Abstract
microRNA (miRNA) plays important roles in regulating various biological processes, including host-pathogen interaction. Recent studies have demonstrated that virus-encoded miRNAs can manipulate host gene expression to ensure viral effective multiplication. Bombyx mori cypovirus (BmCPV), a double-stranded RNA virus with a segmented genome, is one of the important pathogens for the economically important insect silkworm. Our present study indicated that two putative miRNAs encoded by BmCPV could promote viral replication by inhibiting the gene expression of B. mori GTP-binding nuclear protein Ran (BmRan), an essential component of the exportin-5-mediated nucleocytoplasmic transport of small RNAs. BmCPV-miR-1 and BmCPV-miR-3 are two of the BmCPV-encoded miRNAs identified in our previous studies. BmRan is a common target gene of them with binding sites all located in the 3′-untranslated region (3′-UTR) of its mRNA. The expression levels of the two miRNAs in the midgut of larvae infected with BmCPV gradually increased with the advance of infection, while the expression of the target gene BmRan decreased gradually. The miRNAs and the recombinant target gene consisting of reporter gene mCherry and 3′-UTR of BmRan mRNA were expressed in HEK293T cells for validating the interaction between the miRNAs and the target gene. qRT-PCR results revealed that BmCPV-miR-1 and BmCPV-miR-3 negatively regulate target gene expression not only separately but also cooperatively by binding to the 3′-UTR of BmRan mRNA. By transfecting miRNA mimics into BmN cells and injecting the mimics into the body of silkworm larvae, it was indicated that both BmCPV-miR-1 and BmCPV-miR-3 could repress the expression of BmRan in BmN cells and in the silkworm, and the cooperative action of the two miRNAs could enhance the repression of BmRan expression. Furthermore, the repression of BmRan could facilitate the replication of BmCPV genomic RNAs. It is speculated that BmCPV-miR-1 and BmCPV-miR-3 might reduce the generation of host miRNAs by inhibiting expression of BmRan, thus creating a favorable intracellular environment for virus replication. Our results are helpful to better understand the pathogenic mechanism of BmCPV to the silkworm, and provide insights into one of the evasion strategies used by viruses to counter the host defense for their effective multiplication.
Keywords: silkworm, cypovirus, microRNA, target gene, BmRan, virus replication
Introduction
Bombyx mori cypovirus (BmCPV) is one of the important pathogens of the silkworm (Cao et al., 2012). It is a typical RNA virus belonging to the Cypovirus genus of Reoviridae. Its genome consists of 10 segmented double-stranded RNAs with a total length of more than 24 kbp, each encoding a structural or non-structural protein (Hagiwara et al., 2002; Hu et al., 2019). The virus infects only midgut epithelial cells in the silkworm and produces polyhedra in the cytoplasm. The intact virus particles are generally embedded in the polyhedra and must be released in the larval midgut to infect the silkworm. The infection of the virus to the silkworm always causes serious cytoplasmic polyhedrosis disease resulting in big losses to the commercial sericultural production. The mechanism of interaction between the virus and silkworm needs to be further explored for the development of effective strategies to control the occurrence and prevalence of the disease. In the silkworm, major antiviral defense mechanisms such as RNA interference (RNAi), NF-kB-mediated, Imd (immune deficiency), stimulator of interferon gene (STING), and Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathways have been shown to play important roles in antiviral immunity (Jiang, 2021). In contrast, viruses can modulate prophenol oxidase (PPO), phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt), and extracellular signal-regulated kinase (ERK) signaling pathways of the host to elevate their proliferation in the silkworm (Jiang, 2021). Transcriptome studies of silkworm have revealed a complex response against BmCPV infection. While, studies of deep sequencing of viral small RNAs have indicated the importance of the RNAi pathway in the control of cypovirus infection although many functional aspects still need to be elucidated and conclusive evidence is lacking (Swevers et al., 2020).
microRNA (miRNA) is a type of 19–25 nt single-stranded non-coding small RNAs, which is widely found in animals, plants, and nematodes, and regulates the expression of target genes at the post-transcriptional level (Filipowicz et al., 2008). A miRNA can target and regulate multiple target genes, and a target gene may also be regulated by multiple miRNAs. At the same time, studies have shown that the increase in the number of miRNA binding sites in mRNA 3′-untranslated region (3′-UTR) can enhance the translational repression of a target gene (Heiss et al., 2012). Similar studies also showed that co-operativity between two or more miRNA-binding sites enhanced repression of mRNA translation via an unknown mechanism when sites were separated by 13–35 nucleotides (Saetrom et al., 2007). Many viruses also encode miRNAs and 569 miRNAs of virus origin have been registered in the miRBase database version 22.1 (2018)1. Virus-encoded miRNAs play an important role in the intricate interaction between virus and hosts, including regulation of host immune response, evasion from recognition by the host immune system (Liang et al., 2014), inhibition of apoptosis (Zhao et al., 2011), regulation of cell cycle (Gottwein et al., 2007), mimicking host miRNAs (Grimson et al., 2007; Zhao et al., 2009), and so on. At present, most viral miRNAs reported are encoded by DNA viruses, but some RNA viruses can also encode functional miRNAs (Swaminathan et al., 2013; Qiu et al., 2018). Our previous deep sequencing of small RNA in the midgut of silkworm larvae infected by BmCPV virus identified some virus-derived non-coding RNA sequences similar to miRNA. Further study proved that a BmCPV-encoded miRNA can regulate the expression of host genes and affect the replication and proliferation of the virus (Guo et al., 2020).
The present work studies the functions of two putative BmCPV-encoded miRNAs, namely BmCPV-miR-1 and BmCPV-miR-3. Target gene prediction against the silkworm genome identified that the B. mori GTP-binding nuclear protein ran gene (BmRan) is the common target gene of the two miRNAs. Their regulation on the target gene and its influence on BmCPV virus replication were analyzed. Studying the BmCPV-encoded functional miRNAs and revealing their functions in the process of pathogen-host interaction would enrich the miRNA family encoded by viruses and help to reveal the miRNA-mediated new mechanism of regulation on RNA virus replication and proliferation.
Materials and Methods
Silkworm Strain, Cell Lines, and Virus
The domesticated silkworm of strain 4008 used in this study was supplied by Silkworm Germplasm Conservation Center, Chinese Academy of Agricultural Sciences. BmCPV, BmN cell, and HEK293T cell (human embryonic kidney cells) lines were maintained in our laboratory. miRNA mimics, inhibitors, and negative controls (NCs) were synthesized and chemically modified by Shanghai GenePharma Co., Ltd. The pmCherry-N1 plasmid, lentiviral expression vector pLNHX, pLKO.3G, and packaging plasmids pVSV-G, pSPAX2, and pMD2.G were purchased from Wuhan MiaoLingBio Inc. and kept in our laboratory.
Virus Inoculation and Tissue Collection
Bombyx mori cypovirus polyhedra suspension at a concentration of 1.0 × 108 mL–1 was coated on fresh mulberry leaves cut into 5 × 3 cm. The leaves coated with the virus suspension were fed to the 5th instar silkworm larvae and the average amount of virus ingested by each larva was calculated to be about 1.0 × 106 polyhedra. Another group of larvae fed with mulberry leaves coated with sterile water were used as blank control. When the mulberry leaves coated with virus or water were all eaten up (about 6 h), the larvae were given fresh mulberry leaves and reared under the standard condition of 14 h light and 10 h darkness and relative humidity of about 90%.
The larvae were dissected at 12, 24, 48, 72, and 96 h, respectively after inoculation to collect midguts. The collected midgut was rinsed in DEPC water to remove the attached mulberry leaf pieces and put into a cryotube after the extra water was absorbed with tissue paper. The midguts of every five larvae were mixed as one sample and three samples were taken at each time point. Then the samples were stored at −80°C after quick freezing in liquid nitrogen.
RNA Extraction
In this study, the expression level of both the virus-derived miRNAs and the host target gene, and the replication level of viral genome in the midgut of the silkworm larvae needed to be quantitatively detected, respectively. Therefore, total RNA was extracted with a classic manual method of the following steps. The frozen silkworm larval midguts were ground into fine powder with liquid nitrogen, put into an RNase free centrifuge tube. A total of 1 mL of lysate RL was added into the tube, and the mixture was shaken and mixed, then incubated at room temperature for 5 min. It was centrifuged at 4°C, 12000 rpm for 15 min. Then the supernatant was mixed with 200 μL of chloroform in a new tube, shaken sufficiently, and kept at room temperature for 10 min, followed by centrifugation at 4°C, 12000 rpm for 15 min. The upper aqueous phase was mixed with an equal volume of pre-cooled isopropanol in a new RNase free centrifuge tube, kept at 4°C for 10 min, then centrifuged at 4°C, 12000 rpm for 10 min. The supernatant was discarded, the pellet was washed with 1 mL of 75% ethanol and centrifuged at 4°C, 7500 rpm for 5 min. Then, the supernatant was discarded and the pellet was kept at room temperature for 5–10 min to allow the RNA pellet to dry. The RNA precipitate was dissolved with 30–50 μL of RNase-free water, and stored at −80°C after the concentration was measured.
cDNA Synthesis
cDNA was synthesized with a TaKaRa Primer ScriptTM RT reagent kit for reverse transcription (Takara Biomedical Technology Co., Ltd., China) according to the manufacturer’s instructions. For the first step, 2 μL of 5 × gDNA Eraser Buffer, 1 μL of gDNA Eraser, and 1 μg of RNA were mixed in the reaction tube to a total volume of 10 μL by adding ddH2O, then kept at 42°C for reaction for 2 min. Then, 10 μL of the reaction solution, 4 μL of 5 × Prime Script Buffer 2, 1 μL of Prime Script RT Enzyme Mix I, 1 μL of RT Primer, and 4 μL of ddH2O were mixed, and the reaction was performed with the program 37°C, 15 min, 85°C, 5 s, and 4°C + ∞. After the reaction, the synthesized cDNA was stored at −20°C. The cDNA for miRNA detection was synthesized with stem-loop RT primers. The stem-loop primers were designed with reference to literature (Chen et al., 2005) and the sequences are shown in Table 1.
TABLE 1.
Primer sequences for qRT-PCR.
| Primer name | Sequence (5′–3′) |
| Stem-loop reverse-transcribed PCR for BmCPV-miR-1 | |
|
RT: GTCGTATCCAGTGCAGGGTCCGAGGT ATTCGCACTGGATACGACTAGTGT |
|
| F: ACACTCCAGCTGGGGAAATGGACACAGGC | |
| Stem-loop reverse-transcribed PCR for BmCPV-miR-3 | |
|
RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCA CTGGATACGACATCAAGCC |
|
| F: ACACTCCAGCTGGGTAGGAGAATTAGCGCGG | |
| Universal | R: CCAGTGCAGGGTCCGAGGTA |
| mCherry | F: CTCAGTTCATGTACGGCTCCAAGG |
| R: GGAGTCCTGGGTCACGGTCAC | |
| Human β-actin | F: CTCCATCCTGGCCTCGCTGT |
| R: GCTGTCACCTTCACCGTTCC | |
| BmRan | F: GCCGTAACGACTTTGCTTTGGAAC |
| R: TTGCCAGTACCACCATCTCCTACC | |
| Genomic RNA S2 | F: GTTGAGCGTCAGCAGTCAGATCG |
| R: TGTTTACCCTGAGCAGCGTTATCG | |
| Genomic RNA S5 | F: CGCTTACAGGCAGTGGAATAGGAC |
| R: GCTCTAACACATCGCTGGGCTAAG | |
| Genomic RNA S10 | F: ACCGTCAGTGATTGCTCGTGTAAC |
| R: AGCGTCACCCTATCCGAAGACC | |
| Bmβ-actin | F: CCGTATGAGAAAGGAAATCA |
| R: TTGGAAGGTAGAGAGGGAGG | |
qRT-PCR
Primer Premier 5.0 software was used to design quantitative primers (Table 1) for target gene BmRan, reporter gene mCherry, and internal reference gene β-actin (Table 1), and the stem-loop primers for miRNA quantitative detection were designed with reference to literature (Chen et al., 2005). The reaction system was prepared according to the instructions of the SYBR premix Ex TaqTM kit (Takara Biomedical Technology Co., Ltd., China) (2 × SYBR Premix Ex TaqTM: 10 μL, ROX Reference Dye: 0.4 μL, upstream primer: 0.8 μL, downstream primer: 0.8 μL, cDNA: 1 μL, ddH2O: 7 μL, total volume: 20 μL). Three technical replicates were set for each quantitative reaction and the reaction was run on an ABI Prism fluorescence quantitative PCR instrument (Applied Biosystems, Foster City, CA, United States). The reaction program was 95°C for 45 s followed by 45 cycles of 95°C for 5 s, and 60°C for 31 s. Bmβ-actin and BmTIF-4A were used as internal reference genes for detection of target gene expression. The differences in gene expression levels were calculated using the relative quantitative 2–ΔΔCT method (Chang et al., 2009).
Target Gene Prediction
The target genes of BmCPV-miR-1 and BmCPV-miR-3 was predicted against the silkworm genome by the miRanda and Targetscan2 software. The genes that could be predicted by both the two software programs and that mainly participated in immune response, escape of immune recognition, regulation of cell apoptosis, regulation of cell cycle, etc. were selected as the candidate target genes. The minimum free energy of hybridization between miRNA and target gene mRNA was calculated via software RNAhybrid3.
Construction of Lentiviral Expression Vectors and Transfection of HEK293T Cells
To verify the miRNA regulation on target gene expression by binding to its target site in 3′-UTR of mRNA, the lentiviral expression vectors for expression of the target gene and the miRNAs were constructed respectively and transfected into HEK293T cells. The reporter gene mCherry sequence (731 bp) was obtained from the plasmid pmCherry-N1. The 570 bp sequence encoding 3′-UTR of the target gene (BmRan) mRNA was amplified from the midgut cDNA of the experimental silkworm strain 4008. They were ligated into lentiviral vector pLNHX sequentially to construct the expression vectors pLNHX-mCherry-RanUTR, carrying a recombinant target gene consisting of the reporter gene mCherry and the 3′-UTR of BmRan mRNA. The mCherry gene serves not only as the reporter for successful transfection but also as the substituent target gene for miRNA regulation. At the same time, the miRNA binding sites on the target gene mRNA 3′-UTR were mutated by Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech Co., Ltd., China) to construct the expression vector pLNHX-mCherry-RanUTR-mut, in which the miRNA binding sites were destroyed. On the other hand, the miRNA precursor sequence was cloned from the BmCPV genome and inserted into the lentiviral vector pLKO.3G (carrying EGFP reporter gene) to construct the expression vectors pLKO.3G-miR-1 and pLKO.3G-miR-3. The lentiviral vector inserts and primer sequences are shown in Table 2.
TABLE 2.
Sequences for construction of lentivirus expression vectors.
The single-underlined sequence is the restriction site, the double-underlined is the binding site for BmCPV-miR-1, the wavy-underlined is the binding site for BmCPV-miR-3, the boxed sequences are mature miRNAs.
HEK293T cells were inoculated into a 60 mm cell culture dish in which the cell density should be over 60%, and the transfection was conducted 20 h later. In a 1.5 mL EP tube, 5.3 μg of expression plasmid pLNHX-mCherry-RanUTR or pLNHX-mCherry-RanUTR-mut and 2.7 μg of packaging plasmid pVSV-G were mixed to 125 μL with serum-free DMEM medium (Shanghai Thermo Scientific, China). In another EP tube, 20 μL of transfection reagent (EntransterTM-H4000, Beijing Engreen, China) was mixed with 105 μL of serum-free medium and kept at room temperature for 5 min. Then the transfection reagent mixture was mixed thoroughly into the expression plasmid mixture and left to stand for 15 min. For transfection, the medium in the petri dish was removed and the above mixed solution was added to the cells then fresh medium was added up to 5 mL. At 6 h after the transfection, the culture medium was replaced by the fresh medium containing fetal bovine serum (FBS) (Shanghai Thermo Scientific) and the cells were cultured for 24–48 h before the next transfection. When the cells showing red fluorescence accounted for about 80%, they were inoculated into a six-well plate in which the cell density should be over 60%. In a 1.5 mL EP tube, 2 μg of pLKO.3G-miR-1 or pLKO.3G-miR-3, 1.5 μg of packaging plasmid pMD2.G, and 0.5 μg of packaging plasmid pSPAX2 were mixed with serum-free medium to 50 μL. At the same time, 10 μL of transfection reagent (EntransterTM-H4000, Beijing Engreen) and 40 μL of serum-free culture medium were mixed in another EP tube and kept at room temperature for 5 min. Then the solution in the two tubes was mixed thoroughly and left to stand for 15 min. The medium in the cell culture was removed, the mixture was added to the cells, and then serum-containing medium was added up to 2 mL. The cells were collected 24 h and 48 h after transfection for RNA extraction and for detection of changes in the expression of reporter gene mCherry by qRT-PCR with human β-actin as the internal reference.
BmN Cell Transfection
To verify the miRNA regulation on target gene expression in BmN cells, miRNA mimics were transfected into the BmN cells to detect the changes in expression of the target gene. miRNA mimics and NC (sequences shown in Table 3) were synthesized by Shanghai GenePharma Co., Ltd. BmN cells were inoculated into a six-well plate 16 h before transfection in which the cell density should be over 60%, and they were divided into three groups, i.e., blank control, NC, and mimics, each group was set for three replicates. In a 1.5 mL EP tube, 3.33 μg of mimics or NC was mixed with serum-free TC-100 medium (Sangon Biotech Co., Ltd., Shanghai, China) up to 25 μL. In another tube, 5 μL of transfection reagent (EntransterTM-R4000, Beijing Engreen) was mixed with 20 μL of serum-free medium and kept at room temperature for 5 min. The solution in the two tubes was mixed thoroughly and left to stand for 15 min before transfection. For the transfection, the medium in the six-well cell culture plate was removed, the mixture solution was added to the cells, and culture medium was added up to 2.5 mL. Cells were collected at 24, 48, and 72 h after the transfection for RNA extraction and qRT-PCR detection of the changes in target gene expression with Bmβ-actin as internal reference.
TABLE 3.
Sequences of miRNA mimics, inhibitors, and NC.
| Name | Sequence (5′–3′) | |
| BmCPV-miR-1 | Mimic | GAAAUGGACACAGGCACACUA GUGUGCCUGUGUCCAUUUCUU |
| Inhibitor | UAGUGUGCCUGUGUCCAUUUC | |
| BmCPV-miR-3 | Mimic | UAGGAGAAUUAGCGCGGCUUGAU CAAGCCGCGCUAAUUCUCCUAUU |
| Inhibitor | AUCAAGCCGCGCUAAUUCUCCUA | |
|
| ||
| Negative control (NC) | AGAAGCUUAGUCGUGUCGGAUGA | |
Verification of miRNA Function in Silkworm
To study the regulation of miRNAs on target genes and their influence on virus replication and proliferation in silkworm larvae, the synthesized miRNA mimics, inhibitors (sequence shown in Table 3), and NC were respectively injected into the fifth instar larvae of normal silkworm, 2 μg for each larva. The midgut was dissected at 24, 48, 72, and 96 h after the injection. Total RNA was extracted as described above and the expression level of target gene was detected by qRT-PCR with Bmβ-actin as the internal reference gene.
At the same time, the 5th instar larvae of the silkworm were orally infected with BmCPV, 12 h later, miRNA mimics, inhibitors, and NC were injected respectively into the body cavity of the larvae. At 24, 48, 72, and 96 h after the injection, the midgut tissues were dissected for RNA extraction. Then the replication levels of the second, fifth, and tenth segment of BmCPV genomic RNA were detected respectively by qRT-PCR with Bmβ-actin as internal reference.
Statistical Analysis Methods
One-way ANOVA analysis in the GraphPad Prism package was used to analyze the experimental data statistically. The results are shown as the mean ± standard error (SE) of three independent treatments. Asterisks denote significant differences as compared with the control group, as indicated by ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001.
Results
Stem-Loop PCR Identification of BmCPV-miR-1 and BmCPV-miR-3
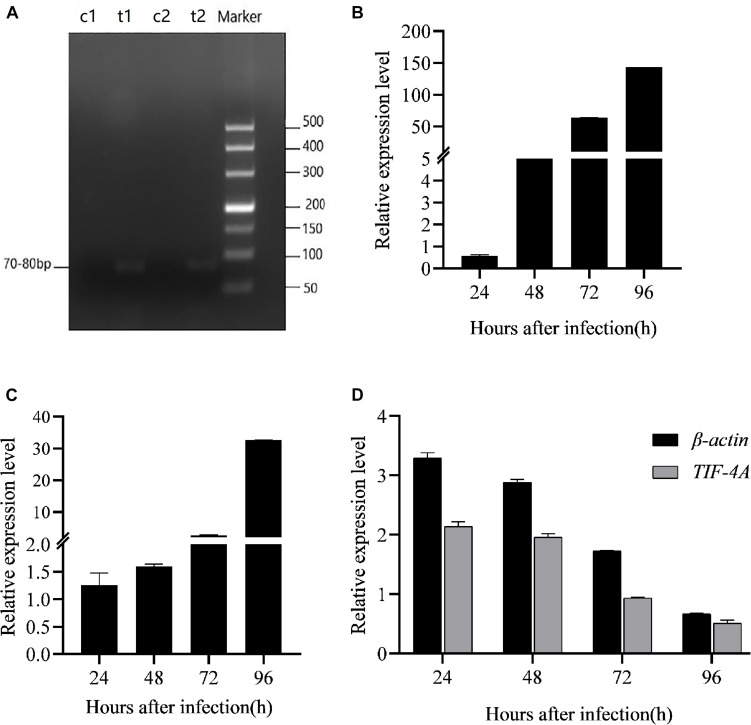
In our previous small RNA sequencing data (data deposited in NCBI Sequence Read Archive Database4, accession number SRP158739) of the midgut of BmCPV-infected silkworm larvae, we found two miRNA-like small RNAs encoded by the first and third segment of BmCPV genomic RNA, with the sequencing abundances 1375 and 2710, respectively. They were named BmCPV-miR-1 (sequence: GAAAUGGACACAGGCACACUA, located at the 5P arm of its precursor sequence) and BmCPV-miR-3 (sequence: UAGGAGAAUUAGCGCGGCUUGAU, located at the 3P arm of its precursor sequence). With stem-loop RT-PCR detection, obvious bands were detected in midgut tissue of the 5th instar silkworm larvae infected with BmCPV (Figure 1A), with a band size of 70–80 bp respectively, while no bands were detected in midgut tissues of normal larvae, indicating that both BmCPV-miR-1 and BmCPV-miR-3 are derived from BmCPV.
FIGURE 1.
Detection of expression patterns of BmCPV-miRNAs and target gene BmRan in the midgut of silkworm larvae infected with BmCPV. (A) BmCPV-miR-1 and BmCPV-miR-3 detected by stem-loop RT-PCR. The bands sized about 70–80 bps were the putative miRNAs. c: control; t: BmCPV infected; t1: BmCPV-miR-1; t2: BmCPV-miR-3. (B) Expression pattern of BmCPV-miR-1. (C) Expression pattern of BmCPV-miR-3. (D) Expression pattern of BmRan with normalization to different internal reference genes. The expression of BmCPV-miRNAs gradually increased, while the expression of the target gene BmRan gradually decreased, as the infection advanced (n = 3 replicate samples, each containing five larvae).
Target Gene Prediction
miRanda and Targetscan software were used to predict the target genes of both BmCPV-miR-1 and BmCPV-miR-3 against the silkworm genome5. As a result, B. mori GTP-binding nuclear protein Ran gene (BmRan, NCBI accession number: NM_001046809.1) was predicted to be the common target gene of these two miRNAs. The binding sites for the two miRNAs are all located in the 3′-UTR region of the BmRan mRNA sequence with an interval of 284 nt. The binding site of BmCPV-miR-1 is located at 11∼34 nt and that of BmCPV-miR-3 is located at 319∼339 nt, both with an individual specific region complementary to the seed sequence of the miRNAs. The binding free energy of the two miRNAs to their respective target sites is lower than −20 kcal/mol (BmCPV-miR-1: −20.8 kcal/mol, BmCPV-miR-3: −22.7 kcal/mol). GTP-binding nuclear protein Ran is a 25 kDa protein and an important component of exportin-5-mediated nucleocytoplasmic transport. It is mainly involved in the transport of small RNA from nucleus to cytoplasm (Bischoff and Ponstingl, 1991). In the process of nuclear export of pre-miRNA, exportin-5 binds pre-miRNA in a Ran-GTP dependent manner. The depletion of Ran protein level leads to a significant reduction in nuclear export of pre-miRNA (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004; Wang et al., 2011). Drosophila exportin-5 can transport pre-miRNA and tRNA (Shibata et al., 2006). However, the mechanism of small RNA transport in the silkworm and its influence factors have not been reported yet. Therefore, the BmRan gene of the silkworm was selected in this study to evaluate the function of the two BmCPV-derived miRNAs.
Expression of BmRan Is Inversely Correlated With Both BmCPV-miR-1 and BmCPV-miR-3 Levels in the Larvae Infected With BmCPV
Fifth-instar silkworm larvae were inoculated per os with BmCPV, and midgut tissues were collected at 12, 24, 48, 72, and 96 h post-infection. Total RNA was extracted for reverse transcription and for detection of the expressional changes of miRNAs and the target gene. The results indicated that the expression level of BmCPV-miR-1 and BmCPV-miR-3 in the virus-infected midgut gradually increased with the advance of infection (Figures 1B,C), while the expression of the target gene BmRan was gradually downregulated as the time advanced after infection (Figure 1D). To ensure the detection objectivity of the target gene expression, two internal reference genes (Bmβ-actin and BmTIF-4A) were used to normalize the qRT-PCR detection data. The overall trend in changes of the target gene expression among the different time points after BmCPV infection was the same, although the calculated expression level of the target genes was slightly different at a specific time point as normalized to different reference genes (Figure 1D). It implies that BmCPV-miR-1 and BmCPV-miR-3 might be inversely correlated with the expression of the target gene BmRan. It can be speculated that BmCPV-miR-1 and BmCPV-miR-3 have negative regulatory effects on BmRan.
BmCPV-miR-1 and BmCPV-miR-3 Negatively Co-regulate Target Gene Expression by Binding to 3′-UTR of BmRan mRNA
The constructed expression vectors pLNHX-mCherry-RanUTR and pLNHX-mCherry-RanUTR-mut, both carrying the recombinant target gene consisting of the 731 bp mCherry reporter gene and the 570 bp sequence encoding the BmRan mRNA 3′-UTR, were confirmed to be correct by restriction enzyme digestion and sequencing of the inserted fragments. The mCherry gene serves here not only as the reporter for successful transfection but also as the substituent target gene for miRNA regulation. On the other hand, the constructed miRNA expression vectors pLKO.3G-miR-1 and pLKO.3G-miR-3 were confirmed by PCR amplification and the specific bands of 87 bp pre-miR-1 and 88 bp pre-miR-3 were detected, then the PCR products were further sequenced as the miRNA precursors of the two miRNAs. These results indicated that the expression vectors were all constructed successfully.
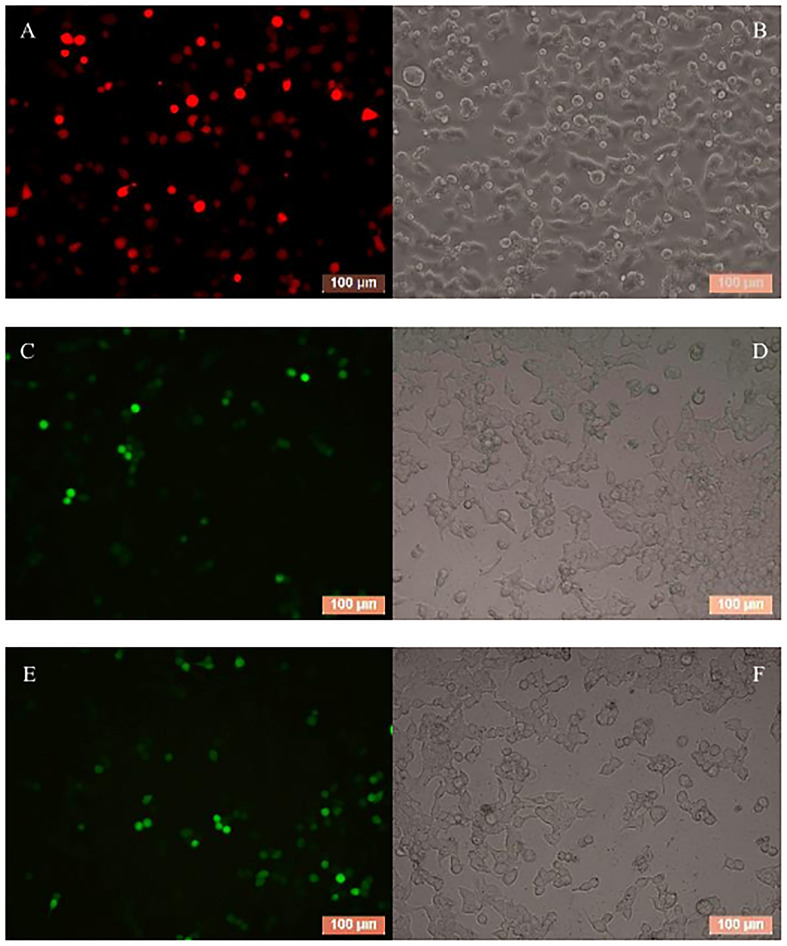
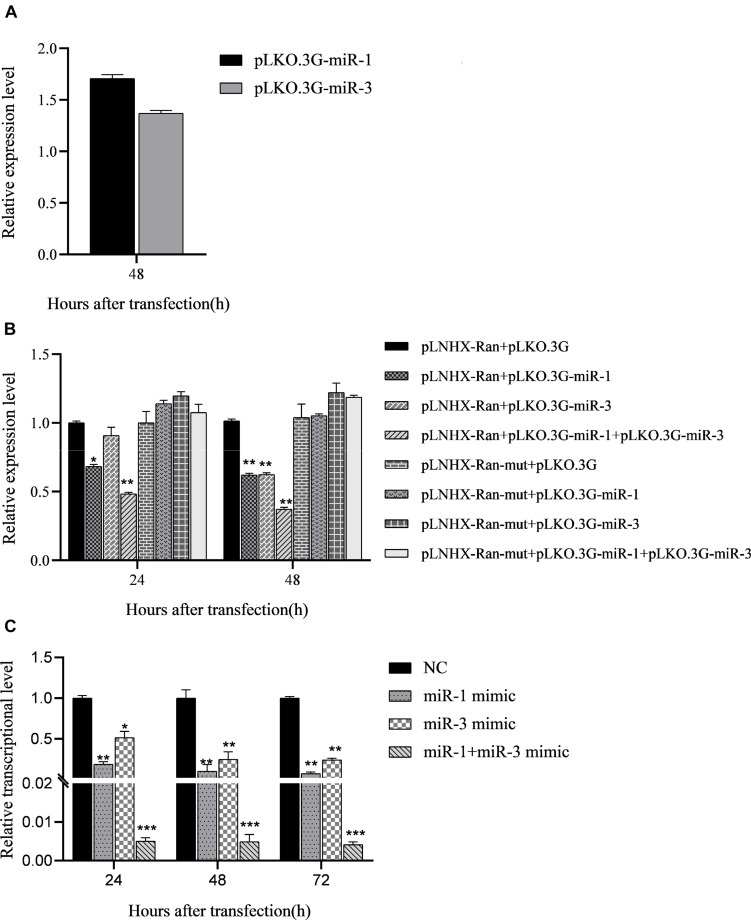
To evaluate the effect of BmCPV-miR-1 and BmCPV-miR-3 bound to the binding sites in 3′-UTR on the expression of the mCherry gene (that served as a substituent target gene), HEK293T cells were transfected successively with the above constructed lentiviral expression vectors for mCherry, BmCPV-miR-1, and BmCPV-miR-3. HEK293T cells were firstly co-transfected with the recombinant plasmid pLNHX-mCherry-RanUTR or pLNHX-mCherry-RanUTR-mut and packaging plasmid pVSV-G. After 48 h, about 80% of cells showed red fluorescence (Figures 2A,B), indicating that the recombinant expression vector was successfully transfected into the cells and the red fluorescent protein was stably expressed. Then, the HEK293T cells were further transfected with the plasmid pLKO.3G-miR-1 and pLKO.3G-miR-3 respectively, meanwhile another group of cells transfected with the pLKO.3G plasmid were used as NC. The transfection efficiency was about 60% (Figures 2C–F) at 20 h post-transfection based on the number of cells showing green fluorescence. Then, cells were collected at 24 and 48 h after transfection of the miRNA expression vectors, and the expression of the miRNAs and the red fluorescent protein gene was quantitatively detected. The expression level of both the miRNAs BmCPV-miR-1 and BmCPV-miR-3 gradually increased with the progression of time after transfection of the miRNA expression vectors as detected by qRT-PCR, indicating that the miRNA expression vectors were not only successfully transfected into HEK293T cells, but the miRNAs were also expressed (Figure 3A). At the same time, the expression of the mCherry gene in the experimental groups transfected with the miRNA expression vectors decreased (Figure 3B), furthermore the downregulated expression of the mCherry gene in the cells co-transfected with both pLKO.3G-miR-1 and pLKO.3G-miR-3 was more significant than that in the cells transfected with only pLKO.3G-miR-1 or pLKO.3G-miR-3 (Figure 4B). While, the expression level of the mCherry gene in the NC cells transfected with pLKO.3G was stable (Figure 3B). However, neither pLKO.3G-miR-1 nor pLKO.3G-miR-3 could downregulate the expression of the mCherry gene after mutation of the miRNA binding sites on 3′-UTR (Figure 3B). These results implied that both pLKO.3G-miR-1 and pLKO.3G-miR-3 could inhibit the expression of the target gene by binding to 3′-UTR of mRNA, and their cooperative action could enhance the repression of the target gene expression.
FIGURE 2.
HEK293T cells transfected with the targe gene expression vector pLNHX-mCherry-RanUTR (A,B), the miRNA expression vectors pLKO.3G-miR-1 (C,D) and pLKO.3G-miR-3 (E,F) respectively. (A,C,E) Observed under fluorescent microscope (A, under green light; C,E, under red light); (B,D,F) Observed under optical microscope. Cells showing red fluorescence indicate successful transfection and transfection efficiency of target gene expression vector (A); cells showing green fluorescence indicate the successful transfection and transfection efficiency of miRNA expression vectors (C,E).
FIGURE 3.
Regulation of BmCPV-miRNAs on the target gene in HEK293T cells and in BmN cells. (A) BmCPV-miRNAs were expressed in the transfected HEK293T cells. (B) BmCPV-miRNAs downregulate the expression of the substituent target gene mCherry in HEK293T cells. pLNHX-Ran: pLNHX-mCherry-RanUTR; pLNHX-Ran-mut: pLNHX-mCherry-RanUTR-mut. (C) Regulation of BmCPV-miRNA mimics on expression of target gene BmRan in BmN cells (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001).
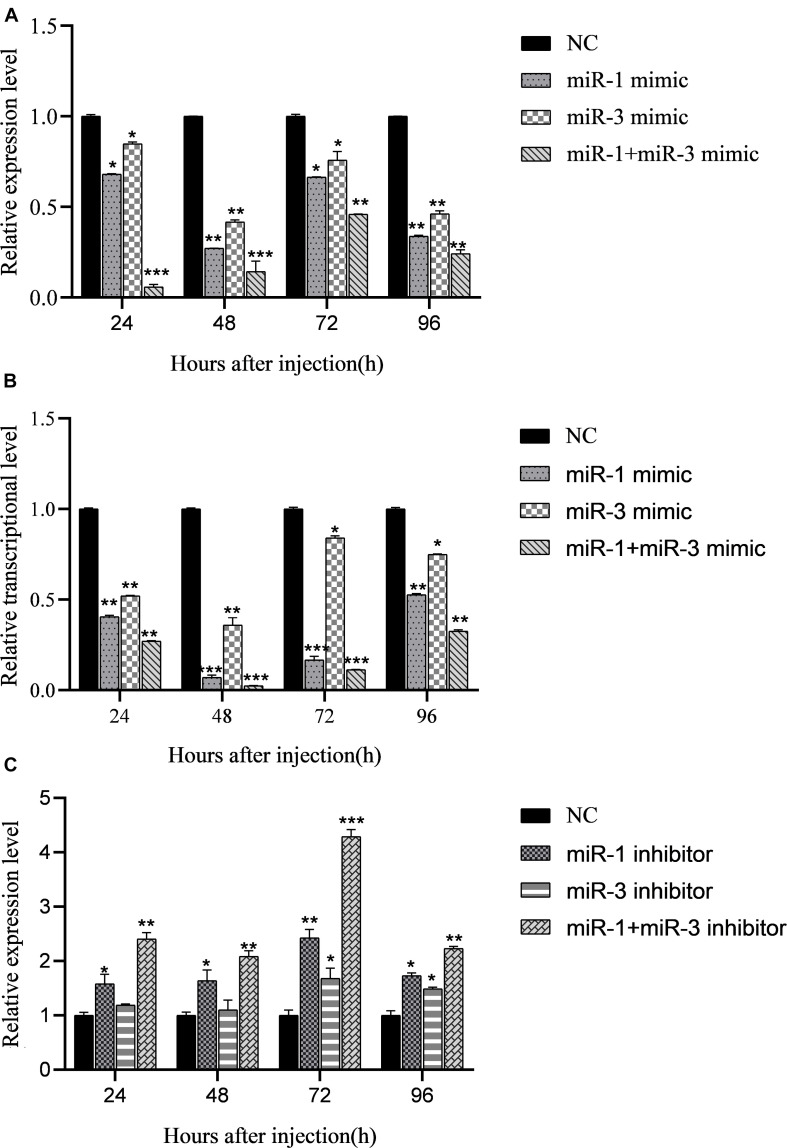
FIGURE 4.
Regulation of miRNA mimics on expression of target gene BmRan in midgut of silkworm larvae. Result shows that BmCPV-miRNAs downregulate the expression of BmRan in vivo in the silkworm. (A) In midgut of normal silkworm larvae. (B) In midgut of silkworm larvae infected with BmCPV. (C) Regulation of BmCPV-miRNA inhibitors on expression of BmRan in midgut of silkworm larvae infected with BmCPV (n = 3 replicate samples, each containing five larvae, *p < 0.05, **p < 0.01, ***p < 0.001).
BmCPV-miR-1 and BmCPV-miR-3 Mimics Negatively Regulate BmRan Expression in BmN Cells
In order to validate the regulation effect of the two BmCPV-miRNAs on target gene BmRan, BmN cells were transfected respectively with BmCPV-miR-1 and BmCPV-miR-3 mimics or NC. Cells were collected at 24, 48, and 72 h after transfection, and the expression changes of the target gene were detected by qRT-PCR. The quantitative detection results (Figure 3C) showed that compared with the cells transfected with NC, the expression of the target gene BmRan was downregulated in the cells transfected with the BmCPV-miR-1 mimics or BmCPV-miR-3 mimics. At the same time, the expression of the target gene BmRan was downregulated more significantly in the cells co-transfected with both the two miRNA mimics (Figure 3C). It indicated that the mimics of BmCPV-miR-1 and BmCPV-miR-3 could effectively repress the expression of the BmRan gene, and furthermore the repression of the expression was enhanced when the mimics of the two miRNAs acted cooperatively. This is consistent with the regulatory effect of BmCPV-miRNAs on the target gene verified by lentiviral expression vectors in HEK293T cells. These results implied the negative regulation of both BmCPV-miR-1 and BmCPV-miR-3 on expression of the target gene BmRan.
BmCPV-miR-1 and BmCPV-miR-3 Downregulate BmRan Expression in the Midgut of Silkworm Larvae
To verify the regulatory effects of BmCPV-miR-1 and BmCPV-miR-3 on the expression of the target gene in vivo in the silkworm, the mimics of BmCPV-miR-1 and BmCPV-miR-3 were injected into the normal fifth instar larvae separately or jointly to detect the expressional changes of BmRan. The larvae injected with NC were used as NC. The results showed that the expression level of the target gene BmRan in the midgut of larvae injected with mimics was lower than that in the NC injection group at all the time points of 24, 48, 72, and 96 h after the injection (Figure 4A), and further lower in the larvae injected with both the two miRNA mimics. The results indicated that BmCPV-miR-1 and BmCPV-miR-3 could also inhibit the expression of the target gene BmRan in silkworm larvae and have a co-operativity in the repression.
At the same time, the mimics and the inhibitors of BmCPV-miR-1 and BmCPV-miR-3 and the NC were injected into the fifth instar larvae infected with BmCPV to further validate the expression changes of the BmRan gene. The results showed that the expression level of the BmRan gene in the mimic injection groups was also lower than that in the NC group, with the largest decrease of 40.9 times at 48 h (Figure 4B), while the expression level in the inhibitor injection groups was higher than that in the NC group (Figure 4C) with the highest increase of 4.28 times at 72 h. Furthermore, in the larvae injected with both the two miRNA mimics, the repression of BmRan expression was more significantly enhanced, while inhibition of both the miRNAs by injecting both miRNA inhibitors resulted in a much higher BmRan expression level. These implied that increasing the level of the BmCPV-miRNAs could inhibit the expression of the BmRan gene, while inhibiting the effect of the miRNAs could effectively increase the expression level of the target gene.
Effect of the BmCPV-miRNAs on BmCPV Replication in the Midgut of Silkworm Larvae
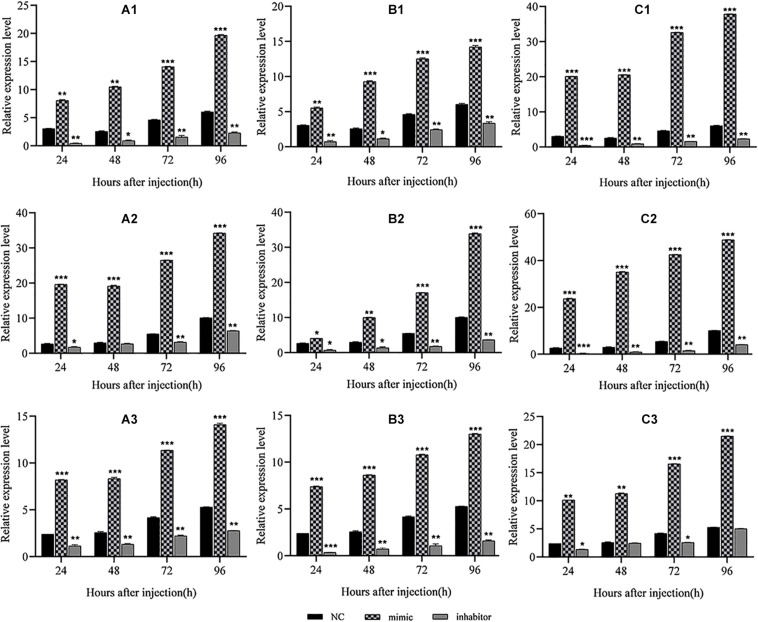
The above results verified the negative regulation of the two BmCPV-miRNAs, BmCPV-miR-1 and BmCPV-miR-3, on the target gene BmRan both in BmN cells and in vivo in the silkworm body. Whether the regulation of BmCPV-miR-1 and BmCPV-miR-3 on the BmRan gene affects the replication of the BmCPV genome was also evaluated by injecting the miRNA mimics, inhibitors, and NC into the fifth instar larvae infected with BmCPV and detecting the replication of the second (S2), fifth (S5), and tenth (S10) segment of BmCPV genomic RNA at the time points of 24, 48, 72, and 96 h post-injection. The results showed that the replication levels of the three RNA segments of the viral genome exhibited the same trend, and they all gradually increased with advanced time (Figure 5), which indicated viral replication. While, compared with the NC group, the replication level of the three RNA segments in the silkworm larvae injected with the miRNA mimics increased much faster (Figures 5A1,B1,A2,B2,A3,B3). However, the replication level of the three RNA segments in the larvae injected with inhibitors was lower than that in the NC group (Figures 5A1,B1,A2,B2,A3,B3). Furthermore, increasing the level of both the miRNAs by injecting the two mimics promoted viral replication more significantly than by any single miRNA, while inhibition of both the two miRNAs resulted in further lower replication than inhibition of any single miRNA (Figures 5C1–C3). This indicated that the replication of BmCPV was enhanced by increasing the level of miRNAs, but repressed by a decrease of the miRNA level. Therefore, it can be speculated that BmCPV-miR-1 and BmCPV-miR-3 could create a favorable intracellular environment and thus promote virus replication by inhibiting the expression of the target gene BmRan.
FIGURE 5.
Regulation of miRNA mimics and inhibitors on replication of the BmCPV genome in midgut of silkworm larvae. Result shows that the BmCPV-miRNAs could enhance the replication of BmCPV genomic RNAs. (A1) BmCPV-miR-1 to S2; (B1) BmCPV-miR-3 to S2; (C1) BmCPV-miR-1 + BmCPV-miR-3 to S2; (A2) BmCPV-miR-1 to S5; (B2) BmCPV-miR-3 to S5; (C2) BmCPV-miR-1 + BmCPV-miR-3 to S5; (A3) BmCPV-miR-1 to S10; (B3) BmCPV-miR-3 to S10; (C3) BmCPV-miR-1 + BmCPV-miR-3 to S10. S2, S5, and S10 represent the 2nd, 5th, and 10th segment of BmCPV genomic RNA, respectively (n = 3 replicate samples, each containing five larvae, *p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
Virus-encoded miRNA plays an important role in the process of virus-host interaction. Its small molecules, non-antigenicity, and target specificity make it a potential strategy for the virus to counter host defense mechanisms. Most of the presently reported viral miRNAs are encoded by DNA viruses, but some RNA viruses can also encode functional miRNAs (Swaminathan et al., 2013; Fani et al., 2018), such as Ebola virus (EBOV) (Liang et al., 2014; Chen et al., 2016; Qiu et al., 2018), hepatitis A virus (HAV) (Shi et al., 2014), bovine leukemia virus (BLV) (Rosewick et al., 2013), and Marek’s disease virus (MDV) (Zhao et al., 2011), etc. Studies have shown that one miRNA can target multiple genes, and a target gene can also be regulated by several miRNAs. Generally, miRNA mainly binds to the 3′-UTR of mRNA to repress the target gene translation, and co-operativity between two or more miRNA-binding sites can enhance repression of the mRNA translation (Saetrom et al., 2007; Fang and Rajewsky, 2011; Trobaugh et al., 2014; Liu et al., 2015). On the other hand, some miRNAs bind to the 5′-UTR of mRNA and upregulate the expression of target genes (Jopling et al., 2005; Ørom et al., 2008; Helwak et al., 2013; Hussain et al., 2013). In addition, some miRNAs can also bind to the CDS region and downregulate the expression of the target gene (Hausser et al., 2013; Pan et al., 2017).
Bombyx mori cypovirus has a larger and double-stranded RNA genome, which makes the virus potentially able to encode functional miRNAs, and the miRNAs encoded by the virus might play an important role in the virus-host interaction and in virus replication. In the present study, two miRNAs namely BmCPV-miR-1 and BmCPV-miR-3, which are encoded by the first and third segment of BmCPV genomic RNA, respectively, and their regulatory effects on target genes and then on virus replication and proliferation were studied. BmCPV-miR-1 and BmCPV-miR-3 share a common target gene, B. mori GTP-binding nuclear protein Ran (BmRan), and their binding sites on BmRan mRNA are all located in the 3′-UTR region. qPCR results showed that the expression levels of BmCPV-miR-1 and BmCPV-miR-3 in the midgut of virus-infected larvae gradually increased with the progression of infection, while the expression level of the target gene BmRan gradually decreased, indicating that both the two miRNAs were negatively correlated with the expression of target gene BmRan. Many virally encoded miRNAs were reported to downregulate the expression of host target genes via binding to the 3′-UTR of their mRNA (Stern-Ginossar et al., 2007; Singh et al., 2012; Skalsky et al., 2012; Fani et al., 2018). Therefore, our results implied that BmCPV-miR-1 and BmCPV-miR-3 might downregulate the expression of the target gene BmRan.
At present, the regulation of miRNAs on target genes is mostly verified using the dual luciferase reporter system, but both miRNA and the target gene can only be expressed transiently. However, the lentivirus expression system has the characteristics of high transfection efficiency and expression stability. Therefore, we employed the lentivirus expression vectors to express the miRNAs and the target gene respectively, and to evaluate the interaction between the BmCPV-encoded miRNAs and their shared target gene. In the system, the mCherry gene and the cDNA sequence encoding 3′-UTR of BmRan mRNA were combined into an expression vector, in which the mCherry gene served as both the reporter gene for successful transfection and the substituent target gene for the miRNAs. The results showed that both BmCPV-miR-1 and BmCPV-miR-3 could downregulate the expression of the target gene, and furthermore they had a co-operativity in the regulation.
At the same time, the negative regulation of BmCPV-miR-1 and BmCPV-miR-3 on the target gene BmRan was also confirmed in the cultured BmN cells by transfection of miRNA mimics, and in vivo in the silkworm by injecting miRNA mimics into both the normal and BmCPV-infected silkworm larvae. Furthermore, injecting miRNA mimics into the larvae enhanced the replication of the tested second, fifth, and tenth segment of the viral genome RNA. This implied that BmCPV-miR-1 and BmCPV-miR-3 encoded by BmCPV might promote the replication and proliferation of the virus by inhibiting the expression of the target gene BmRan.
In plants and invertebrates such as insects, host miRNAs serve as an important antiviral mechanism and degrade viral RNA into siRNAs, which then bind to the virus genome to inhibit viral replication (Ding and Voinnet, 2007; Carl et al., 2013; Bernier and Sagan, 2018). The biological generation of host miRNAs includes the transcription of pri-miRNA containing a stem-loop structure (Lagos-Quintana et al., 2003), which is then digested into pre-miRNA containing a hairpin structure by the Drosha enzyme in the nucleus. Then the pre-miRNA is transported from the nucleus to the cytoplasm, where it is cleaved into mature miRNA by the Dicer enzyme.
GTP-binding nuclear protein Ran is a 25 kDa transporter and serves as an important part of the exportin-5-mediated nucleocytoplasmic transport. It plays an important role in the transport of various non-coding RNAs and proteins from the nucleus to the cytoplasm, and mainly participates in the transport of small RNAs (Bischoff and Ponstingl, 1991). The exportin-5 of Drosophila can transport pre-miRNA and tRNA (Shibata et al., 2006). Research has shown that the nuclear export protein exportin-5 (Exp5) binds specifically to pre-miRNA in a Ran-GTP dependent manner (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004). Then, the pre-miRNA/Exp5/Ran-GTP complex migrates to cytoplasm, where the hydrolysis of Ran-GTP to Ran-GDP induces the release of pre-miRNA. The released pre-miRNA is further processed by the RNase III enzyme called Dicer to release the mature miRNAs (Hutvágner et al., 2001; Ketting et al., 2001). The depletion of Ran results in significant reduction of pre-miRNA export (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004). Other studies have shown that the combination of the pre-miRNA/Exp5/Ran-GTP complex can significantly reduce the efficiency of Dicer cutting pre-miRNA (Kim, 2004; Zeng and Cullen, 2004). In addition, overexpression of the pre-miRNA/Exp5/Ran-GTP complex increased the level of miRNA in the transfected cells, while RNAi-mediated knock-down of the pre-miRNA/Exp5/Ran-GTP complex inhibited the production of mature miRNA. Therefore, the pre-miRNA/Exp5/Ran-GTP complex not only acts as a nuclear export factor for pre-miRNA, but also protects pre-miRNA from degradation, thus promoting the formation of miRNA. Our results indicate that BmCPV-miR-1 and BmCPV-miR-3 may enhance the replication and proliferation of BmCPV by inhibiting the expression of target gene BmRan. The probable reason is that BmCPV-miR-1 and BmCPV-miR-3 bind to the 3′-UTR region of BmRan mRNA, negatively regulate the translation of the BmRan protein, thus inhibit the transport of pre-miRNA from the nucleus to the cytoplasm in host cells, reduce the population of host miRNAs, and consequently create a favorable intracellular environment for BmCPV genome replication and virus multiplication. Based on this speculation, the function mechanism of BmCPV-miR-1 and BmCPV-miR-3 on regulation of target gene BmRan and on replication of BmCPV is proposed (Figure 6). B. mori nuclear polyhedrosis virus (BmNPV) is another important virus pathogen of the silkworm, which is a double-stranded DNA virus belonging to Baculoviridae. The miRNA bmnpv-miR-1 encoded by BmNPV also represses the expression of Ran in the silkworm, leading to the reduction in the host small RNA population, as a consequence, the BmNPV load increases significantly in the infected larvae (Singh et al., 2012). In contrast, blockage of the host miRNA, bmo-miR-8, which targets the immediate-early gene of the virus and whose production was repressed upon bmnpv-miR-1 and Ran dsRNA administration, also resulted in a significant increase in the virus load in the infected silkworm larvae. While inhibition of BmNPV-miR-1 resulted in the significant expression of Ran and the decrease in BmNPV load in the BmNPV-infected larvae (Singh et al., 2012). These results, including those in our present study, provide insights into one of the evasion strategies used by these viruses to counter the host defense for their effective multiplication.
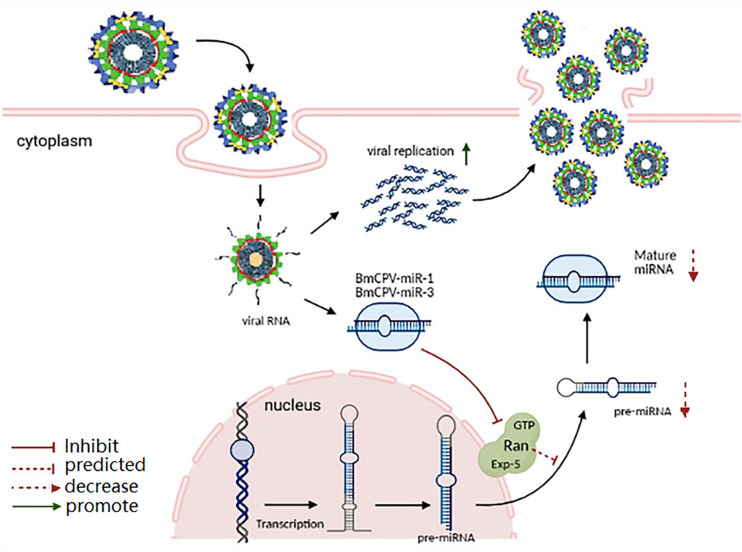
FIGURE 6.
Proposed function mechanism of BmCPV-miR-1 and BmCPV-miR-3 on regulation of the target gene BmRan and on replication of BmCPV. BmCPV-miR-1 and BmCPV-miR-3 negatively regulate the translation of the BmRan protein, thus inhibit the transport of pre-miRNA from the nucleus to the cytoplasm in host cells, thereby the population of host miRNAs decreases, consequently creating a favorable intracellular environment for BmCPV genome replication and virus multiplication is enhanced.
Research on the mechanism of biogenesis of miRNAs by RNA viruses indicated that RNA viruses generate functional miRNAs through non-canonical miRNA biosynthesis pathways. For example, the Drosha enzyme also exists in cytoplasm to cleave pri-miRNA to form pre-miRNA (Shapiro et al., 2012), viruses can encode a protein with the function of the Drosha enzyme to cleave the initial transcripts of miRNA (Kreuze et al., 2005), the tRNase Z in the cytoplasm can cut pri-miRNA into pre-miRNA (Bogerd et al., 2010), and the small stem-loop structure transcripts of the viral genome in the cytoplasm can be directly processed into miRNAs or miRNA-like molecules by the Dicer enzyme (Okamura et al., 2007). Our previous studies identified several miRNAs encoded by BmCPV and have proven that they can regulate the expression of silkworm target genes (Pan et al., 2017; Guo et al., 2020). For example, BmCPV-miR-1 upregulated the expression of another target gene, B. mori inhibitor of apoptosis protein (BmIAP), by binding to the 5′-UTR of its mRNA and then inhibited the apoptosis of the infected cells, thus facilitating the replication of BmCPV (Pan et al., 2017; Guo et al., 2020). The enhanced replication of the BmCPV genome in the present study should include the contribution from the BmCPV-miR-1 upregulation on BmIAP. However, the mechanism with which BmCPV generates the miRNAs requires a further in-depth study in the future.
In summary, the present study revealed the negatively regulatory and co-regulatory function of two BmCPV-encoded putative miRNAs, BmCPV-miR-1 and BmCPV-miR-3, on the host target gene BmRan. Furthermore, repression of BmRan expression by the two BmCPV-miRNAs enhanced replication of the viral genome. The results might imply one of the strategies employed by the insect virus to modulate miRNA-mediated host antiviral defense by generating miRNAs that inhibits Ran, an important component in miRNA generation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SL: conceptualization, methodology, investigation, formal analysis, and writing–original draft and editing. YW, ZZ, WW, and YS: methodology, investigation, and formal analysis. ZDZ, MS, and PW: conceptualization, resources, supervision, and writing–original draft. HQ: resources. XG: conceptualization, methodology, funding acquisition, resources, project administration, supervision, and writing–review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding. This study was supported by the National Natural Science Foundation of China (Grant No. 31572463).
References
- Bernier A., Sagan S. M. (2018). The diverse roles of microRNAs at the host-virus interface. Viruses 10:440. 10.3390/v10080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Ponstingl H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354 80–82. [DOI] [PubMed] [Google Scholar]
- Bogerd H. P., Karnowski H. W., Cai X., Shin J., Pohlers M., Cullen B. R. (2010). A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell 37 135–142. 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack M. T., Czaplinski K., Gorlich D. (2004). Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10 185–191. 10.1261/rna.5167604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Meng X., Xue R., Zhu Y., Zhang X., Pan Z., et al. (2012). Characterization of the complete genome segments from BmCPV-SZ, a novel Bombyx mori cypovirus 1 isolate. Can. J. Microbiol. 58 872–883. 10.1139/w2012-064 [DOI] [PubMed] [Google Scholar]
- Carl J. W., Jr., Trgovcich J., Hannenhalli S. (2013). Widespread evidence of viral miRNAs targeting host pathways. BMC Bioinformatics 14 (Suppl. 2): S3. 10.1186/1471-2105-14-S2-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Chen W., Yang J. (2009). Another formula for calculating the gene change rate in real-time RT-PCR. Mol. Biol. Rep. 36 2165–2168. 10.1007/s11033-008-9430-1 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liang H., Chen X., Ke Y., Zhou Z., Yang M., et al. (2016). An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. 26 380–383. 10.1038/cr.2016.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. W., Voinnet O. (2007). Antiviral immunity directed by small RNAs. Cell 130 413–426. 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Rajewsky N. (2011). The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One 6:e18067. 10.1371/journal.pone.0018067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani M., Zandi M., Rezayi M., Khodadad N., Langari H., Amiri I. (2018). The Role of microRNAs in the viral infections. Curr. Pharm. Des. 24 4659–4667. 10.2174/1381612825666190110161034 [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight?. Nat. Rev. Genet. 9 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W. H., Chi J. T., et al. (2007). A viral microRNA functions as an orthologue of cellular miR-155. Nature 450 1096–1099. 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Wang Y. S., Chen T., Jiang X. X., Wu P., Geng T., et al. (2020). Functional analysis of a miRNA-like small RNA derived from Bombyx mori cytoplasmic polyhedrosis virus. Insect Sci. 27 449–462. 10.1111/1744-7917.12671 [DOI] [PubMed] [Google Scholar]
- Hagiwara K., Rao S., Scott S. W., Carner G. R. (2002). Nucleotide sequences of segments 1, 3 and 4 of the genome of Bombyx mori cypovirus 1 encoding putative capsid proteins VP1, VP3 and VP4, respectively. J. Gen. Virol. 83 1477–1482. 10.1099/0022-1317-83-6-1477 [DOI] [PubMed] [Google Scholar]
- Hausser J., Syed A. P., Bilen B., Zavolan M. (2013). Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 23 604–615. 10.1101/gr.139758.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss B. L., Maximova O. A., Thach D. C., Speicher J. M., Pletnev A. G. (2012). MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J. Virol. 86 5647–5659. 10.1128/jvi.07125-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., Tollervey D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Chen F., Zhu L., Yu L., Zhu M., Liang Z., et al. (2019). Bombyx mori cypovirus encoded small peptide inhibits viral multiplication. Dev. Comp. Immunol. 96 51–57. 10.1016/j.dci.2019.02.017 [DOI] [PubMed] [Google Scholar]
- Hussain M., Walker T., O’Neill S. L., Asgari S. (2013). Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 43 146–152. 10.1016/j.ibmb.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A. E., Bálint E., Tuschl T., Zamore P. D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293 834–838. 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Jiang L. (2021). Insights into the antiviral pathways of the silkworm Bombyx mori. Front. Immunol. 12:639092. 10.3389/fimmu.2021.639092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309 1577–1581. 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., Plasterk R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15 2654–2659. 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N. (2004). MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 14 156–159. 10.1016/j.tcb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Kreuze J. F., Savenkov E. I., Cuellar W., Li X., Valkonen J. P. (2005). Viral class 1 RNase III involved in suppression of RNA silencing. J. Virol. 79 7227–7238. 10.1128/jvi.79.11.7227-7238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. (2003). New microRNAs from mouse and human. RNA 9 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Zhou Z., Zhang S., Zen K., Chen X., Zhang C. (2014). Identification of Ebola virus microRNAs and their putative pathological function. Sci. China Life Sci. 57 973–981. 10.1007/s11427-014-4759-2 [DOI] [PubMed] [Google Scholar]
- Liu G., Zhang R., Xu J., Wu C. I., Lu X. (2015). Functional conservation of both CDS- and 3′-UTR-located microRNA binding sites between species. Mol. Biol. Evol. 32 623–628. 10.1093/molbev/msu323 [DOI] [PubMed] [Google Scholar]
- Lund E., Güttinger S., Calado A., Dahlberg J. E., Kutay U. (2004). Nuclear export of microRNA precursors. Science 303 95–98. 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130 89–100. 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U. A., Nielsen F. C., Lund A. H. (2008). MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30 460–471. 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Pan Z. H., Wu P., Gao K., Hou C. X., Qin G. X., Geng T., et al. (2017). Identification and characterization of two putative microRNAs encoded by Bombyx mori cypovirus. Virus Res. 233 86–94. 10.1016/j.virusres.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Qiu G. H., Weng Z. H., Hu P. P., Duan W. J., Xie B. P., Sun B., et al. (2018). Synchronous detection of ebolavirus conserved RNA sequences and ebolavirus-encoded miRNA-like fragment based on a zwitterionic copper (II) metal-organic framework. Talanta 180 396–402. 10.1016/j.talanta.2017.12.045 [DOI] [PubMed] [Google Scholar]
- Rosewick N., Momont M., Durkin K., Takeda H., Caiment F., Cleuter Y., et al. (2013). Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110 2306–2311. 10.1073/pnas.1213842110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P., Heale B. S., Snøve O., Jr., Aagaard L., Alluin J., Rossi J. J. (2007). Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35 2333–2342. 10.1093/nar/gkm133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. S., Langlois R. A., Pham A. M., Tenoever B. R. (2012). Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 18 1338–1346. 10.1261/rna.032268.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Duan Z., Sun J., Wu M., Wang B., Zhang J., et al. (2014). Identification and validation of a novel microRNA-like molecule derived from a cytoplasmic RNA virus antigenome by bioinformatics and experimental approaches. Virol. J. 11:121. 10.1186/1743-422x-11-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Sasaki M., Miki T., Shimamoto A., Furuichi Y., Katahira J., et al. (2006). Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 34 4711–4721. 10.1093/nar/gkl663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C. P., Singh J., Nagaraju J. (2012). A baculovirus-encoded MicroRNA (miRNA) suppresses its host miRNA biogenesis by regulating the exportin-5 cofactor Ran. J. Virol. 86 7867–7879. 10.1128/jvi.00064-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky R. L., Corcoran D. L., Gottwein E., Frank C. L., Kang D., Hafner M., et al. (2012). The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8:e1002484. 10.1371/journal.ppat.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Elefant N., Zimmermann A., Wolf D. G., Saleh N., Biton M., et al. (2007). Host immune system gene targeting by a viral miRNA. Science 317 376–381. 10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan G., Martin-Garcia J., Navas-Martin S. (2013). RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiol. Genomics 45 1035–1048. 10.1152/physiolgenomics.00112.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swevers L., Feng M., Ren F., Sun J. (2020). Antiviral defense against Cypovirus 1 (Reoviridae) infection in the silkworm, Bombyx mori. Arch. Insect Biochem. Physiol. 103:e21616. [DOI] [PubMed] [Google Scholar]
- Trobaugh D. W., Gardner C. L., Sun C., Haddow A. D., Wang E., Chapnik E., et al. (2014). RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506 245–248. 10.1038/nature12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu X., Ma Z., Huo Y., Xiao Z., Li Y., et al. (2011). Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 17 1511–1528. 10.1261/rna.2732611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I. G., Cullen B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17 3011–3016. 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B. R. (2004). Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32 4776–4785. 10.1093/nar/gkh824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xu H., Yao Y., Smith L. P., Kgosana L., Green J., et al. (2011). Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 7:e1001305. 10.1371/journal.ppat.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yao Y., Xu H., Lambeth L., Smith L. P., Kgosana L., et al. (2009). A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J. Virol. 83 489–492. 10.1128/jvi.01166-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.