Abstract
Background
Studies have reported coinfection of severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), the cause of coronavirus disease‐2019 (COVID‐19), with other viruses that cause respiratory tract infections (RTIs). We investigated the coinfection rate of SARS‐CoV‐2 and other RTI‐causing viruses, and whether the cycle threshold (Ct) value of a real‐time reverse transcriptase PCR (RT‐PCR) differed when the coinfection occurred during the first wave of COVID‐19 in Daegu, Republic of Korea, in 2020.
Methods
After performing PCR for SARS‐CoV‐2, we additionally tested for the presence of RTI‐causing viruses to check for coinfection. Subsequently, we identified the specific coexisting respiratory viruses and calculated the coinfection rate. In addition, based on the coinfection status, we compared the Ct values obtained from RT‐PCR for SARS‐CoV‐2 in patients who tested positive for COVID‐19 PCR.
Results
Of 13,717 patients, 123 had positive results on COVID‐19 PCR testing and six tested positive for an RTI‐causing virus. Thus, the coinfection rate was 4.9%. There were no statistically significant differences in the mean Ct values of SARS‐CoV‐2 RT‐PCR between coinfected and non‐coinfected patients.
Conclusion
This study computed the coinfection rate of SARS‐CoV‐2 and RTI‐causing viruses and revealed that the mean Ct values in SARS‐CoV‐2 real‐time RT‐PCR did not differ according to the coinfection status.
Keywords: COVID‐19, SARS‐CoV‐2, coinfection, real‐time RT‐PCR, cycle threshold
1. INTRODUCTION
Since it was first reported in Wuhan, China, in December 2019, severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), the cause of coronavirus disease‐19 (COVID‐19), quickly spread across the globe; the disease burden is increasing with a global loss of GDP and health. The World Health Organization (WHO) declared the disease a pandemic in March 2020, and as of March 7, 2021, there were 116,166,652 confirmed cases and 2,582,528 deaths.1 Although vaccines against SARS‐CoV‐2 have been developed and are being administered worldwide since December 2020,2, 3, 4 the pandemic remains active. The cycle threshold (Ct) value of a real‐time RT‐PCR SARS‐CoV‐2 test has been associated with disease severity and patient mortality.5 Furthermore, some studies have explored the pathophysiology of SARS‐CoV‐26, 7 as well as the multidisciplinary approaches for COVID‐19 management as it affects most organ systems, including cardiovascular, neurological, and hematological systems.8, 9, 10
Besides SARS‐CoV‐2, other viral respiratory tract infections (RTIs) also have a detrimental impact on human mortality and morbidity and are one of the leading causes of death.11 Although most cases of RTIs do not require additional treatment, Coronaviridae, including SARS‐CoV‐2, Paramyxoviridae, and Picornaviridae, may result in the development of secondary bacterial infections after RTI. Recently, a study promoting an understanding of RTI viruses, including SARS‐CoV‐2, was published for clinicians.12
Coinfection of SARS‐CoV‐2 and other RTI viruses is gaining attention. Several studies have reported the coinfection rates and changes in symptoms, the rate of hospitalization, and the length of hospital stay after coinfection.13, 14, 15, 16, 17, 18, 19, 20, 21, 22
The first wave of COVID‐19 infections occurred in Daegu, Republic of Korea, between February and May 2020. Here, we investigated specific RTI‐causing viruses and their possible interactions with SARS‐CoV‐2. Further, we calculated the coinfection rate and variations in Ct obtained from real‐time RT‐PCR SARS‐CoV‐2 tests during the aforementioned period.
2. MATERIALS AND METHODS
The flow diagram for subject allocation is shown in Figure 1. Here, we enrolled patients who underwent COVID‐19 real‐time RT‐PCR (COVID‐19 PCR) testing between February 28 and May 2, 2020, at the Yeungnam University Medical Center in Daegu. These patients either showed COVID‐19 symptoms or were in close contact with COVID‐19 patients.
FIGURE 1.
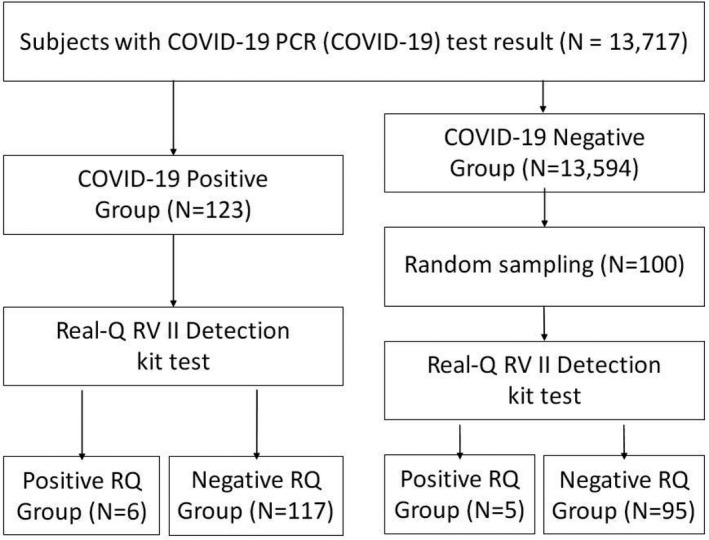
Flow diagram for subject allocation. Abbreviations: COVID‐19, coronavirus disease 2019; PCR, polymerase chain reaction; RQ, Real‐Q RV II Detection kit
This study was approved by the Institutional Review Board of Yeungnam University Medical Center (IRB No. 2020‐03‐115).
2.1. Laboratory test for COVID‐19 and RTI
The SARS‐CoV‐2 must be present for the diagnosis of COVID‐19. At the Yeungnam University Medical Center, real‐time RT‐PCR tests were performed using nasopharyngeal swab samples to confirm COVID‐19. The test kit used was Allplex 2019‐nCoV Assay (Allplex; Seegene Co. Ltd, Seoul, Republic of Korea), which obtained an emergency use authorization (EUA) from the South Korea Ministry of Food and Drug Safety. The samples remaining after COVID‐19 PCR tests were stored in a deep freezer at −70℃ and subsequently analyzed using the Real‐Q RV II Detection kit (RQ; Bioseum Co. Ltd, Seoul, Republic of Korea); the kit simultaneously detects 16 major RTI‐causing respiratory viruses—adenovirus, parainfluenza virus 1/2/3/4, enterovirus, influenza virus A/B, coronavirus 229E/OC43/NL63, rhinovirus, respiratory syncytial virus A/B, metapneumovirus, and bocavirus. The RQ assay was performed in patients who tested positive and 100 randomly selected specimens from those who tested negative on the COVID‐19 PCR test.
2.2. Statistical analysis
The continuous variables were compared using Welch's two‐sample t test; they were presented as median and interquartile range (IQR). The categorical variables were compared using Pearson's chi‐square test or Fisher's exact test; they were presented as total count and percentage. Statistical significance was set at a p‐value of 0.05, and p‐values less than 0.05 were deemed statistically significant. All statistical analyses were performed using R statistical software version 4.0.3, and graphs were generated using the ggplot2 package in R.23
3. RESULTS
3.1. Demographic characteristics of the allocated subjects
During the study period, 13,717 individuals underwent COVID‐19 PCR testing, and 123 tested positive. When the RQ assay was performed on the specimens of these 123 individuals, six (4.9%) tested positive and 117 (95.1%) tested negative (Figure 1).
To represent 13,594 individuals who tested negative on the COVID‐19 PCR test, 100 specimens were randomly selected and analyzed using RQ assay. On analysis, five (5.0%) individuals tested positive and 95 (95.0%) tested negative (Table 1).
TABLE 1.
Demographic characteristics of COVID‐19 PCR‐positive and sampled COVID‐19 PCR‐negative subject groups
|
COVID‐19 PCR Positive |
COVID‐19 PCR Negative |
p‐value | |
|---|---|---|---|
| N | 123 | 100 | |
| Female, median (%) | 70 (56.9) | 47 (47.0) | 0.18 |
| Male, median (%) | 53 (43.1) | 53 (53.0) | |
| Age, median (%) | 60 (47.5, 67) | 71.5 (59.75, 80) | <0.001† |
| Female, median (IQR) | 59 (42, 65.75) | 77 (64.5, 83) | <0.001 |
| Male, median (IQR) | 60 (53, 73) | 66 (57, 75) | 0.53 |
| Real‐Q RV II Detection kit | |||
| Positive, median (%) | 6 (4.9) | 5 (5.0) | 1 |
| Negative, median (%) | 117 (95.1) | 95 (95.0) | |
Abbreviation: IQR, interquartile range.
Comparison using Pearson's chi‐squared test, Welch two‐sample t test, or Fisher's exact test, accordingly.
The percentage of RQ‐positive patients did not significantly differ between COVID‐19 PCR‐positive and ‐negative individuals. Similarly, the male‐to‐female sex ratio also did not significantly differ between the two groups. However, the median age of the male and female individuals differed significantly.
3.2. Clinical characteristics of RQ‐positive subjects
Table 2 shows the clinical characteristics of patients who tested positive on the RQ assay after testing positive (n = 123) or negative (n = 100) on the Allplex test. The sequence information shown in the last row was obtained by searching the website for the National Center for Biotechnology Information using the Basic Local Alignment Search Tool (BLAST).
TABLE 2.
Clinical and laboratory characteristics of the patients with Real‐Q RV II Detection kit‐positive specimens
| Case | Sex | Age, years | Symptoms on arrival | COVID‐19 PCR |
Real‐Q RV II Detection kit Virus group |
Confirmed virus# |
|---|---|---|---|---|---|---|
| 1 | M | 24 | Sore throat, cough, and sputum | Positive | CoV 229E/OC43 | Human coronavirus 229E |
| 2 | F | 26 | . | Positive | AdV (A‐F) | Human mastadenovirus A |
| 3 | F | 60 | . | Positive | HRV (A‐C) | Human rhinovirus B |
| 4 | F | 50 | Sore throat | Positive | FluA | Influenza A virus |
| 5 | F | 56 | Sore throat | Positive | HRV (A‐C) | Human rhinovirus A |
| 6 | M | 61 | Sputum | Positive | AdV (A‐F) | Human mastadenovirus A |
| 1 | M | 54 | General weakness | Negative | CoV 229E/OC43 | Human coronavirus OC43 |
| 2 | F | 72 | Fever, sputum, sore throat, and hemoptysis | Negative | AdV (A‐F) | Human mastadenovirus A |
| 3 | M | 40 | Abdominal pain | Negative | CoV 229E/OC43 | Human coronavirus 229E |
| 4 | M | 85 | Fever | Negative | HRV (A‐C) | Human rhinovirus A |
| 5 | M | 78 | Dyspnea | Negative | HRV (A‐C) | Human rhinovirus B |
Abbreviations: AdV, adenovirus; CoV, coronavirus; FluA, influenza A virus; HRV, rhinovirus.
Sequenced and then NCBI BLAST searched virus names of the Real‐Q RV II Detection kit‐positive specimen.
3.3. Laboratory analysis between Allplex and RQ
The Ct values for the target gene, used to determine positivity/negativity on the Allplex assay, are shown as box plots (Figure 2). The RQ test results for specimens that were positive on the COVID‐19 PCR test are shown on the x‐axis and their E, RdRP, and N gene Ct values, used for the Allplex assay, are shown on the y‐axis. According to the box plots, the differences in the mean Ct values between RQ‐positive (with coinfection) and RQ‐negative (without coinfection) specimens were not statistically significant; P‐values for the E, RdRP, and N genes were 0.91, 0.87, and 0.76, respectively.
FIGURE 2.
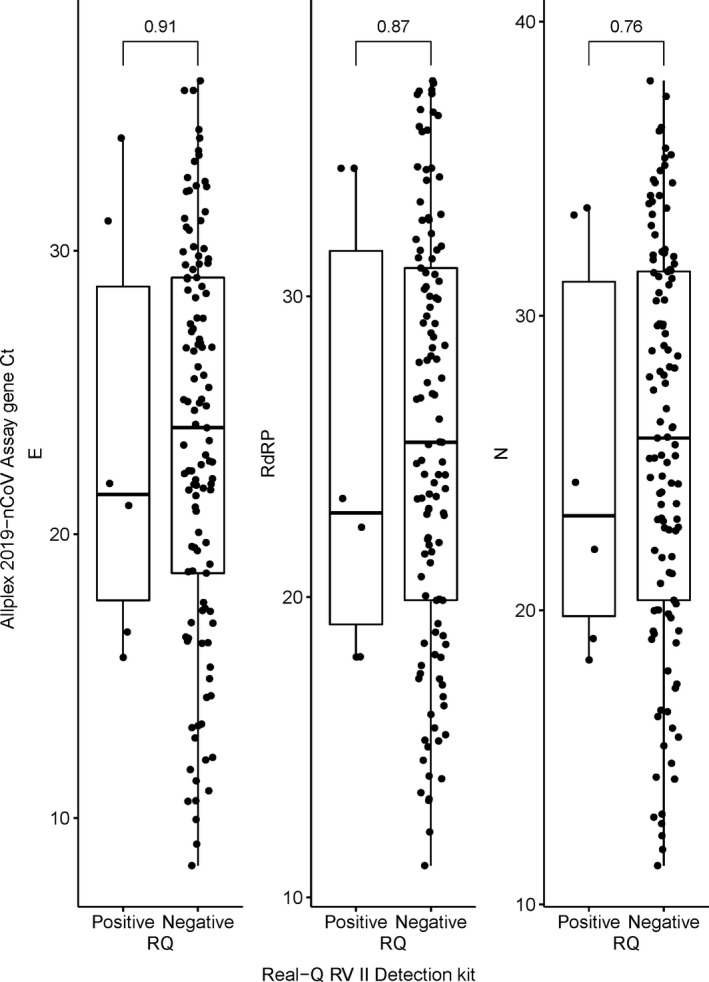
Box plots of E, RdRP, and N gene Ct values of Allplex 2019‐nCoV assay according to positive or negative results of the Real‐Q RV II Detection kit in the COVID‐19 PCR‐positive group. Notes: Comparison using the Wilcoxon test. Abbreviations: Ct, cycle threshold; E, envelope; N, nucleocapsid; RdRP, RNA‐dependent RNA polymerase; PCR, polymerase chain reaction; RQ, Real‐Q RV II Detection kit
4. DISCUSSION
An array of colonized viruses exists in the supposedly sterile pulmonary environment24, 25; however, the presence of these viruses may not cause an infection. Nevertheless, it is still necessary to detect these respiratory tract viruses to verify the assumption that they may cause a coinfection with SARS‐CoV‐2. Therefore, we performed RQ assays on patients who underwent SARS‐CoV‐2 testing for COVID‐19.
Of the 123 patients who tested positive on the SARS‐CoV‐2 PCR test, 4.9% were coinfected with an RTI‐causing virus. This was lower than the coinfection rates reported by Kim et al. (20.7%)13 and Ling Ma et al. (15.6%).26 While examining the coinfection rate, the fact that the time and regions differed across studies should be taken into account. Although we could not find studies confirming coinfection rates in different geographic locations in a single study period, different coinfection rates were found in East/Mid/West Asia, Europe, North Africa, and North/South America.27 Furthermore, as mentioned in previous studies,28, 29 the results could have differed due to the differences in the test kits (such as Allplex and RQ test kits) and obtained specimens (such as nasopharyngeal swabs). Besides, there could be limitations related to the multiplex PCR test method, which simultaneously detects multiple respiratory viruses as opposed to a single virus.
According to a previous study, there was a disagreement between the detection of upper and lower respiratory viruses using nasopharyngeal swabs.28 Karhu et al reported that in only 8 out of 24 patients, the virus which was detected in the trachea was found in the nasopharyngeal swab sample. However, in this study, we only used nasopharyngeal swab samples as the sputum samples were either inadequate or unavailable.
Figure 2 shows the results of the analysis conducted in patients who tested positive on the COVID‐19 PCR test. There were no statistically significant differences in the mean Ct values for E, RdRP, and N genes between patients who tested positive on the RQ test and those who tested negative on the RQ test. However, it should be noted that the analysis was only performed in 123 patients who tested positive on the COVID‐19 PCR test, and only 6 of these patients were coinfected with a virus that caused RTI. Further, the absence of statistically significant parameters might be because we did not conduct a subgroup analysis (e.g., sex and age) in the coinfected patients.
Coinfection and superinfection are possible not only with viruses but also with other pathogens, such as bacteria and fungi. A previous study performed a systematic review and meta‐analysis on coinfection and superinfection of SARS‐CoV‐2 and other microbial pathogens.27 We studied the coinfection of SARS‐CoV‐2 with other respiratory viruses only because we used the remaining specimens from virus transport media (e.g., UTM), which was obtained to confirm the existence of SARS‐CoV‐2. In a future study, we plan to use a protocol encircling virus as well as bacteria and fungi to check species, rate, and Ct difference of the pathogens causing coinfection.
In this study, we could confirm that coinfection of SARS‐CoV‐2 and a respiratory virus did not affect the Ct values of a real‐time RT‐PCR used for SARS‐CoV‐2 detection. The coinfection rates observed in this study differed from those reported in previous studies. However, this study had certain limitations. The study was conducted in a single hospital in a single region. To generalize the findings in terms of time and region, a larger number of specimens from multiple centers, collected at different times, should be analyzed. Further, the failure to use various test kits to detect respiratory viruses may have caused statistical bias. In brief, we computed the coinfection rate of SARS‐CoV‐2 with other RTI viruses and revealed that the mean Ct values for N, RdRP, and E genes in RT‐PCR tests were not significantly affected by coinfection.
CONFLICTS OF INTEREST
There are no potential conflicts of interest relevant to this article.
ACKNOWLEDGMENT
This work was supported by the 2020 Yeungnam University Research Grant.
Kim Z, Lee JH. Coinfection with severe acute respiratory syndrome coronavirus‐2 and other respiratory viruses at a tertiary hospital in Korea. J Clin Lab Anal. 2021;35:e23868. 10.1002/jcla.23868
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.
REFERENCES
- 1.COVID‐19 Weekly Epidemiological Update. World Health Organization. Published May 7, 2021 [Cited 2021 May 14]. Available from: https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20210309_weekly_epi_update_30.pdf?sfvrsn=4e7da248_8&download=true
- 2.Mahase E. Covid‐19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. 2020:m4714. 10.1136/bmj.m4714 [DOI] [PubMed] [Google Scholar]
- 3.Wouters OJ, Shadlen KC, Salcher‐Konrad M, et al. Challenges in ensuring global access to COVID‐19 vaccines: production, affordability, allocation, and deployment. Lancet. 2021;397:1023‐1034. 10.1016/s0140-6736(21)00306-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limb M. Covid‐19: What is happening with the vaccine rollout? BMJ. 2021:n213. 10.1136/bmj.n213 [DOI] [PubMed] [Google Scholar]
- 5.Choudhuri J, Carter J, Nelson R, et al. SARS‐CoV‐2 PCR cycle threshold at hospital admission associated with patient mortality. PLoS One. 2020;15:e0244777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough PA, Kelly RJ, Ruocco G, et al. Pathophysiological basis and rationale for early outpatient treatment of SARS‐CoV‐2 (COVID‐19) infection. Am J Med. 2021;134:16‐22. 10.1016/j.amjmed.2020.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SP, Pritam M, Pandey B, Yadav TP. Microstructure, pathophysiology, and potential therapeutics of COVID‐19: A comprehensive review. J Med Virol. 2021;93:275‐299. 10.1002/jmv.26254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu KK, Cerny J. Managing patients with hematological malignancies during COVID‐19 pandemic. Exp Rev Hematol. 2020;13:787‐793. [DOI] [PubMed] [Google Scholar]
- 9.Whittaker A, Anson M, Harky A. Neurological manifestations of COVID‐19: A systematic review and current update. Acta Neurol Scand. 2020;142:14‐22. 10.1111/ane.13266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolis AS, Manolis AA, Manolis TA, Melita H. COVID‐19 and acute myocardial injury and infarction: Related mechanisms and emerging challenges. J Cardiovasc Pharmacol Ther. 2021;26:107424842110110. 10.1177/10742484211011026 [DOI] [PubMed] [Google Scholar]
- 11.Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non‐influenza‐related viral respiratory tract infection in the United States. Arch Intern Med. 2003;163:487‐494. [DOI] [PubMed] [Google Scholar]
- 12.Yuen E, Gudis DA, Rowan NR, Nguyen SA, Schlosser RJ. Viral infections of the upper airway in the setting of COVID‐19: A primer for rhinologists. Am J Rhinol Allergy. 2021;35:122‐131. 10.1177/1945892420947929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co‐infection between SARS‐CoV‐2 and other respiratory pathogens. JAMA. 2020;323:2085. 10.1001/jama.2020.6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Di, Lu J, Ma X, et al. Coinfection of influenza virus and severe acute respiratory syndrome coronavirus 2 (SARS‐COV‐2). Pediatr Infect Dis J. 2020;39:e79‐e79. 10.1097/inf.0000000000002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touzard‐Romo F, Tapé C, Lonks JR. Co‐infection with SARS‐CoV‐2 and human metapneumovirus. R I Med J. 2013;2020:75‐76. [PubMed] [Google Scholar]
- 16.Alharthy A, Faqihi F, Karakitsos D. SARS‐CoV‐2 Complicated by sinusitis and co‐infection with human metapneumovirus. R I Med J. 2013;2020:23‐24. [PubMed] [Google Scholar]
- 17.Wu X, Cai Y, Huang X, et al. Co‐infection with SARS‐CoV‐2 and influenza A virus in patient with pneumonia, China. Emerg Infect Dis. 2020;26:1324‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak MD, Sordillo EM, Gitman MR, Paniz Mondolfi AE. Coinfection in SARS‐CoV‐2 infected patients: Where are influenza virus and rhinovirus/enterovirus? J Med Virol. 2020;92:1699‐1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang S, Liu P, Xiong G, et al. Coinfection of SARS‐CoV‐2 and multiple respiratory pathogens in children. Clin Chem Lab Med. 2020;58:1160‐1161. 10.1515/cclm-2020-0434 [DOI] [PubMed] [Google Scholar]
- 20.Kondo Y, Miyazaki S, Yamashita R, Ikeda T. Coinfection with SARS‐CoV‐2 and influenza A virus. BMJ Case Rep. 2020;13:e236812. 10.1136/bcr-2020-236812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García‐Martínez FJ, Moreno‐Artero E, Jahnke S. SARS‐CoV‐2 and EBV coinfection. Med Clin (English). 2020;155:319‐320. 10.1016/j.medcle.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuadrado‐Payán E, Montagud‐Marrahi E, Torres‐Elorza M, et al. SARS‐CoV‐2 and influenza virus co‐infection. Lancet. 2020;395:e84. 10.1016/s0140-6736(20)31052-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Team RC . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.R‐project.org/ [Google Scholar]
- 24.Dy R, Sethi S. The lung microbiome and exacerbations of COPD. Curr Opin Pulm Med. 2016;22:196‐202. [DOI] [PubMed] [Google Scholar]
- 25.Costa AN, Costa FMD, Campos SV, Salles RK, Athanazio RA. The pulmonary microbiome: challenges of a new paradigm. J Bras Pneumol. 2018;44:424‐432. 10.1590/s1806-37562017000000209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L, Wang W, Le Grange JM, et al. Coinfection of SARS‐CoV‐2 and Other Respiratory Pathogens. Infect Drug Resist. 2020;13:3045‐3053. 10.2147/idr.s267238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musuuza JS, Watson L, Parmasad V, Putman‐Buehler N, Christensen L, Safdar N. Prevalence and outcomes of co‐infection and superinfection with SARS‐CoV‐2 and other pathogens: A systematic review and meta‐analysis. PLoS One. 2021;16:e0251170. 10.1371/journal.pone.0251170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karhu J, Ala‐Kokko TI, Vuorinen T, Ohtonen P, Syrjala H. Lower respiratory tract virus findings in mechanically ventilated patients with severe community‐acquired pneumonia. Clin Infect Dis. 2014;59:62‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burk M, El‐Kersh K, Saad M, Wiemken T, Ramirez J, Cavallazzi R. Viral infection in community‐acquired pneumonia: a systematic review and meta‐analysis. Eur Respir Rev. 2016;25:178‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


