Abstract
COVID-19 is a severe epidemic affecting the whole world. This epidemic, which has a high mortality rate, affects the health systems and the economies of countries significantly. Therefore, ending the epidemic is one of the most important priorities of all states. For this, automatic diagnosis and detection systems are very important to control the epidemic. In addition to the recommendation of the “reverse transcription-polymerase chain reaction (RT-PCR)” test, additional diagnosis and detection systems are required. Hence, based on the fact that the COVID-19 virus attacks the lungs, automatic diagnosis and detection systems developed using X-ray and CT images come to the fore. In this study, a high-performance detection system was implemented with three different CNN (ResNet50, ResNet101, InceptionResNetV2) models and X-ray images of three different classes (COVID-19, Normal, Pneumonia). The particle swarm optimization (PSO) algorithm and ant colony algorithm (ACO) was applied among the feature selection methods, and their performances were compared. The results were obtained using support vector machines (SVM) and a k-nearest neighbor (k-NN) classifier using the 10-fold cross-validation method. The highest overall accuracy performance was 99.83% with the SVM algorithm without feature selection. The highest performance was achieved after the feature selection process with the SVM + PSO method as 99.86%. As a result, higher performance with less computational load has been achieved by realizing the feature selection. Based on the high results obtained, it is thought that this study will benefit radiologists as a decision support system.
Keywords: COVID-19, X-ray, Deep features, Feature selection, PSO, ACO
1. Introduction
The COVID-19 epidemic, which affected the whole world, was first seen in China in the last months of 2019. The official name of the virus was expressed by the World Health Organization (WHO) as SARS-CoV-2 (Severe Acute Respiratory Syndrome-Coronavirus-2) [1]. It has been declared a “pandemic” by WHO because of the rapid spread of the virus and many people's death. It is known that the most crucial factor in the spread of the virus occurs through air and physical contact. It is stated that this virus, which directly attacks the lungs, causes severe pneumonia [2]. Because it is an RNA virus, it is difficult and time-consuming to detect. As with all epidemics, early diagnosis is crucial to reduce the impact of the COVID-19 virus [3,4]. In this way, it is ensured that people infected with the virus get over the life-threatening situation earlier and get treatment quickly. WHO recommends the “reverse transcription polymerase chain reaction (RT-PCR)” test for all diagnoses of COVID-19 disease. However, this method is troublesome, time consuming, and costly [5]. It is also stated that the real detection sensitivity is low [6]. This test alone is not enough, and medical imaging approaches should also support it. There are two most preferred methods for this. These are X-ray and Computed Tomography (CT) images. These two methods, which have advantages and disadvantages, show high performance [7,8]. CT images, which are more detailed than X-ray images, provide higher accuracy in radiologists' manual examinations. However, this process requires much workload for the radiologist. The most critical aspect of X-ray images is that they are readily available and accessible in all healthcare institutions. However, visible changes in the lungs in X-ray images occur sometime after being infected with the virus. In other words, it was reported that 10–12 days after the transmission of the COVID-19 virus, the disease could be easily diagnosed by radiologists in the X-ray image [9]. This period is long enough to cause a delay in the fight against COVID-19. Therefore, machine learning approaches are widely used to detect of COVID-19 patients [[10], [11], [12]].
Although there are many studies with deep learning approaches in the literature, these methods also include traditional machine learning approaches such as manual feature extraction and feature selection. In Ref. [13] the authors extracted various spatial, frequency and statistical features, such as Texture, Fast Fourier Transform (FFT), Gray-Level Co-Occurrence Matrix (GLCM), Gray Level Difference Method (GLDM), Wavelet transform (WT), Skewness, Kurtosis, Mean, Entropy, Energy, Std, Mean, Median, Max, Min, Mean Deviation, RMS, Range, MeanGradient, StdGradient, and Uniformity, as part of manual feature extraction. A total of 266 features were obtained by the authors thanks to the use of these methods. They achieved COVID-19 detection performances by reducing the size of the feature space with Principal Component Analysis (PCA) method. In Ref. [14], authors used 8196 features, which are 8 first order statistical features (FOSF), 88 GLCM, 8100 histograms of oriented gradients (HOG). Using binary gray wolf optimization (BGWO), they selected the features that showed the best separation between the classes and the feature selection. In another study, In Ref. [12], authors used GLCM, local binary gray level co-occurrence matrix (LBGLCM), gray level run length matrix (GRLLM), and segmentation-based fractal texture analysis (SFTA) features. They used a wide variety of features. To solve the unbalanced dataset problem, they applied SMOTE over-sampling. Size reduction was performed using PCA to identify the most prominent features. In studies using deep learning models, which is another approach, models with different architectures have been used in the literature [[15], [16], [17]]. The absence of manual feature extraction in deep learning and the existence of an end-to-end structure has pushed researchers to further research on this side.
Besides, studies combining deep learning with traditional machine learning approaches known as hybrid systems are encountered rarely [18]. Among these studies, Nour et al. conducted a COVID-19-Normal-Viral Pneumonia, 3-class study using the features they obtained with their Convolutional Neural Network (CNN) models. They used support vector machines (SVM) and k nearest neighbor (k-NN) algorithms as classifiers. They optimized the parameters of the models using the Bayesian optimization algorithm [18]. Sethy et al. performed on 11 different well-known CNN models with very little data set. They presented a 2-class study using the SVM algorithm with the features obtained from these models. In their second study, they performed a similar study by increasing the number of data. Their 3-class study tried to determine their performance with SVM by using the feature maps obtained from 13 CNN models [19]. In another study in the literature, Narin used only the pre-trained ResNet50 CNN model and SVM [20]. Toǧaçar et al. performed a study on a 3-class data set. The feature map sizes obtained using MobileNetV2 and SqueezeNet were reduced by Social Mimic Optimization method, and the detection performances were obtained with SVM [21]. Kassani et al. performed a small number of data using 15 different CNN models and 6 different machine learning algorithms [22]. Finally, Turkoglu used the feature maps obtained from different outputs of the Alexnet-based CNN model. Selecting these features with the help of the Relief feature selection algorithm has detected COVID-19 patients with the SVM algorithm [23].
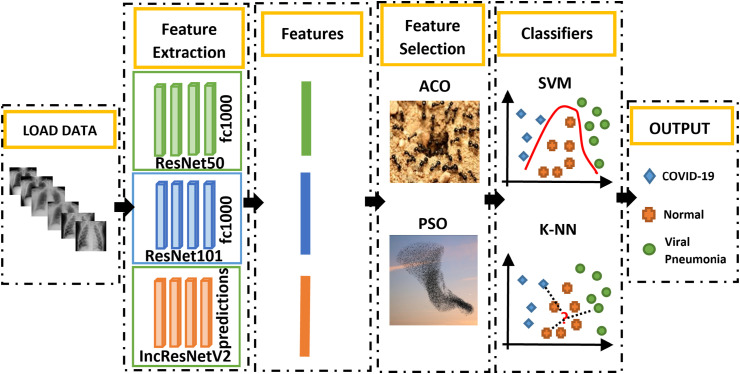
This study, which is carried out as shown in Fig. 1 , generally consists of 4 stages. In the first step, the data were loaded without any pre-processing except for sizing. In the second step, the features were extracted using 3 different CNN models (ResNet50, ResNet101, InceptionResNetV2). In the third step, feature selection has been made. The feature selection was made using Particle Swarm Optimization (PSO) and Ant Colony Algorithm (ACO) methods. In the last stage, the detection performances of COVID-19 and other classes were obtained with two different classifiers, k-NN, and SVM.
Fig. 1.
Flowchart of the proposed systems.
The main contribution of the proposed methods can be summarized as follows:
-
I)
Unlike studies using traditional feature extraction methods, deep features were used over deep learning models.
-
II)
The deep features extracted from three different CNN models have been combined to improve the detection of COVID-19 further.
-
III)
A hybrid study was presented with features derived from three different CNN models, two different metaheuristic feature selection methods and two different classifiers as high performance.
-
IV)
To detect unnecessary and non-informative features, having less computational load and fewer parameters, PSO and ACO were preferred among metaheuristic methods.
-
V)
A high-accuracy decision-making system has been provided to radiologists for detection of COVID-19 and follow-up.
2. Materials and methods
2.1. Dataset
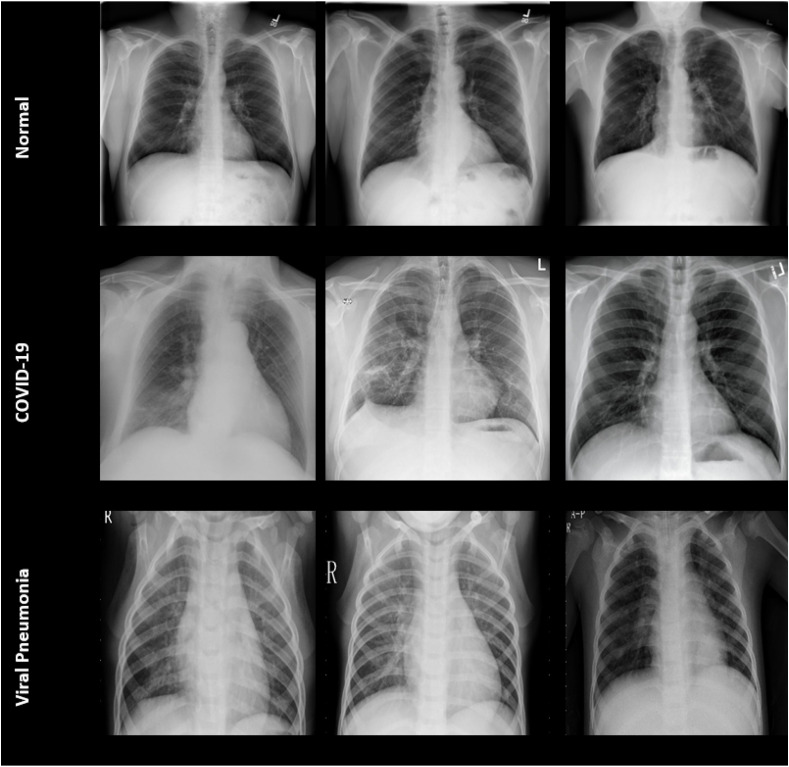
In this study, the X-ray images database known as “COVID-19 radiography database”, which is open to access by Kaggle, was used [24]. There are 4 different datasets in this database: (1) “COVID-19 Database” provided by the Italian Medical and Interventional Radiology Society (SIRM), (2) “Novel Corona Virus 2019 Dataset” provided by Cohen, Morrison and Dao on GitHub, (3) “COVID-19 (+) Chest X-ray Images” compiled from 43 different scientific articles, and (4) “Kaggle chest X-ray database” consisting of Kaggle chest X-ray images [[25], [26], [27]]. In this way, 219 COVID-19 patient images, 1341 normal individuals, and 1345 pneumonia patients were compiled. Fig. 2 shows sample images belonging to these three groups.
Fig. 2.
Samples of X-ray images (Normal (Healthy)) (first row), COVID-19 (second row), Viral Pneumonia (third row) [[25], [26], [27]].
2.2. CNN structure and feature extraction
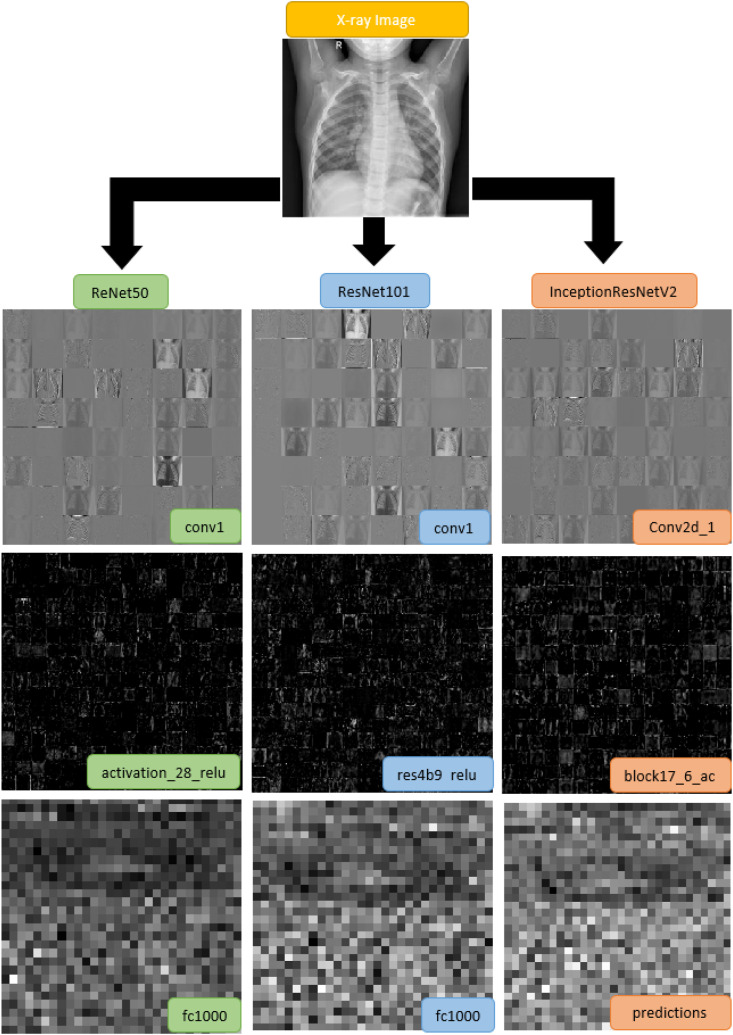
CNN is a sub-branch of deep learning that takes its name from the convolution process. As an end-to-end model, it is understood from the literature that it is highly effective and high performance [28]. Besides this usage, CNNs are also used in feature extraction processes [29,30]. Hybrid approaches are developed by using the extracted features with traditional classifiers. The first reason why CNN models are used in feature extraction is that deep features give very high results in studies in the literature. Secondly, there is a limited number of hybrid approaches using deep attributes to detect COVID-19. CNN networks generally consist of 3 layers: a convolution layer, pooling layer, and fully connected layer [31]. In the convolution layer, it is the most basic layer for which the model is named. The patterns coming here are passed through filters, and feature maps are created. These filters allow a wide variety of features to be detected by shifting them along with the pattern. The higher the number of convolution layers the deeper the properties can be obtained. In the pooling layer, the size of the feature maps and the reduction of the number of network parameters are performed. Finally, the feature maps obtained in the fully connected layer are transformed into one-dimensional vectors. Interconnections of fully connected layers are weighted. In addition to these layers, it is used in layers such as normalization and dropout layers. In the literature, there are famous models created by combining all these structures with different topologies. ResNet50, ResNet101 and InceptionResNetV2 models were used in this study. Unlike classical CNN approaches, the ResNet50 model is created by adding the residual value and residual block model. The ResNet50 architecture, which consists of 50 layers, is a model that won first place in the 2015 ILSVRC (ImageNet Large Scale Visual Recognition Challenge) image classification task on the ImageNet test set and 2015 COCO (Common Objects in Context) competition for COCO detection and COCO segmentation [32]. Convolution processes include 1 x 1, 3 x 3, and 1 x 1 convolution stages. ResNet50 architecture consists of 25.6 million parameters [33]. ResNet101 is another model used. . This model, which also includes 33 residual blocks, consists of 101 layers [34]. The InceptionResNetV2 model, which has a total of 164 deep residual network layers, is used as the third model [35]. In the study, features obtained from fully connected layers were used in the classification stage of all these models. They are fully connected layers from which “fc1000” for ResNet50, “fc1000” for ResNet101 and “predictions” for InceptionResNetV2 features are obtained. In each layer, 1000 feature values were obtained. The total of the features obtained is 3000. Outputs from beginning, middle of 3 different models and layers from which features are taken are shown in Fig. 3 .
Fig. 3.
Output samples of 3 different models.
2.3. Feature selection
Feature selection is used to find the optimum features subset by deleting irrelevant data from large datasets [36]. Among the reasons for doing this, it can be said that decreasing the computational load ratio and increasing the estimation rate (such as accuracy value), and preventing overfitting problem. On the other hand, the feature selection process does not guarantee an increase in estimation rates. However, achieving similar performance even with fewer features is a positive development. Within the scope of this study, using metaheuristic methods such as PSO and ACO, the reduction of too many features and their effects on achievement were investigated. PSO and ACO are used in feature selection because they have a small number of parameters among metaheuristic methods and can quickly reach the result.
2.4. Particle swarm optimization (PSO)
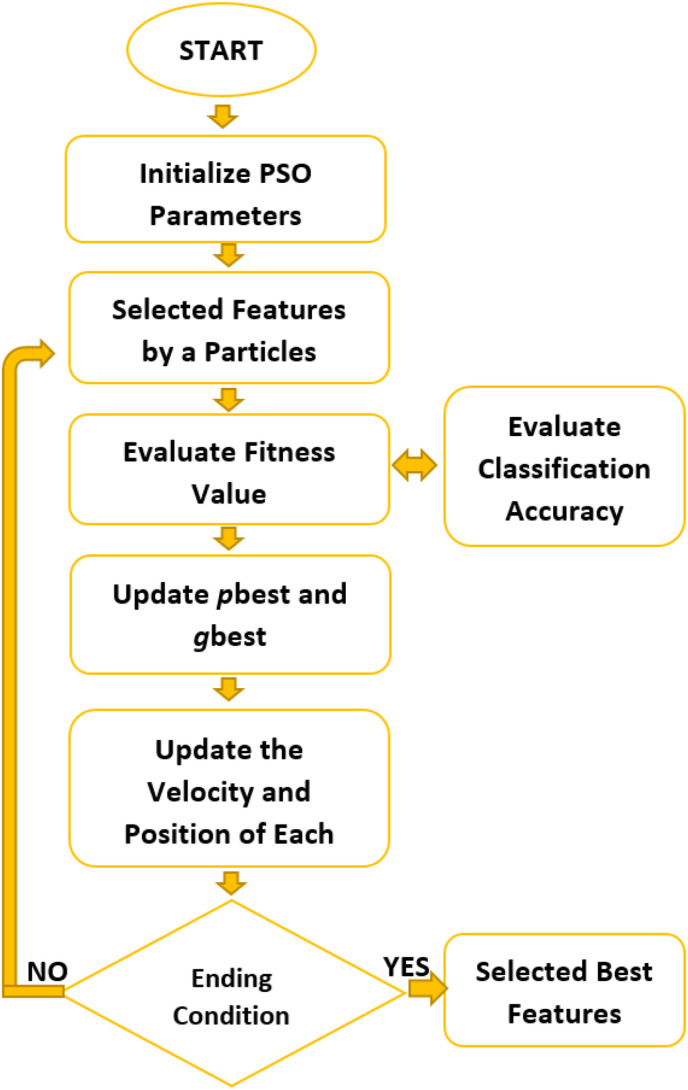
PSO was inspired by the movements of some animals moving in a herd while meeting their basic needs. In this way, it is seen that they reach their goals more quickly. It is an optimization algorithm introduced by Dr. Kennedy and Dr. Eberhart in 1995 [37]. Each individual with velocity and position information is called a “particle,” the group formed by these particles is called a “swarm”. Thanks to mathematically determining fitness functions, the optimization of particles is controlled. While the best state of a particle that comes closest to the solution is called pbest (personnel best), the current state of the particle with which the whole herd is closest to the solution is called gbest (global best). By updating these values, each particle's rate of change and movements are determined [38]. The PSO flow diagram used for feature selection is given in Fig. 4 .
Fig. 4.
Feature selection approach using PSO.
The initial parameters used for PSO are given in Table 1 . In addition, general accuracies for the fitness value were obtained for k-NN (k = 1). The calculated velocity and position equations are as follows:
| (1) |
| (2) |
Where X id and V id are particle velocity and particle position, respectively. t represents the t. iteration in the process, w is the inertia weight. c 1 and c 2 are cognitive and social factors, r 1i and r 2i are random values uniformly distributed in [0, 1]. pbest is best position, gbest is global best.
Table 1.
PSO algorithm initial parameters in used.
| Parameters | Value |
|---|---|
| Number of particles | 10 |
| Maximum number of iterations | 100 |
| Cognitive factor(c1) | 2 |
| Social factor (c2) | 2 |
| Random values (r1, r2) | [0,1] |
| Min-Max bound on inertia weight(w) | 0.4–0.95 |
2.5. Ant colony optimization (ACO)
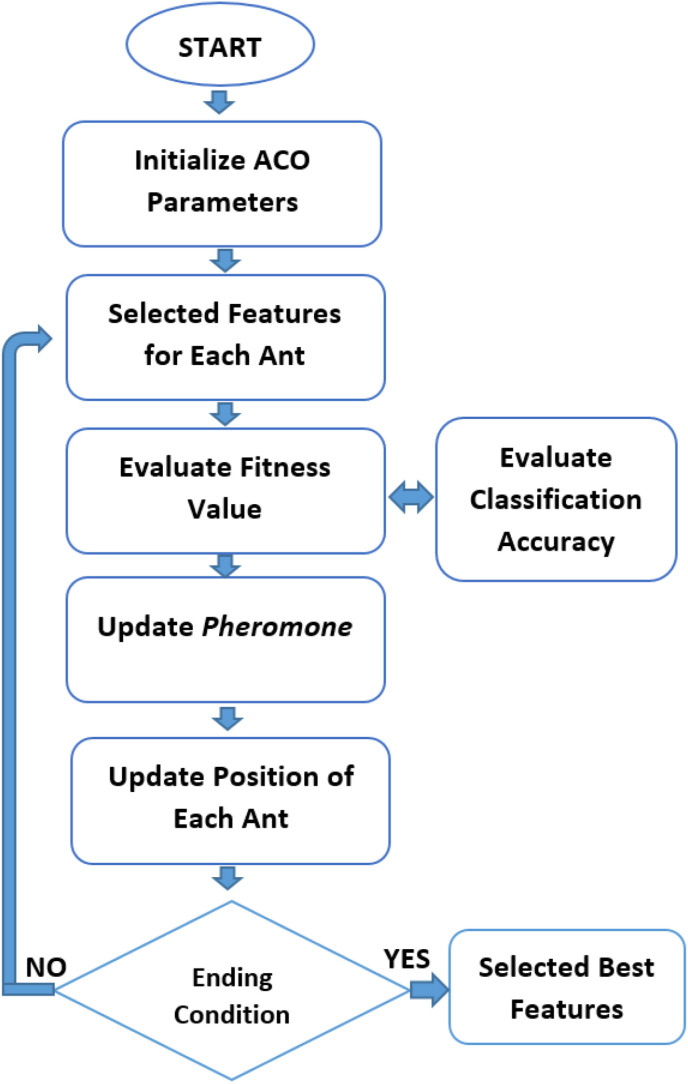
ACO is one of the metaheuristic methods introduced by Dorigo in 1991, inspired by the daily lives of ants [39]. It is known that ants leave a pheromone secretion, which is a chemical substance, on their way to food and move collectively as they communicate between them. There are many possibilities for an ant coming out of the nest looking for food. It decides the direction to go by looking at the pheromone substance density in the environment. Here, the possibility arises that other ants prefer the route with the most pheromone substance. Therefore, the probability of ants choosing this route increases more. In addition, if ants encounter any obstacle or an unusual situation, finding the shortest path between food and their nests as soon as possible reveals the importance of this method in terms of optimization. This method was first applied for the traveling salesman problem, and the results were quite impressive [40]. ACO is also used in feature selection processes. In feature selection with ACO, most ants select a feature that indicates that this feature is more distinctive than other features. As a result, this feature has more pheromone secretion, and other ants in the herd will be more likely to select this feature. According to this approach, the desired number of features can be selected by evaluating the abundance of pheromone secretion. The feature selection stage using ACO is shown in Fig. 5 .
Fig. 5.
Feature selection approach using ACO.
The parameters used for ACO are given in Table 2 . In addition, general accuracy values for the fitness value were obtained for k-NN (k = 1). The pheromone is updated according to the following equation:
| (3) |
Table 2.
ACO algorithm initial parameters in used.
| Parameters | Value |
|---|---|
| Number of ants (n) | 10 |
| Maximum number of iterations | 100 |
| Evaporation rate (p) | 0.2 |
| Pheromone factor (α) | 1 |
| Heuristics factor (β) | 1 |
The mathematical expression of the probability of the ants moving from point i to point j is as follows.
| (4) |
Where τ ij(t) is the pheromone value at the time t, p is the pheromone trail evaporation rate, n is number of ants, η ij(t) is a priori available heuristic information at the time t, α is the weight of pheromone, β is the weight of heuristic information.
2.6. Classification algorithms
SVM and k-NN algorithms, which are not complex and have less computational load, were used to determine the classification performance of the deep features obtained. Detailed information about the algorithms used can be found in the ongoing subtitles.
2.6.1. Support vector machines (SVM)
Vapnik developed SVM for the solution of pattern recognition and classification problems [41]. While classifying the data, it aims to find the closest samples of the classes to each other and to maximize the perpendicular distances of these samples to the separating surface that will separate the two classes. The separator surface can have many different combinations without changing its success on the data set. Thanks to the support vectors, the distance between the classes is maximum. The Lagrange method, which finds the smallest and largest values of a function depending on a constraint, is used to realize this operation. In this study, the results are obtained using the SVM algorithm's linear, quadratic, and cubic kernel functions. The parameter of C was tested from 0.01 to 100 with 0.1 increments. The kernel scale was set to 1 and the box constraint set to 1.
2.6.2. k-Nearest Neighbors (k-NN)
k-NN is a supervised and sample-based classification algorithm. In the k-NN classifier algorithm, the training phase occurs by separating the training set [42]. Therefore, no additional time is wasted for training the model. In this algorithm, a test data is classified by many votes of its neighbors, with the object assigned to the class most common among its k nearest neighbors. In this study, odd numbers with k values from 1 to 11 were used. In order to avoid a tie in voting, only odd numbers were preferred in the selection of the k value. The Euclidean metric was used as the distance criterion.
2.7. Performance metrics
5 metrics were used to evaluate the predictive performance of the classifiers [43]:
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
Here, TP (True positive): To determine the patient as a patient. FP (False positive): To determine the health of a patient. TN (True negative): To determine the healthy as healthy. FN (False negative): To determine the patient as healthy.
In classifier studies, as it is known, the high training performance of the classifier does not mean that its performance on test data will be high. For this reason, the data set is divided into two as training and testing in all classifier studies. The k-fold cross validation (CV) method in which the dataset is divided into k-folds. While the k-1 piece is used for training, one of them is used for testing. This process is repeated until all parts are used for testing. Classifier performance is calculated as classifier training performance and classifier test performance by taking averages separately for training and testing [44]. In this study, the fold value of k is taken as 10.
3. Experimental results
In this study, feature maps obtained from 3 different CNN models, feature selection algorithms, and classification algorithms were implemented using MATLAB2020a. The classifier performances were calculated by applying the 10-fold cross-validation method to the feature maps obtained with 3 different models. Adaptive moment estimation (ADAM) optimization algorithm was used as an optimizer for all models [45]. Learning rate and minibatch size for the CNN models used in feature extraction were chosen as 0.00001 and 10, respectively. In the preliminary trials, the maximum number of iterations was taken as 30 since the overfitting occurred after an average of 30 iterations.
3.1. Performances without feature selection
Table 3 shows the total number of features obtained and all execution times from each CNN model. In terms of time complexity, it can be seen in Table 3 that it takes much longer to extract features from the InceptionResNetv2 model than the other two models. It can be said that the ResNet50 model is the fastest model in feature extraction. The maximum performance values obtained for the linear, quadratic, and cubic kernel functions for SVM using these features are given in Table 4 .
Table 3.
The features obtained in the study and all execution times.
| Model | Total Features |
Execution Times in |
|---|---|---|
| feature extraction (s) | ||
| ResNet50 | 1000 | 484 |
| ResNet101 | 1000 | 925 |
| InceptionResNetV2 | 1000 | 3233 |
| Combined | 3000 | 4642 |
Table 4.
SVM performance results using all features (without feature selection).
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
SVM Type |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 94.52 | 99.04 | 99.93 | 99.52 | 96.73 | SVM Quadratic |
| ResNet101 | 93.15 | 99.51 | 99.96 | 99.45 | 96.23 | SVM Cubic |
| IncResNetV2 | 94.06 | 97.63 | 99.81 | 99.38 | 95.81 | SVM Cubic |
| Combined | 97.72 | 100 | 100 | 99.83 | 98.85 | SVM Quadratic |
According to the results that are shown in Table 4, the general accuracy values are above 99%. It is seen that all performances have increased significantly, especially by using the features obtained from 3 different models (combined features). Among the models, it is seen that the highest performance belongs to the ResNet50 model. It can be stated that cubic and quadratic SVM is more effective than Linear SVM. Considering that working with 3 different classes, it can be noted that the highest performances of those with COVID-19 are 97.72%, while the data belonging to other classes are 100%. If taken for the F1-score value, it was found as 98.85%. For k-NN, the highest results obtained by using (k = 1, 3, 5, 7, 9, 11) values are given in Table 5 .
Table 5.
k-NN performance results using all features (without feature selection).
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
k value (k-NN) |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 88.58 | 99.49 | 99.96 | 99.10 | 93.72 | k = 1 |
| ResNet101 | 92.24 | 98.54 | 99.89 | 99.31 | 95.28 | k = 1 |
| IncResNetV2 | 90.41 | 97.54 | 99.81 | 99.10 | 93.84 | k = 3 |
| Combined | 94.06 | 100 | 100 | 99.55 | 96.94 | k = 1 |
As can be seen from Table 5, the highest overall performance value was obtained in the ResNet101 model among all models. The highest performance has been achieved by combining the features obtained from all models. It can be said that the results are high for small values of k of the k-NN algorithm. It is understood that especially the value of k = 1 stands out. The performance of the k-NN classifier is lower than the SVM algorithm.
3.2. Performances of feature selection
Feature selection performances were investigated by using metaheuristic PSO and ACO optimization algorithms on the features. The number of selected features and all execution times of each model are given in Table 6 . It can be seen from Table 6 that the time complexity value of PSO is much higher than ACO.
Table 6.
Number of features selected by PSO and ACO and all execution times.
| Model | PSO | ACO | Execution Times |
Execution Times |
|---|---|---|---|---|
| for PSO (s) | for ACO (s) | |||
| ResNet50 | 498 | 500 | 2845 | 1275 |
| ResNet101 | 463 | 450 | 2796 | 1201 |
| InceptionResNetV2 | 487 | 420 | 2823 | 1218 |
| Combined | 1490 | 1500 | 5234 | 4210 |
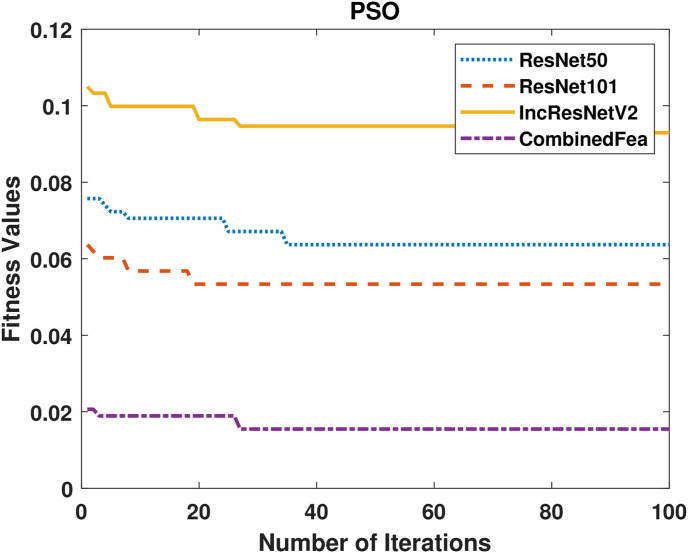
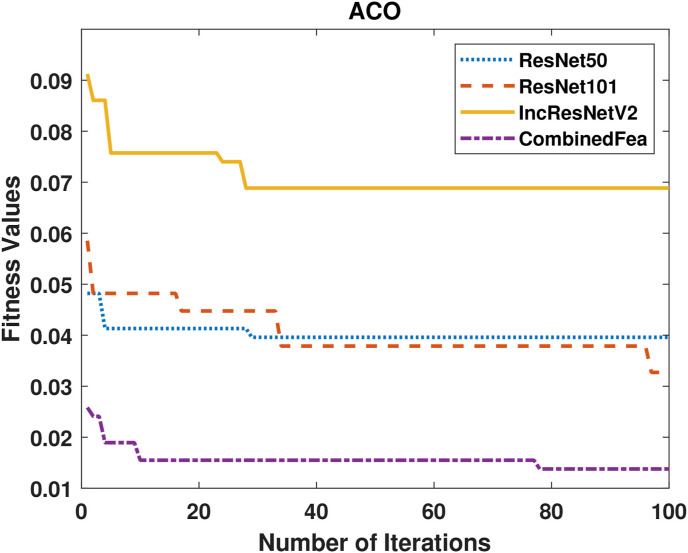
Comparison of convergence curves of the PSO algorithm in the feature selection stage is shown in Fig. 6 . It is clearly seen in the figure that the features obtained from InceptoResNetV2 model show the lowest fitness value for PSO. The best performance is given by the feature group obtained by combining 3 models. Similarly, Comparison of convergence curves obtained for ACO are given Fig. 7 . As in PSO, the lowest performance was achieved with the features obtained from the InceptonResnetV2 model, while the best performance was achieved with the feature group obtained by combining the model.
Fig. 6.
PSO fitness values of 4 different feature groups.
Fig. 7.
ACO fitness values of 4 different feature groups.
Performance results of the feature selection algorithms for both SVM and k-NN are given separately in Table 7 , Table 8 , Table 9 , Table 10 .
Table 7.
SVM performance of features selected with PSO.
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
SVM Type |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 97.26 | 100 | 100 | 99.79 | 98.61 | SVM Quadratic |
| ResNet101 | 92.69 | 99.51 | 99.96 | 99.41 | 95.98 | SVM Cubic |
| IncResNetV2 | 94.98 | 97.20 | 99.78 | 99.41 | 96.07 | SVM Cubic |
| Combined | 98.17 | 100 | 100 | 99.86 | 99.08 | SVM Linear |
Table 8.
k-NN performances of features selected with PSO.
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
k value (k-NN) |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 90.41 | 99.00 | 99.93 | 99.21 | 94.51 | k = 1 |
| ResNet101 | 89.04 | 97.99 | 99.85 | 99.04 | 93.30 | k = 1 |
| IncResNetV2 | 94.52 | 97.18 | 99.78 | 99.38 | 95.83 | k = 1 |
| Combined | 92.24 | 100 | 100 | 99.41 | 95.96 | k = 1 |
Table 9.
SVM performance of features selected with ACO.
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
SVM Type |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 96.35 | 99.06 | 99.93 | 99.66 | 97.69 | SVM Quadratic |
| ResNet101 | 93.15 | 98.55 | 99.89 | 99.38 | 95.77 | SVM Quadratic |
| IncResNetV2 | 95.89 | 97.67 | 99.81 | 99.52 | 96.77 | SVM Quadratic |
| Combined | 98.17 | 99.54 | 99.96 | 99.83 | 98.85 | SVM Quadratic |
Table 10.
k-NN performances of features selected with ACO.
| Methods (Features) | Recall |
Precision |
Specificity |
Accuracy |
F1-score |
k value (k-NN) |
|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | ||
| ResNet50 | 90.87 | 98.03 | 99.85 | 99.17 | 94.31 | k = 1 |
| ResNet101 | 89.04 | 97.99 | 99.85 | 99.04 | 93.30 | k = 1 |
| IncResNetV2 | 91.78 | 97.10 | 99.78 | 99.17 | 94.37 | k = 1 |
| Combined | 91.32 | 100 | 100 | 99.35 | 95.47 | k = 1 |
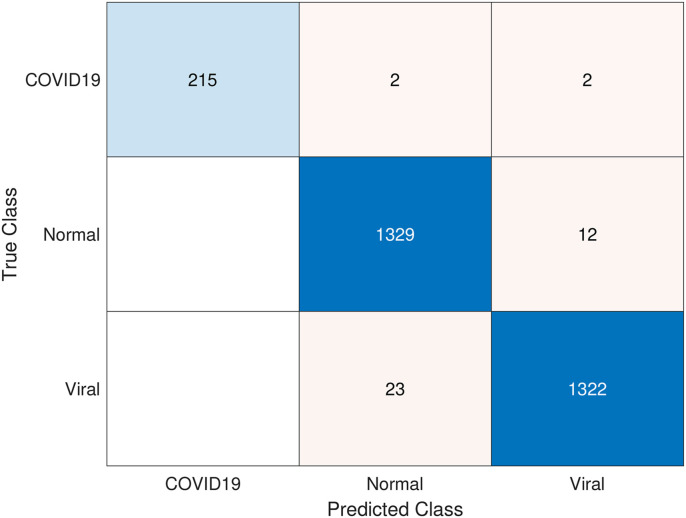
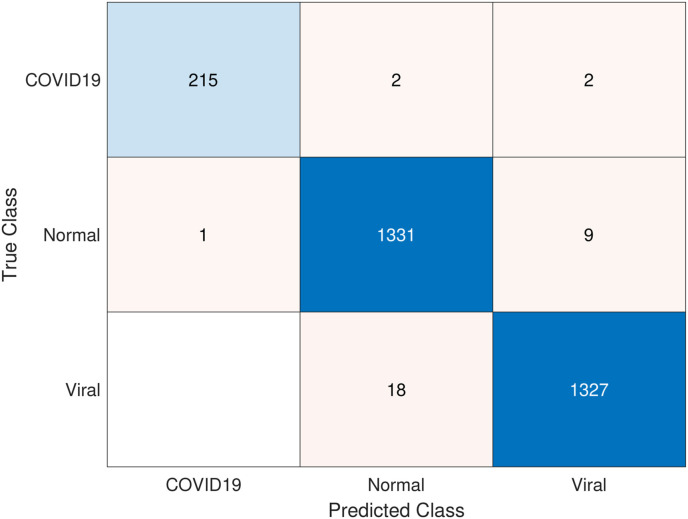
It is clear from Table 7 that the highest performance results were obtained with SVM (linear) and combined features. The overall accuracy value was 99.86% and the F1-score value was 99.08%. In the results given in Table 8, the results obtained by k-NN are lower than SVM. In general, the highest overall accuracy value and F1-score value for k = 1 are 99.41% and 95.96%, respectively. Fig. 8 shows the confusion matrix of the highest COVID-19 detection obtained with PSO. Here, 215 of 219 patients were detected correctly, while 2 were detected to be expected and the other 2 as viral pneumonia.
Fig. 8.
Confusion matrix for the highest performance achieved with PSO.
According to the results obtained with ACO in Table 9, the highest general accuracy and F1-score value were obtained with the combined features. It is seen that the highest classifier performance is achieved with the ResNet50 model. With SVM (quadratic), the overall accuracy and F1-score were achieved at 99.83% and 98.85%, respectively. The results obtained for the k-NN algorithm with the features selected by using ACO are presented in Table 10. It is seen that the highest performances for k-NN are obtained for k = 1. It can be said that k-NN has a lower performance value than SVM. The overall accuracy value and the F1-score values are 99.35% and 95.47%, respectively. In Fig. 9 , the confusion matrix for determining the highest COVID-19 and other classes obtained with ACO is given. Here, 215 of the 219 patients were detected correctly, while 2 were detected to be expected and the other 2 as viral pneumonia.
Fig. 9.
Confusion matrix for the highest performance achieved with ACO.
4. Discussion
As the accurate detection of COVID-19 patients becomes more important day by day, new detection methods have begun to emerge. Many studies in the literature on detecting COVID-19 using CT images, X-ray images and audio signals [15,16]. These two imaging technologies are among the widely used methods in addition to RT-PCR testing in the epidemic. While a large number of daily cases causes excessive density in imaging methods, the workload of radiologists who play a role in the examination and decision-making of these images is increasing. Therefore, the number of false detections may increase due to workloads and human eye vision errors. To minimize these errors, automatic diagnosis and detection systems performed with machine learning algorithms are developed by experts.
Among these detection systems, many studies have been carried out with deep learning algorithms and traditional methods. Some of these studies using lung X-ray images are given in Table 11 . Khuzani et al. performed a detailed study with spatial (Texture, GLDM, GLCM), frequency (Wavelet and FFT), and statistical measurements. Due to the large size of the hand-crafted features obtained, feature selection was performed with PCA. They stated that FFT measurements have a more distinctive feature with their performance, especially in detecting COVID-19. The highest overall accuracy and F1-score values were 95% and 94.3%, respectively. However, these researchers working with a limited number of data (140 COVID-19, 140 Normal, 140 Pneumonia) obtained all the results with the hold-out method [13]. On the other hand, Ozturk et al. studied data from too many classes, unlike many other studies. Since the data in some classes were too small, they increased the data using the SMOTE method. Although they have stated that they have overcome the unbalanced data set problem, the data is still not balanced. In other words, 126 pieces of data in total were increased to 260 with this method. The authors noted that handmade feature extraction would be more appropriate with limited data. In this study, PCA, one of the feature reduction methods, was used to increase performance. They achieved an F1 score of 94.23% and 93.99%, respectively, with very high overall accuracy for small and multi-class datasets. Nevertheless, the researchers obtained performance values using the hold-out method in this study [12].
Table 11.
Performance comparison using deep based features on X-ray images. (CV: cross validation, Acc: Accuracy, F1: F1-score).
| Literature | Method | Number of |
CV | Performance |
|---|---|---|---|---|
| Class | (%) | |||
| Khuzani et al. [13] | Texture,FFT,GLCM,GLDM,WT, and Statistical + PCA + MLP | 3 | Hold-out | Acc = 95.00 F1 = 94.30 |
| Öztürk et l [12]. | GLCM,LBGLCM,GLRLM, and SFTA + PCA + SVM | 6 | Hold-out | Acc = 94.23 F1 = 93.99 |
| Ozturk et al. [46] | DarkNet CNN | 3 | 5-Fold | Acc = 87.02 F1 = − |
| Narin et al. [47] | Pre-trained ResNet50 CNN | 2 | 5-Fold | Acc = 96.01 F1 = 83.50 |
| Ismael and Sengür [48] | ResNet50 Features + SVM | 2 | %50 - %50 | Acc = 94.70 F1 = − |
| Nour et al. [18] | Own deep CNN + SVM, k-NN, decision trees | 3 | %70 - %30 | Acc = 98.97 F1 = 96.72 |
| Sethy et al. [19] | deep CNN models + SVM | 3 | %80 - %20 | Acc = 95.33 F1 = 95.34 |
| Narin [20] | ResNet50+SVM | 3 | 5-Fold | Acc = 99.35 F1 = − |
| Toǧaçar et al. [21] | MobileNetV2, SqueezeNet + SMO + SVM | 3 | 5-Fold | Acc = 99.34 F1 = 99.49 |
| Kassani et al. [22] | DenseNet121+Bagging Trees | 2 | 10-Fold | Acc = 99.07 F1 = 96.00 |
| Turkoǧlu [23] | Pre-Trained CNN model + Relief + SVM | 3 | %90 - %10 | Acc = 99.18 F1 = 99.30 |
| 10-Fold | Acc = 98.08 F1 = − | |||
| This study | (ResNet50+ResNet101+ | 3 | 10-Fold | Acc = 99.83 F1 = 98.85 |
| IncResNetV2 features) + SVM | ||||
| This study | (ResNet50+ResNet101+ | 3 | 10-Fold | Acc = 99.86 F1 = 99.08 |
| IncResNetV2 features)+ PSO + SVM | ||||
| This study | (ResNet50+ResNet101 | 3 | 10-Fold | Acc = 99.83 F1 = 98.85 |
| IncResNetV2 features)+ACO + SVM |
Another fundamental approach in detecting COVID-19 is the studies using end-to-end deep learning models. Ozturk et al. achieved 87.02% accuracy and 87.37% F1-score in their results for multi-class and 5-CV. The most striking aspect of this article is evaluating the results obtained from the model they call DarkCovidNet separately by the radiologist. As a result, it was revealed that the results obtained from DarkCovidNet were successful by an expert. In this study, they used a total of 1125 data (125 COVID-1, 500 Pneumonia, and 500 No-Findings). The only disadvantage of the study is that they have little COVID-19 data [46]. In another study, Narin et al. achieved the highest 96.1% accuracy and 83.5% F1-score with the ResNet50 model among the five different pre-trained CNN models [47]. Nevertheless, all the results of the study were obtained for binary classes. It is known that the results obtained from binary classification problems are higher than in other multi-class studies. Ismael and Sengur presented a versatile study using end-to-end CNN models, deep features approach, and hand-crafted feature extraction. They achieved the highest performance value with 94.70% accuracy, with the features they obtained through the ResNet50 deep learning model. However, in their studies, they obtained performance results by using 50%–50% of the data set as test and training data [48].
In addition to these methods, hybrid studies are using deep learning-based features and traditional classification algorithms. However, these studies are very few. From these studies, Nour et al. performed a 3-class study for the detection of COVID-19. They used the Bayesian algorithm for the optimization of hyper-parameters of machine learning algorithms. Although they obtained the results with the data they reserved for 70% training and 30% testing, they achieved the highest accuracy of 98.97% and F1-score of 96.72%. In this study, they achieved high performance by not using the feature selection method [18]. Sethy et al., one of the first deep features studies on COVID-19 detection, achieved 95.33% accuracy and 95.34% F1-score performance values. They worked with a limited and balanced data set (127 COVID-1, 127 Pneumonia, and 127 Normal). In addition, presenting the performance results of deep features obtained through 13 different deep learning models is one of the prominent aspects of this study. Unfortunately, for this study, the results were obtained by separating 80% of the data for training and 20% for testing [19]. In another study, Narin detected COVID-19 with 3-class and 5-fold CV with ResNet50 deep features. The overall accuracy performance of COVID-19, differing from other classes without feature selection, was found to be 99.35% [20]. Unlike other studies in the literature, Togacar et al. used the fuzzy color technique, one of the pre-treatment methods. They obtained features from two different deep learning models (MobileNetV2, SqueezeNet). Combining the features selected with the Social Mimic optimization (SMO) method reached 99.34% accuracy and 99.49% F1-score values. The results were obtained with a 3-class and 5-fold CV [21]. Kassani et al. presented a comparative study of eight different CNN models. They achieved the highest performance with the help of deep features obtained from DenseNet121 and the Bagging tree classification algorithm. They reported that they achieved 99.07% accuracy and 96.00% F1 score with 10-fold CV. These high results in the study were obtained through binary classification without feature selection [22]. Using the Relief algorithm and feature selection, Turkoglu obtained 99.18% accuracy for 90%–10% training and test data. The same researcher achieved 98.08% accuracy by applying the 10-fold CV method. The researcher used AlexNet, one of the most basic CNN models. Unlike the studies in the literature, deep features obtained from different layers of AlexNet were used [23].
This study was conducted using feature maps obtained from 3 different pre-trained deep learning models and feature maps obtained by combining the features obtained from these methods. Since it is aimed to achieve higher performance with fewer features, feature selection on these feature maps was carried out with PSO and ACO optimization algorithms. It was observed that the features selected with both PSO and ACO slightly increased the performance of the SVM algorithm. When the feature selection strategy based on both PSO and ACO was compared, when SVM was used, the PSO-based feature selection method had higher performance values compared to the ACO method. In the k-NN algorithm, it can be said that the features obtained with PSO and ACO reduce the performance. Considering the model-based features, it can be said that the features obtained from the ResNet50 model have higher overall performance values than other models. It can be clearly stated that SVM is more successful in classification algorithms than the k-NN algorithm, both in this study and in other studies in the literature [49,50]. According to the results obtained, the advantages of the study are as follows:
-
1)
Features have been extracted from pre-trained deep learning models. The results were found to be considerably higher.
-
2)
PSO and ACO metaheuristic feature selection methods were compared on deep features.
-
3)
High results can be obtained when using deep learning approaches and traditional machine learning approaches together.
-
4)
Combining deep features obtained from different CNN models has been shown to improve performance.
The promising results show that deep learning-based models have the potential to assist expert radiologists and medical practitioners in successfully managing the COVID-19 pandemic. We believe that these high-performance approaches will be significant in the epidemic that affects the whole world. The most critical issue limiting the study is the limited number of data. Increasing the data and testing it with data from many different centers will provide more stable systems. Another limitation of the study is that the data used in classes other than COVID-19 data belong to children between the ages of 1–5. There are cases where COVID-19 disease can also be seen in children. However, using adult image classes will provide more stable and generalized results in this disease, generally seen in adults. In the future, features will be extracted using image processing methods on X-ray and CT images. From these extracted features, the features that provide the best separation between classes will be determined and performance values will be measured with different classification algorithms. Apart from this, the results of the study will be tested with data from many different centers. In another study to be carried out in the future, studies will also be conducted to determine the demographic characteristics of patients and the probability of catching COVID-19 with artificial intelligence-based systems.
5. Conclusion
In this study, traditional classification methods and feature selection algorithms are used by using features obtained from the CNN model. The performance of the features obtained from the ResNet50, ResNet101 and InceptionResNetV2 models were tested with two different classification algorithms. It is classified by the high performance in the detection of COVID-19 and other classes. The features selected with PSO and ACO to increase the classification performance also contributed positively to the achievements. As a result, it is seen that the features taken from CNN models show very high performances with traditional classification algorithms and feature selection algorithms.
Funding
There is no funding source for this article.
Ethical approval
This article does not contain any data, or other information from studies or experimentation, with the involvement of human or animal subjects.
Declaration of competing interest
As a result, the authors declare that they have no conflict of interest.
References
- 1.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin. Chim. Acta. 2020;506(March):145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Liu C., Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;87(March):18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu G., Yang Y., Du Y., Peng F., Hu P., Wang R., Yin M., Li T., Tu L., Sun J., Jiang T., Chang C. Clinical pathway for early diagnosis of COVID-19: updates from experience to evidence-based practice. Clin. Rev. Allergy Immunol. 2020;59(1):89–100. doi: 10.1007/s12016-020-08792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos, Solit. Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121(March) doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Tawfiq J.A., Memish Z.A. Diagnosis of SARS-CoV-2 infection based on CT scan vs RT-PCR: reflecting on experience from MERS-CoV. J. Hosp. Infect. 2020;105(2):154–155. doi: 10.1016/j.jhin.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: a deep learning based classification for COVID-19 detection using chest X-ray images. Chaos, Solit. Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imag. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. https://ieeexplore.ieee.org/document/9098956/ [DOI] [PubMed] [Google Scholar]
- 9.Wong H.Y.F., Lam H.Y.S., Fong A.H.T., Leung S.T., Chin T.W.Y., Lo C.S.Y., Lui M.M.S., Lee J.C.Y., Chiu K.W.H., Chung T.W.H., Lee E.Y.P., Wan E.Y.F., Hung I.F.N., Lam T.P.W., Kuo M.D., Ng M.Y. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. 2020;296(2):E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Narin, C. Kaya, Z. Pamuk, Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arxiv 2020, arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- 11.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. 2020;40(3):462–469. doi: 10.1007/s40846-020-00529-4. arXiv:2004.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Öztürk Ş., Özkaya U., Barstuğan M. Classification of coronavirus (covid-19) from x-ray and ct images using shrunken features. Int. J. Imag. Syst. Technol. 2021;31(1):5–15. doi: 10.1002/ima.22469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khuzani A.Z., Heidari M., Shariati S.A. Covid-classifier: an automated machine learning model to assist in the diagnosis of covid-19 infection in chest x-ray images. Sci. Rep. 2021;11(1):1–6. doi: 10.1038/s41598-021-88807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (covid-19) detection in chest x-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoeibi A., Khodatars M., Alizadehsani R., Ghassemi N., Jafari M., Moridian P., Khadem A., Sadeghi D., Hussain S., Zare A., Sani Z.A., Bazeli J., Khozeimeh F., Khosravi A., Nahavandi S., Acharya U.R., Shi P. 2007. Automated Detection and Forecasting of COVID-19 Using Deep Learning Techniques: A Review. arXivarXiv. [Google Scholar]
- 16.Hariri W., Narin A. 2012. Deep Neural Networks for COVID-19 Detection and Diagnosis Using Images and Acoustic-Based Techniques: A Recent Review. arXivarXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman A., Iqbal M.A., Xing H., Ahmed I. Covid-19 detection empowered with machine learning and deep learning techniques: a systematic review. Appl. Sci. 2021;11(8):3414. [Google Scholar]
- 18.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and bayesian optimization. Appl. Soft Comput. 2020;97(xxxx) doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int. J. Math. Eng. Mana. Sci. 2020;5(4):643–651. doi: 10.33889/IJMEMS.2020.5.4.052. [DOI] [Google Scholar]
- 20.Narin A. 2020 Medical Technologies Congress (TIPTEKNO) IEEE; 2020. Detection of covid-19 patients with convolutional neural network based features on multi-class X-ray chest images; pp. 1–4.https://ieeexplore.ieee.org/document/9299289/ arXiv:2012.05525. [Google Scholar]
- 21.M. Toǧaçar, B. Ergen, Z. Cömert, COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches, Comput. Biol. Med. 121 (March). doi:10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed]
- 22.Kassania S.H., Kassasni P.H., Wesolowski M.J., Schneider K.A., Deters R. 2020. Automatic Detection of Coronavirus Disease (COVID-19) in X-Ray and CT Images: A Machine Learning Based Approach; pp. 1–18. arXiv 2019. arXiv:2004.10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Turkoglu, COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble, Appl. Intell.:10.1007/s10489-020-01888-w. [DOI] [PMC free article] [PubMed]
- 24.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Acc. 2020;8:132665–132676. https://ieeexplore.ieee.org/document/9144185/ [Google Scholar]
- 25.Casistica Radiologica Italiana COVID-19 database. https://www.sirm.org/category/senza-categoria/covid-19/
- 26.Cohen J., Morrison P., Dao L. 2020. Covid-19 Image Data Collection. arxiv [preprint] arXiv preprint arXiv:2003.11597. [Google Scholar]
- 27.Mooney P. Chest X-ray images (pneumonia) https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 28.Acharya U.R., Oh S.L., Hagiwara Y., Tan J.H., Adeli H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2018;100(September 2017):270–278. doi: 10.1016/j.compbiomed.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 29.A. M. Hasan, M. M. Al-Jawad, H. A. Jalab, H. Shaiba, R. W. Ibrahim, A. R. Al-Shamasneh, Classification of Covid-19 coronavirus, pneumonia and healthy lungs in CT scans using Q-deformed entropy and deep learning features, Entropy 22 (5). doi:10.3390/E22050517. [DOI] [PMC free article] [PubMed]
- 30.Toǧaçar M., Ergen B., Cömert Z. Classification of white blood cells using deep features obtained from Convolutional Neural Network models based on the combination of feature selection methods. Appl. Soft Comput. J. 2020;97 doi: 10.1016/j.asoc.2020.106810. [DOI] [Google Scholar]
- 31.Goodfellow I., Bengio Y., Courville A., Bengio Y. vol. 1. MIT press Cambridge; 2016. (Deep Learning). [Google Scholar]
- 32.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet large scale visual recognition Challenge. Int. J. Comput. Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. arXiv:1409.0575. [DOI] [Google Scholar]
- 33.Wu Z., Shen C., van den Hengel A. Wider or deeper: revisiting the ResNet model for visual recognition. Pattern Recogn. 2019;90:119–133. doi: 10.1016/j.patcog.2019.01.006. arXiv:1611.10080. [DOI] [Google Scholar]
- 34.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 35.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 36.Abualigah L.M., Khader A.T., Hanandeh E.S. A new feature selection method to improve the document clustering using particle swarm optimization algorithm. J. Comput,. Sci. 2018;25:456–466. [Google Scholar]
- 37.Kennedy J., Eberhart R. vol. 4. IEEE; 1995. Particle swarm optimization; pp. 1942–1948. (Proceedings of ICNN’95-international Conference on Neural Networks). [Google Scholar]
- 38.Xue B., Zhang M., Browne W.N. Particle swarm optimisation for feature selection in classification: novel initialisation and updating mechanisms. Appl. Soft Comput. 2014;18:261–276. [Google Scholar]
- 39.Dorigo M., Birattari M., Stutzle T. Ant colony optimization. IEEE Comput. Intell. Mag. 2006;1(4):28–39. [Google Scholar]
- 40.Dorigo M., Gambardella L.M. Ant colonies for the travelling salesman problem. Biosystems. 1997;43(2):73–81. doi: 10.1016/s0303-2647(97)01708-5. [DOI] [PubMed] [Google Scholar]
- 41.Vapnik V.N. An overview of statistical learning theory. IEEE Trans. Neural Network. 1999;10(5):988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 42.Cover T., Hart P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theor. 1967;13(1):21–27. [Google Scholar]
- 43.Duda R.O., Hart P.E., Stork D.G. PATTERN second edition. 2001. https://cds.cern.ch/record/683166/files/0471056693/TNQDotTNQ/TOC.pdf URL. 654.
- 44.Rodríguez J.D., Pérez A., Lozano J.A. Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE Trans. Pattern Anal. Mach. Intell. 2010;32(3):569–575. doi: 10.1109/TPAMI.2009.187. [DOI] [PubMed] [Google Scholar]
- 45.Kingma D.P., Ba J.L. ICLR 2015 - Conference Track Proceedings. 2015. Adam: a method for stochastic optimization, 3rd international conference on learning representations; pp. 1–15. arXiv:1412.6980. [Google Scholar]
- 46.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021;24:1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismael A.M., Şengür A. Deep learning approaches for covid-19 detection based on chest x-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asghar M.A., Khan M.J., Amin Y., Rizwan M., Rahman M., Badnava S., Mirjavadi S.S., et al. Eeg-based multi-modal emotion recognition using bag of deep features: an optimal feature selection approach. Sensors. 2019;19(23):5218. doi: 10.3390/s19235218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang S., Sabri A.Q.M., Alnajjar F., Loo C.K. Autism spectrum self-stimulatory behaviors classification using explainable temporal coherency deep features and svm classifier. IEEE Acc. 2021;9:34264–34275. [Google Scholar]