Abstract
Objective
To clarify the role of miR‐92a in regulating the malignant progression of cervical cancer and its specific molecular mechanism.
Methods
qRT‐PCR was used to detect the differential expression of miR‐92a in cervical cancer and adjacent tissues. The effects of overexpression of miR‐92a on the proliferation, migration, and invasion of HeLa and SiHa cells were tested. Luciferase assays and rescue experiments were used to investigate the regulatory mechanism of miR‐92a on its downstream gene PIK3R1 and their interaction in the progression of cervical cancer.
Results
miR‐92a was significantly up‐regulated in cervical cancer tissues. Overexpression of miR‐92a significantly increased the ability of cervical cancer cells to proliferate, migrate, and invade. PIK3R1 was identified as a downstream gene of miR‐92a. In cervical cancer tissues, PIK3R1 was found to be down‐regulated and negatively correlated with the level of miR‐92a. Overexpression of PIK3R1 reversed the promotional effect of overexpressed miR‐92a on the proliferation, migration, and invasion of cervical cancer.
Conclusion
miR‐92a is up‐regulated in cervical cancer tissues. miR‐92a promotes the malignant development of cervical cancer by negatively regulating PIK3R1.
Keywords: cervical cancer, miR‐92a, PIK3R1
miR‐92a increases expression in cervical cancer tissues and promotes cervical cancer cell proliferation, invasion, and migration.
1. INTRODUCTION
Cervical cancer is one of the most common malignant tumors in women, and its incidence is second only to breast cancer, colorectal cancer, and lung cancer.1 According to incomplete statistics from the World Health Organization, there are more than 500,000 new cases worldwide each year, with increasing incidence year by year and reduced age of onset.2 Cervical cancer causes approximately 300,000 deaths every year, of which 85% occur in developing countries, and the cancer seriously affects the quality of life of women.3, 4 Therefore, understanding the pathogenesis of cervical cancer and looking for potential prognosis and targeted therapy indicators are of great significance for improving the survival rate of patients with cervical cancer.
MicroRNA molecules (microRNA, miRNA) are small non–protein‐coding RNAs that regulate the expression of target genes at the post‐transcriptional level.5, 6 miRNAs bind to the 3′‐untranslated region (three prime untranslated region, 3′‐UTR) of the target mRNA and regulate gene expression by shearing the mRNA or inhibiting translation. Previous studies have shown that the abnormal expression of miRNA is related to a variety of human malignancies including cervical cancer.7, 8 miR‐92a is a novel miRNA molecule discovered in recent years. It is considered to be a tumor promoter in colorectal cancer,9 liver cancer,10 gastric cancer,11 and renal cell carcinoma12 where it shows up‐regulated expression and appears to regulate cell functions such as tumor cell proliferation, migration, and apoptosis. However, the function of miR‐92a in cervical cancer and its mechanism of action are still unclear.
Phosphoinositide 3‐kinase regulatory subunit 1 (PIK3R1) is significantly reduced in a variety of human tumor tissues compared with normal control tissues, including lung, prostate, liver, kidney, and breast cancers.13, 14 It has been reported that decreased expression of PIK3R1 mRNA and the PIK3R1‐encoding protein p85α is related to reduced metastasis‐free survival in patients with breast cancer.15 In a mouse liver cancer model, the liver‐specific PIK3R1 knockout and an 80%–90% reduction in p85α protein expression lead to PI3K pathway activation and corresponding tumor progression. P85α can stabilize PTEN, and PTEN expression in these PIK3R1 knockout tumors is also reduced.16 These findings indicate that the dysregulation of PIK3R1 expression plays a key role in cancer development. In addition, PIK3R1 has been shown to be a functional target of many miRNAs involved in the regulation of cancer progression.17 However, the role of miRNA and PIK3R1 in cervical cancer requires further study.
In our study, we found that the expression of miR‐92a in cervical cancer tissues was significantly higher than that in adjacent tissues. miR‐92a can promote the proliferation, migration, and invasion of cervical cancer cells. Mechanistically, PIK3R1 was confirmed as a target of miR‐92a. In conclusion, our observations indicate that miR‐92a‐PIK3R1 can regulate the malignant biological characteristics of cervical cancer.
2. MATERIALS AND METHODS
2.1. Cell culture
The cell cryopreservation tube removed from the −80°C refrigerator was placed in a 37°C water bath for 5 min, and immediately resuspended in DMEM (Gibco) containing 10% FBS. Cells were cultured in a constant temperature incubator at 37°C and 5% CO2. The cell culture medium was changed every other day.
2.2. Total RNA extraction
One milliliter of TRIzol was added to the cervical cancer tissue sample, and a tissue homogenizer was used to crush the tissue into a slurry. Two hundred microliters of CHCl3 was then added; the material was vortexed and allowed to stand on ice for 3 min; after centrifugation, the supernatant was retained; and 500 µl IPA was then added. The sample was mixed by inversion on ice for 10 min and centrifuged, the supernatant was discarded, and 1 ml of 75% ethanol was added to the pellet to wash. After recentrifugation, the supernatant was discarded, and the pellet was washed once and allowed to dry naturally. An appropriate amount of RNase‐free water was then added and gently pipetted to fully dissolve the RNA.
2.3. qRT‐PCR
Appropriate amounts of RNA were reverse‐transcribed into cDNA using the reverse transcription kit (Gima). The NovoStart SYBR qPCR SuperMix Plus (Novoprotein) was used for real‐time PCR analysis. The relative expression of miR‐92a in the cells was calculated according to the 2−ΔΔCt formula using U6 as the internal reference (Table S1).
2.4. Cell transfection
Cells were plated in 6‐well plates at a density of 50%–60% per field when viewed under the microscope. A serum‐free culture medium was added, and the cells were transfected with small RNA (Table S2) using Lipofectamine 2000. Cells were incubated at 37°C for 5 h before changing the medium to complete medium and culturing for 24–48 h.
2.5. MTT assay
Cells were plated at 5000 cells per well in 96‐well plates the day before transfection. Cells were divided into three groups for transfection with small interfering RNA. After 0, 24, 48, 72, and 96 h, 20 μl MTT reagent was added to each well and incubated at 37°C for 4 h. The medium was aspirated and 150 μl DMSO added to each well. A microplate reader was used to detect the absorbance (OD) value of each sample at 490 nm.
2.6. Colony formation assay
Logarithmic growth phase cells were inoculated into 6‐well plates one day before transfection and were divided into three groups for transfection with small interfering RNA. After transfection, 500 cells per well were inoculated into a new 6‐well plate and placed in an incubator. After 12 days of culture, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet staining solution, and the visible colonies were counted manually.
2.7. Wound healing assay
Three equidistant lines were drawn on the bottom of each well of a 6‐well plate. Appropriate amounts of logarithmic growth phase well were inoculated into each cell and divided into three groups for siRNA transfection in advance. A pipette tip was used to scratch the cells horizontally, and the marked cells were washed with phosphate‐buffered saline before adding serum‐free medium and culturing at 37°C and 5% CO2 for 0, 24, and 48 h before being photographed.
2.8. Transwell assay
Forty‐eight hours after transfection, the cells in each group were collected by trypsinization and counted; 4 × 104 cells were added to 100 μl serum‐free medium and mixed, and then added to the bottom of the upper chamber of the Transwell chamber. The complete medium containing 10% FBS was added to the lower chamber, and the apparatus was placed in an incubator for 48 h. The cells were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet staining solution. Micrographs were taken to count the number of cells that had penetrated the membrane.
2.9. Western blot
The transfected cells were lysed with RIPA reagent and centrifuged to obtain the protein lysate. The lysate was mixed with an equal volume of SDS loading buffer and boiled to prepare the denatured protein, 50 μg of which was separated on SDS‐PAGE. The electrophoresed proteins were transferred to PVDF membranes by the wet transfer method. The membranes were blocked with 5% skim milk for 1 h and incubated with the primary antibodies overnight at 4°C. The secondary antibodies were incubated for 1 h at room temperature, and the protein bands were exposed by ECL chemiluminescence.
3. RESULTS
3.1. miR‐92a was up‐regulated in cervical cancer tissues
It is well known that miRNAs play important regulatory roles in cervical cancer. Although there are many reports that miRNAs are abnormally expressed in cervical cancer, there are still many miRNAs whose functions in cervical cancer have not been verified. We used cervical cancer microarray data to select miR‐92a as it has significant differences from other candidate miRNAs.18 First, we verified the expression of miR‐92a in cervical cancer tissues. We used qRT‐PCR to detect the expression of miR‐92a in 13 pairs of cervical cancer and adjacent tissues, finding that miR‐92a expression was significantly higher in cervical cancer tissues (Figure 1A and B).
FIGURE 1.
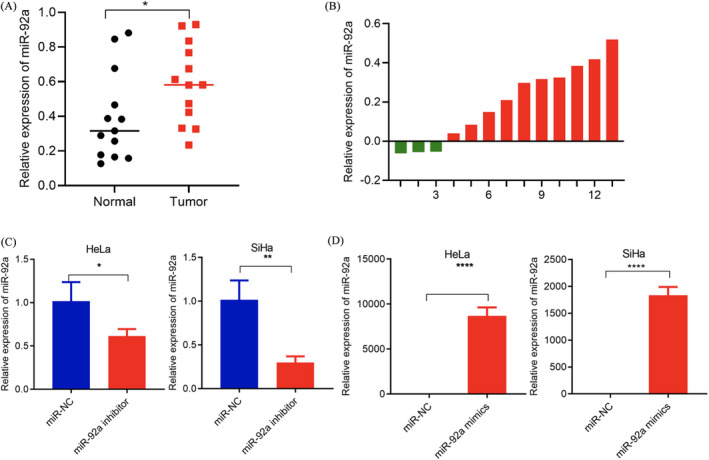
Increased expression of miR‐92a in cervical cancer tissues. (A, B) qRT‐PCR confirmed that the expression of miR‐92a in 13 cervical cancer tissues was higher than that in normal tissues. (C) qRT‐PCR results showed that the miR‐92a inhibitor can specifically reduce the expression of miR‐92a. (D) qRT‐PCR results showed that miR‐92a mimics can specifically up‐regulate the expression of miR‐92a. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001
3.2. miR‐92a promotes cell proliferation, migration, and invasion in vitro
To further explore the effects of miR‐92a on the proliferation, migration, and invasion of cervical cancer cells, the gain of function and loss of function were determined. First, we designed and synthesized miR‐92a inhibitor. We found that the miR‐92a inhibitor can significantly down‐regulate the expression of miR‐92a by 47%–66% (Figure 1C). In addition, we used miR‐92a mimics to perform ectopic expression of miR‐92a and found that miR‐92a may be up‐regulated 2500‐ to 10000‐fold (Figure 1D). MTT and colony formation experiments showed that knocking down miR‐92a inhibited the proliferation of HeLa and SiHa cells. In contrast, overexpression of miR‐92a had an opposite effect on proliferation, indicating that miR‐92a promotes the proliferation of HeLa and SiHa cells (Figure 2A and B). Wound healing and invasion experiments showed that knockout of miR‐92a inhibited the migration and invasion of HeLa and SiHa cells, and overexpression promoted the migration and invasion of HeLa and SiHa cells (Figure 2C and D).
FIGURE 2.
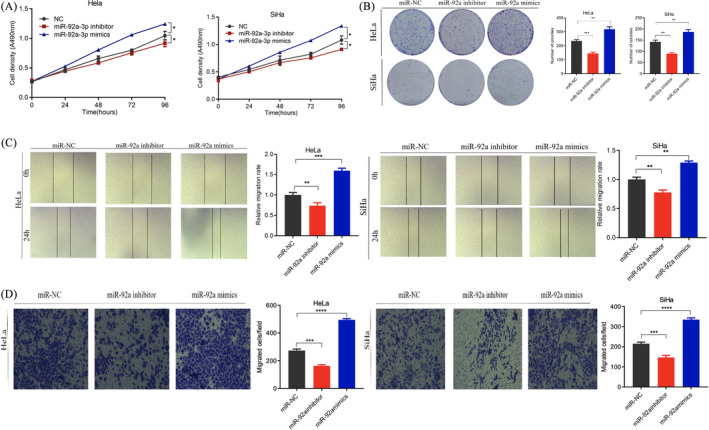
miR‐92a promotes the proliferation, invasion, and migration of cervical cancer cells. (A) Cell viability detected by the MTT method after transfection of miR‐NC, miR‐92a inhibitor, or miR‐92a mimics in HeLa and SiHa cells. (B) Cell proliferation detected by the clone formation method. (C) Cell migration detected by the cell scratch test. (D) Cell invasion detected by the Transwell assay. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001
3.3. PIK3R1 was identified as a direct target of miR‐92a
The cell function experiments demonstrated that miR‐92a can promote the proliferation, invasion, and migration of cervical cancer cells. We suspected that miR‐92a may affect the progression of cervical cancer through a specific molecular mechanism. Many studies have shown that miRNA can specifically bind and regulate the expression of target genes to regulate tumor progression.19 We used StarBase software (http://starbase.sysu.edu.cn/, v3.0) to predict that miR‐92a interacts with PIK3R1 (Figure 3A). To verify this, we knocked down and overexpressed miR‐92a in HeLa and SiHa cells and found that PIK3R1 expression was clearly regulated by miR‐92a (Figure 3B). To further corroborate this interaction, we conducted a dual‐luciferase gene reporter experiment. The results showed that after co‐transfection of miR‐92a mimics with the PIK3R1 wild type, the luciferase activity was significantly reduced, while after co‐transfection of miR‐92a mimics with PIK3R1 mutant type, there was no significant change in luciferase activity (Figure 3C), indicating that miR‐92a can interact with PIK3R1.
FIGURE 3.
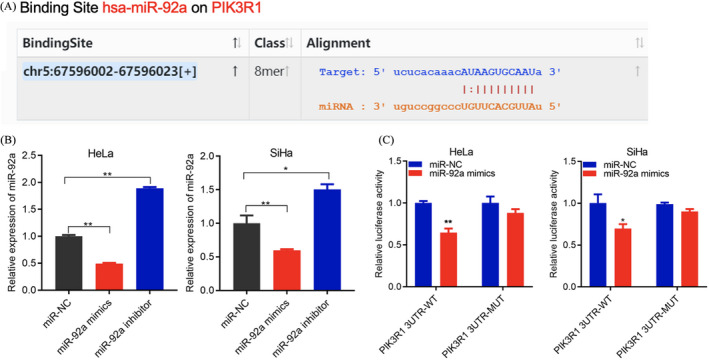
miR‐92a targets PIK3R1. (A) StarBase prediction of miR‐92a and PIK3R1 binding. (B) qRT‐PCR detection of PIK3R1 expression in HeLa and SiHa cells after transfection with miR‐92a mimics and the miR‐92a inhibitor. (C) Luciferase reporter assay of the binding of miR‐92a and PIK3R1. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001
3.4. miR‐92a promotes cell proliferation, migration, and invasion by regulating PIK3R1
To further verify whether miR‐92a promotes the progression of cervical cancer by regulating the expression of PIK3R1, we co‐transfected the miR‐92a inhibitor and PIK3R1 siRNA in HeLa and SiHa cells, and found that compared with PIK3R1 siRNA transfection alone, transfection of the miR‐92a inhibitor significantly inhibited cell proliferation, migration, and invasion (Figure 4A–E). Western blotting results showed that when only PIK3R1 siRNA was transfected, the PIK3R1 protein level was significantly inhibited. After co‐transfection of the miR‐92a inhibitor and PIK3R1 siRNA, the expression level of PIK3R1 was restored to a certain extent (Figure 4F).
FIGURE 4.
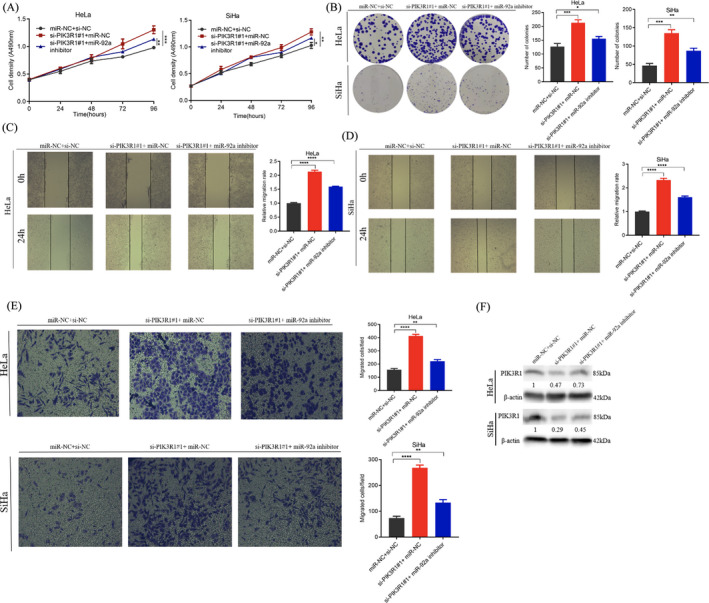
Down‐regulated miR‐92a reverses PIK3R1 promotion of proliferation, migration, and invasion in cervical cancer cells. (A) MTT experiments showed that after co‐transfection of the miR‐92a inhibitor and PIK3R1 siRNA, cell proliferation was partially restored. (B) Clone formation after co‐transfection of miR‐92a inhibitor and PIK3R1 siRNA. (C, D) Wound healing of cell migration after co‐transfection of the miR‐92a inhibitor and PIK3R1 siRNA shown by the scratch experiment. (E) Partial restoration of cell invasion and migration after co‐transfection of miR‐92a inhibitor and PIK3R1 siRNA shown by the Transwell assay. (F) Western blot experiments showed that the expression level of PIK3R1 protein after co‐transfection with miR‐92a inhibitor and PIK3R1 siRNA. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001
4. DISCUSSION
As a type of non‐coding small RNA with a length of about 19–25 oligonucleotide sequences in eukaryotic cells, miRNAs play roles similar to oncogenes or tumor suppressor genes in multiple biological processes such as cell proliferation, apoptosis, and migration.20, 21, 22 Different miRNAs regulate the mRNAs of different target genes forming a complex RNA regulatory network that is frequently both cell‐ and tissue‐specific. The human genome contains thousands of miRNAs. Compared with the large number of siRNA fragments, miRNAs are highly conserved and time‐sequential.23 Their mechanism of action is still not well understood.
Previous studies have demonstrated the involvement of miR‐92a in the occurrence and development of tumors. For example, miR‐92a functions as an oncogenic miRNA in colorectal cancer by regulating the PI3K/AKT pathway mediated by PTEN24 and promotes the proliferation, migration, and invasion of esophageal squamous cell carcinoma by regulating PTEN.25 In contrast, miR‐92a is significantly down‐regulated in Wilms’ tumor, where it inhibits the proliferation, migration, and invasion of Wilms’ tumor cells and induces apoptosis by targeting FRS2.26 In this study, miR‐92a was found to be highly expressed in cervical cancer tissues. In the cell function experiments, we used HeLa and SiHa cells to investigate the relationship between the expression of miR‐92a and the proliferation and invasion of cervical cancer cells. The results suggested that miR‐92a may act as an oncogene in cervical cancer. In addition, we used dual‐luciferase reporter gene detection to demonstrate for the first time that miR‐92a is related to the proliferation, migration, and invasion of cervical cancer cells through targeted regulation of PIK3R1.
The PIK3R1 gene is located on chromosome 5q13.1 and encodes the regulatory subunit of type I PI3K. The PI3K/AKT signaling pathway participates in the regulation of various cell functions including cell proliferation, migration, and invasion.27, 28 The PIK3R1 gene plays an important role in a variety of tumors such as ovarian cancer, colon cancer, hepatocellular carcinoma, and breast cancer.29 Numerous studies have shown that miRNAs can affect tumor cell proliferation, apoptosis, and invasion by regulating the expression of PIK3R1 mRNA. For example, miR‐486‐5p promotes pancreatic cancer cell migration and invasion by directly targeting PIK3R1,30 which is also targeted by miR‐455 to promote the proliferation and migration of kidney cancer cells.31 In addition, miR‐21 directly targets PIK3R1 and activates the PI3K/AKT signaling pathway, thereby promoting breast cancer cell growth, migration, and invasion.32 This study found that knocking down PIK3R1 can promote the proliferation, migration, and invasion of cervical cancer cells, which is consistent with the functional effect of overexpression of miR‐92a. In‐depth studies have found that inhibiting the expression of PIK3R1 can partially reverse the effects of miR‐92a on the proliferation and invasion of HeLa and SiHa cells, indicating that miR‐92a functions by regulating the expression of PIK3R1.
In summary, investigation of the mechanism of PIK3R1 regulation by miR‐92a and its role in the proliferation and metastasis of cervical cancer cells will assist our understanding of miR‐92a's tumor‐promoting mechanism and help clarify the mechanism of cervical cancer occurrence and evolution. This knowledge will provide a new theoretical basis for the diagnosis and treatment of cervical cancer.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Lingjun Zhao designed and performed the experiments, and wrote the manuscript. Yijun Wang, Chenyang Zheng, and Lingjun Zhao contributed to experimental work and data analysis. Yijun Wang and Aner Chen conducted the experiments and revised the manuscript. All authors have read and approved the final manuscript. The authors declare that all data were generated in‐house and that no paper mill was used.
ETHICAL APPROVAL
All samples were obtained with the patient's informed consent and were carried out with the approval of the Ethics Committee of Ningbo Women and Children's Hospital.
Supporting information
Tables S1‐S2
Wang Y, Chen A, Zheng C, Zhao L. miR‐92a promotes cervical cancer cell proliferation, invasion, and migration by directly targeting PIK3R1 . J Clin Lab Anal. 2021;35:e23893. 10.1002/jcla.23893
Funding information
This research was supported by the Public Welfare Science and Technology Plan Project of Ningbo, Major Social Development Projects of Ningbo (Grant No. 2013C51015)
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169‐182. 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235:718‐724. 10.1002/jcp.29009 [DOI] [PubMed] [Google Scholar]
- 4.Zigras T, Lennox G, Willows K, Covens A. Early cervical cancer: current dilemmas of staging and surgery. Curr Oncol Rep. 2017;19:51. 10.1007/s11912-017-0614-5 [DOI] [PubMed] [Google Scholar]
- 5.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202‐1207. 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597‐610. 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Song Y, Mi Y, et al. microRNA‐499a promotes the progression and chemoresistance of cervical cancer cells by targeting SOX6. Apoptosis. 2020;25:205‐216. 10.1007/s10495-019-01588-y [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Li F, Zhu L. Clinical significance and functions of microRNA‐93/CDKN1A axis in human cervical cancer. Life Sci. 2018;209:242‐248. 10.1016/j.lfs.2018.08.021 [DOI] [PubMed] [Google Scholar]
- 9.Yamada NO, Senda T. Circulating microRNA‐92a‐3p in colorectal cancer: a review. Med Mol Morphol. 2021. 10.1007/s00795-021-00282-w. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Yang B, Feng X, Liu H, et al. High‐metastatic cancer cells derived exosomal miR92a‐3p promotes epithelial‐mesenchymal transition and metastasis of low‐metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene. 2020;39:6529‐6543. 10.1038/s41388-020-01450-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao QQ, Chen JJ, Xu WJ, Zhao XZ, Sun X, Zhong L. miR‐92a‐3p promotes the proliferation and invasion of gastric cancer cells by targeting KLF2. J Biol Regul Homeost Agents. 2020;34:1333‐1341. 10.23812/20-209-a [DOI] [PubMed] [Google Scholar]
- 12.Zeng R, Huang J, Sun Y, Luo J. Cell proliferation is induced in renal cell carcinoma through miR‐92a‐3p upregulation by targeting FBXW7. Oncol Lett. 2020;19:3258‐3268. 10.3892/ol.2020.11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Yang Z, Xu A, et al. PIK3R1 negatively regulates the epithelial‐mesenchymal transition and stem‐like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci Rep. 2015;5:8997. 10.1038/srep08997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munkley J, Livermore KE, McClurg UL, et al. The PI3K regulatory subunit gene PIK3R1 is under direct control of androgens and repressed in prostate cancer cells. Oncoscience. 2015;2:755‐764. 10.18632/oncoscience.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cizkova M, Vacher S, Meseure D, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013;13:545. 10.1186/1471-2407-13-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi CM, Winnay J, Kondo T, et al. The phosphoinositide 3‐kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 2010;70:5305‐5315. 10.1158/0008-5472.can-09-3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan X, Hong X, Lai J, et al. Exosomal microRNA‐221‐3p confers adriamycin resistance in breast cancer cells by targeting PIK3R1 . Front Oncol. 2020;10:441. 10.3389/fonc.2020.00441 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Li Z, Zhou X, Gao W, Sun M, Chen H, Meng T. Circular RNA VRK1 facilitates pre‐eclampsia progression via sponging miR‐221‐3P to regulate PTEN/Akt. J Cell Mol Med. 2021. 10.1111/jcmm.16454. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JX, Jia X‐J, Liu Y, et al. Silencing of miR‐17‐5p suppresses cell proliferation and promotes cell apoptosis by directly targeting PIK3R1 in laryngeal squamous cell carcinoma. Cancer Cell Int. 2020;20:14. 10.1186/s12935-020-1096-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203‐222. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 21.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22‐36. 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- 22.Rupaimoole R, Calin GA, Lopez‐Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235‐246. 10.1158/2159-8290.cd-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill M, Tran N. MicroRNAs regulating microRNAs in cancer. Trends Cancer. 2018;4:465‐468. 10.1016/j.trecan.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 24.Ahmadi S, Sharifi M, Salehi R. Locked nucleic acid inhibits miR‐92a‐3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gene Ther. 2016;23:199‐205. 10.1038/cgt.2016.10 [DOI] [PubMed] [Google Scholar]
- 25.Li X, Guo S, Min L, Guo Q, Zhang S. miR‐92a‐3p promotes the proliferation, migration and invasion of esophageal squamous cell cancer by regulating PTEN. Int J Mol Med. 2019;44:973‐981. 10.3892/ijmm.2019.4258 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Li JL, Luo P. MiR‐140‐5p and miR‐92a‐3p suppress the cell proliferation, migration and invasion and promoted apoptosis in Wilms’ tumor by targeting FRS2. Eur Rev Med Pharmacol Sci. 2020;24:97‐108. 10.26355/eurrev_202001_19899 [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Yang L, Yao L, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun. 2018;9:1357. 10.1038/s41467-018-03867-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Lee E, Jung J, et al. microRNA‐155 positively regulates glucose metabolism via PIK3R1‐FOXO3a‐cMYC axis in breast cancer. Oncogene. 2018;37:2982‐2991. 10.1038/s41388-018-0124-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallejo‐Díaz J, Chagoyen M, Olazabal‐Morán M, González‐García A, Carrera AC. The opposing roles of PIK3R1/p85α and PIK3R2/p85β in cancer. Trends Cancer. 2019;5:233‐244. 10.1016/j.trecan.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 30.Kong Y, Li Y, Luo Y, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR‐486‐5p/PIK3R1/VEGF‐C axis in pancreatic cancer. Mol Cancer. 2020;19:82. 10.1186/s12943-020-01205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YD, Sun ZL. Effects of miR‐455 on PIK3R1 gene expression regulation and kidney cancer cell functions. Eur Rev Med Pharmacol Sci. 2017;21:3370‐3376. [PubMed] [Google Scholar]
- 32.Yan LX, Liu Y‐H, Xiang J‐W, et al. PIK3R1 targeting by miR‐21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol. 2016;48:471‐484. 10.3892/ijo.2015.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1‐S2
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


