Abstract
Accumulating evidence indicates that the alternative splicing program undergoes extensive changes during cancer development and progression. The RNA-binding protein QKI-5 is frequently downregulated and exhibits anti-tumor activity in lung cancer. Howeve-r, little is known about the functional targets and regulatory mechanism of QKI-5. Here, we report that upregulation of exon 14 inclusion of cytoskeletal gene Adducin 3 (ADD3) significantly correlates with a poor prognosis in lung cancer. QKI-5 inhibits cell proliferation and migration in part through suppressing the splicing of ADD3 exon 14. Through genome-wide mapping of QKI-5 binding sites in vivo at nucleotide resolution by iCLIP-seq analysis, we found that QKI-5 regulates alternative splicing of its target mRNAs in a binding position-dependent manner. By binding to multiple sites in an upstream intron region, QKI-5 represses the splicing of ADD3 exon 14. We also identified several QKI mutations in tumors, which cause dysregulation of the splicing of QKI targets ADD3 and NUMB. Taken together, our results reveal that QKI-mediated alternative splicing of ADD3 is a key lung cancer-associated splicing event, which underlies in part the tumor suppressor function of QKI.
Keywords: ADD3, alternative splicing, RNA-binding protein, QKI, lung cancer
Introduction
Alternative pre-mRNA splicing generates multiple mRNA variants from a single pre-mRNA through selective recognition of splice sites located at exon/intron boundaries. Recent advances in high-throughput technologies have facilitated the detection of alternative splicing at a genome-wide scale in a variety of organisms, tissues, or cells under different treatments. Alternative splicing is now well recognized as a widespread mechanism for expanding genome complexity and regulating gene expression (Chen and Manley, 2009; Braunschweig et al., 2013; Baralle and Giudice, 2017). Although the functional significance of most alternative splicing events in human genome remains to be determined, numerous examples have illustrated the roles of alternative splicing in a myriad of cellular processes in development and diseases (Chabot and Shkreta, 2016; Scotti and Swanson, 2016; Fiszbein and Kornblihtt, 2017).
Accumulating evidence suggests that the alternative splicing program undergoes dramatic changes during malignant transformation, cancer progression, and anti-cancer drug resistance (Siegfried and Karni, 2017; Song et al., 2017). Defining the molecular mechanisms underlying the misregulation of splicing in cancer and identifying cancer-related splicing targets should facilitate the development of reliable biomarkers for early diagnosis and effective treatment strategies. Somatic mutations, altered copy number and aberrant expression of spliceosomal factors or splicing regulators have been implicated in the dysregulation of the splicing program in cancers (Anczukow and Krainer, 2016; Dvinge et al., 2016). For example, mutations found in the core spliceosomal factors, splicing factor 3B, subunit 1 (SF3B1), and U2 small nuclear RNA auxiliary factor 1 (U2AF1), have been shown to be associated with poor prognosis in patients with haematological malignancies (Papaemmanuil et al., 2013; Haferlach et al., 2014). Most of the known splicing regulators are RNA-binding proteins. Among them, the serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) are two families of extensively studied splicing regulators (Han et al., 2010; Zhou and Fu, 2013). Changes in gene copy number and/or in gene expression level of SR and hnRNP proteins, such as SRSF1, SRSF6, and hnRNP H, have been reported in tumors (Karni et al., 2007; Lefave et al., 2011; Cohen-Eliav et al., 2013). Other RNA-binding proteins, such as RBFOX2, RBM4, and RBM10, also show oncogenic or tumor suppressive activities in cancer-related processes (Venables et al., 2009; Bechara et al., 2013; Wang et al., 2014). However, the underlying molecular mechanisms and functional significance of the cancer-associated splicing changes mediated by trans-acting factors are not well understood.
Lung cancer is a leading cause of cancer-related death world-wide for both men and women (Siegel et al., 2017). About 80%‒85% of lung cancers are non-small-cell lung cancer (NSCLC), which has three main subtypes: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Misregulated splicing changes found in lung cancer have been implicated to drive lung tumorigenesis (Coomer et al., 2019). Large-scale analyses of alternative splicing in lung cancer identified many cancer-associated splicing variants (Langer et al., 2010; de Miguel et al., 2016; West et al., 2019; Zhao et al., 2020). The functions of most lung cancer-related splicing events remain unknown. The RNA-binding protein QKI is a conserved signal transduction and activation of RNA (STAR) family protein that plays an essential role in development as well as in cancers, including lung cancer (Feng and Bankston, 2010; Darbelli and Richard, 2016). QKI contains a STAR domain, which is composed of a maxi-KH RNA binding motif and two flanking QUA motifs (QUA1 and QUA2), several SH3-binding sites, and a tyrosine-rich tail. Both RNA‒protein interaction and structural studies revealed that QKI proteins recognize an RNA binding consensus YUAAY (Galarneau and Richard, 2005; Hafner et al., 2010; Teplova et al., 2013; Fagg et al., 2017; Hayakawa-Yano et al., 2017). The QKI gene generates three main transcripts through alternative splicing, which encode protein isoforms termed QKI-5, QKI-6, and QKI-7 (Kondo et al., 1999). In mouse myoblast, different variants display a network of auto- and cross-regulation of QKI protein isoforms (Fagg et al., 2017). QKI-5 contains a nuclear localization signal in the C-terminus and is localized in the nucleus of cells, while QKI-6 and QKI-7 are found in the cytoplasm and can be translocate to the nucleus (Wu et al., 1999; Pilotte et al., 2001; van der Veer, et al., 2013). Immunohistochemistry staining of QKI was predominantly detected in the nucleus of bronchial epithelium and pneumocytes in non-cancerous lung tissues. QKI staining was weaker in lung adenocarcinoma and squamous cell carcinoma compared to normal tissues (de Miguel et al., 2016; Zhou et al., 2017). We and others have reported previously that QKI-5 is a critical regulator of lung cancer-associated splicing and a prognostic marker for NSCLC at early stage (Zong et al., 2014; de Miguel et al., 2016). We showed that QKI-5 inhibits cell proliferation in part by regulating the splicing of NUMB exon 12 to prevent the inappropriate activation of Notch signaling. Mechanistically, QKI-5 competes with a core splicing factor SF1 for binding to the branchpoint to inhibit exon 12 splicing. A recent study reported that downregulation of QKI increased the stability of an oncogenic miRNA, miR-196b-5p, in lung cancer (Liang et al., 2020). In another work, Wang et al. suggested that QKI is able to promote the biogenesis of circSLC7A6, a circRNA with a tumor repressive activity (Wang et al., 2020). However, the functional targets of QKI in lung cancer remain largely unknown.
In an effort to further explore the role of QKI-5 in lung cancer, in this study, we identified that QKI-5 controls the alternative splicing of Adducin transcripts and that this regulation is important for QKI-5 to regulate cancer cell proliferation and migration. Adducin is a family of membrane-skeletal proteins, encoded by three genes ADD1, ADD2, and ADD3. ADD1 forms heterodimers or heterotetramers with either ADD2 or ADD3 at spectrin–actin junctions. While ADD1 and ADD3 are ubiquitously expressed in all tissues, ADD2 is mainly expressed in the brain and erythropoietic tissues (Gilligan et al., 1999). All these proteins contain an N-terminal globular head domain, a middle neck domain, and a C-terminal protease-sensitive tail domain. ADD proteins are involved in signal transduction, cell migration, and cell-to-cell adhesion (Matsuoka et al., 2000; Luo and Shen, 2017). In our previous study, we identified 25 QKI targets as lung cancer-associated splicing events using paired adjacent and lung tumor tissues (Zong et al., 2014). Among these targets, ADD3 splicing changes displayed the highest prognostic significance. In addition, ADD3 is one of the top-ranked splicing events identified in a large-scale analyses of alternative splicing between normal lung and lung cancer tissues (Langer et al., 2010). However, the role of its splicing change in lung cancer is unknown.
Considering that cytoskeleton genes play critical roles in regulating cancer cell growth and migration, in this study, we focus on the regulation and function of ADD3 alternative splicing. We found that a high level of ADD3 exon 14 inclusion promotes lung cancer cell proliferation and migration. QKI-5 inhibits cell proliferation and migration in part through repressing the inclusion of ADD3 exon 14. By performing individual nucleotide-resolution crosslinking immunoprecipitation coupled to deep sequencing (iCLIP-seq) analysis with a QKI-5-specific monoclonal antibody we generated as well as splicing minigene assays, we demonstrated that QKI-5 regulates alternative splicing in a binding position-dependent manner and controls ADD3 splicing by binding to the 3′ end region of intron 13. In addition, we characterized several QKI mutations which cause aberrant splicing of QKI targets. Our findings provide novel insights into the role and regulation of QKI-mediated alternative splicing in lung tumorigenesis.
Results
Alternative splicing of ADD3 exon 14 is a key lung cancer-related splicing event
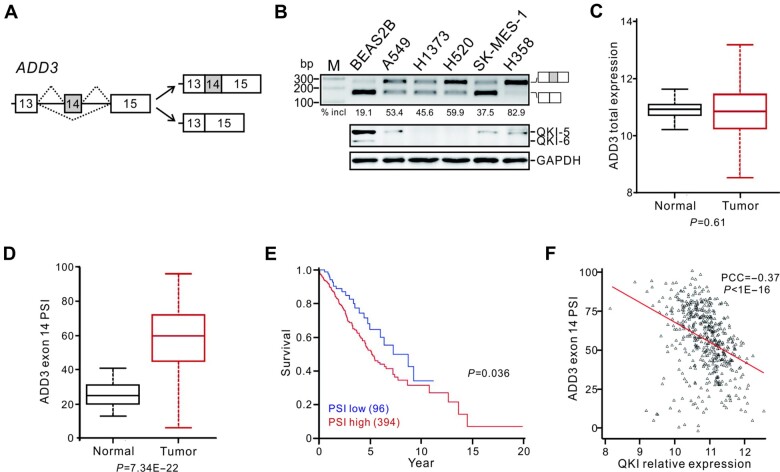
The human ADD3 gene contains 15 exons, among which exon 14 (96 nt in length) can be alternatively spliced (Figure 1A). This alternative exon 14 is conserved in human, mouse, and rat, suggesting a relevant function for the regulated ADD3 splicing (see genome browser at www.ucsc.edu). Previously, we showed that knockdown of QKI induced the inclusion of ADD3 exon 14, and the exon 14 inclusion percentage increased by 1.43‒3.76 folds in 8 out of 9 paired lung adenocarcinoma and squamous cell carcinoma tumor tissues with QKI underexpression (see Figure 3C and D in Zong et al., 2014). Comparing to the normal bronchial epithelial cell line BEAS2B, several lung cancer cell lines including A549, H1373, H520, SK-MES-1, and H358 displayed higher levels of ADD3 exon 14 inclusion, determined by reverse-transcription polymerase chain reaction (RT-PCR) analysis (Figure 1B). We previously also showed that downregulation of QKI in NSCLC patients at stage I is significantly associated with a poor survival rate, suggesting that QKI serves as a prognostic marker for NSCLC at an early stage (Zong et al., 2014). To further investigate the clinical significance of QKI-mediated ADD3 splicing in a large patient cohort, we analyzed TCGA datasets for patients with adenocarcinoma and squamous cell carcinoma at stage I. The results show that the expression level of ADD3 total mRNA in tumors is comparable to that in normal tissues; however, ADD3 exon 14 inclusion is significantly upregulated in tumors (Figure 1C and D). Importantly, the level of exon 14 inclusion inversely correlates with the survival of patients and with the level of QKI mRNA (Figure 1E and F). Our data indicate that the QKI-mediated splicing of ADD3 exon 14 is a lung cancer-related splicing event and that the exon 14 inclusion is a potential prognostic marker for NSCLC at early stage.
Figure 1.
Increased ADD3 exon 14 inclusion ratio represents a poor prognosis for NSCLC at early stage. (A) Schematic representation for the alternative splicing of ADD3 exon 14. Exons are represented by boxes, introns by lines, and splicing pattern by dotted lines. (B) Splicing patterns of ADD3 exons 13‒15 determined by RT-PCR analysis and the expression levels of QKI detected by western blotting in lung cancer cell lines. (C and D) The expression levels of total ADD3 mRNA (C) and the PSI values of ADD3 exon 14 (D) in adjacent normal (n = 53) and NSCLC tissues (n = 490). (E) Kaplan–Meier survival curves for NSCLC patients (n = 490) with high PSI (PSI ≥ 40) or low PSI (PSI < 40). (F) Correlation between QKI mRNA expression level and ADD3 exon 14 PSI in NSCLC patients. Each black triangle represents a lung tumor sample, and the red line depicts the linear regression trend between ADD3 exon 14 PSI values and QKI expression. The Pearson correlation coefficient (PCC) and P-value are shown.
Increased ADD3 exon 14 inclusion promotes lung cancer cell proliferation and migration
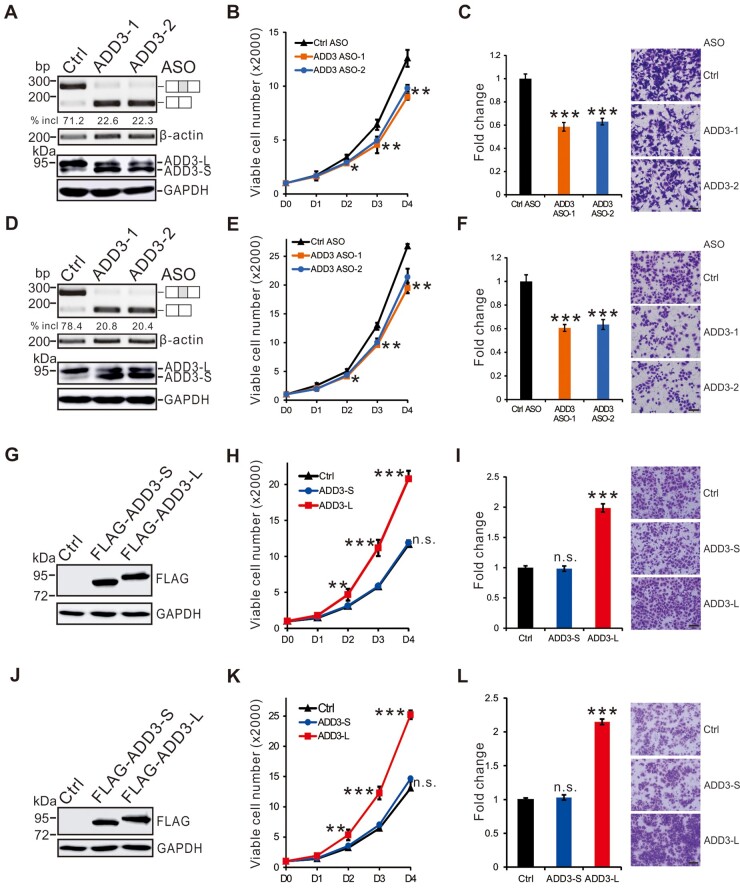
Given its function in cytoskeletal regulation, alternative splicing of ADD3 may affect cell proliferation and migration. To understand the role of ADD3 splicing alterations in lung cancer, we first examined whether the switch between the two ADD3 splicing isoforms affects cell proliferation and migration. To do this, we downregulated exon 14 inclusion ratio in adenocarcinoma A549 or squamous cell carcinoma H520 cells using two antisense oligonucleotides (ADD3-1 and ADD3-2 ASOs) which targeting the upstream 3′ splice site or the downstream 5′ splice site of exon 14, respectively, and then determined the effects on cell proliferation and migration by MTT or transwell assay, respectively (Figure 2A and D). In both cell lines, isoform switch of ADD3 induced by ASOs did not affect the total level of ADD3 expression but decreased cell proliferation (Figure 2B and E) and migration (Figure 2C and F). To dissect the functional difference between the two isoforms of ADD3, we overexpressed the short and long isoforms in A549 or H520 cells, respectively (Figure 2G and J). Interestingly, in both cell lines, the short ADD3 form had no effect on cell proliferation and migration, while the long ADD3 form increased cell proliferation (Figure 2H and K) and migration (Figure 2I and L). Together, these results indicate that upregulation of ADD3 exon 14 inclusion found in lung cancer cells promotes cell proliferation and migration.
Figure 2.
Upregulation of ADD3 long isoform encoding exon 14 promotes cancer cell proliferation and migration. (A‒F) A549 (A‒C) and H520 (D‒F) cells were transfected with control or ADD3-specific ASOs. (A and D) RT-PCR and western blotting analyses of ADD3 splicing patterns and ADD3 expression, respectively. (B and E) MTT analyses of cell proliferation. Error bars represent standard deviations (technical replicates, n = 4). (C and F) Quantitation of results from transwell assays. Error bars represent standard deviation (technical replicates, n = 5). Representative images of transwell membrane are shown on the right. (G‒L) Short and long isoforms of ADD3 were ectopically expressed in A549 (G‒I) or H520 (J‒L) cells. (G and J) Western blotting analyses of the expression of short and long isoforms of ADD3. (H and K) MTT analyses of cell proliferation. Error bars represent standard deviations (technical replicates, n = 4). (I and L) Quantitation of results from transwell assays. Error bars represent standard deviation (technical replicates, n = 5). Student’s t-test, n.s. indicates no significance; *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar, 10 μm in C, F, I, and L.
QKI-5 inhibits lung cancer cell proliferation and migration in part through regulating alternative splicing of ADD3
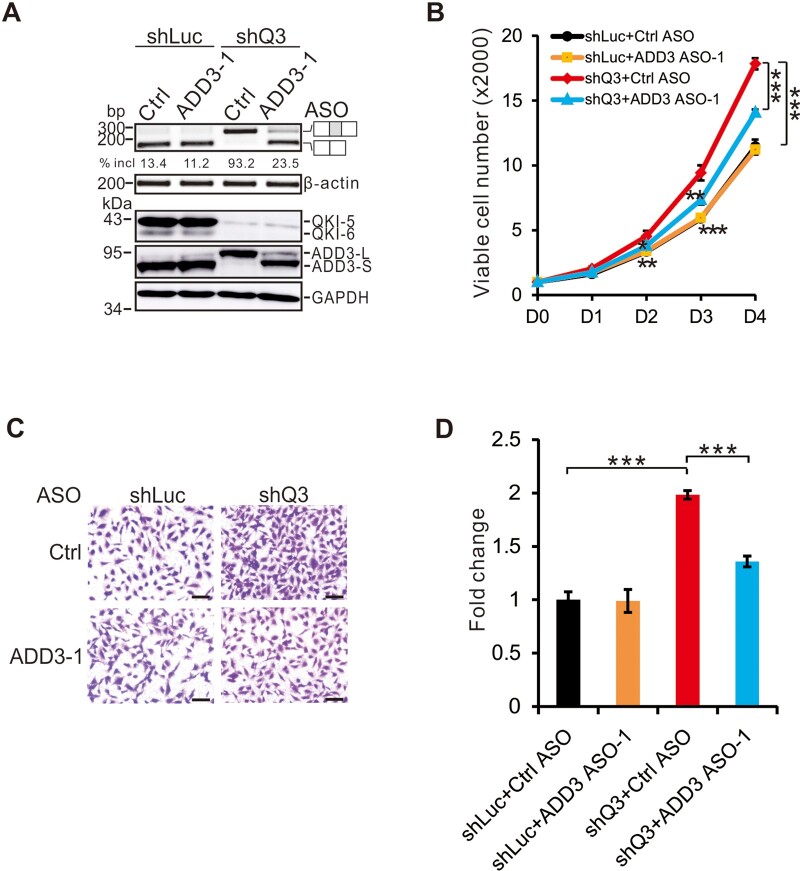
To further determine the potential role of the QKI‒ADD3 axis in lung tumorigenesis, we stably knocked-down QKI in BEAS2B cells using a previously validated shRNA (shQ3, Zong et al., 2014; target sequence is listed in Supplementary Table S1). Control and ADD3-specific ASOs were introduced in QKI-depleted or QKI-undepleted BEAS2B cells by transient transfection. As shown in Figure 3A, upon QKI knockdown, ADD3 exon 14 inclusion increased dramatically at both mRNA and protein levels, which was repressed by the ADD3-specific ASO. Knockdown of QKI promoted cell proliferation and migration. These effects induced by QKI knockdown were partially abrogated by isoform switch of ADD3 (Figure 3B‒D). These data strongly suggest that QKI exerts its tumor suppression function partially through regulating alternative splicing of ADD3 exon 14.
Figure 3.
QKI inhibits cell proliferation and migration in part through regulating ADD3 exon 14 splicing. BEAS2B cells were treated with control or ADD3-specific ASOs in the control (shLuc) or QKI-specific (shQ3) knockdown cells. shLuc is a control shRNA against luciferase gene, while shQ3 is an shRNA specifically targeting QKI mRNA. (A) RT-PCR analysis of ADD3 splicing pattern (upper panel) and western blotting analysis of QKI and ADD3 expression. (B) MTT analyses of cell proliferation. Error bars represent standard deviations (technical replicates, n = 4). (C and D) Transwell assays were performed and the results were quantitated. Scale bar, 10 μm. Error bars represent standard deviations (technical replicates, n = 5). Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
Genome-wide mapping of QKI-5 binding sites at nucleotide resolution through iCLIP-seq analysis
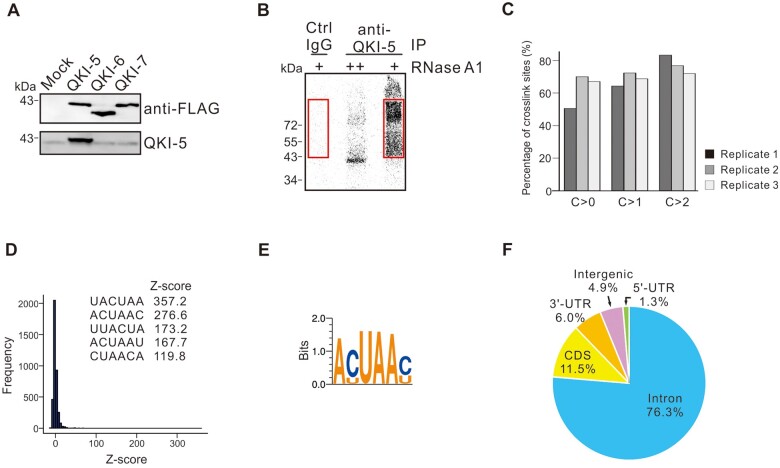
In lung cells, QKI-5 is the dominant isoform of QKI proteins at both mRNA and protein levels and QKI-6 is expressed at a relatively lower level (Figure 1B;Supplementary Figure S1A). Specific knockdown of QKI-5 reduced QKI-6 expression, while siRNA targeting QKI-6 or QKI-7 did not affect QKI-5 expression (Supplementary Figure S1B and C). These data suggest that QKI-5 is required for QKI-6 expression, which is consistent with a recent publication (Fagg et al., 2017). To gain further insights into the splicing regulation by QKI in lung cancer, we mapped the QKI-5‒RNA interactions genome-wide in vivo in BEAS2B cells by performing iCLIP-seq analysis. We generated a monoclonal antibody that recognizes a C-terminal peptide sequence only present in QKI-5. This new antibody successfully recognized FLAG-tagged QKI-5 but not QKI-6 or QKI-7 (Figure 4A). Immunofluorescence analysis using QKI-5 specific antibody showed that QKI-5 is mainly localized in the nucleus of lung cancer cell line A549 (Supplementary Figure S1D). After UV irradiation and RNase partial digestion, anti-QKI-5 monoclonal antibody immunoprecipitated specific QKI-5‒RNA complex, while the control antibody yielded little or almost no signal (Figure 4B). RNAs in the QKI-5 immunoprecipitates or in the control samples were extracted from the gel followed by reverse transcription. The resulting first-strand cDNAs were circularized and the library was subjected to deep sequencing. After the removal of PCR amplification artifacts, QKI-5 immunoprecipitates generated 26.7 million cDNA reads, while the control samples only generated 0.5 million reads, indicating that most of cDNA reads in the QKI-5 samples represent promising QKI-5‒RNA crosslink sites. The cDNA reads were aligned to the human genome by allowing one nucleotide mismatch and single genomic hits. Among QKI-5-specific cDNA reads, 7.7 million crosslink sites were detected, which are highly reproducible among three independent replicates (Figure 4C). Through applying additional algorithms, in total, the iCLIP-seq analysis identified 3.5 million clustered crosslink sites.
Figure 4.
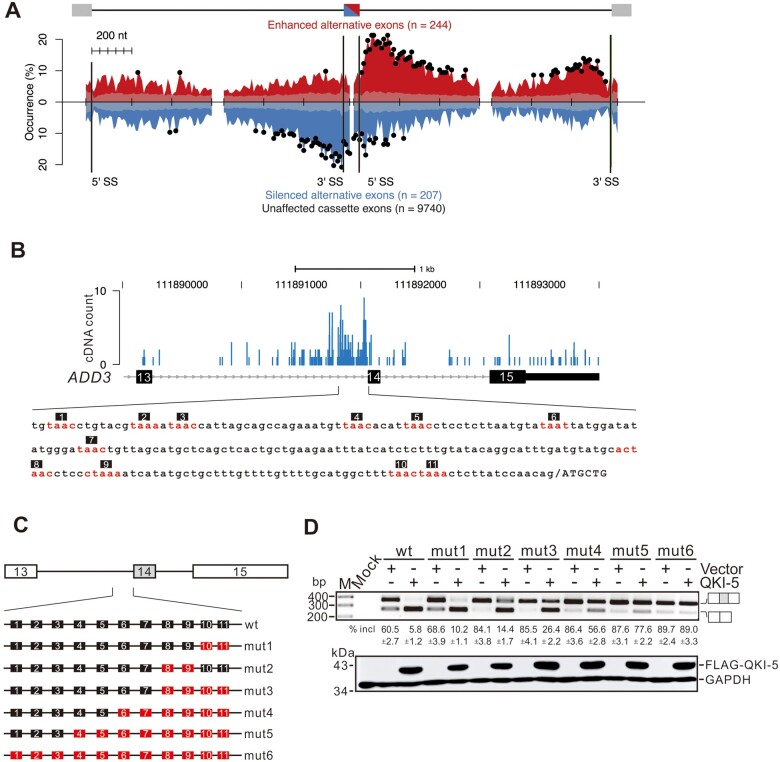
Genome-wide mapping of in vivo QKI-5 binding sites by iCLIP-seq. (A) QKI-5 monoclonal antibody recognizes only FLAG-tagged QKI-5 but not QKI-6 or QKI-7. (B) Phosphor image of QKI‒5-RNA complex fractionated by SDS‒PAGE. Cell extracts were treated with two concentrations of RNase A (high ++; low +) followed by immunoprecipitation with a control or QKI-5-specific antibody. RNAs crosslinked to QKI-5 were 32P-labelled, and RNA‒protein adducts above the position of QKI-5 in control and QKI-5 lanes (marked by red boxes) were purified for iCLIP library preparation. (C) Reproducibility of QKI-5 iCLIP crosslink sites. The crosslink sites in one replicate experiment are grouped according to the cDNA count (C > 0, all sites taken; C > 1, minimal cDNA count of 2; C > 2, minimal cDNA count of 3). The bars in the figure represent the average percentage of crosslink sites in one replicate that are present in at least one of the two other replicates. (D) Histogram of z-scores for hexamers in tags containing 10 nt upstream and 10 nt downstream sequences surrounding the crosslink sites. Inserts are sequences of the top 5 overrepresented hexamers. (E) Deduced QKI-5 binding consensus based on top 20 hexamers. (F) Genomic distribution of QKI-5 crosslink sites.
The RNA-binding specificity of QKI-5 was determined by searching for enriched hexamer motifs in 21 nt sequence tags containing both 10 nt upstream and 10 nt downstream sequences surrounding the crosslink sites. We identified the overrepresented hexamers (Figure 4D) and deduced the QKI-binding consensus as ACUAAC based on top 20 hexamer sequences (Figure 4E), which closely resembles the motif that was defined previously by in vitro systematic evolution of ligands by exponential enrichment (SELEX) as well as in vivo CLIP-seq analyses (Galarneau and Richard, 2005; Hafner et al., 2010; Fagg et al., 2017; Hayakawa-Yano et al., 2017). Consistent with the role of QKI-5 in pre-mRNA splicing and its nuclear localization, QKI-5 crosslink sites (76.3%) are predominantly located in the intron region (Figure 4F).
QKI-5 regulates alternative splicing in a binding position-dependent manner and represses ADD3 exon 14 splicing by binding to multiple sites in the 3′ end of intron 13
To delineate the principles of QKI-5 in splicing regulation, we constructed an RNA splicing map by integrating the iCLIP-seq data in this study with the RNA-seq data in a previous study (Zong et al., 2014). Both iCLIP-seq and RNA-seq data were generated in the same cell line, the normal bronchial epithelial cell line BEAS2B. This map reveals that QKI-5 possesses a binding position-dependent effect on regulating its targets (Figure 5A). As illustrated by cassette exon-type regulation, QKI-5 tends to bind to the 5′ or 3′ end of the introns downstream of QKI-5 activated exons, while represses exon inclusion by binding to the alternative exons or their flanking intronic regions. In addition, we observed two interesting features of the QKI-5 splicing map. First, QKI-5 binding sites are enriched in the 5′ end of the intronic regions downstream of both positively and negatively regulated exons. For the positively regulated exons, QKI-5 binding sites are significantly overrepresented within a 200-nt region proximal to the 5′ splice site and extended to a deep region up to 600 nt in the downstream introns, while for the negatively regulated exons, QKI-5 binding sites are enriched in a narrow region within 200 nt downstream of the 5′ splice site. This feature suggests that QKI-5 binding to the intronic regions immediately downstream of the alternative exons has opposite effects on the outcome of splicing. Second, when QKI-5 activates exon inclusion, it tends to bind to the intronic regions far downstream of the alternative exons. When QKI-5 induces exon skipping, it tends to bind to intronic regions immediately upstream of alternative exons. This feature implies that QKI-5 binding to the regions close to the 3′ end of upstream or downstream intron has distinct effects on the inclusion of the alternative exon.
Figure 5.
QKI-5 regulates alternative splicing in a binding position-dependent manner and represses ADD3 exon 14 splicing by recognizing multiple sites in the 3′ end of intron 13. (A) RNA map of QKI-5-regulated cassette exons. The exon‒intron structure is shown above. The middle cassette exon is in half red and half blue, indicating that its inclusion can be regulated. The black bars represent the exon/intron boundaries. The occurrence (%) indicates the percentage of exons that have at least one crosslink nucleotide within a given window (20 nt). The occurrence of cassette exons with iCLIP cDNA reads located proximal to activated (red curve on positive y-axis), repressed (blue curve on negative y-axis), and unaffected cassette exons (transparent gray on positive and negative y-axis) is shown. The x-axis represents nucleotide position relative to upstream, cassette, and downstream exons. The ticks on the x-axis are 200 nt scale bars. Black dots represent nucleotide positions where QKI-5 binding sites are significantly enriched compared to unaffected cassette exons (P < 0.05, chi-square test). (B) iCLIP cDNA counts for QKI-5 crosslink sites located from exon 13 to exon 15 of ADD3. Sequence immediately upstream of exon 14 is shown in the lower panel. (C) Schematic representation of ADD3 minigene constructs. Sequence elements containing substituted QKI-5 binding sequences shown in B are labelled in red. In these short elements, the conserved motif UAA in the hexamer was substituted by UGA. (D) RT-PCR analyses of the splicing patterns of ADD3 wild-type and mutant minigene constructs transfected into HEK 293 cells together with a vector or a QKI-5 expression construct. The average percentages of exon inclusion with standard deviations are shown below (n = 3).
By examining the in vivo interactions between QKI-5 and ADD3 pre-mRNAs detected by iCLIP-seq, we found that QKI-5 recognizes several cis-elements at the 3′ end of intron 13 that are similar to its binding consensus sequence (Figure 5B). To determine whether QKI-5 inhibits ADD3 splicing through binding to these elements, we made several minigene constructs and carried out in vivo splicing assays by performing cell transfection in HEK 293 cells and RT-PCR analysis. The wild-type minigene construct encompasses exons 13‒15 and the introns in between, whereas the mutant constructs bear substitution of the QKI-5 binding sites at different positions in the minigenes (Figure 5C). Overexpression of QKI-5 significantly repressed exon 14 splicing in the wild-type minigene (Figure 5D, lanes wt). The mutant minigenes carrying mutated QKI-5 binding sites in the vicinity of 3′ splice site in intron 13 exhibited increased inclusion of exon 14 in transfected cells. However, they still responded to QKI-5 overexpression (Figure 5D, lanes mut1, mut2, and mut3), suggesting that other QKI-5-binding elements are functional in splicing inhibition. To test this possibility, we further mutated additional binding sites more upstream of the 3′ splice site to generate mut4, mut5, and mut6 minigenes. Overexpression of QKI-5 inhibited exon 14 inclusion to a less extent in mut4 and mut5 minigenes, and no longer repressed exon 14 splicing in mut6 minigene (Figure 5D, lanes mut4, mut5, and mut6). Taken together, these results indicate that QKI-5 regulates ADD3 exon 14 splicing through recognizing multiple binding sites in the 3′ end of intron 13.
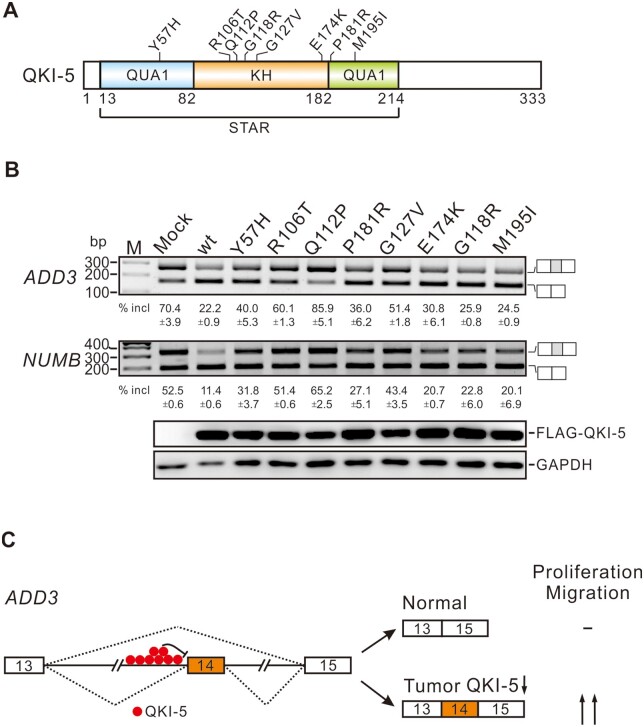
Cancer-associated QKI-5 mutations cause defects in the splicing of ADD3 and NUMB
QKI is frequently downregulated in several types of solid tumors including breast, colon, lung, gastric, and glioblastoma (Yang et al., 2010; Novikov et al., 2011; Chen et al., 2012; Li et al., 2016). By analyzing somatic mutation databases, we identified several QKI mutations in bladder, colon, esophageal adenocarcinoma, endometrial, glioblastoma, and lung adenocarcinoma cancers (Lawrence et al., 2014). To test whether these mutations affect QKI function, we made several constructs expressing QKI mutants containing the cancer-associated mutations, including Y57H, R106T, Q112P, P181R, G127V, E174K, G118R, and M195I. We then transfected these mutants into HEK 293 cells and examined the splicing patterns of endogenous QKI targets ADD3 and NUMB. All these QKI mutants led to defects in inhibiting the splicing of NUMB exon 12 and ADD3 exon 14 to some extents, with Y57H, R106T, and G127V mutations exhibited stronger effects (Figure 6A). Interestingly, Q112P did not show any inhibitory effect on the splicing of NUMB and ADD3, but rather slightly activated exon inclusion, likely because the exogenously expressed Q112P perturbed the function of endogenous QKI-5. To test this possibility, we transfected the wild-type and Q112P QKI-5 constructs into QKI knockdown BEAS2B cells. Introducing the wild-type QKI-5 into QKI knockdown cells recapitulated the repression of exon inclusion in control knockdown cells, while Q112P mutant completely lose the ability to inhibit the splicing of ADD3 and NUMB exons (Supplementary Figure S2). Taken together, our results suggest that both downregulation and mutations of QKI can contribute to tumorigenesis.
Figure 6.
QKI mutations found in cancers affect ADD3 and NUMB splicing. (A) Schematic representation of the domain structure of QKI-5 and its mutations. (B) RT-PCR analyses of the splicing patterns of endogenous ADD3 exon 14 and NUMB exon 12 in HEK 293 cells transfected with wild-type or mutant QKI-5 expression constructs. The average percentages of exon inclusion with standard deviations are shown below (n = 3). (C) A working model of this study. Exons are represented by boxes, introns by lines, and splicing pattern by dotted lines. In normal cells, QKI-5 binds to the intron region upstream of ADD3 exon 14 and inhibits its inclusion, generating the short isoform of ADD3, which has no effect on cell proliferation or migration. In tumors, downregulation of QKI-5 derepresses the inclusion of exon 14, producing the long isoform of ADD3, which is able to promote cell proliferation and migration.
Discussion
In this work, we identified that the splicing target of QKI-5, ADD3, is a potential prognostic marker for NSCLC (Figure 1). Upregulation of ADD3 exon 14 inclusion promotes cancer cell proliferation and migration (Figure 2). QKI-5 exerts its tumor suppressor function in part through suppressing ADD3 exon 14 splicing (Figure 3). Mapping of in vivo QKI-5‒RNA interactions revealed that QKI-5 regulates splicing in a binding position-dependent manner (Figure 5A). It represses ADD3 exon 14 inclusion through recognizing multiple binding sites at the 3′ end of intron 13 (Figures 5D and 6C). Furthermore, we characterized several cancer-associated QKI mutations that cause dysregulation of the splicing of its downstream targets ADD3 and NUMB (Figure 6B;Supplementary Figure S2).
Our results provide novel information about the connection between alternative splicing and cytoskeletal regulation in tumorigenesis. Abnormalities in cytoskeletal network are implicated in the development and progression of many cancers. Changes in splicing of a number of cytoskeletal genes have been implicated in malignancy (Di Modugno et al., 2012; Luo et al., 2015; Wollscheid et al., 2016; de Miguel et al., 2016; Itoh et al., 2017). However, the functional significance of the alternative splicing regulation of cytoskeletal genes remains to be defined. Cytoskeletal protein ADD3 contains an N-terminal globular head domain, a middle neck domain, and a C-terminal tail domain. A myristoylated alanine-rich C kinase substrate (MARCKS)-related domain containing a cluster of lysine residues is located at the very end of the tail domain. ADD3 exon 14 encodes a polypeptide with 32 amino acids between the neck and MARCKS domains. The alternative inclusion of exon 14 in ADD3 provides several potential opportunities to regulate ADD3 function. The ADD3 long isoform containing exon 14 may have a protein-interacting network distinct from the short isoform without exon 14. Moreover, several serine and threonine residues located in this region provide potential phosphorylation sites. Phosphorylation at these sites may impact on the formation of spectrin‒actin meshwork to alter cell proliferation or migration. Recently, using Kras mutant mouse model of lung cancer, we found an increase in ADD3 exon 14 inclusion in the lung of Kras QkL/L mice (data not shown). Relevant to tumorigenesis, Salomonis et al. (2009) reported that the dominant isoform of ADD3 in undifferentiated human embryonic stem cells (hESCs) is the long isoform. The expression pattern of ADD3 shifted from the long isoform to the short one, when hESCs differentiated into cardiac and neural precursors (Salomonis et al., 2009). This finding suggests that the alternative splicing of ADD3 exon 14 may play a role in the self-renewal, pluripotency, and differentiation of embryonic stem cells. The physiological and pathological roles of ADD3 alternative splicing and the underlying mechanisms for differential functions of ADD3 isoforms warrant further investigation.
RNA splicing maps generated through the integration of genome-wide analyses on splicing profiles and RNA‒protein interactions provide global effects and mechanistic insights for the actions of RNA-binding proteins (Witten and Ule, 2011; Fu and Ares, 2014). It has been shown for a number of RNA-binding proteins that can regulate exon inclusion either positively or negatively, depending on the location of their binding sites relative to the regulated exons. In many cases, as has been shown for Nova (Ule et al., 2006), RBFOX (Yeo et al., 2009), MBNL (Wang et al., 2012), and CELF (Ajith et al., 2016), binding of these proteins to intron regions downstream of the regulated exons activates exon inclusion, while binding to the intron regions upstream of the regulated exons or within them represses exon inclusion. Similar to the splicing map of QKI-5 generated in neural stem cells (Hayakawa-Yano et al., 2017), the RNA map of QKI-5 shown in this study indicates that QKI-5 shares these positioning effects. Interestingly, our map also showed that QKI-5 represses exon inclusion when binding to downstream intron regions proximal to the alternative exons, and activates exon inclusion when binding to 3′ end in the downstream introns. In QKI proteins, QUA1 domain has been shown to be critical for forming dimers (Beuck et al., 2012). Dimerization of QKI-5 may facilitate to block exon inclusion via a ‘looping out’ model of repression as proposed for the cases of hnRNP A1 and PTB (Nasim et al., 2002; Oberstrass et al., 2005). It is also possible that the formation of QKI-5 dimer or interaction with other RNA-binding proteins may activate exon inclusion by assisting the 5′ and 3′ splice sites of downstream introns in close vicinity. The positional effects revealed by our QKI-5 RNA map will guide our future mechanistic studies on the alternative splicing of individual mRNA targets, including ADD3.
In support of the role of QKI-5 in regulating alternative splicing in cancer, we found that several cancer-related mutations in QKI cause dysregulation of its key splicing targets ADD3 and NUMB. Among these mutations, Y57H, R106T, Q112P, and G127V exhibited strong effects (Figure 6B). In the crystal structure of the intact QKI STAR domain (residues 7‒214) in complex with RNA (Teplova et al., 2013), the hydroxyl group of Y57 in the QUA1 dimerization domain forms a hydrogen bond with E174 in the KH domain on α6, which may stabilize the interaction between the QUA1 and KH domains (Supplementary Figure S3). The Y57H mutation might diminish intramolecular interactions, resulting in a conformational change. R106 is located in the loop that connects α3 and α4 in the KH domain (Supplementary Figure S3). The side chain of R106 may interact with the phosphodiester group in RNA via electrostatic interactions, which could be disrupted by R106T mutation. Q112 is located in the middle of α4 in the KH domain and stabilizes the conformation of QUA1 through the hydrogen binding with R53 on α2 in the QUA1 domain (Supplementary Figure S3). The Q112P mutation might interrupt α4 structure, leading to a conformational change of QUA1 and the dissociation of QKI dimer. Spatially close to K132, the G127V mutation introduces a bulky hydrophobic group at the position of G127, which may cause a clashing with K132 and change the conformation of the loop connecting β5 and α5 as well as the KH domain (Supplementary Figure S3). Future work is needed to characterize the effects of these mutations on QKI-5 cellular function, splicing of target mRNAs, and tumorigenesis.
Materials and methods
Oligonucleotides
The sequences of all the oligonucleotides purchased from Invitrogen or GenePharma (ASO) for this study are listed in Supplementary Table S1.
ADD3 minigene constructs
The ADD3 minigene constructs consist of three exons and two introns. The genomic sequences of the minigene unit were amplified using genomic DNA isolated from HEK 293 cells as template. To construct the wild-type minigene, most of the intron sequence was deleted from 300 nt downstream of the 5′ splice site to 300 nt upstream of the 3′ splice site. A complete fragment was assembled by overlapping PCRs using oligonucleotides Add3-1‒Add3-6 as primers, and then inserted between HindIII and XhoI sites of the pcDNA3 vector. In the mutant constructs, QKI binding sites in the wild-type minigene construct were mutated using similar overlapping PCRs using primers containing nucleotide substitutions (see Supplementary Table S1 for primer sequences).
Expression constructs of QKI-5 mutants
QKI-5 mutant expression constructs were generated by PCR using pcDNA3-QKI-5-FLAG as template and primers with nucleotide substitution (see Supplementary Table S1 for primer sequences). PCR fragments were inserted between BamHI and XbaI sites of the pcDNA3 vector.
Cell culture
The human bronchial epithelial BEAS2B and HEK 293 cells were grown at 37˚C in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. The human lung cancer cell lines A549, H1373, H520, SK-MES-1, and H358 were cultured at 37˚C in RPMI-1640 medium supplemented with 10% fetal bovine serum.
Cell transfection of ASOs or siRNA
ASOs or siRNA were transiently transfected using Lipofectamine 2000 (Invitrogen). Briefly, the day before transfection, 2 × 105 BEAS2B, A549, or HTB182 cells were seeded in 35-mm culture dishes. According to manufacturer's instruction, the final concentration of ASOs or siRNA was 50 nM. After 48 h, cells were harvested and used for further analysis.
TCGA data analysis
RNA-seq data (BAM files and expression quantification data) and patient survival times for NSCLC lung cancers were obtained from the TCGA projects TCGA-LUAD and TCGA-LUSC (https://gdc.cancer.gov). Percentage splice index (PSI) values in normal and tumor samples were estimated with the splicing analysis tool MISO (Katz et al., 2010). Differential splicing and expression between normal and tumor samples were evaluated with the Wilcoxon signed-rank test. To assess the survival relevance of the ADD3 exon 14 inclusion level, the NSCLC samples were split into PSI-high (PSI ≥ 40) and PSI-low (PSI < 40) groups, fitted to the Kaplan–Meier survival curves, and compared for difference with the log-rank test.
Western blotting
Cell pellets were collected by centrifugation, lysed by sodium dodecyl sulfate (SDS) loading buffer, and separated by SDS‒polyacrylamide gel electrophoresis (SDS‒PAGE) followed by gel transfer to a nitrocellulose membrane (BioRad). The membranes were incubated first with the primary antibodies, and then with secondary antibodies coupled to horseradish peroxidase (HRP). Band signals were detected with an enhanced chemiluminescence (ECL) system (Thermo Scientific) and visualized by image analyzer (Fujifilm). The primary antibodies used for this study are anti-GAPDH (KangCheng Bio-tech), anti-QKI (Sigma), anti-FLAG (Sigma), and anti-ADD3 (Proteintech). The HRP-conjugated secondary antibodies anti-mouse IgG and anti-rabbit IgG were purchased from Promega.
Real-time quantitative PCR
Total RNA was extracted from cells using Trizol (Invitrogen) and reverse-transcribed into first-strand cDNA from random hexamers using MMLV reverse transcriptase (Promega). PCR was then performed using SYBR® Green PCR Master Mix (Applied Biosystems) on a 7500 Fast Real-Time PCR system according to manufacturer’s instruction (Applied Biosystems).
In vivo splicing
The day before transfection, 2 × 105 BEAS2B or HEK 293 cells were seeded in 35-mm culture dishes. Minigene plasmids were transfected using Lipofectamine 2000. After 24 h, total RNAs were isolated using Trizol Reagent. The first-strand cDNAs were reverse-transcribed using BGH-rev as primer and MMLV reverse-transcriptase following the manufacturer’s instruction. PCR was performed using target gene-specific primers. PCR products were loaded and run in 2% agarose gel electrophoresis followed by ethidium bromide staining. Quantification of gel band signals was performed using Gel-Pro Analyzer software according to manufacturer’s instruction.
MTT cell proliferation assay
Cells were seeded at a density of 2000 per well in 24-well culture plates. After 24, 48, 72, 96, and 120 h of incubation, cells were treated with 3-(4,5-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) at a final concentration of 0.5 µg/µl for 4 h. The resulting formazan was solubilized with dimethylsulfoxide (DMSO), and the absorption was measured at 570 nm using a spectrophotometer (Thermo Scientific). The results of MTT assay shown in Figures 2 and 3 were repeated three times.
Cell migration assay
Cell migration was examined in 8-mm transwell chambers with 8-μm pores (Corning Costar). The bottom surface of each membrane was first coated with serum-free medium for 1 h. Approximately 25000 cells were seeded in the upper chambers in 200 μl of serum-free medium. Lower chambers contained 600 μl of 20% serum medium. After the cells were allowed to migrate for 12 h, the medium in the upper chamber was sucked out and cells on the upper side were removed with a cotton swab. Cells on the lower side of the membrane were washed with cold PBS, fixed with methanol, and stained with 0.2% crystal violet in PBS. Cells that migrated through the membrane to the lower surface were counted by a light microscopy. The results of transwell cell migration assay shown in Figures 2 and 3 were repeated three times.
Generating QKI-5-specific monoclonal antibody
Two copies of QKI-5-specific peptide sequence at its C-terminus (GAVATKVRRHDMRVHPYQRIVTADRAATGN) were cloned and expressed in Escherichia coli. The purified protein was immunized into BALB/c mice. Hybridoma cell lines were generated and screened, and the monoclonal antibody against QKI-5 was purified by Abmart.
iCLIP-seq analysis
The iCLIP assay was carried out as described previously (Konig et al., 2010; Wu et al., 2015). Briefly, five 10-cm dishes of BEAS2B cells were irradiated with UV at 150 mJ/cm2. After cell lysis, RNAs were partially fragmented using 10 μl high (1:50) or low (1:5000) dilutions of 5 μg/μl RNase A (QIAGEN) followed by immunoprecipitation with 10 μg anti-QKI5 antibody immobilized on protein A Dynabeads (Life Technologies). RNAs were then ligated at the 3′ end to a DNA adapter and radioactively labelled by T4 polynucleotide kinase (Fermentas), and the protein‒RNA complex was transferred to a nitrocellulose membrane. For iCLIP cDNA library preparation, purified RNAs were reverse-transcribed with a primer containing barcodes. The resulting cDNAs were purified by PAGE, circularized by single-stranded DNA ligase (Epicentre), linearized by restriction enzyme cleavage, and amplified by PCR. The iCLIP cDNA library was processed on an Illumina HiSeq 2000 sequencer. We trimmed the first nine nucleotides at 5′ ends of raw reads, which include a four-nucleotide barcode and five random nucleotides (e.g. NNN[barcode]NN), and removed the 3′ adaptor sequences by cutadapt. The clean reads were mapped against the human genome (version hg19/NCBI37) and transcript sequences allowing one mismatch and single hits using Bowtie. The crosslink sites were identified at one nucleotide upstream of the 5′ end of aligned reads by custom script. iCLIP-seq data and motif enrichment were analyzed as described previously (Wu et al., 2015).
Generation of QKI-5 RNA splicing map
The number of QKI-5 crosslink sites was counted in 40 nt exonic region or 400 nt intronic region surrounding the 5′ or 3′ splice sites with a 20-nt bin for 244 QKI-activated and 207 QKI-repressed events, respectively. For the short exon (<80 nt) and intron (<800 nt), we divided the exon or intron sequence into half. To count the number of QKI-5 crosslink sites for control pre-mRNAs, we randomly selected the same amount of detected cassette-exon events, which are not regulated by QKI (P > 0.95). We repeated this procedure 100 times and calculated the average numbers with 99.9999% confidence interval for each bin. The occurrence (%) was calculated by dividing the number of exons that have at least one crosslink site within a given window by the total number of exons analyzed at this window.
Data availability
The iCLIP-Seq data are available at NCBI’s GEO database under the accession number GSE111133.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We thank Dr Zhongsheng You (Washington University School of Medicine, St. Louis, USA) for critical reading of the manuscript and invaluable suggestions and all the members of Jingyi Hui’s laboratory for technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (31661143035, 31770881, and 32071288) and the National Basic Research Program of China (2017YFA0504400) to J.H.
Conflict of interest: none declared.
References
- Ajith S., Gazzara M.R., Cole B.S., et al. (2016). Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 13, 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczukow O., Krainer A.R. (2016). Splicing-factor alterations in cancers. RNA 22, 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle F.E., Giudice J. (2017). Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara E.G., Sebestyen E., Bernardis I., et al. (2013). RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733. [DOI] [PubMed] [Google Scholar]
- Beuck C., Qu S., Fagg W.S., et al. (2012). Structural analysis of the quaking homodimerization interface. J. Mol. Biol. 423, 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U., Gueroussov S., Plocik A.M., et al. (2013). Dynamic integration of splicing within gene regulatory pathways. Cell 152, 1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B., Shkreta L. (2016). Defective control of pre-messenger RNA splicing in human disease. J. Cell Biol. 212, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.J., Paik J.H., Zhang H., et al. (2012). STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 26, 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Manley J.L. (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Eliav M., Golan-Gerstl R., Siegfried Z., et al. (2013). The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J. Pathol. 229, 630–639. [DOI] [PubMed] [Google Scholar]
- Coomer A.O., Black F., Greystoke A., et al. (2019). Alternative splicing in lung cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194388. [DOI] [PubMed] [Google Scholar]
- Darbelli L., Richard S. (2016). Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 7, 399–412. [DOI] [PubMed] [Google Scholar]
- de Miguel F.J., Pajares M.J., Martínez-Terroba E., et al. (2016). A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer. Mol. Oncol. 10, 1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Modugno F., Iapicca P., Boudreau A., et al. (2012) Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc. Natl Acad. Sci. USA 109, 19280–19285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H., Kim E., Abdel-Wahab O., et al. (2016). RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagg W.S., Liu N., Fair J.H., et al. (2017). Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes Dev. 31, 1894–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.,, Bankston A. (2010) The star family member QKI and cell signaling. Adv. Exp. Med. Biol. 693, 25–36. [PubMed] [Google Scholar]
- Fiszbein A., Kornblihtt A.R. (2017). Alternative splicing switches: important players in cell differentiation. Bioessays 39, doi: 10.1002/bies.201600157. [DOI] [PubMed] [Google Scholar]
- Fu X.D., Ares M. Jr. (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A., Richard S. (2005). Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 12, 691–698. [DOI] [PubMed] [Google Scholar]
- Gilligan D.M., Lozovatsky L., Gwynn B., et al. (1999). Targeted disruption of the β adducin gene (Add2) causes red blood cell spherocytosis in mice. Proc. Natl Acad. Sci. USA 96, 10717–10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T., Nagata Y., Grossmann V., et al. (2014). Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.P., Tang Y.H., Smith R. (2010). Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392. [DOI] [PubMed] [Google Scholar]
- Hayakawa-Yano Y., Suyama S., Nogami M., et al. (2017). An RNA-binding protein, Qki5, regulates embryonic neural stem cells through pre-mRNA processing in cell adhesion signaling. Genes Dev. 31, 1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Radisky D.C., Hashiguchi M., et al. (2017). The exon 38-containing ARHGEF11 splice isoform is differentially expressed and is required for migration and growth in invasive breast cancer cells. Oncotarget 8, 92157–92170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., et al. (2007). The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., et al. (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J., Zarnack K., Rot G., et al. (2010). iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Furuta T., Mitsunaga K., et al. (1999) Genomic organization and expression analysis of the mouse qkI locus. Mamm. Genome 10, 662–669. [DOI] [PubMed] [Google Scholar]
- Langer W., Sohler F., Leder G., et al. (2010). Exon array analysis using re-defined probe sets results in reliable identification of alternatively spliced genes in non-small cell lung cancer. BMC Genomics 11, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Mermel C.H., et al. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefave C.V., Squatrito M., Vorlova S., et al. (2011). Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 30, 4084–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Yi P., Pi J., et al. (2016). QKI5-mediated alternative splicing of the histone variant macroH2A1 regulates gastric carcinogenesis. Oncotarget 7, 32821–32834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Meng W., Huang X., et al. (2020). miR-196b-5p-mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc. Natl Acad. Sci. USA 117, 4347–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Shen J. (2017). Adducin in tumorigenesis and metastasis. Oncotarget 8, 48453–48459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z.L., Cheng S.Q., Shi J., et al. (2015). A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat. Commun. 6, 8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Li X., Bennett V. (2000). Adducin: structure, function and regulation. Cell. Mol. Life Sci. 57, 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim F.U., Hutchison S., Cordeau M., et al. (2002). High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8, 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov L., Park J.W., Chen H., et al. (2011). QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 31, 4244–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass F.C., Auweter S.D., Erat M., et al. (2005). Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Malcovati L., et al. (2013). Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotte J., Larocque D., Richard S. (2001). Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev. 15, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Nelson B., Vranizan K., et al. (2009). Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 5, e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M.M., Swanson M.S. (2016). RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. (2017). Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30. [DOI] [PubMed] [Google Scholar]
- Siegfried Z., Karni R. (2017). The role of alternative splicing in cancer drug resistance. Curr. Opin. Genet. Dev. 48, 16–21. [DOI] [PubMed] [Google Scholar]
- Song X., Zeng Z., Wei H., et al. (2017). Alternative splicing in cancers: from aberrant regulation to new therapeutics. Semin. Cell Dev. Biol. 75, 13–22. [DOI] [PubMed] [Google Scholar]
- Teplova M., Hafner M., Teplov D., et al. (2013). Structure‒function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev. 27, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Stefani G., Mele A., et al. (2006). An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586. [DOI] [PubMed] [Google Scholar]
- doi: 10.1161/CIRCRESAHA.113.301302. van der Veer, E.P., de Bruin, R.G., Kraaijeveld, A.O., et al. (2013). Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ. Res. 113, 1065–1075. [DOI] [PubMed] [Google Scholar]
- van der Veer, E.P.,, de Bruin, R.G.,, Kraaijeveld,A.O., et al , . (2013). Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ. Res. 113, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Venables J.P., Klinck R., Koh C., et al. (2009). Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16, 670–676. [DOI] [PubMed] [Google Scholar]
- Wang E.T., Cody N.A., Jog S., et al. (2012). Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen D., Qian H., et al. (2014). The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 26, 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zheng F., Wang Z., et al. (2020) Circular RNA circ-SLC7A6 acts as a tumor suppressor in non-small cell lung cancer through abundantly sponging miR-21. Cell Cycle 19, 2235–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Kumar S., Batra S.K., et al. (2019). Uncovering and characterizing splice variants associated with survival in lung cancer patients. PLoS Comput. Biol. 15, e1007469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten J.T., Ule J. (2011). Understanding splicing regulation through RNA splicing maps. Trends Genet. 27, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollscheid H.P., Biancospino M., He F., et al. (2016). Diverse functions of myosin VI elucidated by an isoform-specific α-helix domain. Nat. Struct. Mol. Biol. 23, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Zhou L., Tonissen K., et al. (1999). The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 274, 29202–29210. [DOI] [PubMed] [Google Scholar]
- Wu S.L., Fu X., Huang J., et al. (2015). Genome-wide analysis of YB-1‒RNA interactions reveals a novel role of YB-1 in miRNA processing in glioblastoma multiforme. Nucleic Acids Res. 43, 8516–8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Fu H., Zhang J., et al. (2010). RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 138, 231–240.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Coufal N.G., Liang T.Y., et al. (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA‒protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Zhang C., Jiang M., et al. (2020). Survival-associated alternative splicing signatures in non-small cell lung cancer. Aging 12, 5878–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li X., Sun C., et al. (2017). Quaking-5 suppresses aggressiveness of lung cancer cells through inhibiting β-catenin signaling pathway. Oncotarget 8, 82174–82184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Fu X.D. (2013). Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong F.Y., Fu X., Wei W.J., et al. (2014). The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 10, e1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The iCLIP-Seq data are available at NCBI’s GEO database under the accession number GSE111133.