Abstract
The SARS-CoV-2 pandemic has accelerated the study of existing drugs. The mixture of homologs called ivermectin (avermectin-B1a [HB1a] + avermectin-B1b [HB1b]) has shown antiviral activity against SARS-CoV-2 in vitro. However, there are few reports on the behavior of each homolog. We investigated the interaction of each homolog with promising targets of interest associated with SARS-CoV-2 infection from a biophysical and computational-chemistry perspective using docking and molecular dynamics. We observed a differential behavior for each homolog, with an affinity of HB1b for viral structures, and of HB1a for host structures considered. The induced disturbances were differential and influenced by the hydrophobicity of each homolog and of the binding pockets. We present the first comparative analysis of the potential theoretical inhibitory effect of both avermectins on biomolecules associated with COVID-19, and suggest that ivermectin through its homologs, has a multiobjective behavior.
Keywords: SARS-CoV-2, COVID-19, Molecular docking, Molecular dynamic, Ivermectin, Avermectin
Graphical abstract
1. Introduction
SARS-CoV-2 is a novel virus belonging to the β-Coronavirus genus of the 2B group of the Coronaviridae family. This interesting virus contains only 29 proteins, 26 of which have been successfully expressed for in vitro studies to determine targets of interest for drug discovery [1]. For example, the conserved cysteine protease Mpro has been highlighted as an exciting target as it mediates the maturation cleavage of polyproteins during virus replication [2,3].
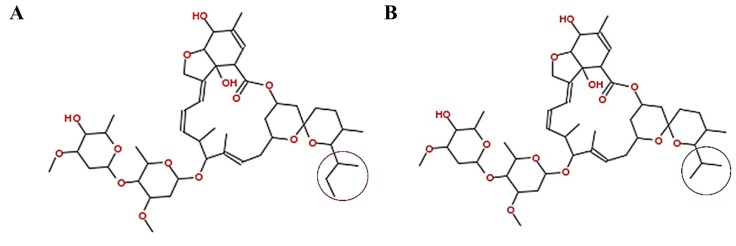
Despite great successes in the production and roll-out of vaccines against SARS-CoV-2, new variants are on the rise and there is still no globally accepted treatment for COVID-19 (https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-1). During the last year many laboratories focused on screening FDA-approved drugs for quick implementation in clincal settings [[4], [5], [6]]. A compound of interest from such work is the racemic mixture ivermectin, typically used to treat helminth infections. Most of the studies on the macrocyclic lactone ivermectin only consider the major constituent B1a in their dockings [7]. However, ivermectin is an approximately 80:20 mixture of two homolog derivatives of the compound avermectin B1, called 22,23-dihydroavermectin B1a (HB1a) and B1b (HB1b) correspondingly, which differ in the presence of a sec-butyl and isopropyl group, at the C25 position, respectively (Fig. 1 ). Interestingly, this mixture has demonstrated in vitro antiviral activity against several single-stranded RNA viruses, such as Zika virus, dengue virus, Chikungunya virus, avian influenza A virus, Porcine Reproductive and Respiratory Syndrome virus, human immunodeficiency virus type 1, among others, including SARS-CoV-2 [8].
Fig. 1.
Molecular structure of the two constituents of ivermectin considered in this study. A) vermectin B1a (HB1a), and B) Avermectin B1b (HB1b). The differential chemical group of each structure is indicated within a circle, For HB1a it is sec-butyl and HB1b is an isopropyl.
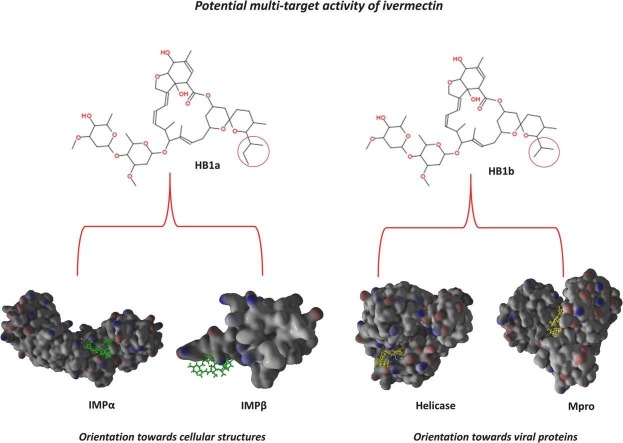
To date, the most accepted mechanism of action for ivermectin against SARS-CoV-2 is inhibition of the nuclear import of viral proteins and RNA, as has been found for HIV-1 and dengue [9]. However, it has also been reported that ivermectin has a potentially inhibitory effect against other viruses such as flaviviruses by blocking the NS3 helicase [10]. Additionally, it has been reported that it can dock in a thermodynamically favorable manner, and with theoretical potential to inhibit other structural and functional proteins associated with SARS-CoV-2 [11]. This diversity of possible targets for ivermectin and the fact that there are very few comparative reports in the SARS-CoV-2 on the behavior of homologs, represents an area of opportunity to contribute with theoretical studies that provide data applicable to the design of drugs and targeted at this new viral strain of interest in global public health.
Therefore, a more in-depth and exhaustive study is necessary on the possible mechanisms of inhibition of the two homologs present in ivermectin (HB1a and HB1b), against biomolecules of interest associated with COVID-19 especially approached from a computational biophysics and chemistry perspective. Here we describe, a rigorous comparative analysis of blind docking and ligand-protein interactions applying various genetic sampling methods, with theoretical calculations of potential inhibitory kinetic activity and analysis of molecular dynamics under controlled environments to study stability and thermodynamic fluctuations of the systems. In addition, we present the first preliminary report of the possible impact of molecular crowding on the activity of avermectin homologs against the structure of the SARS-CoV-2 cysteine protease Mpro.
2. Materials and methods
2.1. Databases and structure selection
As the nuclear import for macromolecules is facilitated by importins, the structures of importin α1 subunit (PDB: 5KLR) from Mus musculus and importin β1 subunit (PDB: 2P8Q) from Homo sapiens were used as a model for the members of the nuclear import superfamily. The host nuclear import system can be bound and sequestered by pathogens such as SARS-CoV-2 allowing transportation of viral proteins to the host nucleus leading to increased viral replication [[12], [13], [14], [15]]. Additionally, we also consider the multi-functional helicase (nsp13) of SARS-CoV-2 responsible for viral replication (PDB: 6ZSL) [[16], [17], [18]], and the main protease (Mpro) of SARS-CoV-2 (PDB: 6LU7) as it is a key enzyme of coronaviruses and has a fundamental role in mediating viral replication and transcription, making it an attractive target for drugs [11,[19], [20], [21], [22]]. All structures were obtained in PDB format from the RCSB protein database (https://www.rcsb.org/). The homologs structures of HB1a (CID_6321424) and HB1b (CID_6321425) that make up ivermectin were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in SDF format, and the SMILES online converter was used (https://cactus.nci.nih.gov/translate/) to get a PDB format. To avoid confusion throughout the study, we only refer to ivermectin when mentioning the mixture of homologs (a mixture of approximately 80:20 of the two homologs derivatives of the compound avermectin B1, called 22,23-dihydroavermectin B1a and B1b), while for the individual study of each homolog, the type of avermectin analyzed (HB1a or HB1b) was always indicated. For a comparison between the ADME profiles of the avermectin homologs, the PubChem databases (https://pubchem.ncbi.nlm.nih.gov/) were used; SwissADME (www.swissadme.ch/) and the Molinspiration Property Engine v2018.10 (https://www.molinspiration.com/).
2.2. Comparative molecular docking
We performed a comparative analysis using five popular molecular docking models for a rigorous prediction of the standard free energy (ΔG) of binding of ligand-protein complexes. The complexes were built in the programs MTiAutoDock (https://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::MTiAutoDock), webina (https://durrantlab.pitt.edu/webina/), DINC 2.0 (http://dinc.kavrakilab.org/), COACH-D (https://yanglab.nankai.edu.cn/COACH-D/) and DockThor (https://dockthor.lncc.br/v2/) were used. They all represent some of the basic, improved, and more advanced versions of molecular docking associated with the efficient AutoDock algorithm either in the sampling stage, blind docking or in the scoring function. To increase accuracy, a minimum of 10 runs per program were performed which implied approximately 106 evaluations per run. The rest of the parameters were considered by default. Additionally, to validate the docking results, the Pose&Rank server (https://modbase.compbio.ucsf.edu/poseandrank/) was used to score the protein-ligand complexes, using the statistical scoring function dependent on the atomic distance RankScore. As usual, all the water molecules were removed and the PDB files were separated into two different files, one containing the protein and the other containing the ligand structure. Only the three runs with the most favorable berth were considered in the sampling of the probabilistically most feasible and thermodynamically most favorable positions in the complexes. This criterion was used to discriminate the complexes that would be subjected to further analysis, including potential theoretical inhibition and molecular dynamics.
2.3. Comparative analysis of the hydrophobic characteristics of the binding sites
We use PockDrug-Server that predicts the drug delivery capacity in the pockets considering the Kyte-Doolittle Pocket Hydrophobicity Scale [23]. The hydrophobic characteristics of the binding sites were also analyzed using Biovia Discovery Studio 2021 [24].
2.4. Comparative analysis of theoretical Inhibition
The binding constant K from the binding free energy was calculated as described [11,25] from the following equation:
| (1) |
And the inhibition constant for binding of ligand to proteins (K i) (in units of M) was obtained as,
| (2) |
where, ∆G is binding affinity (kcal/mol), R is the universal gas constant (1.987 cal/K mol), T is the absolute temperature (298.15 K). The equation states that the higher the K i value, the weaker the binding of the inhibitor to the protein, and, therefore, the protein-inhibitor complex dissociates more easily [[26], [27], [28]]. Additionally, IC50 values were determined using the Dixon Plot, assuming competitive inhibition, as suggested [28]. The Dixon plot provides a good estimate of IC50 values for competitive inhibition of a given substrate. IC50 values were reported by placing the substrate concentration in the same units as IC50 in a subscript in parentheses, as suggested for theoretical predictions. For example, IC50 = 5 μM(10) would indicate an IC50 of 5 μM determined at a substrate concentration of 10 μM [29]. In this study, a hypothetical Substrate-Inhibitor relationship (1:1) was considered to avoid preferential unions favored by concentration, assuming structural similarities between homologs and substrate. Ten concentrations of each avermectin were simulated under a gradient from 0.5 to 5 μM to stimate the inhibitory potential based on plasma concentrations considered safe for ivermectin [30]. A theoretical inhibition potential was also predicted by using the IC50-to-K i web tool, following the considerations in the case of competitive inhibition with the equation,
| (3) |
where P50, is the protein concentration at 50% inhibition; K I, is the affinity constant of the ligand to the protein; [PL50], is the protein concentration by ligand concentration at 50% inhibition; and [L50] [L 50], is the ligand and concentration at 50% inhibition. The IC50-to-Ki web tool was used for all these calculations (https://bioinfo-abcc.ncifcrf.gov/IC50_Ki_Converter/index.php) [11,27].
2.5. Molecular Dynamics (MD) simulations
Simulations for a docking hit were performed for two purposes: 1) to study the relative stability of the ligand residing in the binding pocket; and 2) sampling of the minimum energy conformations to calculate the perturbation of the thermodynamic and structural stability of the complexes. For a protein-ligand complex, the MD system relaxed first through a series of minimization procedures that includes three phases: relaxation, balance and sampling, as recommended [11,31,32]. The MD simulation of the crystal structures was carried out in an explicit water system. Specifically, the solvation of the system was carried out in a solvation box of 8.0 Å. Our MD system also consisted of one copy of each protein system and one copy of the docking ligand. An Amber99SB-ILDN force field was applied to the complex, with TIP3P water model. The whole system was finally neutralized. Water molecules were treated as rigid bodies in all models, allowing a simulation time interval of 2 fs. Periodic boundary conditions were applied, and Berendsen's algorithm for temperature and pressure docking was adopted. After a first steepest descent to 5000 steps and conjugated gradient to 5000 steps energy minimizations with positional restraints on the solute, an initial 100 ps simulation was carried out with the positions of the solute atoms restrained by a force constant of 10 kcal/(mol Å2) to let the water diffuse around the molecule and for equilibration. The method PME was used to calculate the electrostatic contribution to nonbonded interactions with a cutoff of 14.0 Å and a time step of 1 fs. The cutoff distance of the van der Waals interaction was 14.0 Å. After this equilibration run, the NVT (particle numbers, volume, temperature) production run at 300 K was performed with the cell size remaining the same. The SHAKE algorithm was applied to the system, and the time step was set to 2 fs. Ten structures were obtained every 10 ns as target structures extracted from a 100 ns trajectory. For the Root Mean Square Deviations (RMSD) calculations, the equation,
| (4) |
where δi is the distance between atom i and either a reference structure or the mean position of the n equivalent atoms. All MD simulations and additional adjustments were performed using COSGENE/myPresto [11,31,32].
Additionally, to predict conformational changes in each protein after its interaction with the ligands, the NMSim software (https://cpclab.uni-duesseldorf.de/nmsim/main.php) was used, which is a computational technique that uses a three-step including coarse-graining (CG), normal mode analysis (NMA), and elastic network model (ENM) to provide realistic conformations in reasonable simulation time [33]. For this, the minimum energy structure obtained with mypresto at 100 ns was used to also calculate the root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF), using the radius-guided movements (ROG) approach of the NMSim Server [33]. The ROG-guided NMSim simulation is a method to search for a conformation of a given ligand-protein complex, and allows to describe the compactness of a protein. In an ROG-guided NMSim simulation, the trajectory is tailored towards the bound structure by selecting the pathway that leads to a decrease in Radius of gyration (Rg), and conformations are generated by structure distortion along directions of random linear combinations of low-frequency normal modes. For more detailed information on the theory of the method, see [33].
The NMSim web server enables geometric simulations based on NMA approach to explore biologically relevant conformational transitions in proteins, resulting in a trajectory of sampled protein conformations of good stereochemical quality. The NMSim simulation was configured in the “mode 1” to guide the simulation towards the lowest ROG. This allows proteins to be moved without any bias through the conformational space, and to generate the candidate conformation that has the lowest ROG. Therefore, any change in terms of unfolding must be significant. For more detailed information on the individual steps in the NMSim web server, see [33].
Binding affinities based on structural signatures and the force constant relative to the initial frame were also predicted during the 100 ns MD simulation of each complex. Plots of simulated complexes are presented using the CSM-Lig algorithm (http://biosig.unimelb.edu.au/csm_lig/) [34] designed to predict the binding affinity of a protein-small molecule complex based on structural signatures, and WebPSN algorithm (http://webpsn.hpc.unimo.it/wpsn.php) for the force constant using two alternative versions of ENM to calculate the cross-correlation of the motion of the Cα atoms and for the pairwise interactions between the atoms of Cα (linear cutoff-ENM and Kovacs-ENM) [35]. A widely used Molecular Mechanics/Poisson-Boltzmann surface area (MM/PBSA) [[36], [37], [38], [39], [40], [41]] was considered as a thermodynamic integration method, to requalify the complexes, by calculating the free energy binding (ΔGbind) of the MD trajectories. ΔGbind in a solvent medium was calculated as follows:
| (5) |
where Gcomplex, Gprotein and Gligand is the Gibbs free-energy of the ligand-protein complex, protein and the ligand in the presence of the solvent, respectively. The Gibbs free-energy for complex, protein, and ligands was calculated with following equation:
| (6) |
where G comprises the potential energy (EMM) in vacuum and solvation free energy (Gsolv) for each. EMM consists of bonded and non-bonded interactions, whereas Gsolv is the sum of electrostatic and non-polar solvation free energies. The dielectric constant of the biomolecular region (εp) was assigned as 2 and the dielectric constant of the solvent region (εs) was assigned as 80. The Size Modified Poisson-Boltzmann equation (SMPBE) (http://smpbs.math.uwm.edu/index_smpbs.php) and APBS program (https://server.poissonboltzmann.org/) were used to calculate the energetic components and to predict electrostatic solvation and free binding energies [38]. The ΔGbind of the complexes was calculated on frames taken at a 10 ns interval during the 100 ns production run. Molegro Molecular Viewer, version 7.0 was used (MMV_7.0).
2.6. Conformational fluctuations of ligand-protein complexes
The MD-based small-angle and wide-angle X-ray scattering curves (SWAXS) of all explicit solvent atoms were calculated on the WAXSiS server (http://waxsis.uni-goettingen.de) to predict the radius of gyration (Rg), default parameters were also used [42]. We used the HullRad (http://52.14.70.9/index_test.html) method to predict the Rg values, this method uses a convex hull model to estimate of manner fast and exact the Rg of the molecule [43]. To evaluate the conformational quality of each structure, the ProSA algorithm (https://prosa.services.came.sbg.ac.at/prosa.php) was used with which the Z-Score of a specific model was calculated and correlated with this score with the calculated scores of all publicly available structures on PDB websiten. In ProSA the minus negative Z-Score values correspond to conformations closer to native structures [4].
3. Results and discussion
3.1. Molecular Docking of HB1a and HB1b
Most of the computational studies aimed at predicting the best molecular docking for SARS-CoV-2 proteins have focused on the use of algorithms based on AutoDock Vina (ADV). ADV has an improved scoring function of the knowledge-based AutoDock (AD) method that uses a variant of X-Score, with an adjustment using PDBbind, and also considering the inter- and intramolecular contributions, its sampling technique is based on the Iterative Local Search global optimizer, in its Broyden - Fletcher - Goldfarb - Shanno variant (BFGS) [4,[44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]]. The use of the DockThor server was further proposed, which implements a grid-based method that employs a steady-state genetic algorithm for multiple solutions as a search engine and the MMFF94S force field as the scoring function for pose evaluation. This scoring function is obtained by linear regression and more sophisticated machine learning techniques for nonlinear problems using the refined set PDBbind [55]. DockThor has demonstrated a higher success rate than exhibited by AD and ADV-based methods [55,56].
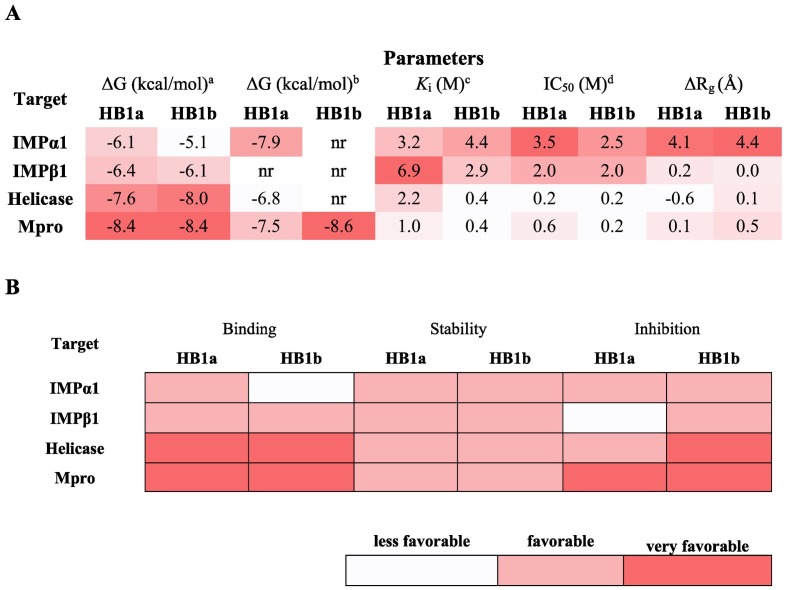
A marked energetic differential docking was observed between each homolog with each of the tested structures. All methods predicted thermodynamically favorable docking between each of the homologs and the proteins considered in this study (Table 1 ). Specifically, and as predicted by highly efficient and discriminatory algorithms used in the literature such as Webina [57], DINC [58,59], COACH-D [60] and DockThor [55,56,61], the existing homolog in the smallest proportion, HB1b, was the compound with the most thermodynamically favorable docking compared to Mpro followed by the helicase, while the thermodynamic union of the majority HB1a was more oriented to Importins (α1 and β1). Ivermectin has been reported to exert its antiviral effect by interfering with nuclear transport after binding with IMPα, affecting the recognition of important substrates, as well as binding to IMPβ [46].
Table 1.
Comparative analysis of the molecular docking of each homolog (HB1a and HB1b) obtained with each of the proteins using the various sampling methods and the RankScore Statistical Scoring Function.
| Target | ΔG (kcal/mol) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AutoDock |
AutoDock Vina (ADV) |
ADV Incremental |
COACH + ADV |
DockThor |
RankScore |
|||||||
| HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | |
| IMPα1 | −8.7 | −8.7 | −3.4 | −1.5 | −6.1 | −5.1 | −3.4 | −1.5 | −8.9 | −8.7 | −31.2 | −29.4 |
| −9.0g | ||||||||||||
| −6.9h | ||||||||||||
| IMPβ1 | −8.5 | −8.7 | −4.4 | −2.5 | −5.7 | −5.6 | −5.1 | −5.2 | −8.4 | −8.9 | −32.0 | −26.4 |
| Helicase | −8.7 | −8.7 | −8.3 | −8.6 | −8.3 | −8.6 | −3.8 | −3.8 | −9.1 | −10.2 | −40.4 | −46.6 |
| −5.1a | −8.1i | |||||||||||
| −7.6j | ||||||||||||
| −5.8k | ||||||||||||
| Mpro | −8.7 | −8.7 | −8.6 | −8.7 | −8.6 | −9.0 | −6.5 | −5.3 | −9.6 | −10.2 | −24.6 | −31.0 |
| −9.3b | −8.3f | −8.3l | −8.2r | −9.3t | ||||||||
| −8.4c | −8.0m | −9.4s | ||||||||||
| −7.7d | −7.2n | |||||||||||
| −5.6e | −6.8o | |||||||||||
| −6.5p | ||||||||||||
| −5.6q | ||||||||||||
Binding energy values reported using complexes and similar algorithms are shown. These are indicated in superscript against the values in this study highlighted in black. HB1a, avermectin B1a; HB1b, avermectin B1b.
[46].
[47].
[46].
[48].
[54].
[47].
[4].
[46].
[46].
[4].
[49].
[4].
[53].
[46].
[49].
[54].
[50].
[51].
[56].
The used genetic algorithm predicted a docking of the HB1b with the Mpro of −10.2 kcal/mol, and with B1a of −9.6 kcal/mol, as for the helicase the free binding energies were − 10.2 kcal/mol and − 9.1 kcal/mol for HB1b and HB1a, respectively (Table 1). These results represented the highest free binding energies predicted under the conditions of this study, and this trend was kept constant between all AD and ADV-based methods without significant difference (p ˃ 0.001). The only difference in this trend was predicted with COACH-D for Mpro and IMPβ1, with a value ≤0.5 kcal/mol. To validate the reproducibility of these results, the best poses were reclassified with PoseScore, especially since the efficient methods used present statistically different magnitudes (p ˂ 0.001). We observe that after reclassification, the same trend predicted above in relation to the affinity of HB1a and HB1b was maintained (Table 1). These results show a greater theoretical individual affinity of the HB1b for Mpro and Helicase, and of the HB1a for Importins (see Table 1).
This corresponds to existing data reported between these structures and HB1a using ADV [4,46]. This result could perhaps be related to the proportion of the HB1a in the ivermectin mixture and to the described mechanism of action of inhibition of import [9]. The binding energy shown by HB1a and HB1b towards Mpro, which in this study exceeds that predicted for Importins by up to ~5 kcal/mol depending on the type of sampling method, allows us to infer the possibility of a more probable docking of this mixture of homologs against Mpro from a statistical-thermodynamic point of view (between −8 kcal/mol and − 10 kcal/mol).
The internal docking control used in this study for Mpro, the alpha-ketoamide type inhibitor 13b [62] presented a less favorable mean binding energy (~7 kcal/mol) compared to ivermectin under conditions of our study (not shown). This trend of a more favorable binding energy (specifically HB1a) for Mpro, followed by IMPα1, has already been reported using ADV [4,46]. This corresponds to our previous report that shows that the constituent homologs of ivermectin present thermodynamically characteristic and differential dockings against various proteins associated with SARS-Cov-2 including Mpro [11]. However, to date there are few studies on ivermectin that consider energetic discrimination between its homologs, some show a more favorable union towards these proteases in the same way but with very narrow energies between both compounds, represented by a difference of ≈ 0.1 kcal/mol using ADV [50], and others studies if they reflect a more favorable energy for the HB1b [51].
In addition, other authors have reported similar binding energies between ivermectin and Mpro using the genetic algorithm tested here but in the same way without considering the discrimination between HB1a and HB1b [46,56] as has already been done [32]. Although the reproducibility of the results as well as the chemical structures offered allow us to suppose that most of the cited studies used the HB1a as a model [63]. This may be because this homolog is the compound with the highest concentration in ivermectin [[64], [65], [66], [67], [68], [69]]. In this sense, this study provides a broader view of the interaction and individual kinetic activity of each homolog, including the compound present in a lower proportion (HB1b).
Table 2 presents a perspective on the ADME profiles of the homologs considered in this study that includes experimental data associated with the plasma half-life of the homologs in healthy volunteers, cell models and animals. Both compounds have similar ADME profiles, and exhibit the same number of Lipinski law violations (MW > 500, HBAC >10). Although compound HB1b has a slightly higher hydrophobicity than HB1a, most of the experimental studies focus on the majority compound HB1a especially because the biological activities in vivo of the two homologs are similar [71].
Table 2.
Comparison between the ADME profiles of the avermectin homologs considered.
| Property | HB1a (ID_6321424)h | HB1b (ID_6321425) |
|---|---|---|
| MWa | 875.1 | 861.1 |
| HBDCa | 3 | 3 |
| HBACa | 14 | 14 |
| RBCa | 8 | 7 |
| ESOLb | −8.73 | −8.49 |
| iLOGPb | 5.74 | 6.44 |
| VOLc | 832.02 | 815.22 |
| VOLgc | 66.77 | 49.97 |
| PHL | 3-5d, h, 3-12e, 12-28f, 18d, h, 25-80g, h | 25-80g, h |
MW, Molecular Weight (g/mol); HBDC, Hydrogen Bond Donor Count; HBAC, Hydrogen Bond Acceptor Count; RBC, Rotatable Bond Count; ESOL, Water Solubility Log S; iLOGP, Lipophilicity (Log PO/W); VOL, Molecular volume based on group contributions (A3); VOLg, Molecular volume of group contribution (A3); PHL, Plasma half-life of Homolog in hours.
PubChem (https://pubchem.ncbi.nlm.nih.gov/).
SwissADME (www.swissadme.ch/).
Molinspiration property engine v2018.10 (https://www.molinspiration.com/).
[70].
[71].
[72].
[73].
No specification.
3.2. Comparative study of molecular docking and interactions of the HB1a and HB1b in the cavity of proteins
The interactions of the homologs with the residues in the binding pockets are presented in Table 3 . Comparison of the hydrophobic characteristics of the binding pockets according to the Kyte-Doolittle pocket hydrophobicity scale showed the most hydrophobic binding pocket in Mpro (−0.01), followed by IMPβ1 (−1.30), Helicase (−1.63) and IMPα1 (−2.59) (Supplementary Table 1). It was found that to IMPα1, 87.5% (14/16) of the interactions of HB1a were hydrophobic and 12.5% (2/16) were of the hydrogen-bond type, while the interactions established by HB1b were distributed in 80% (16/20) and 20% (4/20) for hydrophobic and hydrogen-bond interactions, respectively. The residues with the greatest contribution to the binding of HB1a and HB1b with IMPα1 in terms of hydrophobic interactions and hydrogen bonds were N-188 and W-184; as well as S-149 and N-228, respectively (see Table 3). The main chemical difference of these homologs is given by the presence of a sec-butyl and isopropyl group, in the C25 position, in the HB1a and HB1b, respectively (Table 3 and Fig. 1, Fig. 2 ). It was observed that the differential chemical group (sec-butyl group) of compound HB1a established only hydrophobic interactions with the binding pocket, while the HB1b established both hydrogen bonds and hydrophobic interactions with this protein structure.
Table 3.
Interactions of the HB1a and HB1b with residues in the binding pockets of each protein considered.
| Target | Interactions | Chemical group interactionsa |
|---|---|---|
| IMPα1 |
HB1a: N-188HB, W-184HB, N—228H, E-266H, R-227H, W—184H, W—231H, N—188H, N—146H, D—270H, S—149H, A-269H, L-307H, T-311H, P—308H, W—273H HB1b: S-149HB, N-228HB, R-238HB, E-266HB, S—149H, D—270H, N—235H, G-191H, R-238H, G-150H, D—192H, W—231H, N—188H, E-266H, G-187H, A-148H, G-224H, N—228H, R-227H, W—184H. |
HB1a (sec-butyl): E-266H, R-227H HB1b (isopropyl): S—159H, N—188H. |
| IMPβ1 |
HB1a: K-62HB, K—60H, M-61H, K—62H, R-53H, L-64H, I—54H, D—63H, R-52H HB1b: K-62HB, K—60H, M-61H, K—62H, R-53H, L-64H, I—54H, D—63H, R-52H |
HB1a (sec-butyl): D—63H, K—62H, L-64H, I—54H, M-61H, R-52H HB1b (isopropyl): M-61H, L-64H, D—63H, K—62H, I—54H, ARG-52H. |
| Helicase |
HB1a: S-289HB, A-316HB, H-290HB, E-261HB, R-442HB, E-319H, L-317H, A-316H, S—289H, R-443H, E-540H, G-285H, G-538H, S—264H, D—260H, R-442H, H—290H, F—262H, G-287H, T-286H, E-261H, K—323H, Y—324H, K—320H HB1b: E-261HB, S-289HB, A-316HB, T-286H, G-285H, G-538H, G-287H, R-443H, E-540H, E-319H, S—289H, L-317H, H—290H, A-316H, S—264H, D—260H, R-442H, F—262H, E-261H, K—320H, Y—324H, K-323H |
HB1a (sec-butyl): K—323H, K—320H, Y—324H, E-261H HB1b (isopropyl): K—323H, Y—324H, E-261H, K-320H |
| Mpro |
HB1a: R-298HB, Y-154HB, F—8H, I—152H, F—294H, V—297H, D—153H, G-302H, E-299H, S—301H, R-298H, Y—154H, K—12H, D—155H, P—9H HB1b: Y—154H, D—153H, I—152H, F—264H, F—8H, R-298H |
HB1a (sec-butyl): Y—154H, D—153H, F—8H, I—152H, R-29H. HB1b (isopropyl): no interactions |
The interactions with the differential chemical group of each type of avermectin are also shown. HB: Hydrogen Bonds; H: Hydrophobic Interactions; A: Alanine; R: Arginine; N: Asparagine; D: Aspartic Acid; E: Glutaminic Acid; Q: Glutamine; G: Glycine; H: Histidine; I: Isoleucine; L: Leucine; K: Lysine; M: Methionine; F: Phenylalanine; P: Proline; S: Serine; T: Threonine; W: Tryptophan; Y: Tyrosine; V: Valine.
HB1a is sec-butyl and HB1b is isopropyl group.
Fig. 2.
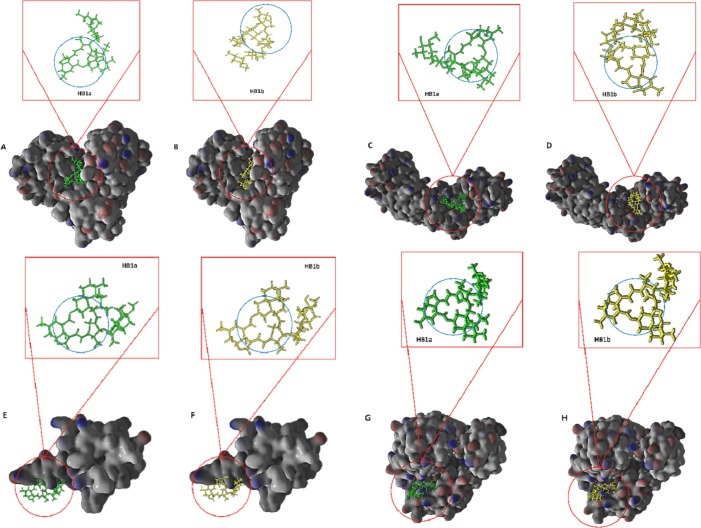
The most stable conformation of each avermectin is shown in the binding pockets of the proteins. Diagram of the orientation in Mpro (A and B), IMPa (C and D), IMPb (E and F), and Helicase (G and H) of HB1a and HB1b and their macrocyclic rings within the respective binding pockets, respectively. HB1a, avermectin B1a; HB1b, avermectin B1b. Macrocyclic rings are shown within a blue circle in the upper boxes; Binding pockets are shown as surface rendering and avermectin is attached as sticks style. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In IMPβ1, a lower number of interactions were observed compared to IMPα1, but in contrast, these were mostly concentrated in the differential chemical groups, approximately 88.8% (8/9) of the interactions between IMPβ1 and HB1a were hydrophobic, followed by 11.2% (1/9) of hydrogen bonding type interactions. In this docking 6/7 of the interactions in the sec-butyl group were hydrophobic in nature, a similar result was observed in the docking of HB1b, with which the same number of hydrophobic interactions was observed, as well as hydrogen bonds. With 4/5 of the hydrophobic type interactions with its isopropyl chemical group (Table 3). The residue with the greatest contribution to the binding of HB1a and HB1b with IMPβ1 in terms of hydrophobic interactions and hydrogen bonds were K-62, this residue established hydrophobic interactions and hydrogen bonds with each homolog and in both cases the union is established in the differential chemical group of each avermectin (see Table 3).
The interactions here predicted between IMPα1 and the HB1a correspond to those reported in the literature using ADV [46]. Analyzes have been performed showing that the binding pocket residues seen in our study Trp184, Arg227, Trp231, Ser149, Asn228, Trp231, and Arg238 appear to be critical in maintaining HB1a docking within the main groove. Trp184, Asn228 and Trp231 residues have already been shown to be vital in providing various intermolecular interactions [46]. No reports of molecular docking specifically between IMPα1 and IMPβ1 with HB1b were found.
In the Helicase protein, 79.2% (19/24) of the interactions presented by HB1a were hydrophobic, followed by 20.8% (5/24) hydrogen bond-type interactions, whereas the HB1b exhibited 86.4% (19/22) hydrophobic interactions and 13.6% (3/22) hydrogen bond-type interactions (Table 3). The differential chemical groups (sec-butyl or isopropyl moieties) of homologs contributed both with hydrophobic interactions and with hydrogen bonds against the Helicase. The residues with the greatest contribution in terms of hydrophobic interactions and hydrogen bonds to the Helicase and HB1a complex were E-261, S-289, H-290, A-316 and R-442; and in the complex with HB1b were E-261, S-289 and A-316. From these residues, only E-261 established hydrophobic interactions and hydrogen bonds with each homolog and in both cases the union is established in the differential chemical group of each avermectin (see Table 3). These results are similar to those reported between the Helicase and HB1a also using ADV on the ADP site [43], and to those reported in similar docking regions [50]. No molecular docking specifically between this protein and HB1b have been reported until now.
In the Mpro protein, interestingly, all the interactions of HB1a were hydrophobic (6/6) and were entirely concentrated around the sec-butyl chemical group, while HB1b showed 86.6% (13/15) of hydrophobic interactions and 13.3% (2/15) of hydrogen bonding type. The residues with the greatest contribution in terms of hydrophobic interactions and hydrogen bonds to the Mpro and HB1a complex were R-298 and Y-154; and in the complex with HB1b were R-298, I-152 and Y-154. Of these residues, Y-154 established hydrophobic interactions and hydrogen bonds with HB1a, as well as hydrophobic interactions with the chemical group (sec-butyl) of HB1a, and only hydrophobic interactions with HB1b (without participation of the chemical group (isopropyl) (Table 3). It was found that both homologs interact with the Arg-298 residue (R-298) (Table 3), which has been reported as key in the catalytic activity of the enzyme [74]. This residue has been described to be involved in dimerization of this protein and plays a key role in the catalytically active conformation of Mpro [75]. This coincides with reports that affecting the Arg-298 residue leads to the alteration of the dimeric structure of Mpro, as well as the irreversible inhibition of the catalytic activity of the enzyme [76,77].
Avermectins established more than 80% of hydrophobic interactions with each of the complexes tested, exhibiting a greater inclination towards this type of interactions, which highlights the influence of the hydrophobic characteristics of the cavities on their union. It is important to point out that the interactions observed with the sec-butyl chemical group were predominantly hydrophobic in the binding pockets of Mpro, while isopropyl exhibited more balanced interactions between hydrophobic and hydrogen bonding types, but at a global and individual level, slightly favored by hydrogen bond. But regardless of the chemical nature of the ivermectin homolog, these can interact with important residues involve in the dimerization of Mpro.
An observation that is important and that requires further study to determine if these interactions can have an impact on the formation of homodimer Mpro. These results differ from those reported by other authors who observed dockings at the catalytic site level, but without considering the differential chemical groups and only evaluating the interaction with HB1a [7]. One of the few works that considered both homologs against Mpro in their theoretical analyzes predicted a favorable but energetically very close docking between HB1a and HB1a using ADV [50]. The predictions have been made in regions other than the one observed in our study [50,78], and differ from other reports using ADV and HB1a with respect to Mpro predicting dockings in the catalytic region [46]. In this sense, this study provides a broader view on the individual interaction of the chemical groups of each homolog, as well as a greater evaluation of each compound through blind docking and a greater number of scoring functions to increase the reproducibility of the results.
3.3. Comparative theoretical study of the inhibition of target proteins by HB1a and HB1b
From the predictions of molecular docking, the theoretical values of both the K i and IC50 of each of the homologs against each of the protein structures considered in this study were determined. In this study we assume a competitive inhibition and constant concentrations of substrate and inhibitor, and we simulate a gradient that includes plasma concentrations considered safe, as will be discussed later, also the theoretical concentration used was placed as a subscript value in μM as suggested [29]. Results showing a more favorable K i compared to Mpro followed by the values for the Helicase protein, especially for the HB1b homolog. Predictions like the values obtained with strong Mpro inhibitors such as dipyridamole (K i = 0.04 μM) have shown promising therapeutic effects in clinical studies conducted for the treatment of patients with COVID-19 [28]. Similarly, compounds such as UAWJ246 and UAWJ247 have been reported to reversibly to Mpro with K i = 0.04 μM [79]. The K i predicted here for both homologs are lower than those reported for drugs such as hydroxychloroquine (K i = 0.36 μM) and chloroquine (K i = 0.56 μM), also calculated using the Dixon plot model and described as inhibitors of Mpro [28]. The results obtained can be seen in the Table 4 .
Table 4.
Comparison of theoretical inhibition kinetic values calculated for HB1a and HB1b with the proteins considered.
| Target |
Ki (M)a |
Ki (M)b |
IC50 (M)c |
IC50 (M)b |
pIC50 (M)b |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | |
| IMPα1 | 3.3 | 4.6 | 3.1 | 4.2 | 5.0 | 2.0 | 2.1 | 3.0 | 6.7 | 6.6 |
| IMPβ1 | 7.5 | 3.0 | 6.4 | 2.8 | 0.1 | 2.0 | 4.0 | 2.0 | 6.4 | 6.7 |
| Helicase | 2.3 | 0.4 | 2.2 | 0.4 | 0.2 | 0.2 | 2.0 | 0.3 | 6.8 | 7.5 |
| Mpro | 1.0 | 0.4 | 1.0 | 0.4 | 0.4 | 0.2 | 0.8 | 0.3 | 7.1 | 7.5 |
Ki, inhibition constant for binding of ligand to proteins in units of M; IC50, maximum mean inhibitory concentration in units of M; pIC50, negative logarithm of IC50. All Ki and IC50 values are in 10−7; HB1a, avermectin B1a; HB1b, avermectin B1b.
Constant predicted from Eq. (2).
Constant predicted from the IC50-to-Ki web server [27].
Constant predicted from the Dixon plot [29].
From the values of the Table 5 can be determined the relationship between the calculated theoretical inhibition constant and the simulated theoretical concentration gradients between 0.5 μM and 5 μM (subscript values) of HB1a and HB1b, according to the Dixon plot, showed that the predicted concentration for the theoretical 50% inhibition of IMPα1 by HB1a could require an IC50 ≈ 0.49 μM(0.5 – 5), with a maximum concentration of IC50 = 1.63 μM(5), while the mean theoretical concentration for HB1b would require an IC50 ≈ 0.69 μM(0.5 – 5), with a maximum concentration under these conditions of IC50 = 2.28 μM(5), this represents a maximum theoretical mean of an IC50 = 2 μM(5) for ivermectin versus IMPα1.
Table 5.
Comparison of the concentration gradient considered for ivermectin to simulate the potential inhibitory kinetics of HB1a and HB1b against the proteins considered.
| IVM [μM]a | IMPα1 |
IMPβ1 |
Helicase |
Mpro |
||||
|---|---|---|---|---|---|---|---|---|
| HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | HB1a | HB1b | |
| IC50 (μM) Dixon plotb | ||||||||
| 0.5 | 0.163 | 0.228 | 0.372 | 0.152 | 0.116 | 0.018 | 0.050 | 0.018 |
| 1.0 | 0.326 | 0.455 | 0.745 | 0.304 | 0.233 | 0.037 | 0.101 | 0.037 |
| 1.5 | 0.488 | 0.683 | 1.117 | 0.457 | 0.349 | 0.055 | 0.151 | 0.055 |
| 2.0 | 0.651 | 0.911 | 1.489 | 0.609 | 0.465 | 0.073 | 0.201 | 0.073 |
| 2.5 | 0.814 | 1.138 | 1.861 | 0.761 | 0.582 | 0.092 | 0.251 | 0.092 |
| 3.0 | 0.977 | 1.366 | 2.234 | 0.913 | 0.698 | 0.110 | 0.302 | 0.110 |
| 3.5 | 1.139 | 1.594 | 2.606 | 1.065 | 0.814 | 0.129 | 0.352 | 0.129 |
| 4.0 | 1.302 | 1.821 | 2.978 | 1.217 | 0.931 | 0.147 | 0.402 | 0.147 |
| 4.5 | 1.465 | 2.049 | 3.351 | 1.370 | 1.047 | 0.165 | 0.452 | 0.165 |
| 5.0 | 1.628 | 2.277 | 3.723 | 1.522 | 1.163 | 0.184 | 0.503 | 0.184 |
| IC50 (μM) Web toolc | ||||||||
| 0.5 | 0.096 | 0.113 | 0.138 | 0.091 | 0.079 | 0.024 | 0.047 | 0.024 |
| 1.0 | 0.130 | 0.159 | 0.204 | 0.123 | 0.103 | 0.027 | 0.057 | 0.027 |
| 1.5 | 0.151 | 0.187 | 0.248 | 0.141 | 0.117 | 0.029 | 0.063 | 0.029 |
| 2.0 | 0.165 | 0.207 | 0.280 | 0.154 | 0.127 | 0.030 | 0.066 | 0.030 |
| 2.5 | 0.176 | 0.223 | 0.305 | 0.164 | 0.134 | 0.031 | 0.069 | 0.031 |
| 3.0 | 0.185 | 0.235 | 0.325 | 0.172 | 0.140 | 0.032 | 0.071 | 0.032 |
| 3.5 | 0.192 | 0.246 | 0.343 | 0.178 | 0.145 | 0.032 | 0.073 | 0.032 |
| 4.0 | 0.198 | 0.255 | 0.358 | 0.184 | 0.149 | 0.033 | 0.074 | 0.033 |
| 4.5 | 0.203 | 0.262 | 0.371 | 0.188 | 0.152 | 0.033 | 0.075 | 0.033 |
| 5.0 | 0.208 | 0.269 | 0.383 | 0.193 | 0.155 | 0.033 | 0.076 | 0.033 |
The hypothetical substrate-inhibitor ratio (1: 1) was considered to avoid preferential binding favored by concentration, assuming a competitive inhibition and structural similarities between homologs and substrate; HB1a, avermectin B1a; HB1b, avermectin B1b.
A 0.5–5 μM gradient was used to speculate the inhibitory potential based on plasma concentrations considered safe for ivermectin (IVM).
Predicted from the Dixon plot [29].
Predicted from the IC50-to-Ki web server [27].
In IMPβ1, the mean for the HB1a was an IC50 ≈ 1.13 μM(0.5 – 5), and the maximum concentration of IC50 = 3.72 μM(5), the mean for HB1b was IC50 ≈ 0.46 μM(0.5 – 5), with a maximum value of IC50 = 1.52 μM(5). This leads to a maximum theoretical mean of an IC50 = 2.6 μM(5) for ivermectin versus IMPβ1 (Table 4, Table 5). These results compared to Importins are related to IC50 values reported in cell culture assays (≈ 2 μM) and therefore, with the potential inhibition of the importation associated mechanism of action as described [80].
It is not clear if the values reported correspond to any of our homologs, or if it is a typical mixture named ivermectin. For the Helicase, a mean of IC50 ≈ 0.35 μM(0.5 – 5) was predicted for HB1a, with a maximum concentration of IC50 = 1.16 μM(5), and to a lesser extent, an average IC50 ≈ 0.06 μM(0.5 – 5), with a maximum concentration of IC50 = 0.18 μM(5) for HB1b, which would represent a maximum theoretical mean of an IC50 ≈ 0.2 μM(5) for ivermectin versus the Helicase (Table 4, Table 5). These results are within the ranges reported of inhibition of activity (IC50 = 0.12–0.5 μM) and kinetics of the Helicase (IC50 = 0.019–0.354 μM), as well as viral replication by 50% (IC50 = 0.0005–4 μM) [81]. Observations that are related to the alteration of viral replication that this compound can induce through this pathway, since ivermectin has also been shown to bind to this non-structural viral protein [82].
On the other hand, compared to Mpro, a mean IC50 ≈ 0.15 μM(0.5 – 5) was predicted for HB1a, with a maximum concentration of IC50 = 0.50 μM(5), and as with the Helicase, an average IC50 ≈ 0.06 μM(0.5 – 5), with a maximum concentration of IC50 = 0.18 μM(5) for the HB1b, which would represent a maximum theoretical mean of an IC50 ≈ 0.1 μM(5) for ivermectin versus Mpro (Table 4, Table 5). The binding affinity of this type of ligand at the submicromolar level has already been reported using algorithms considered here [56]. These concentrations predicted for Mpro inhibition range around concentrations reported as safe in plasma and lung tissue [30], but they differ from those results obtained in in vitro enzyme inhibition assays with isolated protein (IC50 = 20 μM) [7]. This difference could be due to the fact that neither the test with the isolated protein nor our in silico modelling consider the effect of cytoplasmic crowding, an aspect that could explain the differences that exist between the viral inhibition tests in cell culture [80] and enzyme inhibition with isolated proteins [7], because molecular crowding is decisive when it comes to predicting the activity, dynamics and kinetics of biomolecules in a more realistic way [83,84].
It is not clear whether the results reported for isolated Mpro correspond to the HB1a, as evidenced in the graphs of the structure used for molecular docking, or whether it corresponds to a typical ivermectin mixture. An aspect that is important from our results that there exists a clear trend that the HB1b would require a lower concentration for the inhibition of the viral protease, due to the differential energy and kinetic qualities that exist between both homologs. In fact, the only experimental work with cell culture that has reported the use of HB1a is with an IC50 ≈ 50 μM [49]. It is interesting how in the studies that have been carried out to determine the IC50 when working with ivermectin, lower values have been observed than when working with the HB1a. This allows us to suggest that the potency observed when working with the ivermectin mixture may be due to the contribution of HB1b. Which would be in agreement with the results estimated under the conditions of this study.
The results obtained by the Dixon plot were compared with those predicted by the IC50-to-K i web server, and we observed that the same inhibition trend described above is maintained. Therefore, only the concentrations for the theoretical inhibition of Mpro predicted by both the Dixon plot and the IC50-to-K i web server are close to the accepted safe values, followed in second order by the values obtained for the Helicase inhibition (Table 4, Table 5). It has also been suggested that an IC50 ≈ 6–17 μM can convert ivermectin into a non-selective ATPase inhibitor [30,85]. We suggest that the individual activity of each homolog be evaluated independently, especially as the concentrations predicted here have already been suggested for evaluation [86], and because the pharmacokinetics of individual homologs against SARS-CoV-2 have not been reported up to now. Although our results show a tendency and a self-consistency between the binding energy and the constants and/or theoretical inhibition concentrations predicted for the homologs against each of the proteins tested, they should be taken as reference. However, these observations should not be ignored because as many of the cited pharmacokinetic evaluations indicate that ivermectin could be toxic in vitro, so further work is needed to reconcile the observations in vitro with clinical efficacy as suggested [87].
It is important to note that 1% DMSO concentration used for isolated Mpro has been shown to be capable of inducing folding in model proteins [88] which could affect the required concentration of the compound at sites of interest for enzyme inhibition, by favoring the natural folding mechanism and reducing the competitiveness of the drug for the site, as has already been studied [89]. That is why we conducted a preliminary study that represents the first reported evidence of the possible effect of molecular crowding and the addition of 1% of DMSO on the activity of avermectin homologs in structures associated with SARS-CoV-2 such as Mpro. The Mpro in presence and absence of homologs at 100 ns simulation was compared in three media: physiological environment (0.9% NaCl), 1% DMSO and 20% polyethylene glycol (PEG), a common crowding agent.
We consider the synthetic polymer PEG because, although less physiologically relevant, it is often used usually to mimic the interior of the cell as crowding agent and is useful for stabilizing protein-based drugs as recommended [90]. We define ΔRg as the change in Rg in DMSO or PEG of Mpro with and without ligands minus Rg in physiological solution. At which we could observe that the changes at a concentration of 1% DMSO were very discrete, suggesting that no significant effects are expected at the experimental level (ΔRg ≈ 0.1 Å). The ΔRg in an environment crowded with PEG versus the physiological environment did show important changes in terms of Rg of all complexes. We observe that the free Mpro protein and complex with HB1a are little affected by crowding (ΔRg ≈ 0.1 Å), however, the Mpro complex with HB1b is significantly affected, showing great structural compaction (ΔRg ≈ − 0.7 Å) (Supplementary Fig. 1), which thermodynamically favors this system because under crowding the compact structures are more favorable as has been already reported [83]. As in a physiological environment and 1% of DMSO, the energetic stability of the docking favors the HB1b compound despite inducing a conformational unfolding, this allows us to assume that the complex established with HB1b is stable and capable of producing a more favorable minimum energy structure than that generated by HB1a, although both compounds in DMSO destabilize the native (Supplementary Fig. 1). This could explain the differences between the aforementioned in vitro and cell culture results, and demonstrates the challenge of reconciling the often inconsistent observations at the in silico, in vitro and in vivo levels to increase the safety and clinical efficacy of drugs, especially in the early stages of studies with predictive models. Therefore, a more in-depth on the impact of variables such as cell crowding is currently being investigated by our team, while we recommend carrying out this type of study with a greater number of structural and functional proteins associated with SARS-CoV-2, including cellular proteins of the Importins superfamily.
Although the difference between both homologs of ivermectin could imply a non-functionalized part of their structures, and it is very likely that this type of structural difference affects the pharmacokinetic disposition more than the binding of the target. Interestingly, our results suggest that there is a differential behavior to level of the potential inhibitory effect of compounds HB1a and HB1b on the targets considered, and that this inhibitory effect is related to the predicted binding energy values. This kinetic aspect is favored in HB1a and HB1b for the cellular and viral proteins studied, respectively. A differential activity that has already been reported for these compounds both at the energy level and in terms of their particular affinity for related cellular and viral proteins [91].
It is also likely that both the sec-butyl and isopropyl groups contribute to the orientation, conformation, or predicted intermolecular interactions in the target binding pockets, in conjunction with the hydrophobic characteristics of each pocket (Table 3 and Fig. 2) and of each homolog (Table 2), because the steric effect of each chemical group contributes differently to the total volume of the HB1a (832.02 A3) and HB1b (815.22 A3) molecules. In fact, the sec-butyl group contributes 66.77 A3 and the isopropyl 49.97 A3 to the macrolactone molecule according to the Molinspiration server calculations. This method for predicting molecular geometries is very robust and relies on the contributions of each group for the calculation of molecular volume [92]. The efficient molecular docking algorithms used consider the spatial configuration of functional groups, as well as the incidence of molecular interactions by hydrogen bonding, rotation and steric shocks [[55], [56], [57], [58], [59], [60], [61]].
The in vivo biological activities of the two homologs are known to be identical in cell and animal models [71]. However, it is important to note that while the structural similarity of compounds HB1a and HB1b could explain why both homologs bind the various targets with comparable affinity values, the impact of binding energies on predicted inhibition kinetics should not be ignored because comparative analyzes of binding energies have been shown to be an important part in attempts to predict drug-target binding kinetics [93,94]. Emphasizing that the genetic docking algorithm included in this study has shown success rates like that exhibited by other methods used to validate experimental drug affinity results [55,56,94].
3.4. Comparative study on molecular dynamics of HB1a and HB1b
MD simulations in the field of computer-aided drug design have gained substantial importance for the estimation of dynamic and thermodynamic parameters of living systems in specific situations of physiological environments such as those considered here [46]. To investigate the stability and effect of the most favored docking at the energy and kinetic level, MD simulation studies were carried out for both homologs, HB1a and HB1b, against each of the proteins of interest, considering the stability of the systems in terms of total thermodynamic fluctuations and the difference in the atomic distance between structures every 10 ns of simulation in the protein systems in the presence and absence of ligands. The purpose of this study was to determine both the stability of the complexes and the potential changes and disturbances that each of the homologs may be capable of inducing on the proteins of interest.
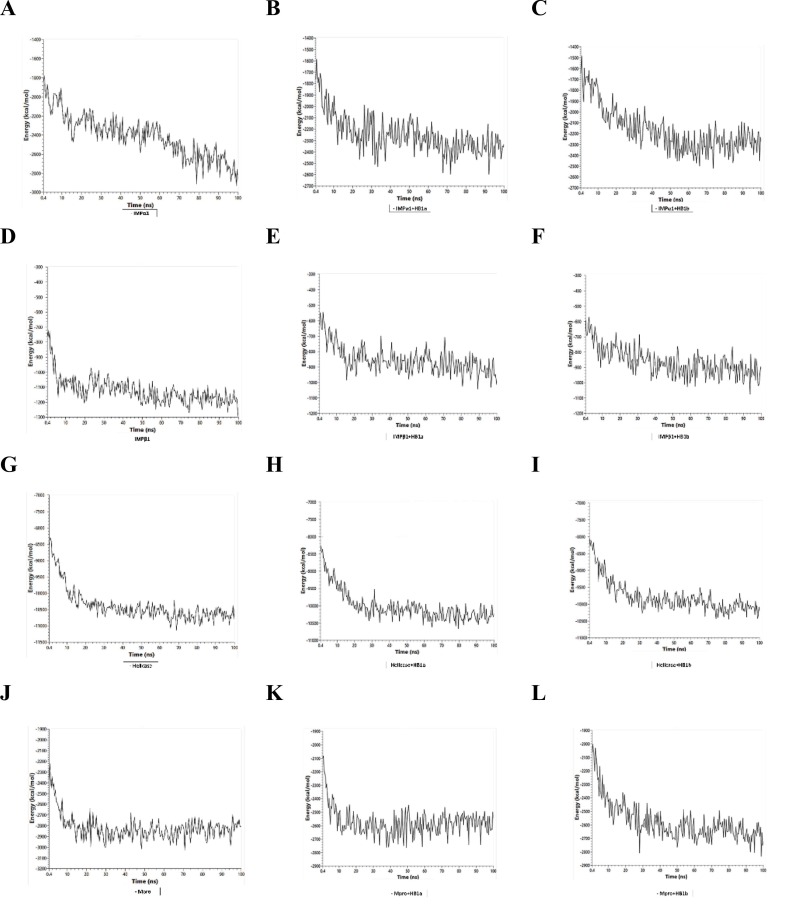
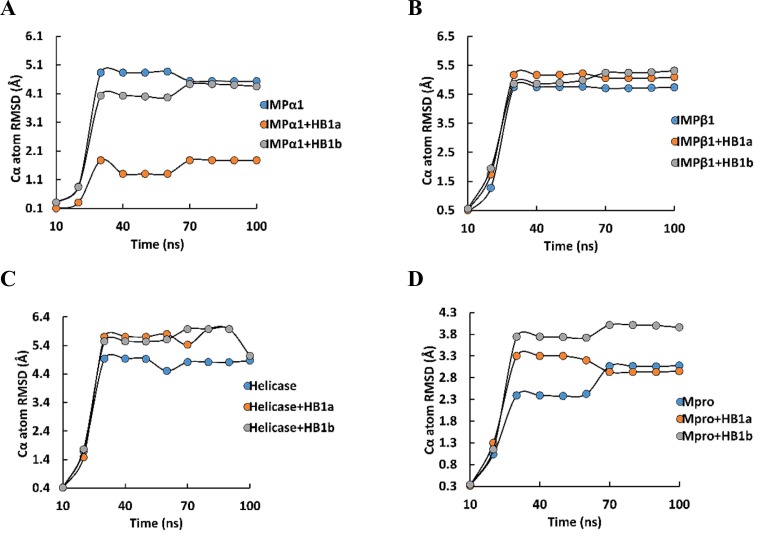
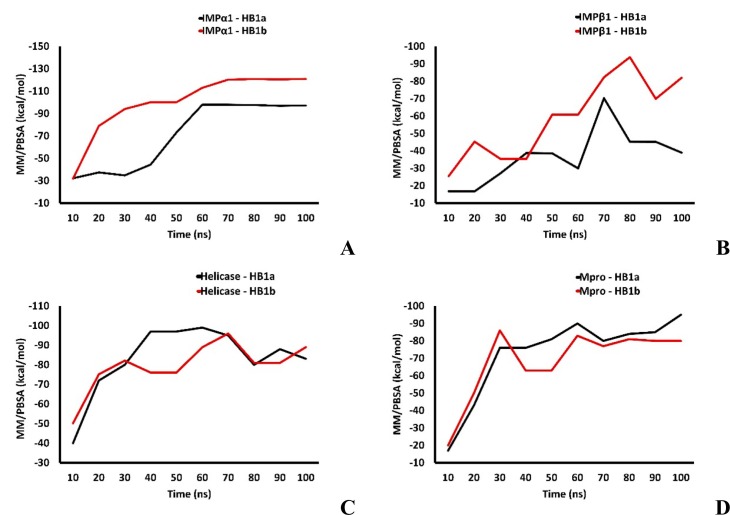
The energetic fluctuations associated with the thermodynamic stability of the ligand-protein complexes showed that both homologs establish thermodynamically stable systems compared to native structures throughout the time considered in this study. As can be seen in Fig. 3 , the ligands affected the energetic contributions and limited the degrees of freedom of the energetic transitions by mediating minimum energy conformations of up to a maximum of −2600 kcal/mol, compared to the native structure that free reached about −2900 kcal/mol at the end of the simulation time. IMPα1 reached favorable minimum energy structures, thermodynamically coupled with each homolog, stabilizing the trajectories from approximately 30 ns with total energy fluctuations in the order of −2300 and − 2500 kcal/mol over time. Avoiding the progressive descent seen of the native structure through the different energetic and probabilistically feasible conformations that it suffered from 15 ns. It is important to highlight that the IMPα1 + HB1a complex reached a more stable minimum energy conformation (−2600 kcal/mol) and in less simulation time (−70 ns) than that observed with the HB1b (−2500 kcal/mol at −75 ns) (Fig. 3A–C).
Fig. 3.
Fluctuation of the thermodynamic stability at 100 ns of proteins in the presence and absence of each homolog. A) IMPα1 free; B) IMPα1 + avermectin B1a (HB1a); C) IMPα1 + avermectin B1b (HB1b); D) IMPβ1 free; E) IMPβ1 + avermectin B1a (HB1a); F) IMPβ1 + avermectin B1b (HB1b), G) Helicase free; H, Helicase + avermectin B1a (HB1a); I) Helicase + avermectin B1b (HB1b); J) Mpro free; K) Mpro + avermectin B1a (HB1a); L) Mpro + avermectin B1b (HB1b).
In IMPβ1, the relationship between the decrease and the thermodynamic stabilization of the minimum energy conformations maintained a similar trend as seen with IMPα1 in the presence and absence of the ligands. Free protein reached a minimum energy conformation of about −1300 kcal/mol at 75 ns, after the initial decline at 5 ns. In presence of the two homologs, the energy fluctuation of the minimum energy conformation ranged from −1100 kcal/mol at approximately 95 ns. It is important to highlight that in the presence of HB1a, IMPβ1 reached stabilization in thermodynamic fluctuation at 15 ns, unlike the HB1b, at which this effect was observed after 30 ns. A stabilization that required more time but was maintained until the end of the simulation (Fig. 3D–F). Which could indicate that the predictions made by genetic algorithms about the most favorable energetic orientation between HB1b and IMPβ1 are thermodynamically feasible and show both a differential energetic between the homologs when docking and when inducing changes in the thermodynamic stability of tested protein systems. More important, unlike general molecular docking methods, MD simulations consider target flexibility and combined with energy calculations of thermodynamic fluctuations in binding, however a more accurate prediction of potential inhibitors can be developed [95].
In the Helicase, the homologs stabilized the thermodynamic fluctuations of the Helicase after 35 ns of the simulation system, this system was the largest studied with about 9407 atoms (≈ 2000 atoms above the closest system in size, represented by IMPβ1), reaching a total energy that ranged about −10,000 kcal/mol, which remained stable until the end of the MD cycle. This energetic behavior is faster with HB1b (at 30 ns). The fluctuations of the native structure showed a decrease in the minimum energy conformations from 25 ns of around −10,000 kcal/mol, until reaching some −11,000 kcal/mol at 70 ns (Fig. 3G–I). The results show that the homologs affected the energy contributions and limited the degrees of freedom of the thermodynamically probable transitions in the Helicase, as with the Importins, which could indicate an increase in the conformational stiffness of the system because of the ligands. The earliest stabilization phenomenon evidenced with HB1b is related to the binding energetics and inhibitory kinetics described above and which are slightly more favorable for this homolog compared to HB1a.
The homologs stabilized the thermodynamic fluctuations of Mpro. Specifically, after 40 ns of the simulation system, the HB1a stabilized the thermodynamic fluctuations constantly throughout the 100 ns of simulation, reaching a total energy that ranged around −2600 kcal/mol. As the HB1b stabilized the system at 30 ns of the simulation. An energetic behavior like that observed against the Helicase and faster than that presented by homolog HB1a. Reaching an energy of −2800 kcal/mol (similar to the energy of the native structure). The fluctuations of the native structure showed a decrease in the minimum energy conformations from 15 ns of −2800 kcal/mol throughout the simulation (Fig. 3J–L). In relation to the behavior of the RMSD of Cα for each protein studied and their complexes with the homologs, in the Fig. 4 can be seen the results obtained in each case. In IMPα1, the values obtained show that the thermodynamic changes induced on IMPα1 by both homologs caused a decrease in the distance of the alpha carbons of the complexes, with a difference at 100 ns with respect to the native RMSD of ≈ 3 Å and ≈0.5 Å approximatly for HB1a and HB1b, respectively (Fig. 4A), which would represent a significantly folded structural state with HB1a and favorable.
Fig. 4.
The Root Mean Square Deviations (RMSD) of Cα relative to the starting frame during 100 ns MD simulation. A, IMPα1 (IMPα1); B, IMPβ1 (IMPβ1); C, Helicase and D, Mpro in the presence and absence of each homolog (HB1a and HB1b).
Unlike the results predicted at 100 ns against IMPβ1 in which no differences were observed in the systems with and without ligand with an RMSD ≈ 5 Å. Results that show that the HB1a exerts a significant differential effect at the thermodynamic and structural level on IMPα1 compared to the induced by HB1b. Neither of the two homologs was able to induce significant differential perturbations against the native structure of IMPβ1 under the conditions of this study (Fig. 4B). The equilibrium of the IMPα system in the presence of HB1a as well as the large fluctuations of the Cα atoms have already been reported with different magnitudes and are considered an indication of large conformational changes in the protein structure during the simulation [4,46].
Against the Helicase, both homologs presented an RMSD ≈ 5 Å at the end of 100 ns. The difference between each Helicase-homolog complex and Helicase was RMSD ≈1 Å at 100 ns of the simulation (Fig. 4C). Results that do not show important and differential perturbations between these complexes compared to the native structure under the simulation conditions. The energetic fluctuations over time, specifically at ≈ 30 ns and ≈ 70 ns have already been described and show the variations in the thermodynamic stability of the Helicase system in the presence of ivermectin [4]. Furthermore, the Helicase values in terms of global RMSD are high due to its large structure, which includes several loops and flexible domains as reported against other types of ligands [16].
On the other hand, the results obtained for Mpro system revealed that the Mpro + HB1a complex has an RMSD ≈ 3.3 Å between 30 ns and 60 ns, and a value of RMSD ≈ 3 Å at 100 ns, with results similar to the native structure (RMSD ≈ 3 Å) at 100 ns. During the same periods of time, the complex with HB1b presented an RMSD ≈ 3.8 Å between 30 ns and 60 ns, and an RMSD ≈ 4 Å at 100 ns. This represents a difference of RMSD ≥1 Å at 100 ns between HB1b and the rest of the structures with and without ligand and show differential perturbations between these compounds compared to Mpro, which favor the HB1b in terms of folding. This shows that the homologs could modify the natural thermodynamic stability of Mpro by different mechanisms (Fig. 4D). There are few reports of DM of ivermectin versus Mpro, however, the results obtained in terms of RMSD of the Cα atoms of Mpro in SARS-CoV-2 are related to those reported during the simulation period of 100 ns versus other compounds that also remained in the steady state at ≈ 3 Å RMSD [16,96,97].
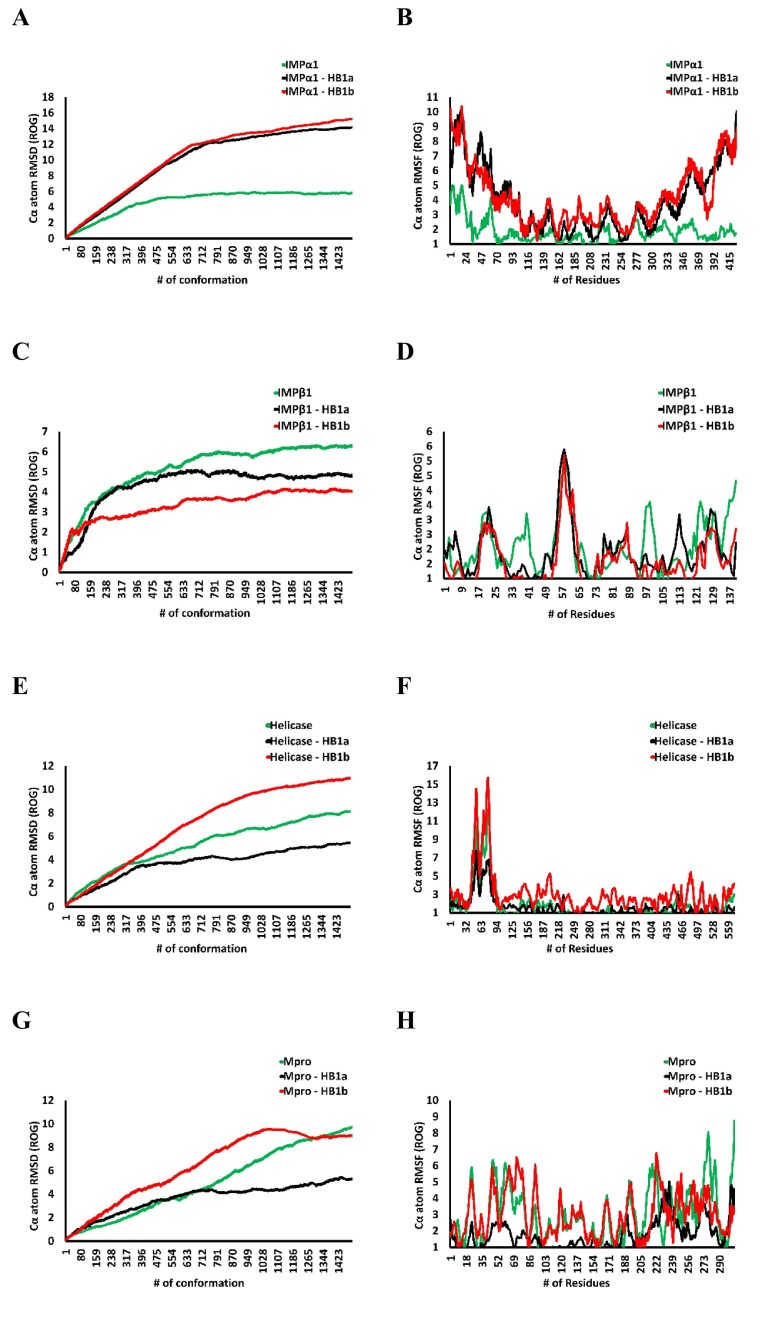
Also, all simulated complex graphs are presented as Radius of Gyration (ROG) guided movements using a geometric simulation based on normal mode. The resulting trajectories of the simulated complexes was inspected for different ROG simulation parameters such as backbone root mean square deviations (RMSD) for all Cα carbon atoms of the protein and the root mean square fluctuations (RMSF) of individual amino acid residues (Fig. 5 ). A behavior similar to those predicted by RMSD was observed after MD. The ROG-based fluctuations are related to the ROG-predicted RMSD values, showing a slightly more unfolded state of the ligand-IMPα1 complexes relative to the free IMPα1 structure (Fig. 5A and Fig. 5B). A similar fluctuation was also predicted in the case of IMPβ1 (Fig. 5C and D), Helicase (Fig. 5E and F) and Mpro (Fig. 5G and H).
Fig. 5.
The Root Mean Square Deviations (RMSD) of Cα and root mean square fluctuations (RMSF) of individual amino acid residues relative to the starting frame during 100 ns MD simulation of each homolog in complex with IMPα1 (A and B), IMPβ1 (C and D), Helicase (E and F), Mpro (G and H).
Variations in terms of RMSF could suggest a stable binding of homologs at their respective binding sites, generating complexes with fluctuations close to the free proteins, specifically, the region between residues N-146 and T-311 represents the binding site for both homologs in IMPα1; the region between R-52 and L-64 for each IMPβ1 homolog; as well as the E-261 and E-540 region for the Helicase with each homolog; observing a greater diversity of residues involved in binding with Mpro for each homolog (region between F-8 and S-301) (see Table 2, Table 3). Furthermore, the binding of each homolog could induce slight conformational perturbations in terms of RMSD, especially since even though the NMSim simulation was configured with “mode 1” to guide the simulation towards the lowest ROG, in the case of IMPα1, a slight protein unfolding was observed with both homologs (Fig. 5A).
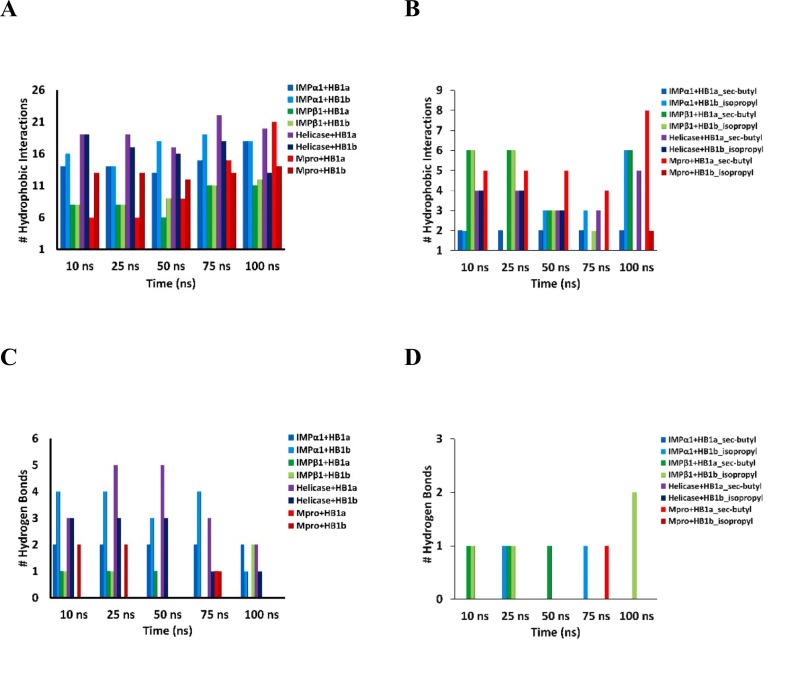
When studying the interactions of each of the homologs attached to their respective pockets in the importins and throughout the simulation time of 100 ns, it was observed that HB1a presented few variations in the interactions initially established throughout the simulation time maintaining a stable binding in place, as described (Table 6 ) [46]. The number of hydrogen bonds throughout the simulation, or the hydrophobic interactions established by its sec-butyl chemical group, were not altered. An increase in the number of hydrophobic interactions was only observed after 75 ns.
Table 6.
Interactions of the HB1a y HB1b with residues in the binding pockets of the proteins at 100 ns of MD.
| Time (ns) | Total interactions | Interactions in chemical groupa |
|---|---|---|
| IMPα1 + HB1a//IMPα1 + HB1b | ||
| 25 | T-184HB, N-188HB, N—228H, D—270H, A-269H, E-266H, T-331H, R-227H, W—184H, S—149H, N—188H, N—146H, W—231H, P—308H, L-307H, W—273H//E-266HB, N-228HB, S-149HB, R-238HB, G-224H, W—184H, N—228H, ARG—227H, E-266H, S—149H, D—270H, G-150H, N—235H, R-238H, D—192H, G-187H, G-191H, W—231H. | E-266H, R-227H//S—159H, N-188H |
| 50 | T-184HB, N-188HB, R-227H, D—270H, N—228H, E-266H, A-269H, P—308H, T-273H, T-184H, N—188H, S—149H, N—146H, L-307H, T-311H//R-238HB, N-228HB, A-148HB, S—149H, G-187H, G-150H, N—235H, G-191H, D—192H, W—231H, R-238H, D—270H, W—273H, N—188H, A-148H, Y—277H, E-266H, N—228H, R-227H, G-224H, W-184H | E-266H, R-227H//S—149H, N—188H, W-231H |
| 75 | W-184HB, N-188HB, R-227H, W—231H, E-266H, D—270H, W—184H, N—188H, S—149H, W—273H, N—146H, T-145H, P—312H, P—308H, A-269H, L-307H, T-311H//R-238HB, A-148HB, G-224HB, W-184HB, G-191H, N—188H, G-187H, R-238H, T-151H, Y—277H, W—231H, N—235H, W—273H, T-155H, D—192H, N—228H, R-227H, W—184H, E-180H, G-224H, S—149H, A-148H, D-270H | R-227H, E-266H//A-148H, S—149H, N—188H, W-231H |
| 100 | W-184HB, N-188HB, W—273H, P—308H, P—312H, T-311H, A-269H, L-307H, D—270H, V—267H, R-227H, E-266H, G-224H, C—223H, W—231H, N—228H, W—184H, N—146H, S—149H, N—188H//E-180HB, R-238H, T-277H, N—235H, D—270H, W—273H, E-266H, W—231H, N—188H, G-191H, T-155H, S—149H, A-148H, G-150H, N—228H, R-227H, E-180H, W—184H, G-224H | E-266H, R-227H//W—231H, N—188H, W—155H, G-191H, G-150H, A-148H, R-238H |
| IMPβ1 + HB1a//IMPβ1 + HB1b | ||
| 25 | K-62HB, K—60H, M-61H, K—62H, L-64H, I—54H, D—63H, R-53H, R-52H//K-62HB, R-52H, K—60H, M-61H, I—54H, K—62H, L-64H, R-53H, D-63H | L-64H, K—62H, MET-61H, I—54H, R-52H, D—63H//K—62H, L-64H, D—63H, M-61H, R-52H, I-54H |
| 50 | K-62HB, K—60H, M-61H, K—62H, L-64H, I—54H, G-56H//L-64H, I—54H, R-53H, L-62H, M-61H, R-52H, L-60H, R-55H, G-56H | K-62H, M-61H, K—60H//K—62H, M-61H, K-60H |
| 75 | R-52H, L-64H, D—63H, K—62H, R-55H, G-56H, M-61H, K—60H, G-57H, D—58H, I—54H//R-55H, G-57H, D—58H, G-59H, G-56H, K—60H, R-52H, K—62H, M-61H, I—54H, R-53H | K-62H//K—62H, I-54H |
| 100 | D-63H, D—58H, K—62H, G-57H, G-56H, R-55H, I—54H, S—65H, R-52H, L-64H, H—19H//Y-132HB, H-19HB, I—54H, K—62H, G-57H, D—58H, D—63H, H—19H, L-64H, Q-18H, R-52H, Y—132H, G-56H, R-55H | D-63H, D—58H, K—62H, G-57H, G-56H, L-64H//H—19H, L-64H, Y132H |
| Helicase + HB1a//Helicase + HB1b | ||
| 25 | E-261HB, A-316HB, S-289HB, R-442HB, H-290HB, E-319H, L-317H, A-316H, S—289H, E-540H, R-443H, G-538H, G-285H, D—260H, S—264H, H—290H, R-442H, G-287H, F—262H, T-286H, E-261H, K—323H, Y—324H, K—320H//A-316HB, S-289HB, E-261HB, G-285H, T-286H, G-287H, R-443H, G-538H, E-319H, E-540H, L-317H, S—289H, H—290H, A-316H, S—246H, D—260H, F—262H, E-261H, K—320H, Y—324H, K-323H | K-323H, K—320H, Y—324H, E-261H//K—323H, Y—324H, E-261H, K-320H |
| 50 | S-289HB, A-316HB, R-442HB, E-261HB, H-290HB, S—289H, K—320H, E-319H, L-317H, A-316H, G-538H, G-287H, G-285H, T-286H, E-261H, D—260H, S—264H, H—290H, K—323H, R-443H, R-442H, E-540H//A-316HB, H-290HB, E-261HB, T-286H, G-287H, G-538H, E-319H, K—320H, S—289H, L-317H, D—260H, A-316H, S—264H, E-261H, H—290H, K—323H, R-442H, E-540H, R-443H | K-323H, K—320H, E-261H//K—323H, E-261H, K-320H |
| 75 | E-261HB, A-316HB, S-289HB, E-540H, R-442H, G-285H, T-286H, H—464H, ARG—443H, LYS-323H, TYR-324H, H—290H, T-440H, G-287H, ASN—265H, ASP—260H, S—264H, A-316H, E-261H, A-312H, L-317H, S—289H, K—320H, S—539H, E-319H//A-316HB, G-285H, R-442H, E-540H, S—539H, K—320H, K—323H, E-319H, A-316H, S—264H, D—260H, E-261H, H—290H, G-287H, H—464H, K—465H, K—569H, R-443H, T-286H | K-323H, Y—324H, E-261H//No interactions detected |
| 100 | K-569HB, E-261HB, Y—541H, N—542H, N—190H, E-319H, K—320H, F—343H, S—539H, E-540H, E-261H, R-442H, C—441H, H—290H, L-438H, R-443H, G-287H, T-286H, D—260H, Y—324H, K—323H, K—569H//R-443HB, G-538H, Y—541H, E-540H, S—539H, D—260H, E-261H, H—464H, K—465H, R-442H, K—569H, T-286H, R-443H, H-290H | K-323H, Y—324H, D—260H, E-261H, R-442H//K-569H |
| Mpro + HB1a//Mpro + HB1b | ||
| 25 | Y-154H, D—153H, I—152H, F—8H, R-298H, F—294H//R-298HB, Y-154HB, I—152H, F—8H, D—153H, F—294H, V—297H, G-302H, S—301H, Q-299H, R-298H, P—9H, K—12H, D—155H, Y-154H | Y-154H, D—153H, I—152H, F—8H, R-298H//No interactions detected |
| 50 | K-12H, D—155H, Y—154H, D—153H, F—8H, F—294H, I—152H, R-298H, G-302H//Y—154H, K—12H, D—155H, P—9H, I—152H, F—294H, F—8H, V—297H, G-302H, S—301H, R-298H, D-153H | Y-154H, D—153H, F—8H, I—152H, D—155H//No interactions detected |
| 75 | Y-154HB, F—8H, F—294H, V—297H, S—301H, D—153H, I—152H, Y—154H, R-298H, S—1H, G-302H, V—303H, C—300H, Q-299H, D—155H, K—12H//Y-154HB, F—294H, S—301H, M-6H, R-298H, V—297H, Q-299H, I—152H, D—153H, F—8H, P—9H, K—12H, D—155H, Y-154H | F-8H, Y—154H, I—152H, D—153H, R-298H//No interactions detected |
| 100 | K-12H, P—9H, D—155H, Y—154H, F—8H, D—153H, I—152H, R-298H, V—297H, F—294H, N—151H, G-302H, S—301H, C—300H, V—303H, S—1H, Q-299H, Y—154H, D—155H, K—12H, P—9H//S—1H, V—303H, C—300H, Q-299H, S—301H, G-2H, G-302H, N—214H, P—9H, R-298H, F—8H, M-6H, A-7H, Y-154H | F-8H, D—153H, I—152H, R-298H, V—297H, N—151H, F—294H//G-302H, V-303H |
The interactions with the differential chemical group of each type of avermectin are also shown.HB: Hydrogen Bonds; H: Hydrophobic Interactions; A: Alanine; R: Arginine; N: Asparagine; D: Aspartic Acid; C: Cysteine; E: Glutaminic Acid; Q: Glutamine; G: Glycine; H: Histidine; I: Isoleucine; L: Leucine; K: Lysine; M: Methionine; F: Phenylalanine; P: Proline; S: Serine; T: Threonine; W: Tryptophan; Y: Tyrosine; V: Valine.
HB1a is sec-butyl and HB1b is isopropyl derivative of ivermectin, respectively.
Results that show a more favorable and stable binding of HB1a against IMPα1, compared to that predicted with HB1b. This variation in docking stability has already been reported but compared only to HB1a [4]. We observed that the HB1b presented more fluctuations in the interactions established over time, showing variations in global hydrophobic interactions and a decrease in the number of hydrogen bonds, as well as in the hydrogen bonds and hydrophobic interactions established by its isopropyl chemical group, however, fluctuations in global hydrophobic interactions were represented by an increase from 50 ns.
Specifically, the HB1a maintained its 2 initial hydrogen bonds and went from having 14 hydrophobic interactions to 18 within 100 ns of the simulation. As HB1b went from establishing 4 hydrogen bridges to 1, with a compensation in the number of hydrophobic interactions from 14 to 18 for 100 ns. In its differential chemical group (isopropyl group), HB1b lost the hydrogen bonds, but increased the number of hydrophobic interactions. Results showing less stable binding of HB1b than predicted for HB1a, and this is consistent with the previously described energetic and kinetic values that favor the binding of the HB1a against IMPα1.
In IMPβ1, the interactions established by HB1a fluctuated at the level of hydrogen bonds and hydrophobic interactions (Table 6), these fluctuations were represented by a decrease in the hydrogen bonds from 75 ns, as well as an increase in the hydrophobic interactions from the same period of time. The same tendency was observed in the interactions established by its sec-butyl chemical group, with a loss of hydrogen bonds from 75 ns and a decrease in hydrophobic interactions between 50 ns and 75 ns. As the HB1b showed an increase in all its interactions, both hydrophobic and hydrogen bonds at a global level, as well as its differential chemical group, which, although it suffered a decrease in the number and hydrophobic interactions, presented a compensatory increase in the number of hydrogen bonds at the end of the 100 ns simulation. It is important to note, that the HB1a after 100 ns lost all its hydrogen bonds and went from having 8 hydrophobic interactions to 11. In contrast, the same time of simulation, HB1b exhibited an increase in the number of global and local hydrogen bonds (in the differential chemical group), as well as an increase from 8 to 12 in terms of hydrophobic interactions at the end of 100 ns. Results that correspond with the observations at the energy and kinetic level, which show that, of the two homologs, HB1b presents a more stable and favorable binding by IMPβ1 (Table 6).
Although the results previously described on the interaction of the Helicase with both homologs showed an earlier stabilization in terms of thermodynamic fluctuation between HB1b and this enzyme over time, the results show a higher binding stability by the HB1a, which maintained the highest number of hydrogen bonds throughout the simulation. Both homologs showed a decrease both in the number of hydrogen bonds at the global level, and in the number of hydrophobic interactions at the local level in each of their differential chemical groups (sec-butyl vs isopropyl group).
The HB1a presented a decrease in the number of hydrogen bonds, offset by an increase in the number of hydrophobic interactions at the global level and in the sec-butyl group from 50 ns, reflected in a slight increase from 19 to 20 global hydrophobic interactions, and from 4 to 5 in their differential chemical group, but with a 5 to 2 loss in their hydrogen bonds at the end of 100 ns. The HB1b presented a significant decrease after 50 ns in all the interactions initially presented (Table 6), but without losing all the interactions as reported [46]. These interactions permit to suppose that under the conditions of this study, the docking of both homologs occurred in a different region than the one reported using similar docking algorithms [63].
Compared to Mpro, both compounds presented stable dockings reflected by an increase in their number of hydrophobic interactions over time, especially in its differential chemical groups. HB1a went from establishing 6 hydrophobic interactions to 21. With an increase of 5–8 hydrophobic interactions in its sec-butyl chemical group. HB1b lost the initial hydrogen bonds, and only increased the number of hydrophobic interactions from 13 to 14 after 100 ns simulation, the isopropyl differential chemical group established 2 hydrophobic interactions not initially predicted during docking. Indicative of stable unions at the end of the simulation (Table 6). Neither of the two compounds lost the interaction with the A-298 residue previously described as associated with dimerization and catalytic activity of the enzyme [76,77]. Time series graphs and/or histograms of ligand-protein interactions are shown in the Fig. 6 as evidence of persistence during MD simulations.
Fig. 6.
Total number of hydrophobic interactions and hydrogen bonding between each homolog (A and C) and its differential chemical group (B and D) with the proteins considered, monitored throughout the simulated trajectory of 100 ns, respectively. The lifespan of the hydrogen bonds and the hydrophobic interactions of the minimum energy structure of each complex were represented as absolute numbers of interactions at each specific time (in nanoseconds) indicated on the X-axis.
An increase in the number of hydrophobic interactions (Fig. 6A) as well as a decrease in the number of hydrogen bonds (Fig. 6C) is observed throughout the simulation of 100 ns. A differential depletion of hydrophobic interactions and hydrogen bonds in the typical chemical groups of each homolog was also predicted as a function of time (Fig. 6B and D).
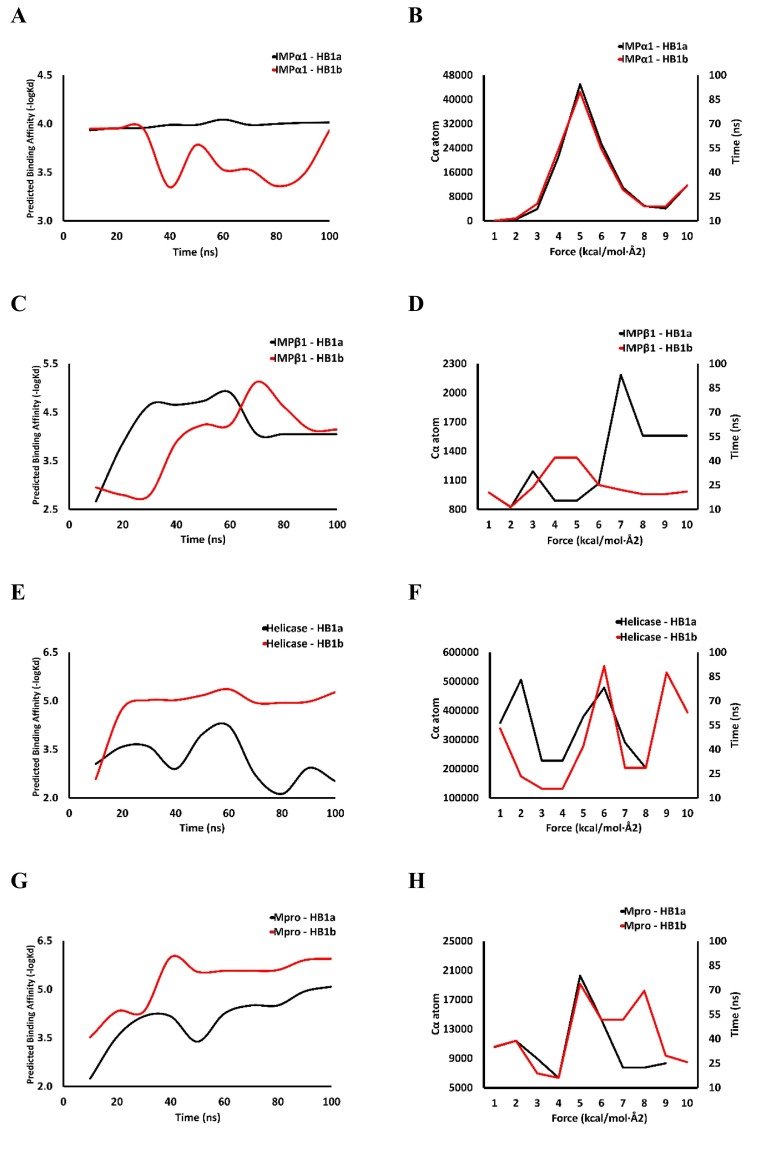
A clearer view of the contact stability between HB1a/HB1b and their target receptors is provided in Fig. 7 . Interaction energies between the ligand and the receptor were calculated and plotted using the predicted binding affinities based on structural signatures and the force constant relative to the initial frame during the 100 ns MD simulation of each homolog in complex with IMPα1 (Fig. 7A and B), IMPβ1 (Fig. 7C and D), Helicase (Fig. 7E and F), Mpro (Fig. 7G and H).
Fig. 7.
Predicted binding affinities based on structural signatures and the force constant relative to the starting frame during 100 ns MD simulation of each homolog in complex with IMPα1 (A and B), IMPβ1 (C and D), Helicase (E and F), Mpro (G and H).
The simulated complex plot is presented using the CSM-Lig algorithm [34] designed to predict the binding affinity of a protein-small molecule complex based on structural signatures (Fig. 7A, C, E and G), and the WebPSN algorithm for force constant (Fig. 7B, D, F and H) using two alternative versions of ENM to calculate the cross-correlation of the movement of Cα atoms and for the pairwise interactions between the Cα atoms (linear cut-ENM and Kovacs-ENM) [35].
The results of affinity and binding energy of the CSM-Lig and WebPSN algorithms (see Fig. 7), respectively, had similar behaviors for each complex, and suggest that the compound HB1a binds more strongly to the two importins studied, while HB1b binds more strongly to viral proteins (Helicase and Mpro), with a binding force of between 4 and 5 kcal/mol*Å2 above 50 ns according to WebPSN. The strength of the interaction is often based on geometric criteria (for example, distance cutoff, in this study was 4.5 Å) [35] and it is well known that a ligand stably bound to its target tends to localize at short distances and energetically favored [98]. In fact, the union between two atoms is determined by the energy produced by the interaction, which is affected by many variables, but mainly by the distance between the two atoms, so the distance used was the one suggested within the limit of 4–6 Å between heavy atoms for optimal performance [99].
As various docking methods were used to provide a consensus result to predict the preferred homologous ivermectin targets, the MD simulations provided an opportunity to potentially produce better estimates of the binding affinity values for these complexes. In this sense, the MM/PBSA prediction (see Fig. 8 ) was considered as a thermodynamic integration method, to requalify the complexes, taking into account multiple structures obtained from the MD trajectories. For the free energy binding (ΔGbind) of the MD trajectory, the widely used molecular mechanics/Poisson-Boltzmann surface area (MM/PBSA) was adapted [[36], [37], [38], [39], [40], [41]].
Fig. 8.
Predicted Binding energy (kcal/mol) from MM/PBSA relative to the starting frame during 100 ns MD simulation of each homolog in complex with IMPα1 (A), IMPβ1 (B), Helicase (C) and Mpro (D).
Stable binding of each homolog was predicted in the complexes according to MM/PBSA (see Fig. 8). The thermodynamic energy mean for IMPα1 + HB1a was −71 kcal/mol, and for IMPα1 + HB1b it was −100 kcal/mol (Fig. 8A). The residues with the highest energy contribution were W-184 (−18.8 kcal/mol) and N-228 (−12.3 kcal/mol) for the union of HB1a and HB1b with IMPα1, respectively. These residues established hydrophobic interactions and hydrogen bonds with each homolog (see Table 3, Table 6). For IMPβ1 + HB1a and IMPβ1 + HB1b it was −36 kcal/mol and − 59 kcal/mol, respectively (Fig. 8B). The residue with the highest energy contribution was K-62 for the union of HB1a (−24.3 kcal/mol) and HB1b (−18.0 kcal/mol) with IMPβ1. This residue established hydrophobic interactions and hydrogen bonds with each homolog and in both cases the union is established in the differential chemical group of each avermectin (see Table 3, Table 6).
For the Helicase + HB1a and Helicase + HB1a complex −83 kcal/mol and − 79 kcal/mol, respectively (Fig. 8C), the residue with the highest energy contribution was E-261 for the union of HB1a (−13.2 kcal/mol) and HB1b (−15.4 kcal/mol) with the Helicase. This residue established hydrophobic interactions and hydrogen bonds with each homolog and in both cases the union is established in the differential chemical group of each avermectin (see Table 3, Table 6). As for Mpro + HB1a and Mpro + HB1b it was −72 kcal/mol and − 68 kcal/mol, correspondingly (Fig. 8D). The residue with the highest energy contribution was Y-154 for the union of HB1a (−24.2 kcal/mol) and HB1b (−25.0 kcal/mol) with Mpro, this residue established hydrophobic interactions and hydrogen bonds with HB1a, and only hydrophobic interactions with HB1b. The differential chemical group of HB1a was the only one that interacted with this residue (see Table 3, Table 6). A stable MM/PBSA-based binding of ivermectin (only for the HB1a) against the proteins considered has already been reported [36,100].
3.5. Comparative study between conformational fluctuations of protein-avermectin complexes
The Z-Score allowed to observe the conformational fluctuation between the native structures and the dynamized ligand-protein complexes, measuring the deviation of the total energy of the structures with respect to their energy distribution derived from the contrast with other native random conformations. Starting from this model, it was observed that all the avermectin-protein complexes presented differences in the distances of their Cα atoms, as well as in their energetics at 100 ns of simulation and with respect to their respective native structures subjected to the same dynamic conditions.
The IMPα1 + homolog complex presented the most distant conformational fluctuation from the native one with a Z-Score more negative than that obtained for protein free of ligand, which suggest unfolding of this protein in both cases, as occurred with the Mpro protein. In contrast, the results obtained for IMPβ1 suggest refolding or compactation of structure in all cases, as for the case of the Helicase, the results suggest that both homologs induced refolding of this protein (Table 7 ; and Supplementary Fig. 2 and Supplementary Fig. 3). The scores obtained by the ProSA server for all structures validate that the predicted complexes, although with values within the graph and close to zero as described for native models [[101], [102], [103]], differ from the native structures without ligands tested here and therefore show conformational changes after simulations, a phenomenon that was observed more consistently between the HB1b with most of the coupled proteins.
Table 7.
Comparison between Z-score and radius of gyration in relation to the minimum energy structure at 100 ns MD simulation for each complex.
| Complexes | Simulation time (100 ns) |
||
|---|---|---|---|
| Z_Score | Rga | Rgb | |
| IMPα1 | −8.50 | 21.89 | 21.93 |
| IMPα1 + HB1a | −12.20 | 28.12 | 23.92 |
| IMPα1 + HB1b | −10.38 | 28.64 | 23.92 |
| IMPβ1 | −5.93 | 14.92 | 14.92 |
| IMPβ1 + HB1a | −4.87 | 15.48 | 14.72 |
| IMPβ1 + HB1b | −4.72 | 15.17 | 14.67 |
| Helicase | −5.27 | 32.02 | 31.97 |
| Helicase + HB1a | −5.03 | 31.34 | 31.32 |
| Helicase + HB1b | −5.24 | 32.14 | 32.09 |
| Mpro | −6.17 | 21.56 | 21.62 |
| Mpro + HB1a | −6.50 | 21.71 | 21.72 |
| Mpro + HB1b | −6.28 | 22.02 | 22.09 |
Radius of gyration (Rg) with WAXSiS server.
Radius of gyration (Rg) with HullRad; HB1a, avermectin B1a; HB1b, avermectin B1b.
The radius of gyration Rg is a measure of the compactness of a protein which allows to understand its folding properties. Small Rg values indicate a tight packing whereas high Rg values show a floppy packing, a relative constant Rg value through time indicates that the ligands hold the folding behavior of the protein whereas abrupt fluctuations of the Rg values denote protein folding instability [95]. All the methods predicted conformational fluctuations in the studied complexes oriented towards the folding or unfolding of the complexes. Observations dependent on both the type of protein system and the homolog considered (Table 7).
For IMPα1, HB1a and HB1b induced an unfolded conformation of the protein, with an average difference between both homologs and the native structure of Rg ≈ 4 Å, with mean values for each homolog of Rg ≈ 4.11 Å and Rg ≈ 4.37 Å, for complex with HB1a and HB1b, respectively (Table 7). Results that would allow us to infer, together with those of the atomic distance in terms of RMSD, that IMPα1 presents an almost “sphero-cylindrical” folded structural state, different from the native globular state (Supplementary Fig. 4). Similar results have already been reported in IMPα systems in the presence of HB1a [4], as the IMPβ1 protein shown a slight unfolded with fluctuating values between each homolog.
The mean difference between both complexes and the native structure was Rg ≈ 0.45 Å, with mean values Rg for the complex with HB1a and HB1b of Rg ≈ 0.54 Å and Rg ≈ 0.36 Å, respectively (Table 7). This corresponds to the observations at the RMSD level at which evidenced that none of the homologs was capable of inducing important differential perturbations compared to the native structure, generating a folded globular-type structural state in both cases. These results show a slightly greater effect of unfolding HB1b on IMPα1 and folding on IMPβ1, compared to HB1a, as well as a differential behavior of each homolog against importins tested in terms of induction of conformational changes.
In the Helicase, the HB1b induced a slightly unfolded conformation of this viral protein, the HB1a induced slightly refolding of this protein (Table 7). This is related to the atomic distance, which show no important or significant variations between these compounds compared to the native structure of the Helicase, inducing a globular folded conformational state with both homologs corresponding to data previously reported [4]. For Mpro the effect of both homologs was oriented towards conformational unfolding, with a mean difference between both homologs and the native structure was Rg ≈ 0.29 Å. Specifically, the mean values of the difference between each homolog were Rg ≈ 0.12 Å and Rg ≈ 0.46 Å, for HB1a and HB1b, respectively (Table 7).
Fig. 9 shows a summary of the main differences in the interaction of avermectin-B1a (HB1a) and avermectin-B1b (HB1b) with the selected targets. The binding of each homolog was stable in all cases over time in terms of RMSD, RMSF, relative binding energy and according to the MM/PBSA term. The differences presented are mainly focused on the discriminant values of the binding energies and the potential theoretical inhibitory effect on the kinetics of the compounds HB1a and HB1b on the cellular and viral proteins studied, respectively. The differential activity of compound HB1b, which is favorable against the viral proteins studied, can be highlighted more appropriately. The fact that there are no theoretical reports of the interaction of compound HB1b with most of the structures considered in this study is also highlighted.
Fig. 9.
Comparison of the observed differential characteristics of each homolog in terms of docking energy, dynamic, and potential theoretical inhibitory activity. aglobal mean binding free energy through AutoDock, AutoDock Vina, AutoDock Vina incremental, COACH-D and DockThor algorithms); breported global mean binding free energy (prediction from energy mean calculated with similar algorithms); nr, no report.
These theoretical predictions could guide the possibility of deriving formulations or analogues that can be administered to achieve relevant therapeutic concentrations, as it has been suggested necessary to do in the case of ivermectin [8,81]. It is important to say that these predictions based on theoretical models cannot predict a relevant clinical outcome and do not consider important variables such as time of exposure to the compound, cytoplasmic crowding (a phenomenon explored preliminarily in this study), among other aspects, which are key to a more adequate prediction of the kinetic qualities of each of the homologs. Furthermore, since it is a superfamily of proteins, the study of a greater number of importins is recommended, as well as a greater number of possible targets associated with SARS-CoV-2 under the same conditions of this study. It would be interesting to compare our results with experimental analyzes that consider each homolog in isolation, or variations in the proportion of each constituent of the ivermectin mixture.
4. Conclusions
There are several studies on possible targets for the controversial ivermectin, but there are few comparative reports on the behavior of avermectin homologs. Our objective was to investigate from a biophysical and computational chemical perspective the ligand-protein interactions, and the effect of these interactions on the kinetics and dynamics of the complexes predicted with each homolog. In this sense, after analysis of molecular dynamics and docking, as well as inhibitory kinetics, hydrophobicity studies, interactions between residues, RMSD, RMSF, Rg, ROG and preliminary results under crowding, we found that each of the homologs of ivermectin could establish thermodynamically favorable dockings with each of the proteins tested in this study. Each homolog produced different changes in the thermodynamic stability of the complexes, affecting the degrees of freedom of energy transitions by mediating minimum energy conformations, with fluctuations different from those of the free proteins. We also found at a global level a theoretical individual affinity of the HB1b for the viral structures and of the HB1a for the host structures. This corresponds to the predicted inhibition kinetics which, in turn, is influenced by the hydrophobicity of the cavities in the binding pockets and by the affinity of the differential chemical groups of each homolog. The theoretical behavior of these homologs could contribute to the possible reported multi-target ivermectin activity. However, it is necessary to carry out experimental demonstrations to corroborate this differential behavior, as well as its clinical relevance.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sector.
Author's Contributions
Lenin González-Paz: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. María Laura Hurtado-León: Investigation. Carla Lossada: Investigation. Francelys V. Fernández-Materán: Investigation. Joan Vera-Villalobos: Supervision, Validation. Marcos Loroño: Writing – original draft. JLPaz: Supervision, Validation, Writing – original draft. Laura Jeffreys: Writing – review & editing. Ysaias J. Alvarado: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bpc.2021.106677.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
References
- 1.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., MacKain A., Miorin L., Moreno E., Naing Z.Z., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Wang G., Ouyang L. Promising inhibitors targeting M pro: an ideal strategy for anti-SARS-CoV-2 drug discovery. Signal Transduct. Target. Therapy. 2020;5(1):1–2. doi: 10.1038/s41392-020-00291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9:622898. doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life sciences. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koulgi S., Jani V., Uppuladinne M., Sonavane U., Nath A.K., Darbari H., Joshi R. Drug repurposing studies targeting SARS-CoV-2: an ensemble docking approach on drug target 3C-like protease (3CLpro) J. Biomol. Struct. Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1792344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam M.T., Murshed R., Gomes P.F., Masud Z.M., Saber S., Chaklader M.A., Khanam F., Hossain M., Momen A.B.I.M., Yasmin N., Alam R.F., Sultana A., Robin R.C. Ivermectin as pre-exposure prophylaxis for COVID-19 among healthcare providers in a selected tertiary hospital in Dhaka – an observational study. Eur. J. Med. Health Sci. 2020;2(6) doi: 10.24018/ejmed.2020.2.6.599. [DOI] [Google Scholar]
- 7.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C., Fortin G.M., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021;4(1):1–10. doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidary F., Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen. J. Antibiot. 2020;73(9):593–602. doi: 10.1038/s41429-020-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudatsir M., Yufika A., Nainu F., Frediansyah A., Megawati D., Pranata A., Mahdani W., Ichsan I., Dhama K., Harapan H. Antiviral activity of ivermectin against SARS-CoV-2: an old-fashioned dog with a new trick–a literature review. Sci. Pharm. 2020;88(3) doi: 10.3390/scipharm88030036. [DOI] [Google Scholar]
- 10.Mastrangelo E., Pezzullo M., De Burghgraeve T., Kaptein S., Pastorino B., Dallmeier K., de Lamballerie X., Neyts J., Hanson A.M., Frick D.N., Bolognesi M., Milani M. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 2012;67(8):1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Paz L.A., Lossada C.A., Fernández-Materán F.V., Paz J.L., Vera-Villalobos J., Alvarado Y.J. Can non-steroidal anti-inflammatory drugs affect the interaction between receptor binding domain of SARS-COV-2 spike and the human ACE2 receptor? A computational biophysical study. Front. Phys. 2020;8:526. doi: 10.3389/fphy.2020.587606. [DOI] [Google Scholar]
- 12.Halder K., Dölker N., Van Q., Gregor I., Dickmanns A., Baade I., Kehlenbach R.H., Ficner R., Enderlein J., Grubmüller H., Neumann H. MD simulations and FRET reveal an environment-sensitive conformational plasticity of importin-β. Biophys. J. 2015;109(2):277–286. doi: 10.1016/j.bpj.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith K.M., Di Antonio V., Bellucci L., Thomas D.R., Caporuscio F., Ciccarese F., Ghassabian H., Wagstaff K.M., Forwood J.K., Jans D.A., Palù G., Alvisi G. Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting. Biochim. Biophys. Acta, Mol. Cell Res. 2018;1865(8):1114–1129. doi: 10.1016/j.bbamcr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Bernardes N.E., Fukuda C.A., da Silva T.D., de Oliveira H.C., de Barros A.C., Dreyer T.R., Bertolini M.C., Fontes M.R.M. Comparative study of the interactions between fungal transcription factor nuclear localization sequences with mammalian and fungal importin-alpha. Sci. Rep. 2020;10(1):1458. doi: 10.1038/s41598-020-58316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin A.J., Jans D.A. Antivirals that target the host IMPα/β1-virus interface. Biochem. Soc. Trans. 2021;9(1):281–295. doi: 10.1042/BST20200568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurakkal L., Singh S., Roy R., Kar P., Sadhukhan S., Porel M. An in-silico study on selected organosulfur compounds as potential drugs for SARS-CoV-2 infection via binding multiple drug targets. Chem. Phys. Lett. 2021;763:138193. doi: 10.1016/j.cplett.2020.138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Culletta G., Gulotta M.R., Perricone U., Zappalà M., Almerico A.M., Tutone M. Exploring the SARS-CoV-2 proteome in the search of potential inhibitors via structure-based pharmacophore modeling/docking approach. Computation. 2020;8(3):77. doi: 10.3390/computation8030077. [DOI] [Google Scholar]
- 18.Kapoor N., Ghorai S.M., Kushwaha P.K., Shukla R., Aggarwal C., Bandichhor R. Plausible mechanisms explaining the role of cucurbitacins as potential therapeutic drugs against coronavirus 2019. Informatics Med. Unlocked. 2020;21:100484. doi: 10.1016/j.imu.2020.100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah S., Chaple D., Arora S., Yende S., Mehta C., Nayak U. Prospecting for Cressa cretica to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2. J. Biomol. Struct. Dyn. 2021;15:1–9. doi: 10.1080/07391102.2021.1872419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panikar S., Shoba G., Arun M., Sahayarayan J.J., Nanthini A.U.R., Chinnathambi A., Alharbie S., Nasif O., Kim H.J. Essential oils as an effective alternative for the treatment of COVID-19: molecular interaction analysis of protease (Mpro) with pharmacokinetics and toxicological properties. J. Infect. Publ. Health. 2021 doi: 10.1016/j.jiph.2020.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chhetri A., Chettri S., Rai P., Sinha B., Brahman D. Exploration of inhibitory action of Azo imidazole derivatives against COVID-19 main protease (Mpro): a computational study. J. Mol. Struct. 2021;1224:129178. doi: 10.1016/j.molstruc.2020.129178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neupane N.P., Karn A.K., Mukeri I.H., Pathak P., Sharma P.K., Singh S., Qureshi I.A., Jha T., Verma A. Molecular dynamics analysis of phytochemicals from Ageratina adenophora against COVID-19 main protease (Mpro) and human angiotensin-converting enzyme 2 (ACE2) Biocatalysis Agric. Biotechnol. 2021;32:101924. doi: 10.1016/j.bcab.2021.101924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussein H.A., Borrel A., Geneix C., Petitjean M., Regad L., Camproux A.C. PockDrug-server: a new web server for predicting pocket druggability on holo and apo proteins. Nucleic Acids Res. 2015;43(W1):W436–W442. doi: 10.1093/nar/gkv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshpande R.R., Tiwari A.P., Nyayanit N., Modak M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur. J. Pharmacol. 2020;5(886):173430. doi: 10.1016/j.ejphar.2020.173430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Rehman M.T., Islam A., Hassan M. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40(6) doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popov M.E., Karlinsky D.M. Search for invisible binding sites of low-molecular-weight compounds on protein molecules and prediction of inhibitory activity. Mol. Biol. 2013;47(4):592–598. doi: 10.1134/S0026893313040122. [DOI] [PubMed] [Google Scholar]
- 27.Cer R.Z., Mudunuri U., Stephens R., Lebeda F.J. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37(2):W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z., Li X., Huang Y.Y., Wu Y., Liu R., Zhou L., Lin Y., Wu D., Zhang L., Liu H., Xu X., Yu K., Zhang Y., Cui J., Zhan C.G., Wang X., Luo H.B. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. 2020;117(44):27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burlingham B.T., Widlanski T.S. An intuitive look at the relationship of Ki and IC50: a more general use for the Dixon plot. J. Chem. Educ. 2003;80(2):214. doi: 10.1021/ed080p214. [DOI] [Google Scholar]
- 30.Momekov G., Momekova D. Ivermectin as a potential COVID-19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol. Biotechnol. Equip. 2020;34(1):469–474. doi: 10.1080/13102818.2020.1775118. [DOI] [Google Scholar]
- 31.Kasahara K., Terazawa H., Itaya H., Goto S., Nakamura H., Takahashi T., Higo J. myPresto/omegagene 2020: a molecular dynamics simulation engine for virtual-system coupled sampling. Biophys. Physicobiol. 2020;17:140–146. doi: 10.2142/biophysico.BSJ-2020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Paz L.A., Lossada C.A., Moncayo L.S., Romero F., Paz J.L., Vera-Villalobos J., Pérez A.E., Portillo E., San-Blas E., Alvarado Y.J. A bioinformatics study of structural perturbation of 3CL-protease and the HR2-domain of SARS-CoV-2 induced by synergistic interaction with ivermectins. Biointerface Research in Applied Chemistry. 2021;11(2) doi: 10.33263/BRIAC112.98139826. [DOI] [Google Scholar]
- 33.Krüger D.M., Ahmed A., Gohlke H. NMSim web server: integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res. 2012;40(Web Server issue):W310–W316. doi: 10.1093/nar/gks478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pires D.E., Ascher D.B. CSM-lig: a web server for assessing and comparing protein-small molecule affinities. Nucleic Acids Res. 2016;44(W1):W557–W561. doi: 10.1093/nar/gkw390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felline A., Seeber M., Fanelli F. webPSN v2. 0: a webserver to infer fingerprints of structural communication in biomacromolecules. Nucleic Acids Res. 2020;48(W1):W94–W103. doi: 10.1093/nar/gkaa397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen Gupta P.S., Biswal S., Panda S.K., Ray A.K., Rana M.K. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1839564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erdogan T. DFT, molecular docking and molecular dynamics simulation studies on some newly introduced natural products for their potential use against SARS-CoV-2. J. Mol. Struct. 2021:130733. doi: 10.1016/j.molstruc.2021.130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Y., Ying J., Xie D. SMPBS: Web server for computing biomolecular electrostatics using finite element solvers of size modified Poisson-Boltzmann equation. J. Comput. Chem. 2017;38(8):541–552. doi: 10.1002/jcc.24703. [DOI] [PubMed] [Google Scholar]
- 39.Kalhor H., Sadeghi S., Abolhasani H., Kalhor R., Rahimi H. Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1824816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qureshi U., Mir S., Naz S., Nur-e-Alam M., Ahmed S., Ul-Haq Z. Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications. J. Biomol. Struct. Dyn. 2021:1–12. doi: 10.1080/07391102.2021.1906750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Oliveira O.V., Rocha G.B., Paluch A.S., Costa L.T. Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1772885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatoki T.H., Ibraheem O., Ogunyemi I.O., Akinmoladun A.C., Ugboko H.U., Adeseko C.J., Awofisayo O.A., Olusegun S.J., Enibukun J.M. Network analysis, sequence and structure dynamics of key proteins of coronavirus and human host, and molecular docking of selected phytochemicals of nine medicinal plants. J. Biomol. Struct. Dyn. 2020;20:1–23. doi: 10.1080/07391102.2020.1794971. [DOI] [PubMed] [Google Scholar]
- 43.Fleming P.J., Fleming K.G. HullRad: fast calculations of folded and disordered protein and nucleic acid hydrodynamic properties. Biophys. J. 2018;114(4):856–869. doi: 10.1016/j.bpj.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagadala N.S., Syed K., Tuszynski J. Software for molecular docking: a review. Biophys. Rev. 2017;9(2):91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azam F., Taban I.M., Eid E.E.M., Iqbal M., Alam O., Khan S., Mahmood D., Anwar M.J., Khalilullah H., Khan M.U. An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1841028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohapatra R.K., Perekhoda L., Azam M., Suleiman M., Sarangi A.K., Semenets A., Pintilie L., Al-Resayes S.I. Computational investigations of three main drugs and their comparison with synthesized compounds as potent inhibitors of SARS-CoV-2 main protease (Mpro): DFT, QSAR, molecular docking, and in silico toxicity analysis. J. King Saud Univ. 2020;33(2):101315. doi: 10.1016/j.jksus.2020.101315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surti M., Patel M., Adnan M., Moin A., Ashraf S.A., Siddiqui A.J., Snoussi M., Deshpande S., Reddy M.N. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: designing, docking and molecular dynamics simulation study. RSC Adv. 2020;10:37707–37720. doi: 10.1039/D0RA06379G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabie A.M. Two antioxidant 2, 5-disubstituted-1, 3, 4-oxadiazoles (CoViTris2020 and ChloViD2020): successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs. New J. Chem. 2021;45(2):761–771. doi: 10.1039/D0NJ03708G. [DOI] [Google Scholar]
- 50.Marak B.N., Dowarah J., Khiangte L., Singh V.P. Step toward repurposing drug discovery for COVID-19 therapeutics through in silico approach. Drug Dev. Res. 2020:1–19. doi: 10.1002/ddr.21757. [DOI] [PubMed] [Google Scholar]
- 51.Oliveira M.D., Oliveira K.M. Comparative docking of SARS-CoV-2 receptors antagonists from repurposing drugs. ChemRxiv Prepr. 2020 doi: 10.26434/chemrxiv.12044538.v4. [DOI] [Google Scholar]
- 52.Cheke R.S., Narkhede R.R., Shinde S.D., Ambhore J.P., Jain P.G. Repurposing of anthelmintic drugs against SARS-CoV-2 (Mpro and RdRp): novel disease, older therapeutics. Letters in Applied NanoBioScience. 2020;10(2):2331–2338. doi: 10.33263/LIANBS102.23312338. [DOI] [Google Scholar]
- 53.Sahu S.N., Mishra B., Sahu R., Pattanayak S.K. Molecular dynamics simulation perception study of the binding affinity performance for main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020;23:1–16. doi: 10.1080/07391102.2020.1850362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adejoro I.A., Babatunde D.D., Tolufashe G.F. Molecular docking and dynamic simulations of some medicinal plants compounds against SARS-CoV-2: an in silico study. J. Taibah Univ. Sci. 2020;14(1):1563–1570. doi: 10.1080/16583655.2020.1848049. [DOI] [Google Scholar]
- 55.Santos K.B., Guedes I.A., Karl A.L., Dardenne L.E. Highly flexible ligand docking: benchmarking of the DockThor program on the LEADS-PEP protein–peptide data set. J. Chem. Inf. Model. 2020;60(2):667–683. doi: 10.1021/acs.jcim.9b00905. [DOI] [PubMed] [Google Scholar]
- 56.Guedes I.A., Costa L.S., Dos Santos K.B., Karl A.L., Rocha G.K., Teixeira I.M., Galheigo M.M., Medeiros V., Krempser E., Custódio F.L., Barbosa H.J., Nicolás M.F., Dardenne L.E. Drug design and repurposing with DockThor-VS web server: virtual screening focusing on SARS-CoV-2 therapeutic targets and their non-synonym variants. Sci. Rep. 2020;11(1):5543. doi: 10.1038/s41598-021-84700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kochnev Y., Hellemann E., Cassidy K.C., Durrant J.D. Webina: an open-source library and web app that runs AutoDock Vina entirely in the web browser. Bioinformatics. 2020;36(16):4513–4515. doi: 10.1093/bioinformatics/btaa579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antunes D.A., Moll M., Devaurs D., Jackson K.R., Lizée G., Kavraki L.E. DINC 2.0: a new protein–peptide docking webserver using an incremental approach. Cancer Res. 2017;77(21):e55–e57. doi: 10.1158/0008-5472.can-17-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhanik A., McMurray J.S., Kavraki L.E. DINC: a new AutoDock-based protocol for docking large ligands. BMC Struct. Biol. 2013;13(1):1–14. doi: 10.1186/1472-6807-13-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Q., Peng Z., Zhang Y., Yang J. COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018;46(W1):W438–W442. doi: 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolcato G., Cuzzolin A., Bissaro M., Moro S., Sturlese M. Can we still trust docking results? An extension of the applicability of DockBench on PDBbind database. Int. J. Mol. Sci. 2019;20(14):3558. doi: 10.3390/ijms20143558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khater S., Das G. Repurposing Ivermectin to inhibit the activity of SARS CoV2 helicase: possible implications for COVID 19 therapeutics. OSF Prepr. 2020 doi: 10.31219/osf.io/8dseq. [DOI] [Google Scholar]
- 64.Kojima K., Yamamoto K., Nakanishi Y., Katae H. Determination of 22,23-dihydroavermectin B1a in dog plasma using solid-phase extraction and high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1987;413:326–331. doi: 10.1016/0378-4347(87)80248-7. [DOI] [PubMed] [Google Scholar]
- 65.Chiu S.H., Sestokas E., Taub R., Smith J.L., Arison B., Lu A.Y. The metabolism of avermectin-H2B1a and-H2B1b by pig liver microsomes. Drug Metab. Dispos. 1984;12(4):464–469. PMID: 6148214. [PubMed] [Google Scholar]
- 66.Miwa G.T., Walsh J.S., VandenHeuvel W.J., Arison B., Sestokas E., Buhs R., Rosegay A., Avermitilis S., Lu A.Y., Walsh M.A., Walker R.W., Taub R., Jacob T.A. The metabolism of avermectins B1a, H2B1a, and H2B1b by liver microsomes. Drug Metab. Dispos. 1982;10(3):268–274. PMID: 6125361. [PubMed] [Google Scholar]
- 67.Chiu S.H., Sestokas E., Taub R., Buhs R.P., Green M., Sestokas R., Vandenheuvel W.J., Arison B.H., Jacob T.A. Metabolic disposition of ivermectin in tissues of cattle, sheep, and rats. Drug Metab. Dispos. 1986;14(5):590–600. PMID: 2876867. [PubMed] [Google Scholar]
- 68.Campbell W.C. An introduction to the avermectins. N. Z. Vet. J. 1981;29(10):174–178. doi: 10.1080/00480169.1981.34836. [DOI] [PubMed] [Google Scholar]
- 69.Campbell W.C., Fisher M.H., Stapley E.O., Albers-Schonberg G., Jacob T.A. Ivermectin: a potent new antiparasitic agent. Science. 1983;221(4613):823–828. doi: 10.1126/science.6308762. [DOI] [PubMed] [Google Scholar]
- 70.Bray M., Rayner C., Noël F., Jans D., Wagstaff K. Ivermectin and COVID-19: a report in antiviral research, widespread interest, an FDA warning, two letters to the editor and the authors’ responses. Antivir. Res. 2020;178:104805. doi: 10.1016/j.antiviral.2020.104805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell W.C. History of avermectin and ivermectin, with notes on the history of other macrocyclic lactone antiparasitic agents. Curr. Pharm. Biotechnol. 2012;13(6):853–865. doi: 10.2174/138920112800399095. [DOI] [PubMed] [Google Scholar]
- 72.Baraka O.Z., Mahmoud B.M., Marschke C.K., Geary T.G., Homeida M.M.A., Williams J.F. Ivermectin distribution in the plasma and tissues of patients infected with Onchocerca volvulus. Eur. J. Clin. Pharmacol. 1996;50(5):407. doi: 10.1007/s002280050131. [DOI] [PubMed] [Google Scholar]
- 73.Peña-Silva R., Duffull S.B., Steer A.C., Jaramillo-Rincon S.X., Gwee A., Zhu X. Pharmacokinetic considerations on the repurposing of ivermectin for treatment of COVID-19. Br. J. Clin. Pharmacol. 2021;87(3):1589–1590. doi: 10.1111/bcp.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22(6):297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 75.Lin P.Y., Chou C.Y., Chang H.C., Hsu W.C., Chang G.G. Correlation between dissociation and catalysis of SARS-CoV main protease. Arch. Biochem. Biophys. 2008;472(1):34–42. doi: 10.1016/j.abb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82(9):4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu C.G., Cheng S.C., Chen S.C., Li J.Y., Fang Y.H., Chen Y.H., Chou C.Y. Mechanism for controlling the monomer–dimer conversion of SARS coronavirus main protease. Acta Crystallogr. D Biol. Crystallogr. 2013;69(5):747–755. doi: 10.1107/S0907444913001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maurya D.K. A combination of ivermectin and doxycycline possibly blocks the viral entry and modulate the innate immune response in COVID-19 patients. ChemRxiv Prepr. 2020 doi: 10.26434/chemrxiv.12630539.v1. [DOI] [Google Scholar]
- 79.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6(50) doi: 10.1126/sciadv.abe0751. eabe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laing R., Gillan V., Devaney E. Ivermectin – old drug, new tricks? Trends Parasitol. 2017;33(6):463–472. doi: 10.1016/j.pt.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wubben J.M., Atkinson S.C., Borg N.A. The role of protein disorder in nuclear transport and in its subversion by viruses. Cells. 2020;9(12):2654. doi: 10.3390/cells9122654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dupuis N.F., Holmstrom E.D., Nesbitt D.J. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. 2014;111(23):8464–8469. doi: 10.1073/pnas.1316039111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Junker N.O., Vaghefikia F., Albarghash A., Höfig H., Kempe D., Walter J., Otten J., Pohl M., Katranidis A., Wiegand S., Fitter J. Impact of molecular crowding on translational mobility and conformational properties of biological macromolecules. J. Phys. Chem. B. 2019;123(21):4477–4486. doi: 10.1021/acs.jpcb.9b01239. [DOI] [PubMed] [Google Scholar]
- 85.Li N., Zhan X. Anti-parasite drug ivermectin can suppress ovarian cancer by regulating lncRNA-EIF4A3-mRNA axes. EPMA Journal. 2020;11(2):289–309. doi: 10.1007/s13167-020-00209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmith V.D., Zhou J., Lohmer L.R. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacolo. Ther. 2020;108(4):762–765. doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kinobe R.T., Owens L. A systematic review of experimental evidence for antiviral effects of ivermectin and an in-silico analysis of ivermectin’s possible mode of action against SARS-CoV-2. Fundam. Clin. Pharmacol. 2021;35(2):260–276. doi: 10.1111/fcp.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roy S., Jana B., Bagchi B. Dimethyl sulfoxide induced structural transformations and non-monotonic concentration dependence of conformational fluctuation around active site of lysozyme. J. Chem. Phys. 2012;136(11):115103. doi: 10.1063/1.3694268. [DOI] [PubMed] [Google Scholar]
- 89.Bergasa-Caceres F., Rabitz H.A. Interdiction of protein folding for therapeutic drug development in SARS CoV-2. J. Phys. Chem. B. 2020;124(38):8201–8208. doi: 10.1021/acs.jpcb.0c03716. [DOI] [PubMed] [Google Scholar]
- 90.Stadmiller S.S., Pielak G.J. Protein-complex stability in cells and in vitro under crowded conditions. Curr. Opin. Struct. Biol. 2021;66:183–192. doi: 10.1016/j.sbi.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 91.Choudhury A., Das N.C., Patra R., Bhattacharya M., Ghosh P., Patra B.C., Mukherjee S. Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach. Futur. Virol. 2021;16(4):277–291. doi: 10.2217/fvl-2020-0342. [DOI] [Google Scholar]
- 92.Unnisa A., Abouzied A.S., Baratam A., Lakshmi K.N.V., Hussain T., Kunduru R.D., Banu H., Fatima S.B., Hussian A., Selvarajan K.K. Design, synthesis, characterization, computational study and in-vitro antioxidant and anti-inflammatory activities of few novel 6-aryl substituted pyrimidine azo dyes. Arab. J. Chem. 2020;13(12):8638–8649. doi: 10.1016/j.arabjc.2020.09.050. [DOI] [Google Scholar]
- 93.Ganotra G.K., Wade R.C. Prediction of drug–target binding kinetics by comparative binding energy analysis. ACS Med. Chem. Lett. 2018;9(11):1134–1139. doi: 10.1021/acsmedchemlett.8b00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Işık M. The binding mechanisms and inhibitory effect of intravenous anesthetics on AChE in vitro and in vivo: kinetic analysis and molecular docking. Neurochem. Res. 2019;44(9):2147–2155. doi: 10.1007/s11064-019-02852-y. [DOI] [PubMed] [Google Scholar]
- 95.Mosquera-Yuqui F., Lopez-Guerra N., Moncayo-Palacio E.A. Targeting the 3CLpro and RdRp of SARS-CoV-2 with phytochemicals from medicinal plants of the Andean Region: molecular docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1835716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lokhande K., Nawani N., Venkateswara K., S., & Pawar, S. Biflavonoids from Rhus succedanea as probable natural inhibitors against SARS-CoV-2: a molecular docking and molecular dynamics approach. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1858165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Al-Karmalawy Ahmed, Alnajjar Radwan, Dahab Mohammed, Metwaly Ahmed, Eissa Ibrahim. Molecular docking and dynamics simulations reveal the potential of anti-HCV drugs to inhibit COVID-19 main protease. Pharmaceutical Sciences. 2021;9:661230. doi: 10.34172/PS.2021.3. [DOI] [Google Scholar]
- 98.Jiang L., Gao Y., Mao F., Liu Z., Lai L. Potential of mean force for protein–protein interaction studies. Proteins Struct. Funct. Bioinforma. 2002;46(2):190–196. doi: 10.1002/prot.10031. [DOI] [PubMed] [Google Scholar]
- 99.Xie Z.R., Hwang M.J. An interaction-motif-based scoring function for protein-ligand docking. BMC Bioinformatics. 2010;11(1):1–16. doi: 10.1186/1471-2105-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eweas A.F., Alhossary A.A., Abdel-Moneim A.S. Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2. Front. Microbiol. 2020;11:592908. doi: 10.3389/fmicb.2020.592908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oany A.R., Pervin T., Moni M.A. Pharmacoinformatics based elucidation and designing of potential inhibitors against Plasmodium falciparum to target importin α/β mediated nuclear importation. Infect. Genet. Evol. 2021;88:104699. doi: 10.1016/j.meegid.2020.104699. [DOI] [PubMed] [Google Scholar]
- 102.Yadav R., Choudhury C., Kumar Y., Bhatia A. Virtual repurposing of ursodeoxycholate and chenodeoxycholate as lead candidates against SARS-Cov2-envelope protein: a molecular dynamics investigation. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1868339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gurung A.B. In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep. 2020;21:100860. doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5