ABSTRACT
Mycobacterium abscessus is a rapidly growing nontuberculous mycobacterial species that comprises three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii. These predominantly environmental microorganisms have emerged as life-threatening chronic pulmonary pathogens in both immunocompetent and immunocompromised patients, and their acquisition of macrolide resistance due to the erm(41) gene and mutations in the 23S rrl gene has dramatically impacted patient outcome. However, standard microbiology laboratories typically have limited diagnostic tools to distinguish M. abscessus subspecies, and the testing for macrolide resistance is often not done. Here, we describe the development of a real-time multiplex assay using molecular beacons to establish a robust, rapid, and highly accurate method to both distinguish M. abscessus subspecies and to determine which strains are susceptible to macrolides. We report a bioinformatic approach to identify robust subspecies sequence targets, the design and optimization of six molecular beacons to identify all genotypes, and the development and application of a 2-tube 3-color multiplex assay that can provide clinically significant treatment information in less than 3 h.
KEYWORDS: Mycobacterium abscessus, macrolide resistance, molecular beacon
INTRODUCTION
Mycobacterium abscessus is a rapidly growing nontuberculous mycobacterial species that comprises three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii (1–3). These predominantly environmental microorganisms are opportunistic pathogens that have been associated with skin and soft tissue infections resulting from contaminated equipment and hospital water supplies; but, more recently, they have emerged as life-threatening chronic pulmonary pathogens in both immunocompetent and immunocompromised patients (1, 3, 4). Among persons with cystic fibrosis (CF), M. abscessus has become a significant cause of chronic lung infections, and treatment is confounded by their intrinsic resistance to antibiotics, including antituberculosis agents. The inability to successfully treat M. abscessus infections with macrolides, clarithromycin, or azithromycin, as a result of the erm(41) gene and mutations in the 23S rrl gene, has dramatically impacted patient outcomes, as cure rates drop to 25 to 40% against clarithromycin-resistant M. abscessus (5–8).
As a result of the increased number of high-risk immunocompromised patient populations, including bone marrow transplant patients, those receiving solid organs, and those with underlying lung disease, such as cystic fibrosis patients, there is an increase in M. abscessus chronic infections and a dire clinical need to improve patient treatment. Macrolide antibiotics remain one of the most effective classes of antibiotics against susceptible M. abscessus strains; however, standard microbiology laboratories typically have limited diagnostic tools to distinguish subspecies of M. abscessus, and the testing for macrolide resistance is often not done (9).
There is significant literature that links subspecies of M. abscessus and genotypic resistance to macrolides, including the important observation that nearly all M. abscessus subsp. massiliense strains have two deletions (one of 2 bp and the other of 274 bp, bases 64 to 65 and 159 to 432, respectively) in the erm(41) gene that lead to susceptibility to clarithromycin (10). Similarly, there are clinical isolates of M. abscessus subsp. abscessus with a single base change from T to C in codon 28 in the erm(41) gene that correlates with clarithromycin susceptibility (11).
The diagnostic challenge of developing a rapid and simple assay to both distinguish M. abscessus subspecies and genotype macrolide resistance revolves around the requirement to sequence the various subspecies and drug susceptibility gene targets. A number of examples, including genetic changes in rpoB, hsp65, and secA1, have been used to distinguish M. abscessus subspecies with relatively high accuracy (12–14), but, in each case, the assay requires PCR amplification followed by DNA sequencing. The same issue applies to defining the resistance mutations in the 23S rrl gene and the erm(41) genetic alterations that are associated with macrolide susceptibility in M. abscessus subsp. abscessus and M. abscessus subsp. massiliense. PCR approaches that require gel electrophoresis have been developed (15, 16); but, these research techniques are labor intensive, are subjective in interpreting the results, and are not amenable to the routine clinical microbiology laboratory.
Here, we describe the development of a real-time multiplex assay using molecular beacons to establish a robust, rapid, and highly accurate method to both distinguish M. abscessus subspecies and determine which strains are susceptible to macrolides. We report a bioinformatic approach to identify robust subspecies sequence targets, the design and optimization of six molecular beacons to identify all genotypes, and the development and application of a 2-tube 3-color multiplex assay that can provide clinically significant treatment information in less than 3 h.
MATERIALS AND METHODS
Clinical isolates and clarithromycin testing.
A collection of 115 M. abscessus clinical isolates, including 100 recovered from patients with cystic fibrosis and catalogued and archived at National Jewish Health, was utilized for this study. The strains included 69 M. abscessus subsp. abscessus strains, 38 M. abscessus subsp. massiliense strains, and 10 M. abscessus subsp. bolletii strains. All strains were analyzed by whole-genome sequencing (BioProject accession no. PRJNA319839 and PRJNA549322). A collection of 22 strains from 20 different nontuberculous Mycobacterium species, including M. aurum, M. avium, M. gallinarum, M. gastri, M. gordonae, M. haemophilum, M. intracellulare, M. marinum, M. scrofulaceum, M. simiae, M. szulgai, M. terrae, M. ulcerans, M. chelonae, M. fortuitum, M. kansasii, M. phlei, M. simiae, plus M. tuberculosis and M. microti, archived at the Center for Discovery and Innovation, was used to evaluate the specificity of the assay.
A panel of six test strains, which includes all three subspecies and genotypic diversity associated with clarithromycin resistance, was analyzed for response to clarithromycin. Susceptibility and resistance were assessed according to the CLSI recommendations (17, 18). Constitutive clarithromycin resistance was defined by a MIC of ≥8 μg/ml at day 5. Inducible resistance was defined by an increase in clarithromycin MIC from ≤2 μg/ml at day 5 to ≥8 μg/ml at day 14 (17, 18).
Design of the molecular probes and PCR primers.
For the identification and development of subspecies-specific sequence targets that are amenable for PCR and molecular beacon probe detection, we interrogated 33 completely sequenced M. abscessus genomes from the NCBI (www.ncbi.nlm.nih.gov/assembly, as of January 2020), including 12 M. abscessus subsp. abscessus, 19 M. abscessus subsp. massiliense, and 2 M. abscessus subsp. bolletii genomes. The genomes were aligned by Parsnp from the Harvest suite, and subspecies-specific single nucleotide polymorphisms (SNPs) were extracted. A core genome analysis was conducted using Roary (19) to identify conserved genes across different M. abscessus subspecies. Core SNP numbers within a 20-nucleotide (nt) sliding window across M. abscessus core genes were calculated using Bedtools (20) to identify putative regions for molecular beacon probe design.
We next used blastn to examine the in silico sensitivity and specificity of putative molecular beacon probes and their neighboring sequences against 1,930 Mycobacterium abscessus complex (MABC) draft genomes in the NCBI assembly database (www.ncbi.nlm.nih.gov/assembly) and among our in-house sequenced genomes (21). In addition, the specificity of the molecular beacons and primers was also examined against 476 representative or completely sequenced genomes of nontuberculous mycobacterial species in the NCBI Refseq database (www.ncbi.nlm.nih.gov/refseq/), including genomes from 119 unique mycobacterial species or subspecies.
Acquired macrolide resistance in M. abscessus is largely associated with the point mutations A2058 and A2059 in the rrl gene encoding the peptidyltransferase domain of the 23S rRNA (22, 23), and inducible macrolide resistance is linked to the ribosomal methylase gene erm(41) (10). The erm(41) gene with a T-to-C polymorphism at nucleotide 28 is associated with macrolide susceptibility. Currently, most clinical isolates of M. abscessus subsp. abscessus and M. abscessus subsp. bolletii express inducible macrolide resistance; but, in contrast, M. abscessus subsp. massiliense is nearly always associated with clarithromycin susceptibility due to two deletions (bases 64 to 65 and 159 to 432) within erm(41). This association correlates with a favorable clinical outcome with macrolide treatment (24). In this study, we designed and validated three sets of primer pairs and molecular beacon probes to detect the rrl macrolide susceptible (wild-type) gene, the erm(41) deletion, and the erm(41) T28C mutation in order to specifically and rapidly genotype macrolide resistance in different M. abscessus subspecies. A region in gene MAB2248 and two regions in MAB2830 were used to design three primer pairs and molecular beacon probes to specifically identify M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense, respectively (see below).
PCR primers were designed utilizing Primer-BLAST (25). Molecular beacon probes were designed to identify genes or species-specific single nucleotide polymorphisms using guidelines described by Vet and Marras (26). PCR primers and molecular beacon probes were evaluated with Integrated DNA Technologies (IDT)’s OligoAnlyzer tool software (https://www.idtdna.com/calc/analyzer) for compatibility in multiplex PCR assays. PCR primers were obtained from Integrated DNA Technologies (Coraville, IA), and molecular beacon probes were obtained from LGC Biosearch Technologies (Novato, CA). The sequences of the PCR primers and molecular beacon probes are listed in Table 1.
TABLE 1.
Molecular beacon probe and primer sequences
| Oligonucleotidea | Sequence (5′–3′)b |
|---|---|
| MAB2248 for | GTGACCGGTCTCGATCAGTT |
| MAB2248 rev | CATTGTCTGGCGTCAATGGC |
| MAB2248 bol MB | CFR610-CCGTGC CTCGGGAACCGGATGACT GCACGG-BHQ-2 |
| MAB2830 for | CCTCATCGAGGACGGTCAGA |
| MAB2830 rev | CACGAATCCGGGCAGCAATA |
| MAB2830 abs MB | FAM-CCGTCG CGAGGCCGGCATCGGCGCA CGACGG-BHQ-1 |
| MAB2830 mas MB | Q670-CCGTCG GAAGCTGGTATCGGTCCAGG CGACGG-BHQ-2 |
| 23S rrl for | CGGCGAAATTGCACTACGAG |
| 23S rrl rev | CCGAACCAAACGCCAATACC |
| 23S rrl WT MB | FAM-CGCTGC CAGGACGAAAAGACCCC GCAGCG-BHQ-1 |
| Δerm(41) for | TTCAGGGGAGTTCGTTGTGA |
| Δerm(41) rev | AAAGTGCTTCGCGGCAATG |
| Δerm(41) MB | CFR610-CACGG TGGACGCTCCGGGC CCGTG-BHQ-2 |
| erm(41) T28C for | GCATGCCCCGATATCTTTGG |
| erm(41) T28C rev | ATCCACAACGAACTCCCCTG |
| erm(41) T28C MB | Q670-CGCTG ACGCCAGCGGGGCTG CAGCG-BHQ-2 |
Forward (for) and reverse (rev) primers and molecular beacon probe (MB) sequences for each of the target genes. abs, M. abscessus subsp. abscessus; bol, M. abscessus subsp. bolletii; mas, M. abscessus subsp. massiliense; WT, wild-type.
BHQ-1, Blackhole Quencher 1; BHQ-2, Blackhole Quencher 2; CFR610, Cal Fluor Red 610; FAM, fluorescein; Q670, Quasar 670; underlined nucleotides represent arm sequences of the molecular beacon probe.
Denaturation profile analysis.
Molecular beacon probes were characterized in a denaturation profile analysis to determine their specificity. For each molecular beacon probe, dependent on the number of DNA target sequences they have to discriminate (wild-type gene sequences versus mutant gene sequences or multiple species to distinguish), three or four 20-μl reaction volumes were prepared containing 1× Platinum Hot Start PCR master mix (Thermo Fisher Scientific, Waltham, MA) and 400 nM molecular beacon probe. To one tube, 1,000 nM final concentration of the matched reverse complement target oligonucleotide was added, and 1,000 nM final concentration of mismatched reverse complement targets was added to other tubes (corresponding to the DNA sequences to be discriminated against). As a control, the target oligonucleotide was left out of one tube. The denaturation profile analysis was performed in 200-μl white polypropylene PCR tubes (USA Scientific, Ocala, FL) in a CFX96 Touch real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). The hybridization mixtures were incubated for 2 min at 95°C to denature the oligonucleotides and molecular beacon probes, followed by a decrease in temperature from 90°C to 30°C in −1°C steps and holding each step for 10 s while monitoring the fluorescence intensity of the molecular beacon probes.
Assay design.
A two-tube real-time PCR assay was developed to be able to both distinguish the three M. abscessus subspecies and identify genetic changes in the erm(41) and 23S rrl genes that predict macrolide resistance. In PCR Assay I, molecular beacons were targeted to the wild-type 23S rrl sequence, the subspecies M. bolletii-specific sequence, and the erm(41) T28C mutation and labeled with fluorophores: (FAM) fluorescein, (CFR610) Cal Fluor Red 610, and (Q670) Quasar 670, respectively. In Assay II, molecular beacons were designed for M. abscessus subspecies abscessus, the erm(41) deletion specific for the M. massiliense subspecies, and the sequence-specific target that distinguishes the M. massiliense subspecies. The three molecular beacons were labeled with fluorophores: (FAM) fluorescein, (CFR610) Cal Fluor Red 610, and (Q670) Quasar 670, respectively. The molecular beacon probes and the PCR primers for the three subspecies are listed in Table 1 as well as the primers and molecular beacon probes to identify the T28C mutation in erm(41), the unique deletion of erm(41) found in M. abscessus subsp. massiliense, and the wild-type target sequence in the 23S rrl gene.
Real-time polymerase chain reactions (RT-PCR).
For each DNA sample, two RT-PCR assays were performed. One assay (Assay I) contained PCR primers and molecular beacon probes to determine the presence of M. abscessus subsp. bolletii, the wild-type gene for 23S rrl 2058-2059, and the erm(41) T28C mutation. The second assay (Assay II) contained PCR primers and molecular beacon probes to determine the presence of either M. abscessus subsp. abscessus or M. abscessus subsp. massiliense and the erm(41) deletion. Each RT-PCR assay was carried out in 20-μl volumes that contained 1× Platinum Hot Start PCR master mix (Thermo Fisher Scientific, Waltham, MA). For Assay I, 250 nM MAB2248 forward (for), 250 nM MAB2248 reverse (rev), 500 nM MAB2248 M. abscessus subsp. bolletii (bol) molecular beacon probe (MB), 250 nM 23S rrl for, 250 nM 23S rrl rev, 500 nM 23S rrl MB, 100 nM erm(41) T28C for, 1,000 nM erm(41) T28C rev, and 500 nM erm(41) T28C MB were added. For Assay II, 250 nM MAB2830 for, 250 nM MAB2830 rev, 500 nM MAB2830 M. abscessus subsp. abscessus (abs) MB, 250 nM MAB2830 M. abscessus subsp. massiliense (mas) MB, 250 nM Δerm(41) for, 250 nM Δerm(41) rev, and 500 nM Δerm(41) MB was added. Each assay was initiated with a 5-μl DNA template. The PCR assays were performed in 200-μl white polypropylene PCR tubes (USA Scientific, Ocala, FL) in a CFX96 Touch real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). The reaction mixtures were incubated for 2 min at 95°C to activate the DNA polymerase, followed by 40 thermal cycles that consisted of denaturation at 95°C for 15 s and annealing and chain elongation at 60°C for 60 s. Molecular beacon fluorescence intensity was monitored during the 60°C annealing and chain elongation stage of each thermal cycle.
RESULTS
Target selection and in silico sensitivity and specificity.
Genomic analysis of 33 whole-genome sequenced (WGS) M. abscessus genomes identified candidate genetic regions that were amenable to distinguish M. abscessus subspecies isolates using molecular beacon probes. Two targets, MAB2248 and MAB2830, were selected based on their conservation and specificity to differentiate the three subspecies. MAB2248 encodes a putative peptide synthetase MbtE protein, and MAB2830 is the dihydroorotase PyrC gene. An 18-bp sequence within the MAB2248 gene, specific to M. abscessus subsp. bolletii, was targeted for the molecular beacon probe to distinguish this subspecies. The MAB2248 beacon differs from the sequences of M. abscessus subsp. abscessus or M. abscessus subsp. massiliense by three SNPs (3/18, 16.7% variation), providing the specificity to detect M. abscessus subsp. bolletii in an allele-specific RT-PCR assay (Table 1). MAB2830 homologous sequences in M. abscessus subsp. abscessus and M. abscessus subsp. massiliense were used to design the two subspecies-specific molecular beacons, and the target regions can be amplified by the same PCR primer pairs in the two subspecies. An 18-bp MAB2830 sequence specific for M. abscessus subsp. abscessus was used as a molecular beacon probe, and this sequence is distinguishable from M. abscessus subsp. massiliense and M. abscessus subsp. bolletii by 4 to 5 SNPs (22.2 to 27.8% variations), respectively. Similarly, a 19-bp MAB2830 homolog sequence specific to M. abscessus subsp. massiliense was targeted for the design of the molecular beacon probe and differed from the MAB2830 sequences in M. abscessus subsp. abscessus and M. abscessus subsp. bolletii by 5 and 3 SNPs (16.7 to 27.8% variations), respectively (Table 1).
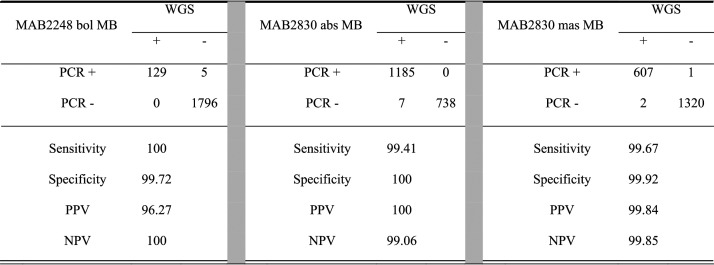
We then examined the in silico sensitivity and specificity of the three designed molecular beacon probes and primers against 1,930 M. abscessus genomes from NCBI and from our own collection. The 1,930 genomes were assigned to three subspecies, and they include 1,192 M. abscessus subsp. abscessus, 609 M. abscessus subsp. massiliense, and 129 M. abscessus subsp. bolletii genomes. MAB2248 and MAB2830 sequences were found in all 1,930 MABC genomes. The in silico sensitivities were 100%, 99.41%, and 99.67% for M. abscessus subsp. bolletii-, M. abscessus subsp. massiliense-, and M. abscessus subsp. abscessus-specific molecular beacons, and the specificities were 99.72%, 100%, and 99.92% for the three subspecies, respectively (Table 2).
TABLE 2.
In silico sensitivity and specificity of PCR probes used to identify subspeciesa
abs, M. abscessus subsp. abscessus; bol, M. abscessus subsp. bolletii; mas, M. abscessus subsp. massiliense; MB, molecular beacon probe; NPV, negative predictive value; PPV, positive predictive value; WGS, whole-genome sequencing.
With the intent of developing this assay to ultimately diagnose primary patient specimens, we checked the in silico specificity of the three subspecies molecular beacon probes and two primer pairs against 476 representative or completely sequenced Mycobacterium genomes deposited in the NCBI Refseq database. The results showed that the primers and probes are highly specific to M. abscessus strains, showing no or low sequence identities to other Mycobacterium species. Among the most clinically relevant nontuberculous Mycobacterium species listed in the Materials and Methods, the MAB2248 primer and probe sequences were absent in all but the five species that belong to the M. chelonae complex (M. chelonae, M. saopaulense, M. immunogenum, M. salmoniphilum, and M. franklinii), and, when present, the molecular beacon target region only showed 61 to 82% (4 to 7 mismatches) similarities. Similarly, the MAB2830 target was absent in most species and revealed 70 to 80% (4 to 6 mismatches) nucleotide identity against only genomes from M. chelonae, M. saopaulense, M. immunogenum, M. salmoniphilum, and M. franklinii. In addition, the two targets were absent in other CF- and lung-associated pathogens, including Staphylococcus aureus, Haemophilus influenzae, Pseudomonas aeruginosa, Burkholderia cepacia complex, and Stenotrophomonas maltophilia. The in silico specificity examination of the primers and probes for the erm(41) T28C mutation and the deletion were specific for the M. abscessus genomes, and there are no sequence matches against other queried genomes. The 23S rrl primers and molecular beacon were designed to target the wild-type alleles (A2058 and A2059), and this region showed 100% identity to other species of M. chelonae, M. saopaulense, M. immunogenum, M. salmoniphilum, and M. franklinii.
Denaturation profile analysis.
Each molecular beacon design used in this assay was characterized in a denaturation profile analysis to determine that they only elicit a fluorescent signal if their intended perfect-matched target is present at the annealing temperature of the PCR and remain nonfluorescent if a nonintended mismatched target is present at the annealing temperature of the PCR. For each of the three molecular beacon probes that distinguish the M. abscessus subspecies present in a sample, three oligonucleotide targets were designed (Table 3): one oligonucleotide is identical to the target region of the probe to M. abscessus subsp. abscessus, one to the target region of the M. abscessus subsp. massiliense probe, and one to the target region of the probe for M. abscessus subsp. bolletii. Each analysis also included a control in which no oligonucleotide target was added, and this control shows at which temperature the molecular beacon probe stem opens, resulting in an increase in the background fluorescence of the probe.
TABLE 3.
Denaturation profile analysis target oligonucleotides
| Oligonucleotidea | Sequence (5′–3′) |
|---|---|
| MAB2248 abs | CCGATCGGAGTCAACCGGTTCGCGGGCATCGAAC |
| MAB2248 bol | CCGATCGGAGTCATCCGGTTCCCGAGCATCGAAC |
| MAB2248 mas | CCGATCGGAGTCAACCGGTTCGCGGGCATCGAAC |
| MAB2830 abs | TCGATGCCTGCGCCGATGCCGGCCTCGATGCGGG |
| MAB2830 bol | TCGATGCCTGGGCCGATACCGGCCTCGATGCGGG |
| MAB2830 mas | TCGATGCCTGGACCGATACCGGCTTCGATGCGGG |
| 23S rrl 2058-2059 wt | AAGGTCCCGGGGTCTTTTCGTCCTGCCGCGCGT |
| 23S rrl 2058-2059 AG mut | AAGGTCCCGGGGTCTCTTCGTCCTGCCGCGCGT |
| 23S rrl 2058-2059 GA mut | AAGGTCCCGGGGTCTTCTCGTCCTGCCGCGCGT |
| erm(41) T28C wt | GCGGATACCAGCCCCACTGGCGTCGCGACCG |
| erm(41) T28C mt | GCGGATACCAGCCCCGCTGGCGTCGCGACCG |
| Δerm(41) wt | TGATTCCGGCCCGTAGCGTCCAATGG |
| Δerm(41) mut | TGATTCCGGCCCGGAGCGTCCAGCGG |
abs, M. abscessus subsp. abscessus; bol, M. abscessus subsp. bolletii; mas, M. abscessus subsp. massiliense; mut, mutant; wt, wild-type.
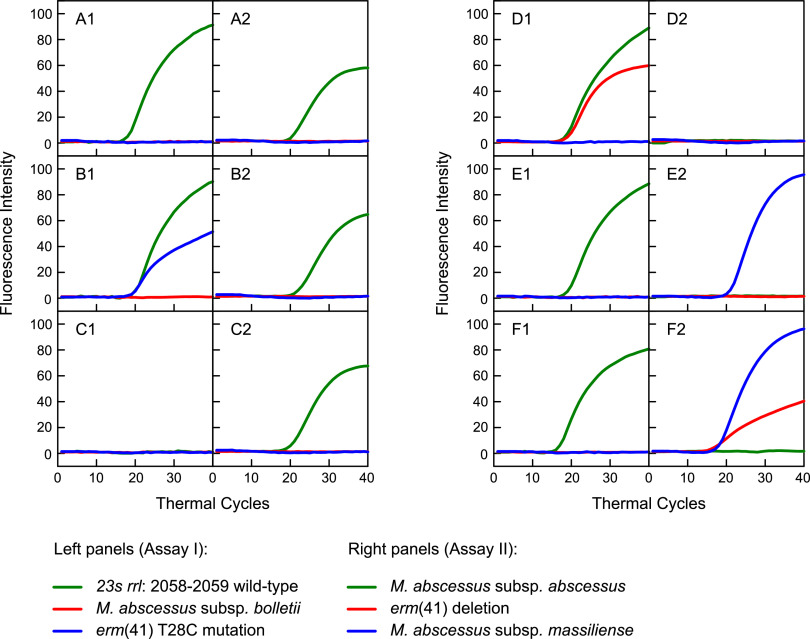
The top row in Fig. 1 shows the denaturation profiles of the three subspecies molecular beacon probes. The plots show that at the annealing temperature of the PCR assay (60°C, represented by a dashed line), a fluorescent signal only arises from the subspecies-specific molecular beacon probe and its intended perfect-matched target oligonucleotide, and that, at this temperature, the probes do not elicit a fluorescent signal from the other two mismatched targets. Furthermore, the stem of the molecular beacon probes is closed at this temperature, and there is no increased background fluorescence from the probes. Similar denaturation profile analyses were carried out for the three molecular beacon probes designed to identify the wild-type rrl gene, the erm(41) T28C mutation, and the erm(41) deletion. For these probes, oligonucleotide targets were designed as a perfect match for the intended target and with mismatches compared to nonintended targets. The bottom row of Fig. 1 shows the denaturation profiles of these three molecular beacon probes. For example, the left plot shows the denaturation profile for the wild-type rrl gene molecular beacon probe in the presence of its intended wild-type target (green curve), in the presence of the two possible mismatched targets, a GA mutant (red curve) or AG mutant (blue curve), and in the absence of any target (black curve) as a control to determine the stability of the stem hybrid of the probe. The plot shows that at the 60°C annealing temperature, a fluorescent signal from the probe only arises when its intended perfect-matched, wild-type target is present. At this temperature, the molecular beacon probe does not show any fluorescence in the presence of the two nonintended mutant targets, indicating the perfect discrimination of this molecular beacon probe between the wild-type and mutant targets. This is also the case for the other molecular beacon probes in this assay.
FIG 1.
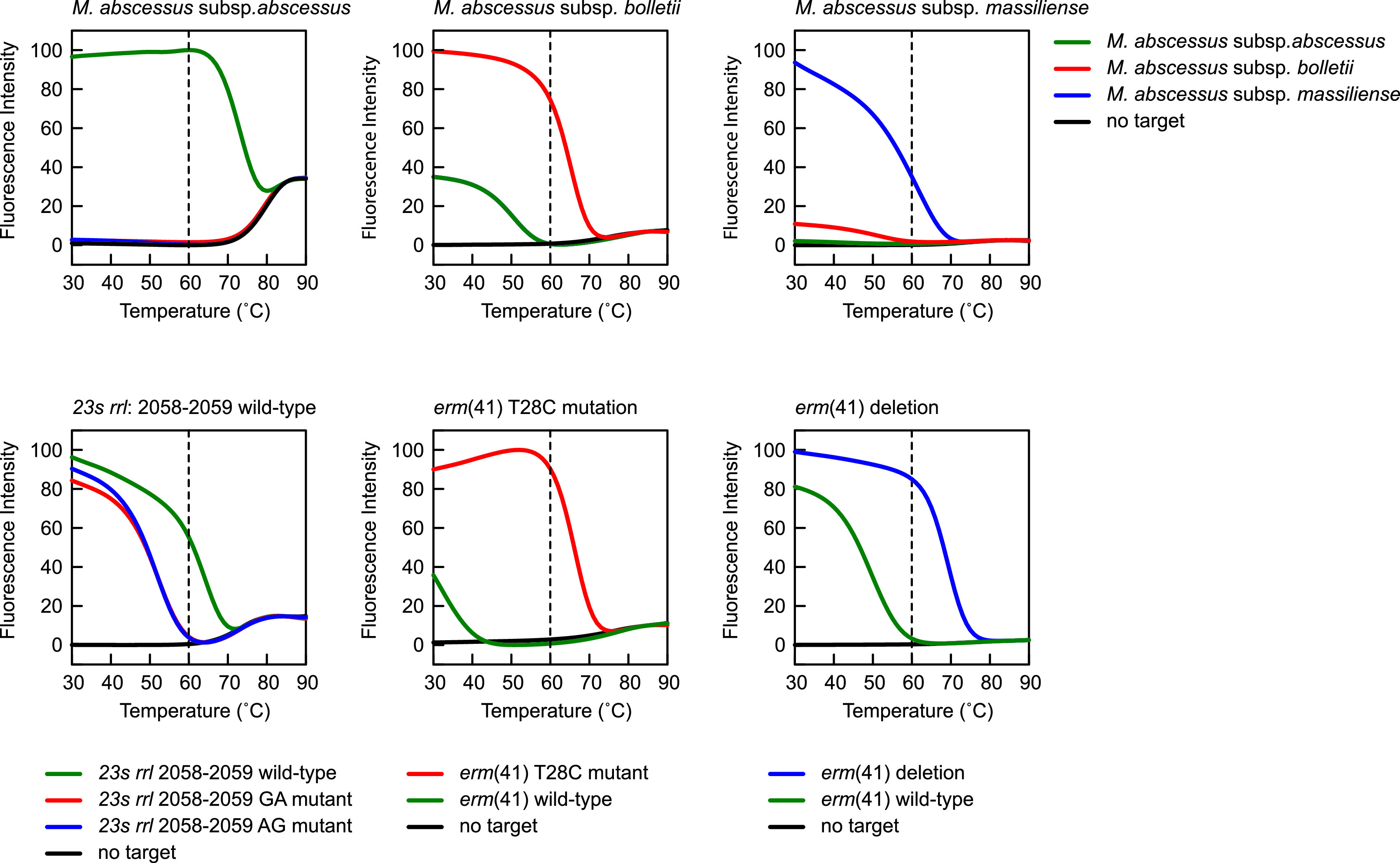
Denaturation profile analysis of the six molecular beacon probes utilized in this assay. For each molecular beacon, a denaturation profile analysis was carried out in the presence of its intended perfectly matched target sequence, its mismatched target sequence, and a no target control. The dashed vertical line in each plot shows that at the 60°C annealing temperature step in the PCR assay, where the fluorescence intensity of the molecular beacon probes is recorded, a fluorescent signal is only generated from the molecular beacon probe hybridized to its intended target, and no fluorescent signal is generated in the presence of a mismatched target or when no target is present.
Assay analysis.
Analysis of the real-time PCR results combines the information for both multiplex assays to predict the M. abscessus subspecies and to determine whether the strain is macrolide resistant or susceptible. The assay is designed with the understanding that all isolates have the erm(41) gene, and its wild-type structure correlates with inducible macrolide resistance. Therefore, those strains that do not have the T28C mutation or the 274-bp deletion are predictively resistant. Similarly, the wild-type 23S rrl gene is associated with macrolide susceptibility, and rare strains have a mutation that correlate with constitutive macrolide resistance. In the current assay, the absence of the rrl-positive molecular beacon predicts resistance.
The multiplex real-time PCR results of Assays I and II are shown in Fig. 2, and the genotypes of 6 representative clinical strains are described in Table 4. For each strain, the real-time PCR profiles of Assay I are shown in the left panel and the profiles of Assay II are shown in the right panel. The results for strain A reveal the presence of the wild-type rrl gene and the identification of M. abscessus subsp. abscessus. The interpretation is that this strain has the wild-type erm(41) gene, wild-type rrl gene, and, consequently, it has inducible macrolide resistance. The results for strain B show the wild-type rrl gene but also the presence of the erm(41) T28C mutation, indicating that this strain is susceptible to macrolides, and the positive FAM molecular beacon profile in the right panel indicates it is an M. abscessus subsp. abscessus strain. The absence of a positive molecular beacon profile for strain C in the left panel indicates that it has the rrl gene mutation, predictive of constitutive macrolide resistance, and the positive FAM molecular beacon profile in the right panel indicates it is an M. abscessus subsp. abscessus strain. The results for strain D reveal the wild-type rrl gene and a positive CFR610 molecular beacon fluorescence in the left panel, predictive of M. abscessus subsp. bolletii. The absence of any molecular beacon profile in the right panel confirms the subspecies identification, and the overall interpretation is that this is an M. abscessus subsp. bolletii strain with inducible resistance to macrolides as a result of having the wild-type erm(41) gene. The results for strains E and F reveal the wild-type rrl gene and a positive Q670 fluorescence in the right panel, predictive of M. abscessus subsp. massiliense. For strain E, the erm(41) gene shows a wild-type genotype, indicating that this strain has inducible resistance to macrolides, whereas strain F has the definitive deletion that is predictive of a macrolide-susceptible strain.
FIG 2.
Two-tube real-time PCR assay. Assay I, left plots (A1 to F1), determines if the sample contains the wild-type sequences for the 23S rrl gene at position 2058-2059 (green curves), if the sample contains M. abscessus subsp. bolletii (red curves), and if the sample contains the erm(41) T28C mutation (blue curves). Assay II, right plots (A2 to F2), determines if the sample contains M. abscessus subsp. abscessus (green curves) or M. abscessus subsp. massiliense (blue curves) and if the sample contains the erm(41) deletion (red curves). The following six different samples are shown: (A) M. abscessus subsp. abscessus, with a wild-type 23S rrl gene at position 2058-2059, no erm(41) T28C mutation, and no erm(41) deletion; (B) M. abscessus subsp. abscessus, with a wild-type 23S rrl gene at position 2058-2059, but with an erm(41) T28C mutation and no erm(41) deletion; (C) M. abscessus subsp. abscessus, with a mutation in the 23S rrl gene at position 2058-2059, no erm(41) T28C mutation, and no erm(41) deletion; (D) M. abscessus subsp. bolletii with a wild-type 23S rrl gene at position 2058-2059, no erm(41) T28C mutation, and no erm(41) deletion; (E) M. abscessus subsp. massiliense with a wild-type 23S rrl gene at position 2058-2059, no erm(41) T28C mutation, and no erm(41) deletion; (F) M. abscessus subsp. massiliense with a wild-type 23S rrl gene at position 2058-2059, no erm(41) T28C mutation, but with a deletion in the erm(41) gene.
TABLE 4.
Interpretation of real-time PCR results for six MABC isolates
| Strains | Real-time PCR Assay Ia |
Real-time PCR Assay IIa |
Results interpretation |
|||||
|---|---|---|---|---|---|---|---|---|
|
MAB2248 bol (MB) |
23S rrl WT (MB) |
erm(41) T28C (MB) |
MAB2830 abs (MB) |
MAB2830 mas (MB) |
Δerm(41) (MB) |
Subspecies | Macrolide phenotype | |
| A | − | + | − | + | − | − | abscessus | Inducible resistance |
| B | − | + | + | + | − | − | abscessus | Susceptible |
| C | − | − | − | + | − | − | abscessus | Constitutive resistance |
| D | + | + | − | − | − | − | bolletii | Inducible resistance |
| E | − | + | − | − | + | − | massiliense | Inducible resistance |
| F | + | + | + | + | + | + | massiliense | Susceptible |
+, positive amplification for the molecular beacon; −, negative amplification for the molecular beacon; abs, M. abscessus subsp. abscessus; bol, M. abscessus subsp. bolletii; mas, M. abscessus subsp. massiliense; MB, molecular beacon probe; WT, wild-type.
Technical sensitivity and specificity.
The sensitivity of the two-tube multiplex assay was determined by initiating PCR assays with different quantities of DNA obtained from strains B, D, and F. These strains were chosen because together they contain target regions for all six molecular beacon probes utilized in this assay. The amount of DNA added as a template to each of the reactions was calculated to be equivalent to 1,000, 100, 10, and 1 copies of genomic DNA. The results show that Assays I and II, for each strain, were able to detect and identify 1 or 10 copies of genomic DNA (data not shown).
We tested the specificity of the assay using DNA from 19 different Mycobacterium species. No positive amplifications were observed for MAB2248 and MAB2830 molecular beacon probes, which is consistent with our above in silico data-mining results. The bioinformatic and experimental approaches confirm that the assay is highly specific for the detection of M. abscessus. In addition, none of the 19-sample panel were positive for the erm(41) T28C mutation or erm(41) 274-bp deletion. For the 23S rrl molecular beacon, only DNA from M. chelonae gave a positive amplification with a threshold cycle (CT) value of <30 cycles, while other species only showed very late amplification signals (CT > 33). It is significant to note that the 23S rrl primer and molecular beacon regions are 100% identical between M. abscessus and M. chelonae, and, in agreement with the in silico data, the rrl RT-PCR result was positive.
Clinical isolate testing.
The two-tube multiplex assay was used to evaluate a large collection of genetically characterized clinical M. abscessus isolates from National Jewish Health, representing nontuberculous mycobacterial (NTM) isolates from CF centers across the United States, and from selected strains archived at the Center for Discovery and Innovation. Blinded DNA samples were assayed and compared to the subspecies and the macrolide genotype of each strain as determined by whole-genome sequencing. A summary of each of the molecular beacon results is shown in Table 5 for a total of 115 strains that were assayed. The results showed 100% specificity and sensitivity in determining the subspecies and the macrolide genotype, and the individual results are shown in Table S1 in the supplemental material.
TABLE 5.
Performance of the real-time PCR in 115 clinical MABC isolates in comparison to the whole-genome sequencing results
| Whole-genome sequencing analysis resultsa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abs |
Mas |
Bol |
23s rrl: 2058-2059 mutantsb |
erm(41) truncation |
erm(41) T28C mutation |
||||||||
| Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | ||
| MB-real-time PCR results | MAB2830 abs MB pos | 69 | 0 | ||||||||||
| MAB2830 abs MB neg | 0 | 46 | |||||||||||
| MAB2830 mas MB pos | 36 | 0 | |||||||||||
| MAB2830 mas MB neg | 0 | 79 | |||||||||||
| MAB2248 bol MB pos | 10 | 0 | |||||||||||
| MAB2248 bol MB neg | 0 | 105 | |||||||||||
| 23S rrl MB pos | 4 | 0 | |||||||||||
| 23S rrl MB neg | 0 | 111 | |||||||||||
| Δerm(41) MB pos | 35 | 0 | |||||||||||
| Δerm(41) MB neg | 0 | 80 | |||||||||||
| erm(41) T28C MB pos | 18 | 0 | |||||||||||
| erm(41) T28C MB neg | 0 | 97 | |||||||||||
abs, M. abscessus subsp. abscessus; bol, M. abscessus subsp. bolletii; mas, M. abscessus subsp. massiliense; MB, molecular beacon probe; neg, negative; pos, positive.
The 23S rrl MB was designed to detect wild-type 2058-2059 alleles, and mutations in 2058 or 2059 will fail to show an amplification curve. The negative amplification curve for 23S rrl MB is regarded as a 23S rrl 2058-2059 mutation result.
DISCUSSION
In addressing the need for a simple, yet comprehensive, assay to improve the treatment of M. abscessus, we have developed a robust and rapid molecular beacon diagnostic that can determine both the identification of the M. abscessus subspecies and the genotypic profile associated with macrolide resistance. Several sequencing-based methods using regular PCR followed by Sanger sequencing to characterize sequence variations among some housekeeping genes, including hsp65, rpoB, secA, and 16S, either individually or in combination, were developed previously to identify the three M. abscessus subspecies (12, 27, 28). Additionally, multiplex PCR-based targeted next-generation sequencing (29) and Cas12a/single guide RNA (sgRNA)-based nucleic acid detection (30) platforms were recently developed for distinguishing M. abscessus subspecies. Commercial nucleic acid amplification tests, such as the line probe GenoType NTM-DR assay, which distinguishes the M. abscessus subspecies and macrolide resistance, are also available (31, 32); however, all of these approaches involve post-PCR procedures, which require longer turnaround times. Due to the high level of genetic relatedness, it is a challenge to differentiate strains from M. abscessus based on a single gene sequence (33).
To facilitate our ability to select alternative targets for M. abscessus detection and identification of subspecies, we analyzed 1,930 M. abscessus genomes and selected two targets for SNP-based real-time PCR design. The subspecies identification PCR targets are highly specific for M. abscessus based on both in silico analysis and comparison of the public nontuberculous mycobacterial genomes and our experimental testing of representative isolates from clinically frequently encountered mycobacterial species, supporting their future use in testing primary specimens where other confounding species could be present.
The molecular epidemiology of the three subspecies varies significantly, and infections caused by M. abscessus subsp. bolletii are rarer than the other two subspecies (3, 34, 35). However, clinical disease and outbreaks caused by M. abscessus subsp. bolletii have been documented (34). In contrast to M. abscessus subsp. bolletii, M. abscessus subsp. abscessus and M. abscessus subsp. massiliense postsurgical wound infections and respiratory disease are frequently reported globally (36–40), and differences in clinical outcomes commonly correlate to the strain’s susceptibility to macrolides (41). Infections due to M. abscessus subsp. massiliense respond more favorably to macrolide therapy. However, as shown in the current study and in other publications, not all M. abscessus subsp. massiliense harbor the truncated erm(41), and M. abscessus subsp. massiliense isolates with a wild-type erm(41) have been described (15). The erm(41) C28 sequevar M. abscessus subsp. abscessus is associated with significantly higher culture conversion rates with treatment, since these isolates do not exhibit inducible resistance to macrolides (5). Finally, M. abscessus subsp. massiliense with the truncated erm(41) gene has been identified with mutations in the rrl 2058 and 2059 sites, making them fully resistant to macrolides (42). Collectively, identification of subspecies and genotyping macrolide susceptibility are necessary to guide “precise treatment” for clinical M. abscessus infections (41, 43). It is noted that our assay was designed to detect the most common mutations associated with macrolide resistance in MABC with the understanding that rare resistance mechanisms (44) could be targeted in the future and added to the assay if they become more prevalent among clinical isolates.
Besides the identification of subspecies, our assay has strong predictive ability to determine whether to use a macrolide in the patient’s treatment regimen. Compared to the 3 to 14 days needed for clarithromycin phenotypic susceptibility testing, our real-time PCR method can be completed in 2 to 3 h and provides timely and definitive laboratory results. Results from current and other studies (5, 45) also suggested that the erm(41) gene truncation is not a reliable marker to distinguish M. abscessus subsp. massiliense. Importantly, our assay showed 100% specificity and sensitivity among 115 tested M. abscessus strains from multiple health care facilities in the United States, further supporting its clinical application. The ability for health care facilities to extend their diagnostic capability in the treatment of M. abscessus infections would dramatically improve patient care by providing appropriate antibiotic treatment and establishing epidemiological markers to monitor and track M. abscessus infections.
ACKNOWLEDGMENTS
This work was in part supported by grants from the National Institutes of Health (R01AI141805) and the Cystic Fibrosis Foundation to B.N.K. C.L.D., R.M.D., and M.S. thank the Cystic Fibrosis Foundation for funding pertaining to the National Jewish Health NTM biobank and genomics. S.A.E.M. is partly supported by a grant from the National Cancer Institute, National Institutes of Health (number R01CA227291).
Footnotes
Supplemental material is available online only.
Contributor Information
Barry N. Kreiswirth, Email: Barry.Kreiswirth@hmh-cdi.org.
Melissa B. Miller, UNC School of Medicine
REFERENCES
- 1.Lee MR, Sheng WH, Hung CC, Yu CJ, Lee LN, Hsueh PR. 2015. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis 21:1638–1646. 10.3201/2109.141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tortoli E, Kohl TA, Brown-Elliott BA, Trovato A, Leao SC, Garcia MJ, Vasireddy S, Turenne CY, Griffith DE, Philley JV, Baldan R, Campana S, Cariani L, Colombo C, Taccetti G, Teri A, Niemann S, WallaceRJ, Jr., Cirillo DM. 2016. Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. nov. Int J Syst Evol Microbiol 66:4471–4479. 10.1099/ijsem.0.001376. [DOI] [PubMed] [Google Scholar]
- 3.Johansen MD, Herrmann JL, Kremer L. 2020. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 18:392–407. 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- 4.Cowman S, van Ingen J, Griffith DE, Loebinger MR. 2019. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J 54:1900250. 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 5.Koh WJ, Jeong BH, Kim SY, Jeon K, Park KU, Jhun BW, Lee H, Park HY, Kim DH, Huh HJ, Ki CS, Lee NY, Kim HK, Choi YS, Kim J, Lee SH, Kim CK, Shin SJ, Daley CL, Kim H, Kwon OJ. 2017. Mycobacterial characteristics and treatment outcomes in Mycobacterium abscessus lung disease. Clin Infect Dis 64:309–316. 10.1093/cid/ciw724. [DOI] [PubMed] [Google Scholar]
- 6.Koh WJ, Stout JE, Yew WW. 2014. Advances in the management of pulmonary disease due to Mycobacterium abscessus complex. Int J Tuberc Lung Dis 18:1141–1148. 10.5588/ijtld.14.0134. [DOI] [PubMed] [Google Scholar]
- 7.Smibert O, Snell GI, Bills H, Westall GP, Morrissey CO. 2016. Mycobacterium abscessus complex - a particular challenge in the setting of lung transplantation. Expert Rev Anti Infect Ther 14:325–333. 10.1586/14787210.2016.1138856. [DOI] [PubMed] [Google Scholar]
- 8.Guo Q, Chu H, Ye M, Zhang Z, Li B, Yang S, Ma W, Yu F. 2018. The clarithromycin susceptibility genotype affects the treatment outcome of patients with Mycobacterium abscessus lung disease. Antimicrob Agents Chemother 62:e02360-17. 10.1128/AAC.02360-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffith DE. 2014. Mycobacterium abscessus subsp abscessus lung disease: 'trouble ahead, trouble behind…'. F1000Prime Rep 6:107. 10.12703/P6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nash KA, Brown-Elliott BA, WallaceRJ, Jr. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother 53:1367–1376. 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maurer FP, Castelberg C, Quiblier C, Bottger EC, Somoskovi A. 2014. Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J Antimicrob Chemother 69:1559–1563. 10.1093/jac/dku007. [DOI] [PubMed] [Google Scholar]
- 12.Nie W, Duan H, Huang H, Lu Y, Bi D, Chu N. 2014. Species identification of Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii using rpoB and hsp65, and susceptibility testing to eight antibiotics. Int J Infect Dis 25:170–174. 10.1016/j.ijid.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, WallaceRJ, Jr., Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng A, Sun HY, Tsai YT, Chang SY, Wu UI, Hsueh PR, Sheng WH, Chen YC, Chang SC. 2019. Comparing the utilities of different multilocus sequence typing schemes for identifying outbreak strains of Mycobacterium abscessus subsp. massiliense. J Clin Microbiol 58:e01304-19. 10.1128/JCM.01304-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shallom SJ, Gardina PJ, Myers TG, Sebastian Y, Conville P, Calhoun LB, Tettelin H, Olivier KN, Uzel G, Sampaio EP, Holland SM, Zelazny AM. 2013. New rapid scheme for distinguishing the subspecies of the Mycobacterium abscessus group and identifying Mycobacterium massiliense isolates with inducible clarithromycin resistance. J Clin Microbiol 51:2943–2949. 10.1128/JCM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mase A, Yamaguchi F, Funaki T, Yamazaki Y, Shikama Y, Fukuchi K. 2020. PCR amplification of the erm(41) gene can be used to predict the sensitivity of Mycobacterium abscessus complex strains to clarithromycin. Exp Ther Med 19:945–955. 10.3892/etm.2019.8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI. 2018. Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes; 1st ed. CLSI M62. Wayne, PA. [PubMed] [Google Scholar]
- 18.CLSI. 2018. Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes; 3rd ed. CLSI M24. Wayne, PA. [PubMed] [Google Scholar]
- 19.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey R, Chen L, Manca C, Jenkins S, Glaser L, Vinnard C, Stone G, Lee J, Mathema B, Nuermberger EL, Bonomo RA, Kreiswirth BN. 2019. Dual beta-lactam combinations highly active against Mycobacterium abscessus complex in vitro. mBio 10:e02895-18. 10.1128/mBio.02895-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 23.Hanson KE, Slechta ES, Muir H, Barker AP. 2014. Rapid molecular detection of inducible macrolide resistance in Mycobacterium chelonae and M. abscessus strains: a replacement for 14-day susceptibility testing? J Clin Microbiol 52:1705–1707. 10.1128/JCM.03464-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux AL, Catherinot E, Soismier N, Heym B, Bellis G, Lemonnier L, Chiron R, Fauroux B, Le Bourgeois M, Munck A, Pin I, Sermet I, Gutierrez C, Véziris N, Jarlier V, Cambau E, Herrmann JL, Guillemot D, Gaillard JL. 2015. Comparing Mycobacterium massiliense and Mycobacterium abscessus lung infections in cystic fibrosis patients. J Cystic Fibros 14:63–69. 10.1016/j.jcf.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vet JA, Marras SA. 2005. Design and optimization of molecular beacon real-time polymerase chain reaction assays. Methods Mol Biol 288:273–290. 10.1385/1-59259-823-4:273. [DOI] [PubMed] [Google Scholar]
- 27.Adékambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 41:5699–5708. 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adékambi T, Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol 54:2095–2105. 10.1099/ijs.0.63094-0. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Xu L, Guo Q, Chen J, Zhang Y, Huang W, Zhang Z, Han L, Xu X, Chu H. 2020. GenSeizer: a multiplex PCR-based targeted gene sequencing platform for rapid and accurate identification of major Mycobacterium species. J Clin Microbiol 59:e00584-20. 10.1128/JCM.00584-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao G, Zhang S, Liang Z, Li G, Fang M, Liu Y, Zhang J, Ou M, He X, Zhang T, Zeng C, Liu L, Zhang G. 2020. Identification of Mycobacterium abscessus species and subspecies using the Cas12a/sgRNA-based nucleic acid detection platform. Eur J Clin Microbiol Infect Dis 39:551–558. 10.1007/s10096-019-03757-y. [DOI] [PubMed] [Google Scholar]
- 31.Huh HJ, Kim SY, Shim HJ, Kim DH, Yoo IY, Kang OK, Ki CS, Shin SY, Jhun BW, Shin SJ, Daley CL, Koh WJ, Lee NY. 2019. GenoType NTM-DR performance evaluation for identification of Mycobacterium avium complex and Mycobacterium abscessus and determination of clarithromycin and amikacin resistance. J Clin Microbiol 57:e00516-19. 10.1128/JCM.00516-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mougari F, Loiseau J, Veziris N, Bernard C, Bercot B, Sougakoff W, Jarlier V, Raskine L, Cambau E, French National Reference Center for Mycobacteria. 2017. Evaluation of the new GenoType NTM-DR kit for the molecular detection of antimicrobial resistance in non-tuberculous mycobacteria. J Antimicrob Chemother 72:1669–1677. 10.1093/jac/dkx021. [DOI] [PubMed] [Google Scholar]
- 33.Macheras E, Roux AL, Ripoll F, Sivadon-Tardy V, Gutierrez C, Gaillard JL, Heym B. 2009. Inaccuracy of single-target sequencing for discriminating species of the Mycobacterium abscessus group. J Clin Microbiol 47:2596–2600. 10.1128/JCM.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rollet-Cohen V, Roux AL, Le Bourgeois M, Sapriel G, El Bahri M, Jais JP, Heym B, Mougari F, Raskine L, Véziris N, Gaillard JL, Sermet-Gaudelus I. 2019. Mycobacterium bolletii lung disease in cystic fibrosis. Chest 156:247–254. 10.1016/j.chest.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Benwill JL, WallaceRJ, Jr. 2014. Mycobacterium abscessus: challenges in diagnosis and treatment. Curr Opin Infect Dis 27:506–510. 10.1097/QCO.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 36.Leão SC, Viana-Niero C, Matsumoto CK, Lima KV, Lopes ML, Palaci M, Hadad DJ, Vinhas S, Duarte RS, Lourenço MC, Kipnis A, das Neves ZC, Gabardo BM, Ribeiro MO, Baethgen L, de Assis DB, Madalosso G, Chimara E, Dalcolmo MP. 2010. Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil. Future Microbiol 5:971–980. 10.2217/fmb.10.49. [DOI] [PubMed] [Google Scholar]
- 37.Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, Tonelli MR, Cangelosi GA, Dirac MA, Olivier KN, Brown-Elliott BA, McNulty S, WallaceRJ, Jr. 2012. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med 185:231–232. 10.1164/ajrccm.185.2.231. [DOI] [PubMed] [Google Scholar]
- 38.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker AW, Lewis SS, Alexander BD, Chen LF, WallaceRJ, Jr., Brown-Elliott BA, Isaacs PJ, Pickett LC, Patel CB, Smith PK, Reynolds JM, Engel J, Wolfe CR, Milano CA, Schroder JN, Davis RD, Hartwig MG, Stout JE, Strittholt N, Maziarz EK, Saullo JH, Hazen KC, WalczakRJ, Jr., Vasireddy R, Vasireddy S, McKnight CM, Anderson DJ, Sexton DJ. 2017. Two-phase hospital-associated outbreak of Mycobacterium abscessus: investigation and mitigation. Clin Infect Dis 64:902–911. 10.1093/cid/ciw877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peralta G, Tobin-D'Angelo M, Parham A, Edison L, Lorentzson L, Smith C, Drenzek C. 2016. Notes from the field: Mycobacterium abscessus infections among patients of a pediatric dentistry practice–Georgia, 2015. MMWR Morb Mortal Wkly Rep 65:355–356. 10.15585/mmwr.mm6513a5. [DOI] [PubMed] [Google Scholar]
- 41.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis 71:905–913. 10.1093/cid/ciaa1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haworth CS, Banks J, Capstick T, Fisher AJ, Gorsuch T, Laurenson IF, Leitch A, Loebinger MR, Milburn HJ, Nightingale M, Ormerod P, Shingadia D, Smith D, Whitehead N, Wilson R, Floto RA. 2017. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax 72:ii1–ii64. 10.1136/thoraxjnl-2017-210927. [DOI] [PubMed] [Google Scholar]
- 44.Lipworth S, Hough N, Leach L, Morgan M, Jeffery K, Andersson M, Robinson E, Smith EG, Crook D, Peto T, Walker T. 2019. Whole-genome sequencing for predicting clarithromycin resistance in Mycobacterium abscessus. Antimicrob Agents Chemother 63:e01204-18. 10.1128/AAC.01204-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline: executive summary. Clin Infect Dis 71:e1–e36. 10.1093/cid/ciaa241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JCM.00455-21-s0001.xlsx, XLSX file, 14 KB (13.1KB, xlsx)