ABSTRACT
For the control of immunity in COVID-19 survivors and vaccinated subjects, there is an urgent need for reliable and rapid serological assays. Based on samples from 63 COVID-19 survivors up to 7 months after symptom onset, and on 50 serum samples taken before the beginning of the pandemic, we compared the performances of three commercial immunoassays for the detection of SARS-CoV-2 IgA and IgG antibodies (Euroimmun SARS-COV-2 IgA/IgG, Mikrogen recomWell SARS-CoV-2 IgA/IgG, and Serion ELISA agile SARS-CoV-2 IgA/IgG) and three rapid lateral flow (immunochromatographic) tests (Abbott PanBio COVID-19 IgG/IgM, Nadal COVID-19 IgG/IgM, and Cleartest Corona 2019-nCOV IgG/IgM) with a 50% plaque-reduction neutralization test (PRNT50) representing the gold standard. Fifty-seven out of 63 PCR-confirmed COVID-19 patients (90%) showed neutralizing antibodies. The sensitivity of the seven assays ranged from 7.0% to 98.3%, and the specificity ranged from 86.0% to 100.0%. Only one commercial immunoassay showed a sensitivity and specificity of greater than 98%.
KEYWORDS: COVID-19, ELISA, SARS-CoV-2, flow cytometry bead-based surrogate test, lateral flow assay, plaque-reduction neutralization test
INTRODUCTION
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing coronavirus disease (COVID-19) was first reported in China at the end of 2019 and has subsequently caused a worldwide pandemic. As of 7 February 2021, more than 105 million cases and 2.2 million deaths have been reported to be attributed to COVID-19 worldwide (1).
SARS-CoV-2 is mainly transmitted from person to person through respiratory droplets and aerosols, although indirect transmission via contaminated objects or surfaces may also be possible (2, 3).
According to the recommendations of the CDC (4) and European Centre for Disease Prevention and Control (ECDC) (5), antibody testing is not recommended to test for acute SARS-CoV-2 infections. However, antibody testing is useful in determining exposure to SARS-CoV-2, especially in patients with potential post-COVID-19 symptoms, as well as for epidemiological investigations. In patients with low viral RNA load detected by PCR, antibody testing helps to distinguish between the beginning of a SARS-CoV-2 infection, when no antibodies are detectable and infectivity is expected to increase, and the persistent PCR positivity after an undiagnosed SARS-CoV-2 infection, when antibodies should be detectable and no further infectivity is expected (6). Antibody testing also allows the identification of suitable convalescent plasma donors and may become important for the determination of postvaccination immunity. For these purposes, a high specificity is especially important to avoid the assumption of immunity in individuals who are still susceptible to SARS-CoV-2.
Virus neutralization assays remain the gold standard for determining antibody efficacy but are complex, difficult to standardize, and time-consuming. In contrast, enzyme-linked immunosorbent assays (ELISAs) are feasible in a routine laboratory setting and inexpensive. To support decision making on the employment of antibody testing for either diagnostic or population screening, we present a detailed comparison of serological COVID-19 assays to virus neutralization assays.
Several publications have assessed sensitivities of various ELISAs or chemiluminescence immunoassays for laboratory use and lateral flow assays for point-of-care use as proportion of previously PCR-positive patients at least 14 days after the PCR and found sensitivities between 15% and 100% (7–10), but only a few have evaluated these assays against tests that directly measure virus neutralization (11–14).
The aim of this study was to evaluate the performance and utility for routine diagnostic testing of three commercially available immunoassays (Euroimmun SARS-COV-2 IgA and IgG [CE marked], Mikrogen recomWell SARS-CoV-2 IgA and IgG [CE marked], and Serion ELISA agile SARS-CoV-2 IgA and IgG [CE marked]), three rapid lateral flow (immunochromatographic) tests (Abbott PanBio COVID-19 IgG/IgM rapid test device [CE marked], Nadal COVID-19 IgG [CE marked], and Cleartest Corona 2019-nCOV IgG [CE marked]), and a noncommercial bead-based surrogate test, which was designed to also estimate antibody affinity and predict SARS-CoV-2-neutralizing activity as read out by a pseudotype-based virus (micro)neutralization assay (15). It was recently reported that this pseudotype-based surrogate test accurately measures protective immunity against SARS-CoV-2 and correlates well with neutralizing activity using SARS-CoV-2 in the neutralization assay (16).
MATERIALS AND METHODS
Study protocol.
Patients participating in the study were asked to answer a questionnaire about their previous COVID-19 disease, and serum specimens were collected from the participants. Inclusion criteria were a PCR-confirmed COVID-19 disease with a first positive SARS-CoV-2 PCR at least 14 days before serum collection, age of at least 18 years, and written informed consent. In patients with more than one available serum specimen, the first specimen fulfilling the inclusion criteria was included in the main analysis.
Questionnaire.
All participants filled out a questionnaire including questions about age, gender, date of symptom onset, date of first positive PCR test, symptoms (fever, cough, sore throat, dyspnea, rhinorrhea, headache, joint pain, diarrhea, olfactory or gustatory dysfunction, other), and comorbidities (chronic pulmonary disease, ischemic heart disease, autoimmunological disease) as well as immunodeficiencies, intake of corticosteroids (systemically or inhaled), and other immunosuppressants.
Serum samples comprising the negative panel.
All serological assays were performed on 50 serum samples from 50 different individuals. All of these samples were collected in spring 2018, before the circulation of SARS-CoV-2 in Europe. Use of these samples was approved by the local ethics committee.
Pseudotyping of VSV with SARS-CoV-2 Spike protein.
Pseudotyping of vesicular stomatitis virus (VSV) with the SARS-CoV-2 Spike protein was performed as previously described (17). Briefly, VSVΔG-enhanced green fluorescent protein (eGFP)/luciferase was pseudotyped on BHK cells inducibly expressing the VSV G protein (kindly provided by Gert Zimmer) to generate VSVΔG-G. Next, 293T cells were transfected with a plasmid expressing SARS-CoV-2 Spike (kindly provided by Markus Hoffmann and Stefan Pöhlmann [17]) using TransIT-X2 (Mirus) according to the manufacturer’s instructions. At 24 h posttransfection, cells were infected with VSVΔG-G. In order to neutralize residual input virus, inoculated cells were washed twice with phosphate-buffered saline (PBS) 2 h postinfection, and new medium containing 1:1,000 anti-VSV-G antibody (8G5F11; Kerafast) was added. Supernatant containing replication-deficient VSVΔG pseudotyped with SARS-CoV-2 Spike protein (VSVΔG-S) was harvested 18 h postinfection, clarified by centrifugation, and stored at −80°C. Viral titers were determined on Vero E6 cells quantifying green fluorescence.
Pseudovirus neutralization assay.
For pseudovirus neutralization assays, Vero cells (ATCC CCL-81) were seeded in 96-well plates (20,000 cells/plate) in culture medium and were incubated for 4 h at 37°C. Sera were serially diluted 1:3 in infection medium starting with a 1:10 dilution. VSVΔG-S pseudoparticles were diluted in infection medium to obtain ∼200 GFP+ cells/well in the assay. Serum dilutions were mixed 1:1 with pseudoparticles and incubated for 45 min at 37°C prior to addition to the preplated Vero cells and incubation for 16 h at 37°C. The number of GFP+ viral foci was counted using a fluorescence microscope. The 50% pseudovirus neutralization titer (pVNT50) was reported as the interpolated reciprocal of the dilution yielding a 50% reduction in fluorescent viral foci. Neutralization against GFP-expressing recombinant VSV was assayed in parallel to exclude unspecific inhibition of infection. In case of an inhibition greater than 30% in the inhibition control, 50% plaque-reduction neutralization test (PRNT50) titers were read in relation to the PFU in the inhibition control. Neutralization testing was repeated two times in sera with a SARS-CoV-2-independent neutralization greater than 50% as well as in sera negative in the neutralization testing but with at least one borderline or positive ELISA. The median titer was then selected for further analysis. PRNT50s of ≥20 were considered positive.
Immunoassays for anti-SARS-CoV-2 IgA and IgG.
ELISA kits from three manufacturers were included in the study: Euroimmun SARS-COV-2 IgA and IgG (CE marked; Euroimmun Medizinische Labordiagnostika, Lübeck, Germany), Mikrogen recomWell SARS-CoV-2 IgA and IgG (CE marked; Mikrogen, Neuried, Germany), and Serion ELISA agile SARS-CoV-2 IgA and IgG (CE marked; Institut Virion/Serion, Wuerzburg, Germany). The Euroimmun assay is based on S1 antigen, Mikrogen recomWell is based on the nucleocapsid (N) protein as antigen, and Serion ELISA agile SARS-CoV-2 IgA is based on a mixture of recombinant highly purified N protein and whole spike protein (S1/S2 ectodomain), whereas Serion ELISA agile SARS-CoV-2 IgG is based exclusively on the S1/S2 ectodomain. The assays were performed following the manufacturer’s instructions. The following thresholds were used according to the manufacturer’s instructions. In the Mikrogen assays, samples with a concentration of <20 U/ml were assessed as negative, samples with a concentration of ≥20 U/ml but <24 U/ml were counted as borderline, and samples with a concentration of ≥24 U/ml were counted positive. For Euroimmun’s IgA and IgG assays, extinction ratios were calculated out of specimen extinction and calibrator extinction. Ratio values of <0.8 were considered negative, those with a ratio of ≥1.1 were considered positive, and those with a ratio of ≥0.8 to <1.1 were recorded as borderline according to the manufacturer’s recommendations. The thresholds in the Serion assay were <10 U/ml for negative results, ≥10 U/ml but <14 U/ml for IgA and ≥10 U/ml but <15 U/ml for IgG antibodies as borderline, and >14 U/ml for IgA and >15 U/ml for IgG as positive.
Rapid lateral flow tests.
The serum samples were also used to evaluate three rapid lateral flow antibody tests: Nadal COVID-19 IgG/IgM test (whole blood, serum, or plasma) from New Art Laboratories (NAL)-von Minden (Moers, Germany) targeting S1 receptor binding domain (RBD), PanBio COVID-19 IgG/IgM rapid test device from Abbott (Chicago, IL, USA) (9), and Cleartest Corona 2019-nCOV IgM/IgG (Servoprax, Wesel, Germany), the latter two both targeting the N antigen, according to the manufacturers’ information. All three tests are based on immunochromatography for the qualitative detection of IgM and IgG antibodies specific to SARS-CoV-2 in human serum, plasma, and fingerstick and venipuncture whole blood. The tests were performed following the manufacturers’ instructions. Readout (positive/negative) was interpreted by two independent operators. As the volume of the negative sera was limited, not all negative sera could be tested with all lateral flow tests.
Bead-based surrogate test for the detection of neutralizing anti-SARS-CoV-2 antibodies.
M-270 epoxy Dynabeads (ThermoFisher, Waltham, MA, USA) were coated according to the manufacturer’s instructions with different concentrations, to be able to measure relative affinity (18) of either commercially available full-length SARS-CoV-2 S1-Spike protein (His tagged; Sino Biological, Beijing, China) expressed and purified from HEK cells or SARS-CoV-2 RBD protein expressed and purified from Expi293F cells (19–23). Importantly, the receptor binding domain of the viral spike protein has been found to be an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients (24). The coated beads were resuspended in PBS-0.1% bovine serum albumin (BSA) and stored at 4°C. For detection of anti-SARS-CoV-2 antibodies, beads were first blocked with PBS-10% BSA followed by incubation with a 1:100 dilution of serum in PBS-0.1% BSA-0.02% NaN3. After extensive washing, bead-bound antibodies were detected with goat anti-human IgG Alexa 647 (Dianova, Hamburg, Germany). A mouse/human chimeric monoclonal anti-SARS-CoV-2-Spike-S1 antibody (Sino Biological, Beijing, China) was used as a positive control for all measurements. Samples were measured on a FACSCalibur flow cytometer (Becton, Dickinson, Franklin Lakes, NJ, USA), and FlowJo software version 10 (TreeStar, Ashland, OR, USA) was used for data analysis. The flow cytometric data were converted into scores taking into account binding and affinity of the bound antibodies. The test was calibrated in a discovery cohort comprising samples of 72 healthy blood donors (50% male/50% female) aged 18 to 67 years taken more than a year before the onset of the pandemic and 25 samples taken during the pandemic, of which 13 contained neutralizing antibodies. Using the scoring system detailed below, the test detected neutralizing antibodies with a sensitivity of 85.7% and 100% specificity.
RBD score was defined as follows: (a) for binding, MFI(sample)/MFI(negative control) ratio of ≤1 = score 0; ratio of >1 to ≤1.19 = score 1; ratio of ≥1.2 to ≤1.99 = score 2; ratio of >1.99 = score 3; (b) for affinity, MFI(sample − beads with low Ag coating)/MFI(sample − beads with high Ag coating) value of ≤39.99% = score 1; >39.99% = score 1.5; total score(RBD) = score(binding) × score(affinity). MFI is mean fluorescence intensity; Ag is antigen.
S1 score was defined as follows: (a) for binding, MFI(sample)/MFI(negative control) ratio of ≤1.19 = score 0; ratio of >1.19 to ≤1.99 = score 1; ratio of >1.99 = score 2; (b) for affinity, MFI(sample − beads with low Ag coating)/MFI(sample − beads with high Ag coating) value of ≤74.99% = score 1; >74.99% = score 2; total score(S1) = score(binding) × score(affinity); total score = score(RBD) × score(S1).
Statistical analysis.
Data analysis was performed using Microsoft Excel 2016, IBM SPSS Statistics 26, and GraphPad Prism 8. Differences in symptoms between patients with and without neutralizing antibodies as well as differences in test sensitivity were analyzed using two-tailed Fisher’s exact test. Differences in the time interval between PCR result and serum collection were compared using Student’s t test. Spearman’s rank correlation coefficient rho was calculated to determine the correlation of different tests as well as the correlation between titers and time between first positive PCR and serum sampling. Two-tailed significance was calculated using exact permutation-based probabilities. For the calculation of specificity and sensitivity, borderline results of the serological tests were counted as positive. Specificity was calculated as the proportion of negatively detected samples in the negative sera from 2018, and sensitivity was calculated as proportion of samples with neutralizing activity in the pseudovirus neutralization assay. Confidence intervals (CI) were calculated using the Wilson score method without continuity correction (25) using a confidence interval calculator sheet (26). A significance level α of 0.05 was used.
Ethics declaration.
The study protocol was approved by the local ethics committee in accordance with the Declaration of Helsinki (file no. 84/20).
RESULTS
Patient selection.
Out of 68 participating patients, 63 were included in the study. Five patients did not meet the inclusion criteria. Three of these patients reported that their diagnosis was not based on a PCR result but on SARS-CoV-2 antibody rapid testing. Neutralizing antibodies were not found in these three patients, but various commercial assays were positive for SARS-CoV-2 antibodies. The serum samples of two patients were taken at 3 and 13 days, respectively, and thus too early after their first positive PCR result.
Demographic data.
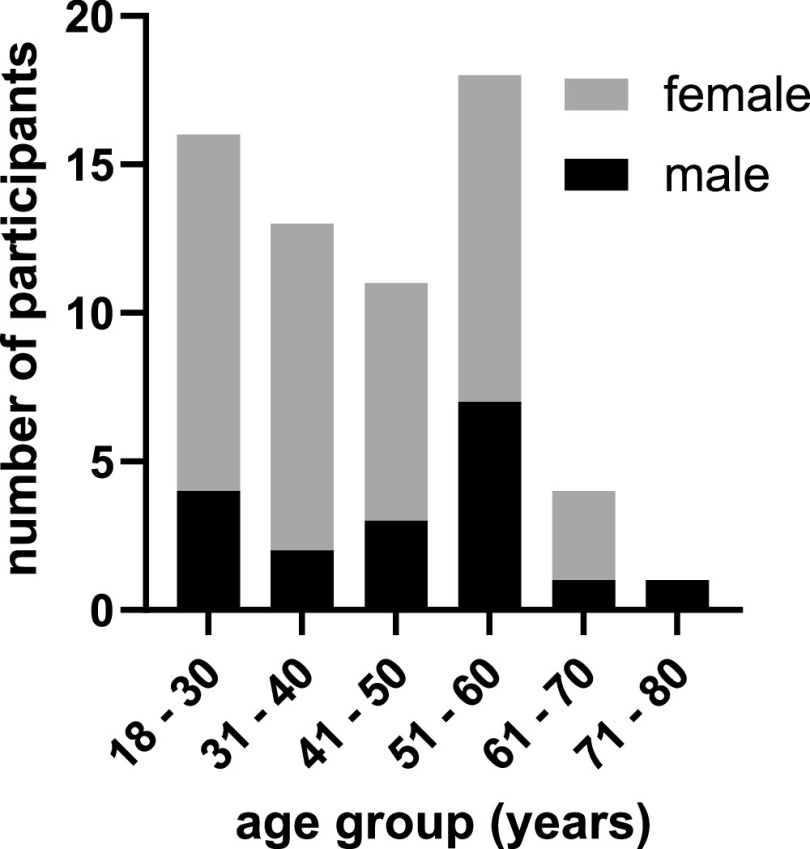
Forty-five of the 63 patients, mostly health care workers, assigned themselves to a female gender (71%), 18 assigned themselves to a male gender (29%), and no participants felt themselves to belong to diverse gender. The gender and age distribution of the participants is shown in Fig. 1. The median time span between the first positive PCR result and serum collection was 91 days (range: 14 to 226 days), and the median time span between the onset of symptoms and serum collection was 94 days (range: 17 to 228 days).
FIG 1.
Participants’ demographics (n = 63).
Clinical characteristics of the participants of the study.
Sixty-two of 63 participants answered questions on their course of disease, comorbidities, and immunosuppression. Sixty of 62 participants reported symptoms, and two did not report any symptoms. The most common symptoms were olfactory or gustatory dysfunction (69%, 43/62), headache (55%, 34/62), cough (52%, 32/62), joint pain (44%, 27/62), rhinorrhea (42%, 26/62), sore throat (39%, 24/62), dyspnea (29%, 18/62), and diarrhea (21%, 13/62). Other reported symptoms were fatigue (16%, 10/62), nausea (5%, 3/62), angina pectoris (5%, 3/62), and dizziness (3%, 2/62). Mucosal dryness, erythema, back pain, shivering, inappetence, vision disorder, and fine motor difficulties were reported by one participant each. Fever was reported by 31 of 62 patients (50%). In 4% (2/57) of cases, no symptoms were reported. Eight patients (13%) reported chronic pulmonary disease, five (8%) reported autoimmune disease, and one (2%) reported ischemic heart disease as comorbidities. One participant had an immunodeficiency (2%), nine (15%) had inhaled corticosteroids, and three participants had taken systemic corticosteroids during the last year. None of the participants had taken other immunosuppressant or immunomodulatory substances during the last year.
Pseudovirus neutralization assay.
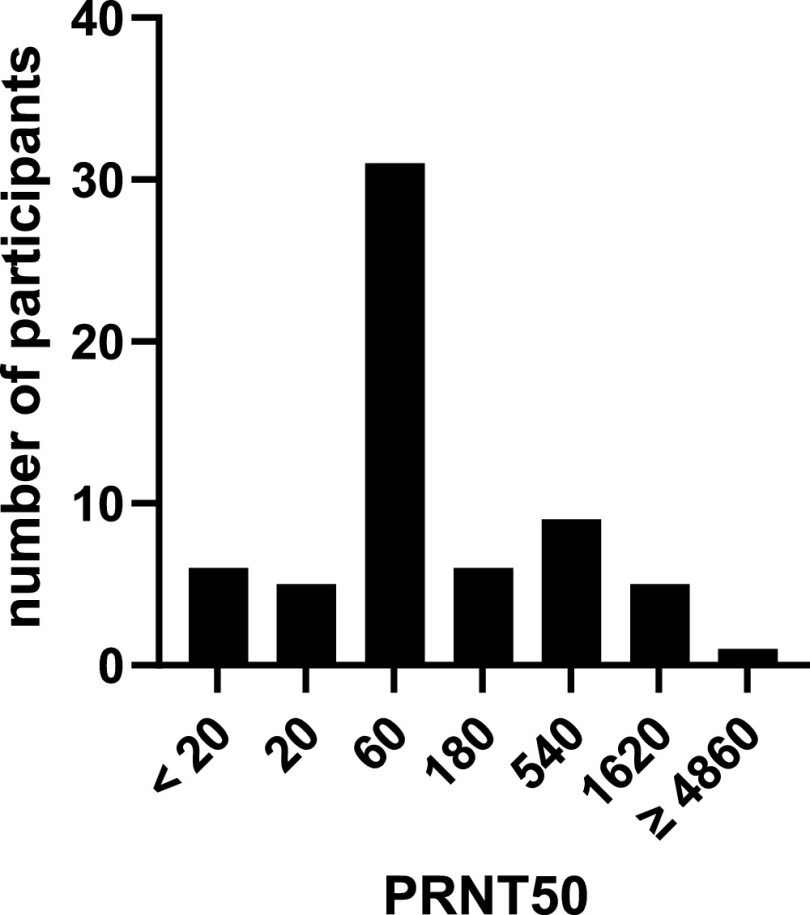
Neutralizing SARS-CoV-2 antibodies could be detected in 57 of the 63 patients. Neutralization assay results could be reproduced in all cases except one where the initial test showed no neutralization at all but both repeated measures showed neutralization (50% plaque-reduction neutralization test [PRNT50] titer: 540). The distribution of the PRNT50 titers is shown in Fig. 2. Both asymptomatic participants tested negative (P < 0.01). None of the patients without neutralizing antibodies reported fever, compared to 55% (31/56) of the patients with neutralizing antibodies (P = 0.02). No other significant differences in symptoms could be determined between patients with and without neutralizing antibodies. The median time between first PCR and serum collection (median, 82 days; minimum, 26 days; maximum, 194 days) in patients without neutralizing antibodies did not differ significantly from the patients with neutralizing antibodies (P = 0.79). One of the participants without neutralizing antibodies inhaled corticosteroids (P = 1.00), and no participant without neutralizing antibodies had taken other immunosuppressant or immunomodulatory substances during the last year or had an immunodeficiency. Neutralizing antibodies could not be detected in any of the 50 sera from 2018.
FIG 2.
Distribution of the PRNT50 titers of the included patients (n = 63). PRNT50, 50% plaque-reduction neutralization test. Method described in Materials and Methods. PRNT50 values of <20 were considered negative.
Comparison of commercial immunoassays with PRNT50.
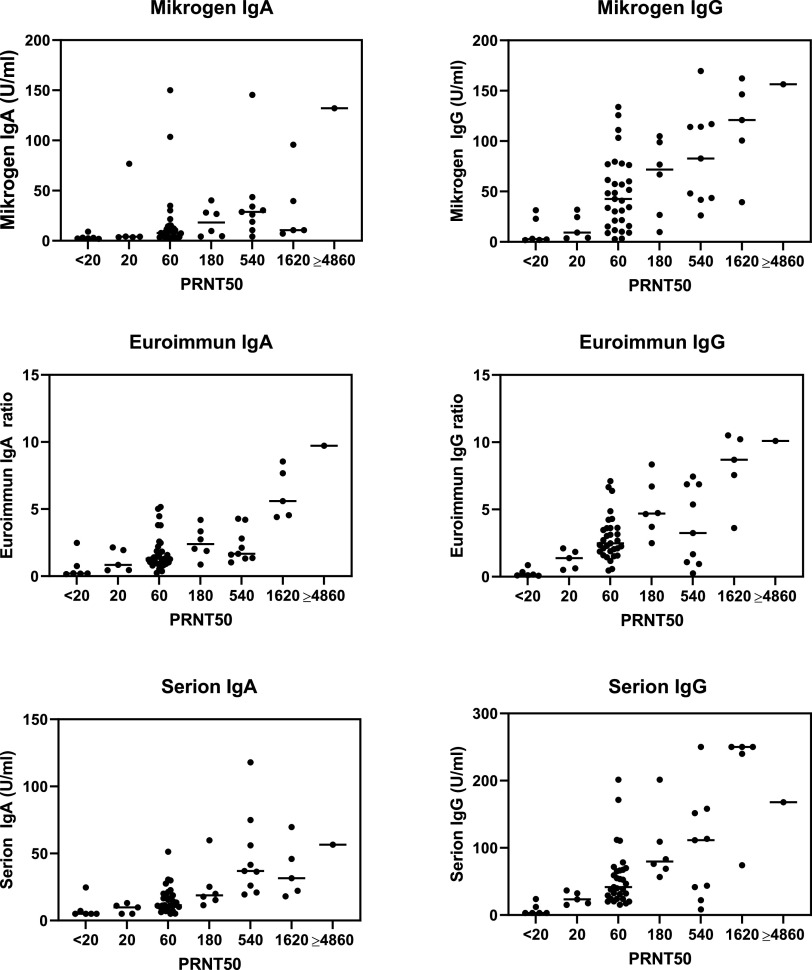
Results of Euroimmun SARS-COV-2 IgA/IgG, Mikrogen recomWell SARS-CoV-2 IgA/IgG and Serion ELISA agile SARS-CoV-2 IgA/IgG were plotted against the titers observed in the neutralization assays (Fig. 3). All ELISA results correlated significantly with PRNT50 titers (P < 0.001). Serion ELISA agile SARS-CoV-2 IgA correlated most strongly with PRNT50 titers (rho = 0.715) followed by Serion ELISA agile SARS-CoV-2 IgG (rho = 0.685), Euroimmun SARS-COV-2 IgG (rho = 0.645), Mikrogen recomWell SARS-CoV-2 IgG (rho = 0.630), Euroimmun SARS-COV-2 IgA (rho = 0.597), and Mikrogen recomWell SARS-CoV-2 IgA (rho = 0.562). The strongest interassay correlation was detected between Euroimmun SARS-COV-2 IgG and Serion ELISA agile SARS-CoV-2 IgG (rho = 0.860; P < 0.001), while correlation was weakest between Euroimmun SARS-COV-2 IgG and Mikrogen recomWell SARS-CoV-2 IgA (rho = 0.362; P = 0.004).
FIG 3.
Results of the ELISAs plotted against the titers from the neutralization assay (n = 63). Values above the maximum or below the minimum in a test were plotted at this maximum or minimum value, respectively (Mikrogen IgA, >150.12 U/ml; Mikrogen IgG, >133.93 U/ml; Euroimmun IgA, >8.54; Serion IgA, <5.0 U/ml; Serion IgG, <3.0 U/ml and >250.0 U/ml). Mikrogen, Mikrogen recomWell SARS-CoV-2 IgA/IgG (Mikrogen, Neuried, Germany), based on the nucleocapsid; Euroimmun, Euroimmun SARS-COV-2 IgA/IgG (Euroimmun, Lübeck, Germany), based on S1 antigen; Serion, Serion ELISA agile SARS-CoV-2 IgA/IgG (Institut Virion/Serion, Wuerzburg, Germany), based on the S1/S2 ectodomain and the nucleocapsid (IgA) or S1/S2 ectodomain antigen (IgG); PRNT50, 50% plaque-reduction neutralization test. Method described in Materials and Methods. PRNT50 values of <20 were considered negative.
The sensitivities of the IgA assays to detect neutralizing antibodies ranged from 31.6% (Mikrogen) to 91.2% (Euroimmun), and the specificities ranged from 86.0% (Euroimmun) to 100.0% (Serion). The sensitivities of the IgG assays ranged from 79.0% (Mikrogen) to 100.0% (Serion), and the specificities ranged from 94.0% (Mikrogen) to 100.0% (Serion). Detailed data and confidence intervals are shown in Table 1.
TABLE 1.
Sensitivity and specificity data of the six ELISAs included in the studya
| Assay | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|
| Mikrogen IgA | 31.6% (21.0–44.5%) | 94.0% (83.8–97.9%) |
| Mikrogen IgG | 79.0% (66.7–87.5%) | 94.0% (83.8–97.9%) |
| Euroimmun IgA | 91.2% (81.1–96.2%) | 86.0% (73.8–93.1%) |
| Euroimmun IgG | 91.2% (81.1–96.2%) | 96.0% (86.5–98.9%) |
| Serion IgA | 82.5% (70.6–90.2%) | 100.0% (92.9–100.0%) |
| Serion IgG | 98.3% (90.7–99.7%) | 100.0% (92.9–100.0%) |
Sensitivity data were collected on 57 samples positive in the neutralization assay. Specificity data were collected on 50 negative sera. Confidence intervals were calculated using the Wilson score method without continuity correction (25) using a confidence interval calculator sheet (26). Mikrogen, Mikrogen recomWell SARS-CoV-2 IgA/IgG (Mikrogen, Neuried, Germany); Euroimmun, Euroimmun SARS-COV-2 IgA/IgG (Euroimmun, Lübeck, Germany); Serion, Serion ELISA agile SARS-CoV-2 IgA/IgG (Institut Virion/Serion, Wuerzburg, Germany); 95% CI, 95% confidence interval.
While the sensitivities of Mikrogen IgA (P < 0.001), Mikrogen IgG (P = 0.002), and Serion IgA (P = 0.008) were significantly lower than those of the best-performing assay, Serion IgG, the differences between Euroimmun IgA/IgG and Serion IgG were not significant (P = 0.21). All ELISA values correlated significantly with the PRNT50 values (P < 0.001). The rho values ranged from 0.562 (Mikrogen IgA) to 0.685 (Serion IgG) and 0.715 (Serion IgA).
A combination of IgG and IgA (IgA or IgG) positivity increases the sensitivity of Serion to 100.0% (95% CI, 92.9 to 100.0%) without specificity losses. The combined sensitivity for Euroimmun IgG and IgA increases to 96.5% (95% CI, 88.1 to 99.0%) with a loss of specificity to 86.0% (95% CI, 73.8 to 93.1%). For the Mikrogen test, no increase in sensitivity can be reached combining IgG and IgA, but specificity decreases to 88.0% (95% CI, 76.2 to 84.4%).
Rapid antibody testing (lateral flow assays).
The sensitivities of the IgM assays to detect neutralizing antibodies ranged from 7.0% (Abbott) to 66.7% (Nadal), and the specificities ranged from 91.1% (Cleartest) to 100.0% (Abbott and Nadal). The sensitivities of the IgG assays ranged from 84.2% (Abbott) to 89.5% (Cleartest), and the specificities ranged from 95.6% (Cleartest) to 100.0% (Abbott and Nadal). Detailed data and confidence intervals are shown in Table 2. Regarding sensitivity, all rapid antibody tests were significantly inferior to the Serion IgG ELISA regarding sensitivity (P = 0.03 and lower) except for the Cleartest IgG, where statistical significance was not reached (P = 0.11).
TABLE 2.
Sensitivity and specificity data of the six lateral flow immunochromatographic assays included in the studya
| Assay | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|
| Abbott IgM | 7.0% (2.8–16.7%) | 100.0% (92.4–100.0%) |
| Abbott IgG | 84.2% (72.6–91.5%) | 100.0% (92.4–100.0%) |
| Nadal IgM | 66.7% (53.7–77.5%) | 100.0% (92.1–100.0%) |
| Nadal IgG | 86.0% (74.7–92.7%) | 100.0% (92.1–100.0%) |
| Cleartest IgM | 21.1% (12.5–33.3%) | 91.1% (79.3–96.5%) |
| Cleartest IgG | 89.5% (78.9–95.1%) | 95.6% (85.2–98.8%) |
Sensitivity data were collected on 57 samples positive in the neutralization assay. Specificity data were collected on 47 negative sera (Abbott) or 45 negative sera (Nadal and Cleartest). Confidence intervals were calculated using the Wilson score method without continuity correction (25) using a confidence interval calculator sheet (26). Abbot, Abbott PanBio COVID-19 IgM/IgG (Abbott, Chicago, IL, USA); Nadal, Nadal COVID-19 IgM/IgG (NAL von Minden, Moers, Germany); Cleartest, Cleartest Corona 2019-nCOV IgM/IgG (Servoprax, Wesel, Germany); 95% CI, 95% confidence interval.
Bead-based surrogate test.
The sensitivity of the bead-based surrogate test for neutralizing antibodies was 71.9% (95% CI, 59.2 to 81.9%), and the specificity was 100% (95% CI, 92.9 to 100.0%) (see Fig. S1 in the supplemental material). The total bead score correlated with the inhibition-corrected PRNT50 (rho = 0.414; P = 0.001).
Survivors of a PCR-confirmed COVID-19 disease without neutralizing antibodies.
In three of the six patients without detectable neutralizing antibodies against the full-length Spike protein or the Spike protein receptor-binding domain, no IgM, IgA, or IgG antibodies could be detected in any of the six commercial assays. In one patient, Mikrogen and Serion showed borderline results in the IgG assay and the Abbott IgG showed a positive result, and in one patient only Nadal IgM/IgG was positive. Interestingly, one patient had positive IgG antibodies in the Mikrogen, Serion, and Abbott assays as well as positive IgM and borderline IgG in the Euroimmun assay. The bead-based surrogate test was negative in all of these six patients.
Antibody dynamics.
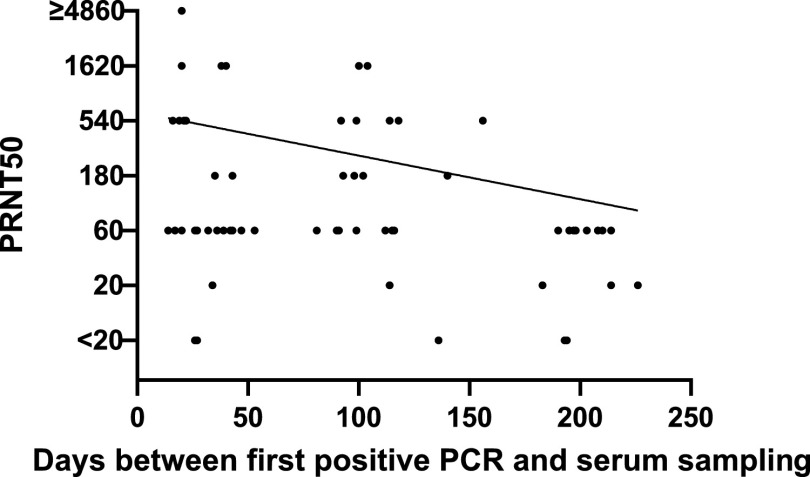
While the IgA antibodies determined by the Mikrogen ELISA were significantly negatively correlated with the time between the first positive PCR and the serum sampling (rho = −0.313; P = 0.01), this was not the case for antibodies measured by other ELISAs and neutralization testing (rho = −0.234; P = 0.06) (Fig. 4).
FIG 4.
Correlation of the PRNT50 titers and the time (in days) between first positive PCR and serum sampling of the included patients (n = 63). PRNT50, 50% plaque-reduction neutralization test. Method described in Materials and Methods. PRNT50 values of <20 were considered negative and plotted as 6 for drawing the logarithmic correlation line. A PRNT50 value of ≥4,860 was plotted as 4,860 for drawing the logarithmic correlation line.
We followed the seroconversion against SARS-CoV-2 in one representative patient with moderate symptoms using the neutralization assay, all six ELISA systems, and the bead-based surrogate test (Table 3). Positive IgA levels were detected from day 8 after onset of symptoms with the Serion IgA ELISA, whereas the Euroimmun IgA assay and Mikrogen IgA assay detected positive IgA levels from day 11 after onset of symptoms. Seroconversions for IgG occurred later than that for IgA, and positive IgG levels were detected from day 11 on with the Serion IgG ELISA and the Mikrogen IgG assay, whereas the Euroimmun IgG assay failed to detect IgG antibody levels until day 24. Interestingly, both IgA and IgG levels decreased after day 17 in all but the Euroimmun IgG assay, where the levels of detected antibodies even continued to increase. Neutralizing antibodies could be detected from day 11 on with a peak on day 14. The flow cytometry bead-based assay gave ambiguous results.
TABLE 3.
Antibody testing results in one individual in chronological sequencea
| Days after onset of symptoms | PRNT50 titer | Mikrogen IgA (U/ml) | Mikrogen IgG (U/ml) | Euroimmun IgA (ratio) | Euroimmun IgG (ratio) | Serion IgA (U/ml) | Serion IgG (U/ml) | BBST (total bead score) |
|---|---|---|---|---|---|---|---|---|
| 2 | <20 | Neg (3.25) | Neg (3.02) | Neg (0.48) | Neg (0.20) | Neg (<5.00) | Neg (<3.00) | Neg (0) |
| 5 | <20 | Neg (3.12) | Neg (3.40) | Neg (0.47) | Neg (0.20) | Neg (<5.00) | Neg (<3.00) | Neg (0) |
| 8 | <20 | Neg (4.75) | Neg (3.63) | Neg (0.55) | Neg (0.22) | Pos (29.21) | Neg (<3.00) | Neg (0) |
| 11 | 60 | Pos (48.64) | Pos (25.84) | Pos (1.34) | Neg (0.37) | Pos (131.54) | Pos (17.84) | Neg (0) |
| 14 | ≥4,860 | Pos (56.86) | Pos (39.12) | Pos (1.48) | Neg (0.57) | Pos (148.26) | Pos (22.50) | Pos (3) |
| 17 | 540 | Pos (43.59) | Pos (41.68) | Pos (1.32) | Bor (0.94) | Pos (117.97) | Pos (22.20) | Neg (0) |
| 24 | 540 | Pos (25.93) | Pos (44.72) | Bor (0.78) | Pos (1.16) | Pos (66.21) | Pos (18.37) | Pos (6) |
| 121 | 60 | Neg (5.13) | Neg (18.95) | Pos (1.16) | Pos (2.87) | Pos (22.78) | Pos (17.55) | Pos (9) |
| 174 | 60 | Neg (4.46) | Neg (10.81) | Bor (0.92) | Pos (2.79) | Pos (20.44) | Pos (17.42) | Neg (0) |
The first positive PCR sample was taken the day after symptom onset. PRNT50, 50% plaque-reduction neutralization test. Method described in Materials and Methods. Euroimmun, Euroimmun SARS-COV-2 IgA/IgG (Euroimmun, Lübeck, Germany); Mikrogen, Mikrogen recomWell SARS-CoV-2 IgA/IgG (Mikrogen, Neuried, Germany); Serion, Serion ELISA agile SARS-CoV-2 IgA/IgG (Institut Virion/Serion, Wuerzburg, Germany); BBST, bead-based surrogate test. Method described in Materials and Methods. Pos, positive; Neg, negative; Bor, borderline.
DISCUSSION
A large variety of serological assays for the detection of anti-SARS-CoV-2 antibodies has recently become available. Thorough assay validation is required to inform decisions on the use of these assays and the interpretation of test results for specific clinical and public health care purposes. In this study, we determined the specificity and sensitivity of three commercially available immunoassays and three rapid lateral flow tests for the detection of SARS-CoV-2 antibodies in comparison with a neutralization test. Neutralization assays remain the gold standard for determining antibody efficacy. Ninety percent of the PCR-confirmed COVID-19 patients investigated in this study showed neutralizing antibodies against the SARS-CoV-2 Spike protein up to 7 months after symptom onset. This is in line with literature data ranging from 77% (14) to 96% (27). Neutralizing antibody production seems to be correlated with severity of disease (27), and one study found neutralizing antibodies in only 12.5% of asymptomatic SARS-CoV-2-positive patients (28). In line with this report, both asymptomatic patients included in our study did not show neutralizing antibodies either, but it is also conceivable that the original PCR results were false positive. The duration of persistence of IgG antibodies has yet to be defined. However, IgG levels seem to be maintained for at least 7 months, as detected with different commercial assays as well as the bead-based surrogate test and the pseudovirus neutralization assay. In comparison, SARS-CoV-1 patients were shown to maintain IgG antibodies for an average of 2 years, and significant reduction of IgG levels and titers occurred in the third year (29).
Sensitivity of the seven assays to detect sera with neutralizing activity ranged from 7.0% to 98.3%. Specificity varied between 86.0% and 100.0%. Sensitivity was higher in IgG assays than in IgM and IgA assays, and IgA levels were only negatively correlated with time from symptom onset in one assay. Based on the currently available data, IgM and IgG seroconversion occurs within 10 to 12 days and 12 to 14 days, respectively, after onset of symptoms (15, 30–34). A combination of IgA and IgM assays in SARS-CoV-2 diagnostics may increase sensitivity compared to a single IgG assay at the cost of a lower specificity depending on the assay. ELISAs were more sensitive than the lateral flow assays with lower sensitivity values for anti-SARS-CoV-2 IgG in our cohort than in previously published data (9). The low sensitivities of IgM lateral flow assays remain unclear as some samples acquired only a few weeks after infection were negative while other samples acquired more than 6 months after the infection were positive despite average PRNT50 titers. In line with a published study (14), only the Serion ELISA agile SARS-CoV-2 IgG showed a satisfactory sensitivity and specificity of more than 98%. The bead-based surrogate test showed 100% specificity and was the only test in this study to produce no false-positive results regarding neutralizing antibody activity, but as expected, sensitivity was lower than the IgG ELISAs.
The symptoms most commonly described in this study were olfactory or gustatory dysfunction, which occurred in 69% of the participants. These data are in line with previous studies on this topic (35, 36).
The limitations of the study are as follows. The severity of disease was not queried, and one participant suffered from an immunodeficiency. Recruiting participants among health care workers resulted in a gender imbalance toward the female gender. Although there are interassay differences in sensitivity, the number of participants was not high enough to assess a significant superiority. It is noteworthy that the three immune assays incorporated in this study are based on different antigen components. Whereas the Euroimmun assay is based on S1 antigen, Mikrogen recomWell is based on the nucleocapsid protein as antigen and Serion ELISA agile is coated with S1 and S2 ectodomain for IgG testing and S1/S2 ectodomain as well as the nucleocapsid for IgA testing. In the one patient followed over time, IgG dynamics differ between the different assays. While IgG assays were available from all manufacturers, IgM and IgA could be tested only with selective assays. As antibody assays are not recommended for primary SARS-CoV-2 diagnosis (4, 5), the role of IgA and IgM antibody determination remains unclear.
None of the three participants who were excluded from the main analysis because their COVID-19 disease was not confirmed by PCR but by an unknown antibody assay showed neutralizing antibodies. This observation highlights the danger of diagnosing COVID-19 based on antibody detection with apparently insufficient assays.
In this study, only one assay (Serion ELISA agile SARS-CoV-2 IgG) showed a sensitivity and specificity of greater than 98% and may hence be suitable for a large-scale assessment of sera for neutralizing activity against SARS-CoV-2, for the clarification of doubtful PCR results, and for a retrospective diagnosis of COVID-19 disease in patients with typical post-COVID-19-symptoms as well as for epidemiological investigations. Future studies are important to determine immunoassay cutoffs in correlation with immunity to reinfection as antibody testing may then be used as a decision criterion for (re)vaccinations.
ACKNOWLEDGMENTS
We thank Evgeny Golubtsov for sample collection support. We thank Florian Krammer, Icahn School of Medicine, Mount Sinai Hospital, New York, NY, for providing the RBD expression plasmid; Gert Zimmer, Markus Hoffmann, and Stefan Pöhlmann for providing reagents and protocols for pseudovirus generation; and Franziska Seifert for expert technical assistance with the bead-based assay. We thank Lukas B. Krone, Department of Physiology, Anatomy & Genetics, Oxford University, for careful language editing.
This study was funded by the Federal Ministry for Education and Science (BMBF) within the program InfectControl (project COVMon, grant no. 03COV26A) and the Free State of Bavaria with COVID research funds provided to the University of Wuerzburg, Germany.
M. Krone, T.-T. Lâm, C. Schoen, U. Vogel, A. Stich, O. Kurzai, and A. Schubert-Unkmeir carried out the sample collection. J. Gütling carried out the immunoassays and collected the questionnaire data. S. Klingler, S. Backes, and G. Gasteiger established and carried out the (micro)neutralization tests. N. Beyersdorf and T. Kerkau established the bead-based test. F. Wedekink and J. Wischhusen produced recombinant RBD protein. J. Wagener, L. Dölken, O. Kurzai, and A. Schubert-Unkmeir contributed to the conception and design of the study. M. Krone and A. Schubert-Unkmeir analyzed the data. O. Kurzai raised study funding. O. Kurzai and A. Schubert-Unkmeir supervised the study. M. Krone and A. Schubert-Unkmeir wrote the first draft of the manuscript. J. Gütling, J. Wagener, T.-T. Lâm, C. Schoen, U. Vogel, A. Stich, F. Wedekink, J. Wischhusen, T. Kerkau, N. Beyersdorf, S. Klingler, S. Backes, L. Dölken, G. Gasteiger, and O. Kurzai reviewed and modified the manuscript and approved its final version.
None of the authors has any conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Alexandra Schubert-Unkmeir, Email: aunkmeir@hygiene.uni-wuerzburg.de.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.World Health Organization. 2021. WHO coronavirus disease (COVID-19) dashboard. World Health Organization, Geneva, Switzerland. https://covid19.who.int/. Accessed 7 February 2021.
- 2.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kampf G, Todt D, Pfaender S, Steinmann E. 2020. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect 104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2021. Interim guidelines for COVID-19 antibody testing. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html. Accessed 17 March 2021.
- 5.European Centre for Disease Prevention and Control. 11June2020. Diagnostic testing and screening for SARS-CoV-2. European Centre for Disease Prevention and Control, Solna, Sweden. https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing.
- 6.Krüger S, Leskien M, Schuller P, Prifert C, Weißbrich B, Vogel U, Krone M. 2021. Performance and feasibility of universal PCR admission screening for SARS-CoV-2 in a German tertiary care hospital. J Med Virol 93:2890–2898. doi: 10.1002/jmv.26770. [DOI] [PubMed] [Google Scholar]
- 7.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma SA, Lyons AM, Li S, Wong AW-Y, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington LA, Loudermilk RP, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol 38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieri M, Nuccetelli M, Nicolai E, Sarubbi S, Grelli S, Bernardini S. 2021. Clinical validation of a second generation anti-SARS-CoV-2 IgG and IgM automated chemiluminescent immunoassay. J Med Virol 93:2523–2528. doi: 10.1002/jmv.26809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batra R, Olivieri LG, Rubin D, Vallari A, Pearce S, Olivo A, Prostko J, Nebbia G, Douthwaite S, Rodgers M, Cloherty G. 2020. A comparative evaluation between the Abbott Panbio™ COVID-19 IgG/IgM rapid test device and Abbott Architect™ SARS CoV-2 IgG assay. J Clin Virol 132:104645. doi: 10.1016/j.jcv.2020.104645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawandi A, Danner RL. 2020. Antibody tests have higher sensitivity at ≥15 days after symptom onset and 99% specificity for detecting SARS-CoV-2. Ann Intern Med 173:JC57. doi: 10.7326/ACPJ202011170-057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jääskeläinen AJ, Kuivanen S, Kekäläinen E, Ahava MJ, Loginov R, Kallio-Kokko H, Vapalahti O, Jarva H, Kurkela S, Lappalainen M. 2020. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 129:104512. doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padoan A, Bonfante F, Pagliari M, Bortolami A, Negrini D, Zuin S, Bozzato D, Cosma C, Sciacovelli L, Plebani M. 2020. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine 62:103101. doi: 10.1016/j.ebiom.2020.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdivia A, Torres I, Latorre V, Francés-Gómez C, Albert E, Gozalbo-Rovira R, Alcaraz MJ, Buesa J, Rodríguez-Díaz J, Geller R, Navarro D. 2021. Inference of SARS-CoV-2 spike-binding neutralizing antibody titers in sera from hospitalized COVID-19 patients by using commercial enzyme and chemiluminescent immunoassays. Eur J Clin Microbiol Infect Dis 40:485–494. doi: 10.1007/s10096-020-04128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strömer A, Rose R, Grobe O, Neumann F, Fickenscher H, Lorentz T, Krumbholz A. 2020. Kinetics of nucleo- and spike protein-specific immunoglobulin G and of virus-neutralizing antibodies after SARS-CoV-2 infection. Microorganisms 8:1572. doi: 10.3390/microorganisms8101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaplin JW, Chappell CP, Clark EA. 2013. Targeting antigens to CD180 rapidly induces antigen-specific IgG, affinity maturation, and immunological memory. J Exp Med 210:2135–2146. doi: 10.1084/jem.20130188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. 2020. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notz Q, Schmalzing M, Wedekink F, Schlesinger T, Gernert M, Herrmann J, Sorger L, Weismann D, Schmid B, Sitter M, Schlegel N, Kranke P, Wischhusen J, Meybohm P, Lotz C. 2020. Pro- and anti-inflammatory responses in severe COVID-19-induced acute respiratory distress syndrome-an observational pilot study. Front Immunol 11:581338. doi: 10.3389/fimmu.2020.581338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlesinger T, Weißbrich B, Wedekink F, Notz Q, Herrmann J, Krone M, Sitter M, Schmid B, Kredel M, Stumpner J, Dölken L, Wischhusen J, Kranke P, Meybohm P, Lotz C. 2020. Biodistribution and serologic response in SARS-CoV-2 induced ARDS: a cohort study. PLoS One 15:e0242917. doi: 10.1371/journal.pone.0242917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Asthagiri Arunkumar G, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer D, Amarilis Lugo L, Milunka Kojic E, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, Garcia-Sastre A, Caplivski D, Cheng A, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, Cornaby C, Bartelt L, Weiss S, Park Y, Edwards CE, Weimer E, Scherer EM, Rouphael N, Edupuganti S, Weiskopf D, Tse LV, Hou YJ, Margolis D, Sette A, Collins MH, Schmitz J, Baric RS, de Silva AM. 2020. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcombe RG. 1998. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med 17:857–872. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Herbert R. 24March2011, last update. Confidence interval calculator. https://www.pedro.org.au/wp-content/uploads/CIcalculator.xls. Accessed 2020 January 14.
- 27.Chen W, Zhang J, Qin X, Wang W, Xu M, Wang LF, Xu C, Tang S, Liu P, Zhang L, Liu X, Zhang Y, Yi C, Hu Z, Yi Y. 2020. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomed Pharmacother 130:110629. doi: 10.1016/j.biopha.2020.110629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshita M, Nishina N, Moriyama S, Takahashi Y, Uwamino Y, Nagata M, Aoki W, Masaki K, Ishii M, Saya H, Kondo Y, Kaneko Y, Suzuki K, Fukunaga K, Takeuchi T, Keio Donner project . 2021. Incomplete humoral response including neutralizing antibodies in asymptomatic to mild COVID-19 patients in Japan. Virology 555:35–43. doi: 10.1016/j.virol.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu LP, Wang NC, Chang YH, Tian XY, Na DY, Zhang LY, Zheng L, Lan T, Wang LF, Liang GD. 2007. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis 13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, Yu F, Ge SX, Zou QD, Yuan Q, Lin S, Hong CM, Yao XY, Zhang XJ, Wu DH, Zhou GL, Hou WH, Li TT, Zhang YL, Zhang SY, Fan J, Zhang J, Xia NS, Chen Y. 2020. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J 56:2000763. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, et al. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. 2020. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58:e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong JY, Wong A, Zhu D, Fastenberg JH, Tham T. 2020. The prevalence of olfactory and gustatory dysfunction in COVID-19 patients: a systematic review and meta-analysis. Otolaryngol Head Neck Surg 163:3–11. doi: 10.1177/0194599820926473. [DOI] [PubMed] [Google Scholar]
- 36.Lechien JR, Chiesa-Estomba CM, De Siati DR, Horoi M, Le Bon SD, Rodriguez A, Dequanter D, Blecic S, El Afia F, Distinguin L, Chekkoury-Idrissi Y, Hans S, Delgado IL, Calvo-Henriquez C, Lavigne P, Falanga C, Barillari MR, Cammaroto G, Khalife M, Leich P, Souchay C, Rossi C, Journe F, Hsieh J, Edjlali M, Carlier R, Ris L, Lovato A, De Filippis C, Coppee F, Fakhry N, Ayad T, Saussez S. 2020. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol 277:2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download JCM.00319-21-s0001.pdf, PDF file, 27 KB (26.2KB, pdf)