ABSTRACT
We investigated the molecular epidemiology of 21 carbapenem-resistant Acinetobacter baumannii isolates from Libya and assessed their relative fitness. Core genome multilocus sequence typing (MLST) revealed five interhospital transmission clusters. Three clusters were associated with the international clones (IC) IC1, IC2, and IC7. Carbapenem-resistance was associated with blaOXA-23, blaGES-11, or blaNDM-1. Compared to that of A. baumannii DSM 30008, the doubling time was similar over 10 h, but after 16 h, half the isolates grew to higher densities, suggesting a fitness advantage.
KEYWORDS: Acinetobacter baumannii, antimicrobial resistance, carbapenems, fitness cost
INTRODUCTION
Acinetobacter baumannii causes life-threatening hospital-acquired infections among immunocompromised and critically ill patients (1). In addition to being a common nosocomial pathogen, rising rates of A. baumannii infections correlate with armed conflicts, especially during periods of heightened combat intensity (2–6). Libya has experienced a protracted civil war since 2011. The aim of this study was to investigate the molecular epidemiology of A. baumannii isolates from patients hospitalized in Tripoli, Libya, to identify their carbapenem resistance mechanisms and to assess the fitness cost to these isolates.
A total of 23 A. baumannii isolates, confirmed using the gyrB multiplex PCR (7), were collected from patients at three hospitals in Tripoli, Libya (Table 1). Table 2 summarizes antimicrobial susceptibility testing results by broth microdilution according to CLSI guidelines (8). One isolate was resistant to colistin, 12 were resistant to amikacin and tetracycline, 16 to trimethoprim-sulfamethoxazole, 21 to meropenem and imipenem (91%), and 22 were resistant to levofloxacin. All were resistant to ceftazidime, cefepime, gentamicin, ciprofloxacin, and piperacillin-tazobactam. In a previous study from Libya, 107 isolates of 167 (64%) were carbapenem-resistant A. baumannii (CRAB) (9).
TABLE 1.
Summary of the 23 A. baumannii isolates and sources
| Isolate | Hospitala | Genderb | Age | Patientc | Wardd | Samplee | TCf | STg |
|---|---|---|---|---|---|---|---|---|
| N22 | BPSH | M | 43 yr | In | ICU | Wound | A | ST25 |
| N34 | TMC | F | 60 yr | In | RHM | Urine | ||
| N46 | BPSH | M | 32 yr | In | MICU | W/S | ||
| N98 | TMC | M | 70 yr | In | GSICU | Wound | ||
| N33 | BPSH | M | 37 yr | Out | ICU | W/S | ||
| N3 | BPSH | M | 35 yr | In | ICU | Wound | B | ST136 |
| N60 | BPSH | M | 32 yr | In | ICUB | W/S | ||
| N103 | BPSH | M | 25 yr | In | ICUB | W/S | ||
| N19 | BPSH | M | 32 yr | In | ICU | Wound | C | ST1 |
| N20 | BPSH | M | 27 yr | In | ICU | Wound | ||
| N44 | TMC | F | 40 yr | In | GSICU | FC tip | ||
| N13 | BPSH | M | 35 yr | In | ICU | Wound | Singleton | |
| N51 | TMC | M | 12 yr | In | PO | Blood | Singleton | |
| N54 | BPSH | M | 23 yr | In | ICUB | Blood | E | ST2 |
| N57 | TMC | M | 61 yr | In | GSICU | Sputum | ||
| N99 | TMC | F | Newborn | In | SCBU | Vaginal swab | ||
| N27 | BPSH | M | 25 yr | In | ICU | Wound | Singleton | |
| N52 | BPSH | M | 35 yr | In | ICUB | Blood | Singleton | |
| N25 | BPSH | M | 37 yr | In | ICU | Wound | Singleton |
BPSH, burn and pediatric surgery hospital; TMC, Tripoli Medical Center; CHT, Central Hospital of Tripoli.
M, male; F, female.
IN, inpatient; OUT, outpatient.
ICU, intensive care unit; RHM, rheumatology; MICU, medical ICU; GSICU, general surgery ICU; ICUB, ICU unit B; SCBU, special care baby unit; PO, Prenatal oncology.
W/S, wound swab; Throat/s, throat swab; FC tip, Foley catheter tip.
TC, transmission cluster.
ST, sequence type.
TABLE 2.
Antimicrobial susceptibility testing results of all the 23 A. baumannii isolates
| Antibiotic | No. of isolates resistant (%) | MIC (μg/ml) |
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Meropenem | 21 (91) | <1 to 128 | 32 | 64 |
| Imipenem | 21 (91) | 2 to 128 | 64 | 128 |
| Ceftazidime | 23 (100) | 32 to >1,024 | >1,024 | >1,024 |
| Cefepime | 23 (100) | 32-1,024 | 256 | 512 |
| Gentamicin | 23 (100) | 8 to >1,024 | 256 | >1,024 |
| Ciprofloxacin | 23 (100) | 8 to 256 | 32 | 64 |
| Levofloxacin | 22 (96) | 2 to 64 | 8 | 16 |
| Colistin | 1 (4) | <1 to 8 | <1 | <1 |
| Amikacin | 12 (52) | 2 to >1,024 | 32 | 64 |
| Piperacillin-tazobactam | 23 (100) | 128 to 1,024 | 512 | 1,024 |
| Trimethoprim-sulfamethoxazole | 16 (70) | 0.5 to >64 | 4 | >64 |
| Tetracycline | 12 (52) | <1 to 1,024 | 16 | 1,024 |
Whole-genome sequencing and core genome multilocus sequence typing (cgMLST) were performed as previously described (10). Pasteur sequence types were determined using the pubMLST website (https://pubmlst.org/abaumannii/). Resistomes were identified using ResFinder. The ADC and OXA variants were identified using the BLAST function of the beta-lactamase database (http://www.bldb.eu/). The novel insertion (IS) element ISAba58 was identified using ISfinder (https://www-is.biotoul.fr/). Mutations in gyrA and parC associated with fluoroquinolone resistance were detected manually.
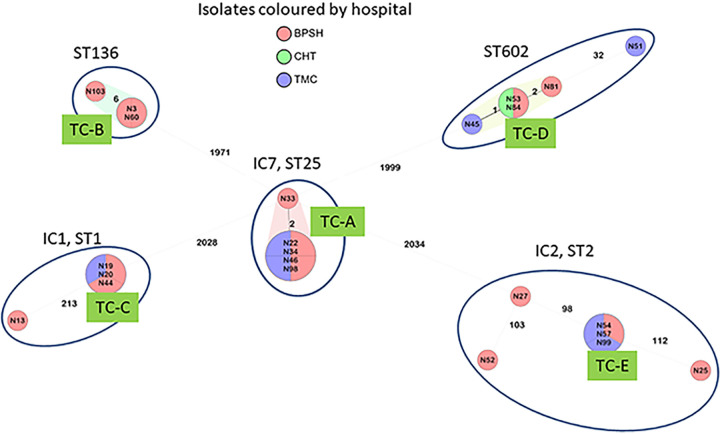
The isolates were assigned to five sequence types (STs) (Table 1), three of which are associated with international clones (IC): ST1 (IC1), ST2 (IC2), and ST25 (IC7). cgMLST results are summarized in Fig. 1. Transmission clusters (TCs) suggest interhospital transmissions, with TC-D involving all three hospitals. The five IC7 isolates formed a TC from five different wards at two of the three hospitals, suggesting patient movement or an endemic clone circulating between the hospitals. Interhospital TCs were also seen within the IC1, IC2, and the ST136 groups. However, not all isolates within a ST were part of a TC. Several studies have highlighted a wide diversity of STs associated with CRAB in Libya (11–13). Notably, ST25 was missing from previous studies, possibly reflecting a recent importation into Libya.
FIG 1.
cgMLST of the 23 A. baumannii isolates.
Table 3 summarizes the antimicrobial resistance determinants. Generally, isolates that clustered together had similar resistomes, with a few exceptions. The four identical isolates in TC-A differ from isolate N33 by 2 alleles, and N33 is also missing the sulfonamide resistance gene sul2. Isolate N45 (TC-D), while only one allele distant from N53 and N84, was carbapenem susceptible and was missing blaOXA-23 as well as aminoglycoside and macrolide resistance determinants.
TABLE 3.
Antibiotic resistance determinants and GyrA/ParC substitutionsa
| Isolate | Beta-lactam gene(s) | Aminoglycoside gene(s) | Sulphonamide gene(s) | FQ/AMG geneb | Phenicol gene | TET genec | TMP gened | Macrolide gene(s) | GyrA substitution | ParC substitution |
|---|---|---|---|---|---|---|---|---|---|---|
| N22 | blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, aph(3′)-VIa-like, strA, strB | sul2 | S83-L | S80-L | |||||
| N34 | blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, aph(3′)-VIa-like, strA, strB | sul2 | S83-L | S80-L | |||||
| N46 | blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, aph(3′)-VIa-like, strA, strB | sul2 | S83-L | S80-L | |||||
| N98 | blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, aph(3′)-VIa-like, strA, strB | sul2 | S83-L | S80-L | |||||
| N33 | blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, aph(3′)-VIa-like, strA-like, strB | S83-L | S80-L | ||||||
| N3 | blaGES-11, blaOXA-23, blaOXA-378 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N60 | blaGES-11, blaOXA-23, blaOXA-378 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N103 | blaGES-11, blaOXA-23, blaOXA-378 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N19 | blaGES-11, blaOXA-23, blaOXA-69 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N20 | blaGES-11, blaOXA-23, blaOXA-69 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N44 | blaGES-11, blaOXA-23, blaOXA-69 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | sul1 | aac(6′)Ib-cr-like | cmlA1-like | dfrA7 | S83-L | S80-L | ||
| N13 | blaCARB-8-like, blaOXA-23, blaOXA-69, blaTEM-1D | aac(3)-Ia-like, aadA1, aph(3′)-Ic, aph(3′)-VIa-like | sul1 | catA1-like | tet(A) | S83-L | S80-A | |||
| N45 | blaOXA-317-like, blaPER-1 | aadB-like, strA-like, strB | sul2 | tet(B) | S83-L | S80-L | ||||
| N53 | blaOXA-317-like, blaPER-1, blaOXA-23 | aadB-like, aph(3′)-VIa-like, aph(3′)-VIb, strA-like, strB-like | sul2 | tet(B) | mph(E), msr(E) | S83-L | S80-L | |||
| N81 | blaOXA-317-like, blaPER-1, blaOXA-23 | aadB-like, aph(3′)-VIa-like, aph(3′)-VIb-like, strA-like, strB-like | sul2 | tet(B) | mph(E), msr(E) | S83-L | S80-L | |||
| N84 | blaOXA-317-like, blaPER-1, blaOXA-23 | aadB-like, aph(3′)-VIa-like, aph(3′)-VIb, strA-like, strB | sul2 | tet(B) | mph(E), msr(E) | S83-L | S80-L | |||
| N51 | blaOXA-317-like, blaOXA-23, blaPER-1 | aadB-like, aph(3′)-VIa-like, strA-like, strB-like | sul2 | tet(B) | mph(E), msr(E) | S83-L | S80-L | |||
| N54 | blaNDM-1, blaOXA-66, blaTEM-1D | aac(6′)-Il-like, aph(3′)-VIa-like, strA, strB | sul1, sul2 | tet(B)-like | S83-L | S80-L | ||||
| N57 | blaNDM-1, blaOXA-66, blaTEM-1D | aac(6′)-Il-like, aph(3′)-VIa-like, strA, strB | sul1, sul2 | tet(B)-like | S83-L | S80-L | ||||
| N99 | blaNDM-1, blaOXA-66, blaTEM-1D | aac(6′)-Il-like, aph(3′)-VIa-like, strA, strB | sul1, sul2 | tet(B)-like | S83-L | S80-L | ||||
| N27 | blaOXA-23, blaOXA-66, blaTEM-1D | aph(3′)-Ic-like, aph(3′)-VIa-like | mph(E), msr(E) | S83-L/E87-V | S80-L | |||||
| N52 | blaOXA-23, blaOXA-66, blaTEM-1D | aph(3′)-Ic, armA, strA, strB | tet(B)-like | mph(E), msr(E) | S83-L | S80-L | ||||
| N25 | blaOXA-66, blaTEM-1D | aac(6′)-Il-like, strA, strB | sul1, sul2 | tet(B)-like | S83-L | S80-L |
Isolates are ordered in accordance with Table 1.
FQ/AMG, fluoroquinolone/aminoglycoside.
TET, tetracycline.
TMP, trimethoprim.
All isolates possessed the intrinsic cephalosporinase blaADC (not shown). There was a good correlation between the STs/IC and the OXA-51-variant: IC1 (blaOXA-69), IC2 (blaOXA-66), IC7 (blaOXA-64), and ST136 (blaOXA-378). All of the ST602 isolates had the novel ISAba58 inserted in their intrinsic blaOXA-51-like. All CRAB in this study carried an acquired carbapenemase. The most common was blaOXA-23 (n = 18), six isolates in addition also possessed blaGES-11. Three isolates possessed blaNDM-1 as their sole carbapenemase. The two carbapenem-susceptible isolates had only their intrinsic blaOXA. ISAba1 was not associated with the intrinsic blaOXA in any isolate. In two previous studies assessing CRAB in Libya, blaOXA-23 was the predominant carbapenemase in both, with the second most common being blaOXA-24 (11) or blaNDM-1 (12), the latter being similar to our findings.
All isolates had a large array of aminoglycoside resistance genes. Isolates with tetracycline MICs of ≥256 μg/ml (n = 11) carried tetB or tetA. One isolate with a tetracycline MIC of 16 μg/ml (N27) had no known tetracycline resistance determinant. Ciprofloxacin MICs of ≥16 μg/ml were associated with a Ser83-Leu substitution in GyrA and Ser80-Leu in ParC, while isolate N27 with a ciprofloxacin MIC of 256 μg/ml had an additional substitution in GyrA, Glu87-Val. One isolate with a ciprofloxacin MIC of 8 μg/ml had the GyrA substitution, but in ParC, Ser80-Ala. The so-called fluoroquinolone resistance gene aac(6′)Ib-cr was found in some isolates but was not associated with higher ciprofloxacin or levofloxacin MICs.
To determine the relative fitness of the isolates, fresh cultures were incubated overnight on MacConkey agar at 37°C and used the following day to inoculate 10 ml of sterile cation-adjusted Mueller-Hinton broth. After incubation overnight at 37°C, each culture was diluted 1:1,000 in fresh broth, and 200-μl aliquots were transferred into 4 separate wells of a 96-well microtiter plate. The growth rate was measured for 16 h with reads at 30-min intervals using a densitometer (optical density at 600 nm [OD600]). The results were averaged, normalized, and plotted against the reference strain A. baumannii (DSM 30008) (14). The results showing the growth curves are summarized in Fig. S1 to S5 in the supplemental material, where the isolates are grouped according to their IC/ST. Most isolates grew at a similar rate as the control strain for the first 10 h. However, after 10 h, differences emerged. For example, five of the six IC2 isolates all grew to higher densities at 16 h, suggesting these isolates have an increased fitness (Fig. S2). Furthermore, in TC-D, both N53 (carbapenem resistant) and N45 (carbapenem susceptible) grew to a higher density than the control, suggesting that the acquisition of carbapenem resistance had no effect on fitness as measured by growth rates (Fig. S5). Kumar et al. showed that CRAB were fitter than carbapenem-susceptible isolates (15). Interestingly, some isolates that were identical by cgMLST and their resistomes exhibited different growth rates; TC-C comprised three isolates, with N19 growing to a lower density, N44 to a higher density, and N20 to the same density as the control (Fig. S1). Taken together, these data suggest that growth rates, while variable between isolates, did not necessarily correlate with clustering, and the acquisition of a carbapenemase was not necessarily detrimental as has sometimes been suggested (16, 17).
Our study demonstrates persistently high rates of CRAB in three hospitals in Tripoli, Libya, a country still in the midst of a decade-long civil war. Rather than a single clone predominating, our results demonstrate, more worryingly, that a diverse population of A. baumannii have acquired carbapenem resistance genes at no fitness cost and are readily spread between hospitals. The lack of fitness cost may explain why no single clone has outcompeted the others. Further research assessing hospital practices, antibiotic use, and infection control protocols, combined with country-level and patient-level data such as periods of active combat and care trajectories of patients as well as isolates from other major cities, would likely shed more light on the underlying dynamics affecting the spread of polyclonal CRAB in Libya.
Data availability.
Raw sequencing reads were submitted to the European Nucleotide Archive under BioProject accession number PRJEB42042.
ACKNOWLEDGMENTS
We thank Julia Wille and Kai Lucaβen for technical support.
This work was funded by the Medical Practice Plan at the American University of Beirut Medical Center.
Footnotes
Supplemental material is available online only.
Contributor Information
Ghassan M. Matar, Email: gmatar@aub.edu.lb.
Paul G. Higgins, Email: paul.higgins@uni-koeln.de.
REFERENCES
- 1.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matar GM, Gay E, Cooksey RC, Elliott JA, Heneine WM, Uwaydah MM, Matossian RM, Tenover FC. 1992. Identification of an epidemic strain of Acinetobacter baumannii using electrophoretic typing method. Eur J Epidemiol 8:9–14. 10.1007/BF02427385. [DOI] [PubMed] [Google Scholar]
- 3.Calhoun JH, Murray CK, Manring MM. 2008. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin Orthop Relat Res 466:1356–1362. 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott P, Deye G, Srinivasan A, Murray C, Moran K, Hulten E, Fishbain J, Craft D, Riddell S, Lindler L, Mancuso J, Milstrey E, Bautista CT, Patel J, Ewell A, Hamilton T, Gaddy C, Tenney M, Christopher G, Petersen K, Endy T, Petruccelli B. 2007. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis 44:1577–1584. 10.1086/518170. [DOI] [PubMed] [Google Scholar]
- 5.Turton JF, Kaufmann ME, Gill MJ, Pike R, Scott PT, Fishbain J, Craft D, Deye G, Riddell S, Lindler LE, Pitt TL. 2006. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J Clin Microbiol 44:2630–2634. 10.1128/JCM.00547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitman TJ, Qasba SS, Timpone JG, Babel BS, Kasper MR, English JF, Sanders JW, Hujer KM, Hujer AM, Endimiani A, Eshoo MW, Bonomo RA. 2008. Occupational transmission of Acinetobacter baumannii from a United States serviceman wounded in Iraq to a health care worker. Clin Infect Dis 47:439–443. 10.1086/589247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins PG, Wisplinghoff H, Krut O, Seifert H. 2007. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin Microbiol Infect 13:1199–1201. 10.1111/j.1469-0691.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 8.CLSI. 2019. EM100 Connect. CLSI M100 ED29. CLSI, Wayne, PA. [Google Scholar]
- 9.Ziglam H, Elahmer O, Amri S, Shareef F, Grera A, Labeeb M, Zorgani A. 2012. Antimicrobial resistance patterns among Acinetobacter baumannii isolated from burn intensive care unit in Tripoli, Libya. Int Arab J Antimicrob Agents 2:716. [Google Scholar]
- 10.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathlouthi N, Areig Z, Al Bayssari C, Bakour S, El Salabi AA, Ben Gwierif S, Zorgani AA, Ben Slama K, Chouchani C, Rolain JM. 2015. Emergence of carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates collected from some Libyan hospitals. Microb Drug Resist 21:335–341. 10.1089/mdr.2014.0235. [DOI] [PubMed] [Google Scholar]
- 12.Mathlouthi N, El Salabi AA, Ben Jomàa-Jemili M, Bakour S, Al-Bayssari C, Zorgani AA, Kraiema A, Elahmer O, Okdah L, Rolain JM, Chouchani C. 2016. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents 48:46–50. 10.1016/j.ijantimicag.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Kieffer N, Ahmed MO, Elramalli AK, Daw MA, Poirel L, Álvarez R, Nordmann P. 2018. Colistin-resistant carbapenemase-producing isolates among Klebsiella spp. and Acinetobacter baumannii in Tripoli, Libya. J Glob Antimicrob Resist 13:37–39. 10.1016/j.jgar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Mu X, Wang N, Li X, Shi K, Zhou Z, Yu Y, Hua X. 2016. The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front Microbiol 7:1715. 10.3389/fmicb.2016.01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Singhal L, Ray P, Gautam V. 2020. In vitro and in vivo fitness of clinical isolates of carbapenem-resistant and -susceptible Acinetobacter baumannii. Indian J Med Microbiol 38:52–57. 10.4103/ijmm.IJMM_19_468. [DOI] [PubMed] [Google Scholar]
- 16.Göttig S, Riedel-Christ S, Saleh A, Kempf VAJ, Hamprecht A. 2016. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int J Antimicrob Agents 47:430–435. 10.1016/j.ijantimicag.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Fernández A, Pérez A, Ayala JA, Mallo S, Rumbo-Feal S, Tomás M, Poza M, Bou G. 2012. Expression of OXA-type and SFO-1 β-lactamases induces changes in peptidoglycan composition and affects bacterial fitness. Antimicrob Agents Chemother 56:1877–1884. 10.1128/AAC.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures. Download AAC.00277-21-s0001.pdf, PDF file, 324 KB (323.9KB, pdf)
Data Availability Statement
Raw sequencing reads were submitted to the European Nucleotide Archive under BioProject accession number PRJEB42042.