ABSTRACT
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with concerning phenotypic mutations is of public health interest. Genomic surveillance is an important tool for a pandemic response, but many laboratories do not have the resources to support population-level sequencing. We hypothesized that a nucleic acid amplification test (NAAT) to genotype mutations in the viral spike protein could facilitate high-throughput variant surveillance. We designed and analytically validated a one-step multiplex allele-specific reverse transcriptase PCR (RT-qPCR) to detect three nonsynonymous spike protein mutations (L452R, E484K, N501Y). Assay specificity was validated with next-generation whole-genome sequencing. We then screened a large cohort of SARS-CoV-2-positive specimens from our San Francisco Bay Area population. Between 1 December 2020 and 1 March 2021, we screened 4,049 unique infections by genotyping RT-qPCR, with an assay failure rate of 2.8%. We detected 1,567 L452R mutations (38.7%), 34 N501Y mutations (0.84%), 22 E484K mutations (0.54%), and 3 (0.07%) E484K plus N501Y mutations. The assay had perfect (100%) concordance with whole-genome sequencing of a validation subset of 229 specimens and detected B.1.1.7, B.1.351, B.1.427, B.1.429, B.1.526, and P.2 variants, among others. The assay revealed the rapid emergence of the L452R variant in our population, with a prevalence of 24.8% in December 2020 that increased to 62.5% in March 2021. We developed and clinically implemented a genotyping RT-qPCR to conduct high-throughput SARS-CoV-2 variant screening. This approach can be adapted for emerging mutations and immediately implemented in laboratories already performing NAAT worldwide using existing equipment, personnel, and extracted nucleic acid.
KEYWORDS: COVID-19, genotyping, RT-qPCR, SARS-CoV-2, variant
INTRODUCTION
High transmission rates during the coronavirus disease 2019 (COVID-19) pandemic have facilitated the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants. The underlying genomic diversity defining these variants is indicative of natural selection of mutations with phenotypic significance (1–3). In particular, variants associated with increased transmission, virulence, or resistance to host, vaccine, or antiviral immunity are of significant public health concern (4–17). Other mutations may also impact diagnostic sensitivity (18). Genomic surveillance that is representative of the newly infected population is therefore an important tool for the pandemic response.
As of 6 May 2021, the U.S. Centers for Disease Control and Prevention (CDC) and the European Centre for Disease Prevention and Control (ECDC) have identified multiple SARS-CoV-2 lineages as variants of concern or variants of interest (VOC/VOI) (19–22). Many share the spike protein mutations E484K (B.1.351, B.1.525, B.1.526, B.1.620, B.1.621, P.1, P.2, P.3), N501Y (B.1.1.7, B.1.351, P.1, P.3, B.1.621), and/or L452R (B.1.427, B.1.429, B.1.617.1, B.1.617.2, B.1.617.3), among others. Such variants have been identified primarily by targeted or whole-genome sequencing (WGS) (23, 24). However, due to the required resources, technical expertise, and equipment for sequencing, few public health systems have successfully coordinated timely large-scale sequencing efforts. Among individual institutions that have developed sequencing programs, there is tremendous variability in throughput, selection criteria, and methodology that confounds accurate estimates of individual variant prevalence. Because the vast majority of laboratories worldwide do not have the resources required to develop and support large-scale sequencing initiatives, a complementary surveillance strategy is desirable.
Upper respiratory nucleic acid amplification tests (NAATs) are the mainstay of SARS-CoV-2 diagnosis. Unlike with sequencing, nearly all laboratories offering NAATs have the necessary equipment and trained personnel to perform nucleic acid extraction and real-time PCR. We therefore aimed to develop a highly accessible spike-genotyping NAAT for high throughput and rapid screening of positive SARS-CoV-2 samples. We hypothesized that this assay would facilitate large-scale VOC/VOI screening and track the emergence of mutations in our population. This approach enables population-based triaging of samples to centers performing sequencing analysis for lineage determination and increases access to variant surveillance.
MATERIALS AND METHODS
Assay design.
In January 2021, the B.1.1.7, B.1.351, B.1.1.28 descendant P.1, B.1.1.28 descendant P.2, and B.1.427/B.1.429 variant lineages were of primary global and local epidemiologic significance. Informed by recent studies describing mutation phenotypic significance and convergent evolution, we elected to begin screening our population for the spike N501Y (B.1.1.7, B.1.351, P.1), E484K (B.1.351, P.1, P.2), and L452R (B.1.427/B.1.429) mutations.
We therefore designed a one-step multiplex allele-specific reverse transcriptase quantitative PCR (RT-qPCR) to detect these three nonsynonymous single-nucleotide variants in the spike gene (N501Y [RefSeq accession no. NC_045512], 23063A>T; E484K, 23012G>A; L452R, 22917 T>G). We selected the wild-type N501 allele (23063A) to serve as a control for those samples that lack any of the targeted mutations.
To permit single-reaction multiplexing, we designed primer sets and dually labeled hydrolysis probes to maximize each probe’s mismatch melting temperature (ΔTm) while maintaining probe specificity and a sufficiently high annealing temperature (see Table S1 in the supplemental material). This 2-to-7°C decrease in each probe’s annealing temperature when binding SARS-CoV-2 cDNA with a single-nucleotide mismatch distinguishes mutant from wild-type nucleic acid templates. Given the proximity between the E484K and N501Y mutations, two primer sets (flanking L452R and E484K/N501Y, respectively) were designed with annealing temperatures similar to or above the probe annealing temperatures.
Recognizing the potential for mutations in the targets, on 26 January 2021, we downloaded all available SARS-CoV-2 sequences from the Global Initiative on Sharing Avian Influenza Data (GISAID) repository annotated as B.1.1.7 (n = 7,864), B.1.351 (n = 341), or B.1.427/B.1.429 variants (n = 622), as well as all available sequences from the National Center for Biotechnology Information (NCBI) Nucleotide Database collected prior to December 2020 (n = 31,016) (25). Primer sequences were conserved in >98.8% of all sequences; mutant probe sequences were conserved in >99.7% of variant-specific sequences and were present in only 0.1% of pre-December 2020 NCBI sequences. The wild-type N501 probe sequence was conserved in 99.2% of pre-December 2020 NCBI sequences.
Additional details regarding synthetic controls, oligonucleotides, cycling conditions, fluorescence thresholds, analytical validation, and assay interpretation for reporting are provided in the supplemental methods and Tables S1 to S6.
Clinical specimens.
This study included upper respiratory tract swab specimens collected between 1 December 2020 and 3 March 2021 from patients as part of routine clinical care. All specimens testing positive for SARS-CoV-2 by a commercial or laboratory-developed NAAT with an RT-qPCR threshold cycle (Ct) of ≤30 or transcription-mediated amplification relative light units (RLU) of ≥1,100 during this period were subjected to multiplex allele-specific genotyping RT-qPCR. All tests were conducted at the Stanford Clinical Virology Laboratory, which serves tertiary-care academic hospitals and affiliated outpatient facilities in the San Francisco Bay Area. After development and analytical validation of our genotyping RT-qPCR, we retrospectively screened all eligible positive specimens dating back to 1 December 2020 and then began prospective genotyping by RT-qPCR on 19 February 2021, for a total of 4,049 unique infections. This study was conducted with Stanford institutional review board approval (protocol 48973), and individual consent was waived.
Prior to the genotyping RT-qPCR, an initial respiratory SARS-CoV-2 NAAT was conducted on a variety of platforms (Table 1; supplemental methods) (26–28) All assays were conducted according to manufacturer and emergency authorization instructions (29; https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2#individual-molecular [accessed 31 March 2021]).
TABLE 1.
Patient characteristics and aggregate genotyping RT-qPCR screening results during the study period
| Characteristic(s) | Valuea in: |
||
|---|---|---|---|
| All SARS-CoV-2 infection group | RT-qPCR population | WGS validation subset | |
| Patients | 11,636 | 4,049 | 229 |
| Age at first positive NAAT (yr [range]) | 36 [23–52] | 37 [23–53] | 37 [23–52] |
| Female | 6,009 (51.64) | 2,065 (51.00) | 128 (55.90) |
| Initial diagnostic specimen | |||
| Month of diagnosis | |||
| December 2020 | 6,074 (52.20) | 344 (8.50) | 18 (7.86) |
| January 2021 | 4,381 (37.65) | 3,050 (75.33) | 170 (74.24) |
| February 2021 | 1,119 (9.62) | 640 (15.80) | 37 (16.16) |
| March 2021 | 62 (0.53) | 15 (0.37) | 4 (1.75) |
| Specimen collection site | |||
| Outpatient COVID-19 screening site | 6,187 (53.18) | 2,393 (59.10) | 138 (60.26) |
| Emergency department | 2,633 (22.62) | 704 (17.39) | 34 (14.85) |
| Outpatient laboratory | 1,190 (10.23) | 469 (11.58) | 24 (10.48) |
| Occupational health | 628 (5.40) | 123 (3.04) | 6 (2.62) |
| Outpatient clinic | 564 (4.85) | 210 (5.19) | 20 (8.73) |
| Urgent care | 220 (1.89) | 75 (1.85) | 6 (2.62) |
| Inpatient floor | 154 (1.32) | 55 (1.36) | 1 (0.43) |
| Perioperative/periprocedural | 28 (0.24) | 11 (0.27) | 0 (0.00) |
| Intensive care unit | 20 (0.17) | 4 (0.10) | 0 (0.00) |
| Infusion center | 10 (0.09) | 3 (0.07) | 0 (0.00) |
| Home health care | 2 (0.02) | 2 (0.05) | 0 (0.00) |
| Specimen source | |||
| Nasopharyngeal swab | 6,607 (56.78) | 1,965 (48.51) | 115 (50.22) |
| Mid-turbinate nasal swab | 4,893 (42.05) | 2,077 (51.30) | 111 (48.48) |
| Other | 136 (1.17) | 7 (0.17) | 3 (1.30) |
| NAAT platform | |||
| PerkinElmer-based RT-qPCR | 7,004 (60.19) | 2,798 (69.10) | 157 (68.56) |
| Lab-developed RT-qPCR | 2,418 (20.78) | 610 (15.07) | 44 (19.21) |
| Hologic Panther | 1,219 (10.48) | 353 (8.72) | 25 (10.92) |
| Cepheid GeneXpert | 573 (4.92) | 147 (3.63) | 1 (0.44) |
| Roche Liat | 320 (2.75) | 97 (2.40) | 0 (0.00) |
| Genmark Eplex | 102 (0.88) | 44 (1.09) | 2 (0.87) |
| RT-qPCR genotype | |||
| L452, E484, N501 | — | 2,308 (57.00) | 120 (52.40) |
| L452R, E484, N501 | — | 1,567 (38.70) | 70 (30.57) |
| L452, E484K, N501 | — | 22 (0.54) | 14 (6.11) |
| L452, E484, N501Y | — | 34 (0.84) | 22 (9.61) |
| L452, E484K, N501Y | — | 3 (0.07) | 3 (1.30) |
| Unable to genotypeb | — | 115 (2.84) | 0 (0.00) |
Values are numbers (percentages) unless otherwise indicated. —, not applicable.
Low viral load, amplification failure, or reaction setup failure despite repeated testing.
Assay validation with next-generation sequencing.
A subset of 120 wild-type and 109 mutant specimens genotyped by RT-qPCR were assessed by WGS with a validation data set. We adapted an existing WGS pipeline for poliovirus genotyping to conduct SARS-CoV-2 whole-genome amplicon-based sequencing of specimens meeting selection criteria (supplemental methods) (30–32) Pangolin was used for PANGO lineage assignment, while Nextclade v0.14.2 and auspice.us 0.5.0. were used to assign Nextstrain clades and perform phylogenetic placement (2, 32, 33). PANGO lineage names and Nextstrain clade names were assigned as of 5 April 2021.
Statistical analysis.
Baseline continuous data were compared with t tests or Wilcoxon rank sum tests, whereas categorical data were compared with Fisher exact tests. Positive percent agreement (PPA) and negative percent agreement (NPA) were reported with Clopper-Pearson score 95% binomial confidence intervals (CI) using next-generation sequencing as the reference method. Analyses were conducted using the R statistical software package. This study was reported in accordance with Standards for the Reporting of Diagnostic Accuracy Studies (STARD) guidelines (34).
Data availability.
Sequencing quality measures and GISAID accession numbers are provided in Tables S7 and S8 in the supplemental material. GISAID accession numbers may be publicly accessed at https://www.epicov.org/epi3/frontend#.
RESULTS
Population-level screening.
During the period between 1 December 2020 and 3 March 2021, our clinical virology laboratory reported 165,501 negative and 13,083 positive SARS-CoV-2 NAATs among 119,154 unique patients. These 13,083 positive NAATs corresponded to 11,635 patients (Table 1). We assumed that this reflected approximately 11,635 unique infections, presuming no reinfection during the study period.
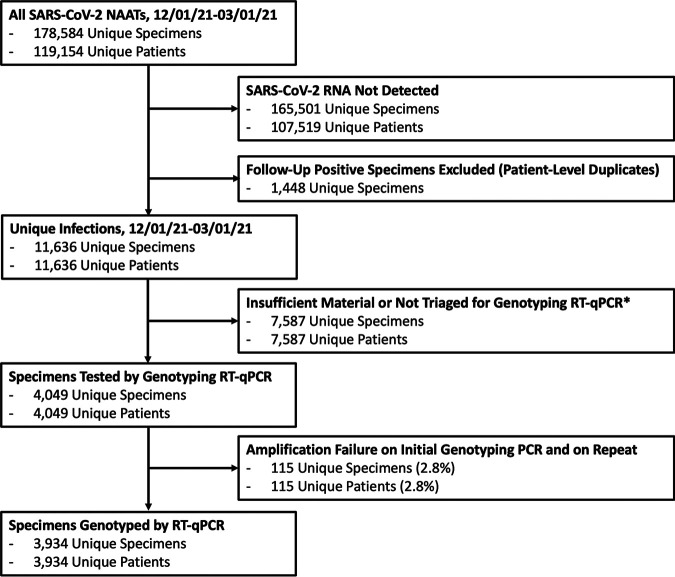
After triaging specimens with higher viral loads and sufficient remaining material, we screened 4,049 specimens from unique patients by genotyping RT-qPCR (Table 1; Fig. 1). Amplification curves from RT-qPCR were interpreted according to Table S4 in the supplemental material, with example cases presented in Fig. 2. A total of 174 specimens had amplification failure on initial RT-qPCR (defined as the absence of both N501-Cy3.5 and N501Y–6-carboxyfluorescein [FAM] amplification). Among these specimens, 59 were successfully genotyped on repeat RT-qPCR and are included in the analysis data set, while the remaining 115 were unable to genotype due to persistent amplification failure, corresponding to an assay failure rate of 2.8%. Among initial diagnostic specimens tested by RT-qPCR with available Ct values (3,142), median viral load was significantly lower among samples unable to genotype (Ct of 28.8 versus 18.8, P < 0.001).
FIG 1.
Flow chart of specimens sent for nucleic acid amplification tests (NAATs) during the study period and numbers of specimens ultimately genotyped by multiplex RT-qPCR. *, predominantly specimens with insufficient material or a low viral load (Ct > 30, RLU < 1,100) or positive specimens in December prior to universal institution-wide genotyping.
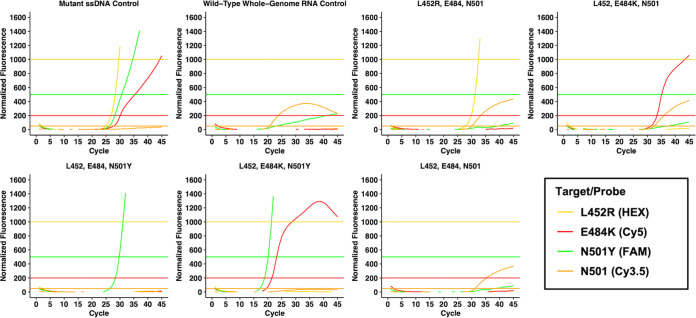
FIG 2.
Example multiplex genotyping RT-qPCR amplification curves for the four hydrolysis probes: L452R (HEX), E484K (Cy5), N501Y (FAM), and N501 (Cy3.5). Horizontal lines represent each probe’s fluorescence threshold in the corresponding color. For specimens without N501Y mutations, N501 (Cy3.5) serves as an internal control in these known SARS-CoV-2-positive specimens. In addition to the single-stranded DNA (ssDNA) mutant control and wild-type whole-genome synthetic RNA control, the assay differentiates among at least five distinct genotypes depicted in this figure.
Among the 3,934 genotyped specimens, we identified 1,567 (38.7%) with isolated L452R mutations, 34 (0.84%) with isolated N501Y mutations, 22 (0.54%) with isolated E484K mutations, and 3 (0.07%) with E484K and N501Y mutations.
Assay validation with next-generation sequencing.
In addition to validating the assay’s analytical performance (Tables S5 and S6), we assessed clinical positive percent agreement (PPA) and negative percent agreement (NPA) of genotyping RT-qPCR using WGS as a reference method. Among 229 specimens from unique patients assessed by both RT-qPCR and WGS (Table 1), the PPAs of RT-qPCR for L452R, E484K, and N501Y were 100% (95% CI, 95 to 100%), 100% (95% CI, 80 to 100%), and 100% (95% CI, 86 to 100%), respectively (Table 2). There were no false negatives by RT-qPCR for L452R, E484K, or N501Y. Furthermore, there were no false positives for E484K (NPA, 100%; 95% CI, 98 to 100%) or N501Y (NPA, 100%; 95% CI, 98 to 100%).
TABLE 2.
Genotyping RT-qPCR validation against whole-genome next-generation sequencing
| RT-qPCRa | Whole-genome next-generation sequencing finding |
|||||
|---|---|---|---|---|---|---|
| L452R |
E484K |
N501Y |
||||
| Detected | Not detected | Detected | Not detected | Detected | Not detected | |
| No. detected | 70 | 0 | 17 | 0 | 25 | 0 |
| No. not detected | 0 | 159 | 0 | 212 | 0 | 204 |
| PPA (95% CI)b | 100 (95–100) | 100 (80–100) | 100 (86–100) | |||
| NPA (95% CI)b | 100 (98–100) | 100 (98–100) | 100 (98–100) | |||
RT-qPCR relative to next-generation sequencing.
PPA, positive percent agreement; NPA, negative percent agreement; CI, confidence interval.
There were three specimens positive for L452R by RT-qPCR but initially negative for L452R by WGS. These had L452R Ct values of 20.0, 22.8, and 31.2, with appropriate amplification curves and appropriate N501 wild-type (Cy3.5) internal control amplification (Cts, 21.0, 18.4, 30.0). All three specimens were reextracted and subjected to repeated RT-qPCR, with the same result. The specimens were then resequenced, and each confirmed the L452R mutation of lineages B.1.427, B.1.427, and B.1.429. These initially discordant results were traced to a pipetting transposition error following long-range PCR. Final L452R NPA after resolution of these discrepancies was 100% (95% CI, 98 to 100%).
Among the 229 specimens tested by both RT-qPCR and WGS, the distribution of PANGO lineages is presented in Table 3, and a phylogenetic tree of Nextstrain clades is presented in Fig. 3. The 120 specimens which were wild type by RT-qPCR were most commonly assigned lineages of B.1.2 (34%) and B.1 (12%). Among the 70 specimens with isolated L452 mutations by RT-qPCR, 43 (60%) and 23 (33%) were B.1.427 and B.1.429, respectively. Among the 22 specimens with isolated N501Y mutations by RT-qPCR, 17 (77%) were B.1.1.7. Among the 14 specimens with isolated E484K mutations by RT-qPCR, 11 (79%) were P.2, 2 (14%) were B.1, and 1 (7%) was B.1.526 (Fig. 4). The three specimens with E484K and N501Y mutations were all of the B.1.351 lineage.
TABLE 3.
PANGO lineage distribution of screen-detected variants by RT-qPCR
| PANGO lineage | RT-qPCR phenotypea |
||||
|---|---|---|---|---|---|
| L452R E484 N501 | L452 E484K N501 | L452 E484 N501Y | L452 E484K N501Y | L452 E484 N501 | |
| B.1 | 2 (3) | 2 (14) | 1 (5) | 0 (0) | 14 (12) |
| B.1.1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.1.174 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.1.222 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.1.228 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| P.2b | 0 (0) | 11 (79) | 0 (0) | 0 (0) | 0 (0) |
| B.1.1.291 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.1.368 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.1.416 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (3) |
| B.1.1.432 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.1.517 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.1.519 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 8 (7) |
| B.1.1.7c | 0 (0) | 0 (0) | 17 (77) | 0 (0) | 0 (0) |
| B.1.139 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.2 | 0 (0) | 0 (0) | 3 (14) | 0 (0) | 41 (34) |
| B.1.232 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (4) |
| B.1.234 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (3) |
| B.1.239 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.240 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (3) |
| B.1.243 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (5) |
| B.1.265 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.311 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (5) |
| B.1.323 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.351c | 0 (0) | 0 (0) | 0 (0) | 3 (100) | 0 (0) |
| B.1.361 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.369 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.375 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.427c | 44 (63) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| B.1.429c | 24 (34) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| B.1.453 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.480 | 0 (0) | 0 (0) | 1 (5) | 0 (0) | 0 (0) |
| B.1.526b | 0 (0) | 1 (7) | 0 (0) | 0 (0) | 0 (0) |
| B.1.561 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (3) |
| B.1.565 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.568 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.577 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| B.1.596 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) |
| B.1.599 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| Total | 70 | 14 | 22 | 3 | 120 |
Data are presented as number (column percent).
CDC-classified variant of interest as of 6 May 2021.
CDC-classified variant of concern as of 6 May 2021.
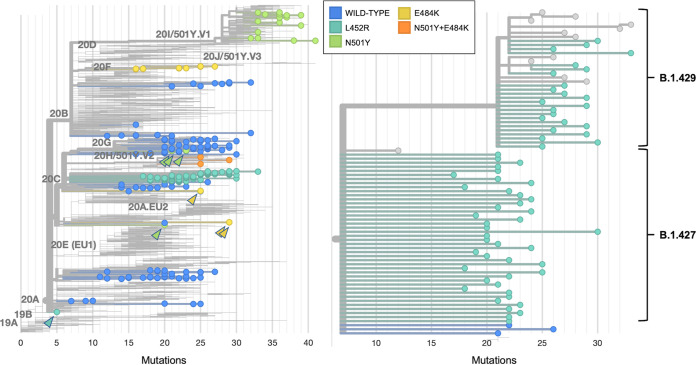
FIG 3.
Nextclade phylogenetic tree of 2,130 SARS-CoV-2 genomes, including 225 of the 229 sequenced genomes from this study and 1,905 genomes from the Nextstrain global reference tree (left). There was one genome excluded because the quality was insufficient to assign a clade. The 225 included genomes are colored by RT-qPCR genotyping result, with each circle representing a sequenced genome. Branch length corresponds to nucleotide divergence. Arrowheads point to mutations identified either sporadically or in rare lineages. Isolates with N501Y mutations belong predominantly to clade 20I/501Y.V1 and lineage B.1.1.7, though sporadic mutations are noted in clade 20G lineage B.1.2 (top three green arrowheads) and clade 20A lineage B.1.480 (bottom green arrowhead). In addition to identifying the clade 20B lineage P.2 variant of concern, E484K RT-qPCR also identified a clade 20C lineage B.1.526 variant of interest (top two yellow arrowheads), as well as a clade 20A proposed lineage B.1 (bottom yellow arrowhead). Isolates with the L452R mutation (turquoise) belong predominantly to clade 20C and lineages B.1.429 and B.1.427 (right). A sporadic L452R mutation was noted within one sample belonging to clade 20A lineage B.1 (turquoise arrowhead). PANGO lineage names and Nextstrain clade names were assigned as of 5 April 2021.
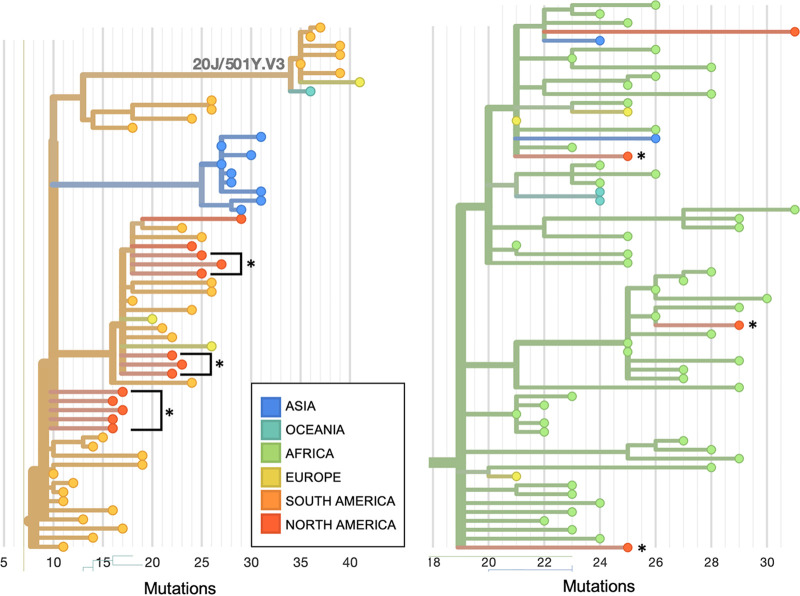
FIG 4.
Nextclade phylogenetic tree from Fig. 3 magnified to visualize isolates with E484K mutations in clade 20B lineage P.2 (left) and isolates with the combined N501Y and E484K mutations, all of which belong to clade 20H/501Y.V2 lineage B.1.351 (right). Isolates from this study are denoted with asterisks. They are shown next to their nearest neighbors from the Nextstrain global reference tree, with branch length corresponding to nucleotide divergence. Isolates are colored by region in which they were collected. PANGO lineage names and Nextstrain clade names were assigned as of 5 April 2021.
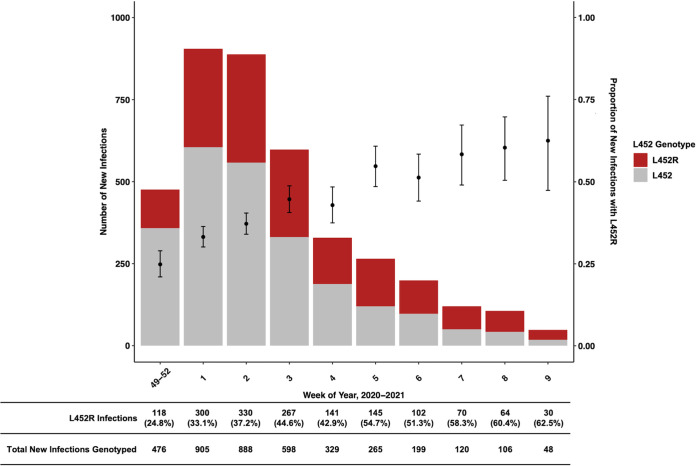
Epidemiologic emergence of L452R.
During the study period, we observed the rapid emergence of the L452R mutation in our population (Fig. 5). The majority of such specimens assessed by WGS were B.1.427/B.1.429 variants (93%). Although the number of new infections in our population decreased between January and March 2021, the proportion of new infections harboring the L452R mutation increased from 24.8% in December 2020 to 62.5% in March 2021.
FIG 5.
Local epidemiologic emergence of L452R mutations in our genotyped population of 3,934 unique infections from December 2020 to March 2021. The bar plot and left y axis represent numbers of new unique genotyped infections by week grouped by RT-qPCR-defined genotype. The scatterplot and right y axis represent proportions of new unique genotyped infections with the L452R genotype, with the 95% Clopper-Pearson confidence interval. Rows under the main figure correspond to numbers and percentages of infections with the L452R genotype among all genotyped strains during each time interval. Infections are grouped by the week in which the initial diagnostic SARS-CoV-2 nucleic acid amplification test was positive. Weeks 48 to 52 are grouped due to fewer tested specimens in December of 2020.
DISCUSSION
Rapid, accessible, and high-throughput epidemiologic surveillance for SARS-CoV-2 variants is of public health interest. We describe a one-step multiplex RT-qPCR assay that facilitated screening of approximately 4,000 unique infections during a period of high transmission in the San Francisco Bay Area. This assay had excellent concordance with WGS and revealed the rapid emergence of variants harboring L452R. Unlike with WGS, the widely available equipment, resources, and trained personnel make this RT-qPCR assay accessible to nearly all laboratories already offering SARS-CoV-2 NAATs. All current CDC-classified variants of concern (B.1.1.7, P.1, B.1.351, B.1.427, B.1.429) and most variants of interest (P.2, B.1.525, some but not all B.1.526 strains) harbor at least one of the L452R, E484K, and N501Y spike protein mutations, enabling population-level triaging by RT-qPCR to centers performing WGS for lineage determination. This approach has implications for outbreak surveillance, diagnostic testing, transmissibility, virulence, and vaccine and/or antiviral efficacy.
The ECDC and the U.S. CDC have identified multiple SARS-CoV-2 lineages as variants of concern or variants of interest (VOC/VOI) (19–22). Many share the spike protein mutations E484K (B.1.351, B.1.525, B.1.526, B.1.620, B.1.621, P.1, P.2, P.3), N501Y (B.1.1.7, B.1.351, P.1, P.3, B.1.621), and/or L452R (B.1.427, B.1.429, B.1.617.1, B.1.617.2, B.1.617.3). Previously reported studies guided by large-scale genomic surveillance have suggested increased transmissibility and virulence or reduced vaccine/antiviral efficacy/neutralization corresponding to the phenotypic significance of these spike protein mutations (4–17). Moreover, mutations in the spike gene (S gene) may lead to false-negative NAAT results (18).
While whole-genome sequencing is required to confirm infection with a specific variant, not all centers have the required resources or personnel to conduct large-scale variant genotyping on a high proportion of incident cases. Allele-specific NAAT-based methods are an alternative to large-scale sequencing programs, but few methods have been described. Korukluoglu et al. recently described a one-step RT-qPCR assay to detect N501Y and HV69-70del using allele-specific forward primers, reserving open reading frame 1ab (ORF1ab) as an internal control (35). This assay is conducted in two separate reactions, with the first targeting N501, ORF1ab, and HV69–70del and the second amplifying N501Y, ORF1ab, and HV69–70del. The authors described high concordance (100%) with results of Sanger and next-generation sequencing. A disadvantage of this method is the absence of screening for E484K and the requirement for two separate reactions. More recently, Banada et al. reported an RT-qPCR method for the detection of N501Y using melt curve analysis (36). This assay includes allele-specific molecular beacon probes for N501 and N501Y conjugated to different fluorophores, as in our assay. Twenty patient specimens were tested, and a subset were confirmed by Sanger sequencing.
Vogels et al. have also described an RT-qPCR assay to detect ORF1a SGF3675-3677del and spike HV69-70del (37). The deletions are detected via ORT1ab-Cy5 and spike-HEX probe dropout, with reservation of the CDC N1 primer/probe set as an internal control. This assay was concordant with 76 sequenced specimens. Limitations of this assay, as described by the authors, are N1-FAM autofluorescence leading to false B.1.1.7 dropout and the absence of the ORF1ab 3675 to 3677 deletion in select B.1.351 isolates. While these deletions are surrogates for B.1.1.7, B.1.351, and P.1, screening for N501Y and E484K may be more specific to variants of concern/interest and appear to be causally associated with concerning phenotypes. Another genotyping NAAT utilized a two-step approach combining the CDC-based laboratory-developed RT-qPCR and the ThermoFisher TaqPath COVID-19 RT-PCR (38). With the intent of identifying B.1.1.7 variants, the authors screened 1,035 samples for TaqPath S gene dropout and genotyped two specimens by multiplex droplet digital PCR (ddPCR), amplifying HV69–70del, N501Y, del145, and S982A in two reactions, which were positive for these four mutations. Next-generation sequencing confirmed B.1.1.7 lineage. Limitations of this genotyping approach include dependence upon S gene dropout followed by more labor-intensive ddPCR.
The largest NAAT-based genotyping series thus far was a national effort in France recently reported by Haim-Boukobza et al (39). The authors used two separate assays to screen for the HVdel69–70 and N501Y mutations in 35,208 samples, with an uninterpretable assay rate of 19.0% and sequencing concordance kappa coefficient of 0.87 to 0.88.
To our knowledge, the present study is the largest NAAT-based SARS-CoV-2 genotyping initiative in the United States. While large multi-institutional programs, such as the United Kingdom COVID-19 genomics consortium, have sequenced more than 250,000 specimens, many public health systems or private institutions are unable to conduct high-throughput genotyping due to the required cost, equipment, and expertise. There are several major advantages to variant screening using NAAT-based genotyping. The RT-qPCR assay that we describe is accessible to any laboratory conducting RT-qPCR SARS-CoV-2 testing and shares much of the same equipment, reagents, and trained personnel. Specimens positive for SARS-CoV-2 by initial NAAT can rapidly be genotyped within hours using the same extracted nucleic acid, and the cost of this assay’s custom oligonucleotides is less than $1 (USA) per reaction. The included interpretation comments may be adapted to individual laboratories and medical records for clinical and epidemiologic reporting, while the described validation procedures serve as a template for laboratorians. As a proof of concept, we have validated this assay on another qPCR instrument (ABI7500 RT-PCR system; Applied Biosystems, Foster City, CA) and with another master mix (NEB Luna Universal one-step RT-qPCR kit; New England BioLabs, Ipswich, MA) to demonstrate that individual laboratories may adapt this assay to existing NAAT workflows (data not shown). Finally, the assay may be further adapted to incorporate emerging mutations of interest (e.g., E484Q).
As evidence of the feasibility of high-throughput testing, we rapidly implemented reflex genotyping RT-qPCR for positive specimens and screened specimens from more than 4,000 infections, thereby facilitating local surveillance of L452R mutation emergence. Our results are consistent with a large-scale whole-genome sequencing initiative of 2,172 specimens from 44 counties in California (12). This series suggested a 19-to-24% increase in transmissibility relative to that of wild-type circulating strains and was the basis for L452R surveillance in our study. Due to the local prevalence of circulating L452R mutations, our screening initiative identified approximately 1,500 unique infections harboring L452R mutations.
This assay has several limitations. Sporadic isolated L452R, E484K, and N501Y mutations are well described and are not necessarily indicative of VOC/VOI. For example, in our series of 229 sequenced specimens genotyped by RT-qPCR, isolated E484K and N501Y mutations by PCR were VOC/VOI in 86% and 77% of cases, respectively, while the remaining were uncommon variants. Although our assay does not screen for several other mutations of interest (HVdel69–70, K417N, P681H, Y453F), it is advantageous that such variants are screen-detected via N501Y (B.1.1.7, del69–70, P681H), E484K (P.1 and B.1.351, K417N), and/or L452R. An additional limitation is the lack of wild-type probes for L452 and E484. Although this choice was intentional to maximize efficiency, it introduces the possibility that synonymous or alternate nonsynonymous mutations (e.g., E484Q) within the probe binding site will lead to an erroneous result. Lastly, variants may develop new mutations in primer and/or probe sites, potentially leading to false negatives. As such, these primer/probe sequences should be regularly checked against all available GISAID sequences, and new VOC/VOI should be monitored for potential assay interference. An additional limitation is exclusion of samples with lower viral loads in an effort to successfully genotype. Although our assay failure rate was low (2.8%), analytical sensitivity might be improved by increasing template volume. While assay failure may have been due to RNA degradation after storage of residual nucleic acid or transport media, it is critical to prospectively monitor for circulating mutations in this assay’s oligonucleotides. Although the present series of 4,049 infections is a subset of the 11,635 unique consecutive infections between December 2020 and March 2021, the genotyped population was similar to the overall population (Table 1). Nevertheless, our results may not be necessarily generalizable to all new infections.
We developed and rapidly implemented a one-step multiplex allele-specific RT-qPCR assay to conduct rapid and high-throughput SARS-CoV-2 variant screening in the San Francisco Bay Area. The assay had near-perfect concordance with the results of whole-genome next-generation sequencing and facilitates detection of all common and uncommon variants of concern/interest harboring the L452R, E484K, or N501Y mutation. This approach can be adapted for emerging mutations and implemented in laboratories already conducting SARS-CoV-2 NAAT using existing resources and extracted nucleic acid.
ACKNOWLEDGMENTS
We sincerely thank the Stanford Clinical Virology Laboratory staff for their dedication and commitment to patient care in the face of unprecedented challenges presented by the COVID-19 pandemic. We also thank the Stanford Protein and Nucleic Acid Facility for oligonucleotide synthesis.
This study was not funded by any grants, scholarships, fellowships, or commercial entities. The authors report no conflicts of interest related to this work.
Footnotes
Supplemental material is available online only.
Contributor Information
Benjamin A. Pinsky, Email: bpinsky@stanford.edu.
Alexander J. McAdam, Boston Children's Hospital
REFERENCES
- 1.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day T, Gandon S, Lion S, Otto SP. 2020. On the evolutionary epidemiology of SARS-CoV-2. Curr Biol 30:R849–R857. 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) Consortium, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. 2021. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12. MMWR Morb Mortal Wkly Rep 70:95–99. 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muik A, Wallisch A-K, Sänger B, Swanson KA, Mühl J, Chen W, Cai H, Maurus D, Sarkar R, Türeci Ö, Dormitzer PR, Şahin U. 2021. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 371:1152–1153. 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kissler SM, Fauver JR, Mack C, Tai CG, Breban MI, Watkins AE, Samant RM, Anderson DJ, Ho DD, Grubaugh ND, Grad YH. 2021. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. medRxiv 10.1101/2021.02.16.21251535. [DOI] [Google Scholar]
- 8.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, The COVID-19 Genomics UK (COG-UK) consortium, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. 2021. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. https://doi.org/2020.12.30.20249034. [Google Scholar]
- 9.Resende PC, Gräf T, Paixão ACD, Appolinario L, Lopes RS, da Fonseca Mendonça AC, da Rocha ASB, Motta FC, Neto LGL, Khouri R, de Oliveira CI, Santos-Muccillo P, Bezerra JF, Teixeira DLF, Riediger I, do Carmo Debur M, Ribeiro-Rodrigues R, Leite AB, do Santos CA, Gregianini TS, Fernandes SB, Bernardes AFL, Cavalcanti AC, Miyajima F, Sachhi C, Mattos T, da da Costa CF, Delatorre E, Wallau GL, Naveca FG, Bello G, Siqueira MM. 2021. A potential SARS-CoV-2 variant of interest (VOI) harboring mutation E484K in the Spike protein was identified within lineage B.1.1.33 circulating in Brazil. bioRxiv 10.1101/2021.03.12.434969. [DOI] [PMC free article] [PubMed]
- 10.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, Xia H, Swanson KA, Cutler M, Cooper D, Menachery VD, Weaver SC, Dormitzer PR, Shi P-Y. 2021. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 27:620–621. 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 11.Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, Costa A, Duarte D, Pessoa K, Goncalvez L, Brandao M, Jesus M, Fernandes C, Pinto R, Silva M, Mattos T, Wallau G, Siqueira M, Resende P, Delatorre E, Graf T, Bello G. 2021. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new variant of concern P.1. Research Square 10.21203/rs.3.rs-275494/v1. [DOI] [Google Scholar]
- 12.Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP, Martin CSS, Federman S, Cheng J, Balcerek J, Taylor J, Streithorst JA, Miller S, Kumar GR, Sreekumar B, Chen P-Y, Schulze-Gahmen U, Taha TY, Hayashi J, Simoneau CR, McMahon S, Lidsky PV, Xiao Y, Hemarajata P, Green NM, Espinosa A, Kath C, Haw M, Bell J, Hacker JK, Hanson C, Wadford DA, Anaya C, Ferguson D, Lareau LF, Frankino PA, Shivram H, Wyman SK, Ott M, Andino R, Chiu CY. 2021. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv https://doi.org/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, Padayachee SD, Dheda K, Barnabas SL, Bhorat QE, Briner C, Kwatra G, NGS-SA, Wits-VIDA COVID Team, Ahmed K, Aley P, Bhikha S, Bhiman JN, Bhorat AE, Du Plessis J, Esmail A, Groenewald M, Horne E, Hwa S-H, Jose A, Lambe T, Laubscher M, Malahleha M, Masenya M, Masilela M, McKenzie S, Molapo K, Moultrie A, Oelofse S, Patel F, Pillay S, Rhead S, Rodel H, Rossouw L, Taoushanis C, Tegally H, Thombrayil A, van Eck S, Wibmer CK, Durham NM, Kelly EJ, Villafana TL, Gilbert S, Pollard AJ, de Oliveira T, Moore PL, Sigal A, Izu A. 2021. Safety and efficacy of the ChAdOx1 nCoV-19 (AZD1222) Covid-19 vaccine against the B.1.351 variant in South Africa. medRxiv https://doi.org/2021.02.10.21251247. [Google Scholar]
- 14.Wang P, Wang M, Yu J, Cerutti G, Nair MS, Huang Y, Kwong PD, Shapiro L, Ho DD. 2021. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. bioRxiv https://doi.org/2021.03.01.433466. [DOI] [PMC free article] [PubMed]
- 15.Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus BJ, Bibi S, Blane B, Bonsall D, Cicconi P, Charlton S, Clutterbuck E, Collins AM, Cox T, Darton T, Dold C, Douglas AD, Duncan CJA, Ewer K, Flaxman A, Faust SN, Ferreira DM, Feng S, Finn A, Folegatti PM, Fuskova M, Galiza E, Goodman AL, Green CM, Green CA, Greenland M, Hallis B, Heath PT, Hay J, Hill HC, Jenkin D, Kerridge S, Lazarus R, Libri V, Lillie PJ, Ludden C, Marchevsky NG, Minassian AM, McGregor AC, Farooq Mujadidi Y, Phillips DJ, Plested E, Pollock KM, Robinson H, Smith A, Song R, Snape MD, et al., COVID-19 Genomics UK (COG-UK) Consortium, Oxford COVID Vaccine Trial Group. 2021. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 VOC 202012/01 (B.1.1.7). ID 3779160. Social Science Research Network, Rochester, NY. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3779160. [Google Scholar]
- 16.Collier DA, Marco AD, Ferreira IATM, Meng B, Datir R, Walls AC, Bassi J, Pinto D, Fregni CS, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A, The CITIID-HIHR BioResource COVID-19 Collaboration, Elmer A, Kingston N, Graves B, McCoy LE, Smith KG, Bradley JR, Temperton N, L LC-G , Barcenas-Morales G, The COVID-19 Genomics UK (COG-UK) consortium, Harvey W, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RK. 2021. SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv https://www.medrxiv.org/content/10.1101/2021.01.19.21249840v4. [Google Scholar]
- 17.Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, Yoon H, Li D, Haynes BF, Sanders KO, Gnanakaran S, Hengartner N, Pajon R, Smith G, Dubovsky F, Glenn GM, Korber B, Montefiori DC. 2021. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. bioRxiv https://www.biorxiv.org/content/10.1101/2021.01.27.428516v2. [DOI] [PMC free article] [PubMed]
- 18.Center for Devices and Radiological Health. 2021. Genetic variants of SARS-CoV-2 may lead to false negative results with molecular tests for detection of SARS-CoV-2—letter to clinical laboratory staff and health care providers. FDA, Silver Spring, MD. https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2. Accessed 29 March 2021. [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2020. SARS-CoV-2 variant classifications and definitions. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html. Accessed 6 May 2021. [Google Scholar]
- 20.Thompson CN. 2021. Rapid emergence and epidemiologic characteristics of the SARS-CoV-2 B.1.526 variant—New York City, New York, January 1–April 5, 2021. MMWR Morb Mortal Wkly Rep 70:712–716. https://www.cdc.gov/mmwr/volumes/70/wr/mm7019e1.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Centre for Disease Prevention and Control. 2021. SARS-CoV-2 variants of concern as of 6 May 2021. European Centre for Disease Prevention and Control, Solna, Sweden. https://www.ecdc.europa.eu/en/covid-19/variants-concern. Accessed 6 May 2021. [Google Scholar]
- 22.Webb LM. 2021. Identification of and surveillance for the SARS-CoV-2 variants B.1.427 and B.1.429—Colorado, January–March 2021. MMWR Morb Mortal Wkly Rep 70:717–718. 10.15585/mmwr.mm7019e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. 2021. Emergence of a novel SARS-CoV-2 variant in southern California. JAMA 325:1324–1326. 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, Scott R, Sconza R, Price J, Margaritis M, Bergstrom M, Spyer MJ, Miralhes PB, Grant P, Kirk S, Valerio C, Mangera Z, Prabhahar T, Moreno-Cuesta J, Arulkumaran N, Singer M, Shin GY, Sanchez E, Paraskevopoulou SM, Pillay D, McKendry RA, Mirfenderesky M, Houlihan CF, Nastouli E. 12April2021. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 1:33–46. 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulterys PL, Garamani N, Stevens B, Sahoo MK, Huang C, Hogan CA, Zehnder J, Pinsky BA. 2020. Comparison of a laboratory-developed test targeting the envelope gene with three nucleic acid amplification tests for detection of SARS-CoV-2. J Clin Virol 129:104427. 10.1016/j.jcv.2020.104427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan CA, Sahoo MK, Pinsky BA. 2020. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 323:1967–1969. 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanford Health Care Clinical Virology Laboratory. 2020. Emergency use authorization (EUA): summary SARS-CoV-2 RT-PCR assay. FDA, Silver Spring, MD. https://www.fda.gov/media/136818/download. Accessed 28 March 2021. [Google Scholar]
- 30.Sahoo MK, Holubar M, Huang C, Mohamed-Hadley A, Liu Y, Waggoner JJ, Troy SB, Garcia-Garcia L, Ferreyra-Reyes L, Maldonado Y, Pinsky BA. 2017. Detection of emerging vaccine-related polioviruses by deep sequencing. J Clin Microbiol 55:2162–2171. 10.1128/JCM.00144-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12:1261–1276. 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagulenko P, Puller V, Neher RA. 2018. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol 4:vex042. 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HCW, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, Cohen JF, STARD Group . 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology 277:826–832. 10.1148/radiol.2015151516. [DOI] [PubMed] [Google Scholar]
- 35.Korukluoglu G, Kolukirik M, Bayrakdar F, Ozgumus GG, Altas AB, Cosgun Y, Ketre CZ. 2021. 40 minutes RT-qPCR assay for screening spike N501Y and HV69-70del mutations. bioRxiv 10.1101/2021.01.26.428302. [DOI]
- 36.Banada P, Green R, Banik S, Chopoorian A, Streck D, Jones R, Chakravorty S, Alland D. 2021. A simple RT-PCR melting temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. medRxiv 10.1101/2021.03.05.21252709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogels CBF, Breban MI, Alpert T, Petrone ME, Watkins AE, Ott IM, de Jesus JG, Claro IM, Magalhães Ferreira G, Crispim MAE, Singh L, Tegally H, Anyaneji UJ, NGS-SA, Hodcroft EB, Mason CE, Khullar G, Metti J, Dudley JT, MacKay MJ, Nash M, Wang J, Liu C, Hui P, Murphy S, Neal C, Laszlo E, Landry ML, Muyombwe A, Downing R, Razeq J, de Oliveira T, Faria NR, Sabino EC, Neher RA, Fauver JR, Grubaugh ND. 2021. PCR assay to enhance global surveillance for SARS-CoV-2 variants of concern. medRxiv 10.1101/2021.01.28.21250486. [DOI] [Google Scholar]
- 38.Perchetti GA, Zhu H, Mills Mg Shrestha L, Wagner C, Bakhash SM, Lin M, Xie H, Huang M-L, Mathias P, Bedford T, Jerome KR, Greninger AL, Roychoudhury P. 2021. Specific allelic discrimination of N501Y and other SARS-CoV-2 mutations by ddPCR detects B.1.1.7 lineage in Washington State. medRxiv 10.1101/2021.03.10.21253321. [DOI] [PMC free article] [PubMed]
- 39.Haim-Boukobza S, Roquebert B, Trombert-Paolantoni S, Lecorche E, Verdurme L, Foulongne V, Selinger C, Michalakis Y, Sofonea MT, Alizon S. 2021. Detecting rapid spread of SARS-CoV-2 variants, France, January 26–February 16, 2021. Emerg Infect Dis 27:1496–1499. 10.3201/eid2705.210397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods and Tables S1 to S8. Download JCM.00859-21-s0001.pdf, PDF file, 306 KB (305.1KB, pdf)
Data Availability Statement
Sequencing quality measures and GISAID accession numbers are provided in Tables S7 and S8 in the supplemental material. GISAID accession numbers may be publicly accessed at https://www.epicov.org/epi3/frontend#.