Abstract
Since the first reports of a novel severe acute respiratory syndrome (SARS)-like coronavirus in December 2019 in Wuhan, China, there has been intense interest in understanding how severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in the human population. Recent debate has coalesced around two competing ideas: a “laboratory escape” scenario and zoonotic emergence. Here, we critically review the current scientific evidence that may help clarify the origin of SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, origins, evolution, zoonosis
A review of the current literature supports a zoonotic origin for SARS-CoV-2 and emphasizes the need to focus on identifying the exact animal host through collaborative and carefully coordinated studies. This will enable the community to understand the roots of the COVID-19 pandemic as well as better prepare us for future pandemics
Evidence supporting a zoonotic origin of severe acute respiratory syndrome coronavirus 2
Coronaviruses have long been known to present a high pandemic risk. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the ninth documented coronavirus that infects humans and the seventh identified in the last 20 years (Lednicky et al., 2021; Vlasova et al., 2021). All previous human coronaviruses have zoonotic origins, as have the vast majority of human viruses. The emergence of SARS-CoV-2 bears several signatures of these prior zoonotic events. It displays clear similarities to SARS-CoV that spilled over into humans in Foshan, Guangdong province, China in November 2002, and again in Guangzhou, Guangdong province in 2003 (Xu et al., 2004). Both these SARS-CoV emergence events were associated with markets selling live animals and involved species, particularly civets and raccoon dogs (Guan et al., 2003), that were also sold live in Wuhan markets in 2019 (Xiao et al., 2021) and are known to be susceptible to SARS-CoV-2 infection (Freuling et al., 2020). Animal traders working in 2003, without a SARS diagnosis, were documented to have high levels of immunoglobulin G (IgG) to SARS-CoV (13% overall and >50% for traders specializing in civets) (Centers for Disease Control and Prevention, 2003). Subsequent serological surveys found ∼3% positivity rates to SARS-related coronaviruses (SARSr-CoV) in residents of Yunnan province living close to bat caves (Wang et al., 2018), demonstrating regular exposure in rural locations. The closest known relatives to both SARS-CoV and SARS-CoV-2 are viruses from bats in Yunnan, although animals from this province have been preferentially sampled. For both SARS-CoV and SARS-CoV-2, there is a considerable geographic gap between Yunnan and the location of the first human cases, highlighting the difficulty in identifying the exact pathway of virus emergence and the importance of sampling beyond Yunnan.
SARS-CoV-2 also shows similarities to the four endemic human coronaviruses: human coronavirus-OC43 (HCoV-OC43), human coronavirus-HKU1 (HCoV-HKU1), human coronavirus-229E (HCoV-229E), and human coronavirus NL63 (HCoV-NL63). These viruses have zoonotic origins, and the circumstances of their emergence are unclear. In direct parallel to SARS-CoV-2, HCoV-HKU1, which was first described in a large Chinese city (Shenzhen, Guangdong) in the winter of 2004, has an unknown animal origin, contains a furin cleavage site in its spike protein and was originally identified in a case of human pneumonia (Woo et al., 2005).
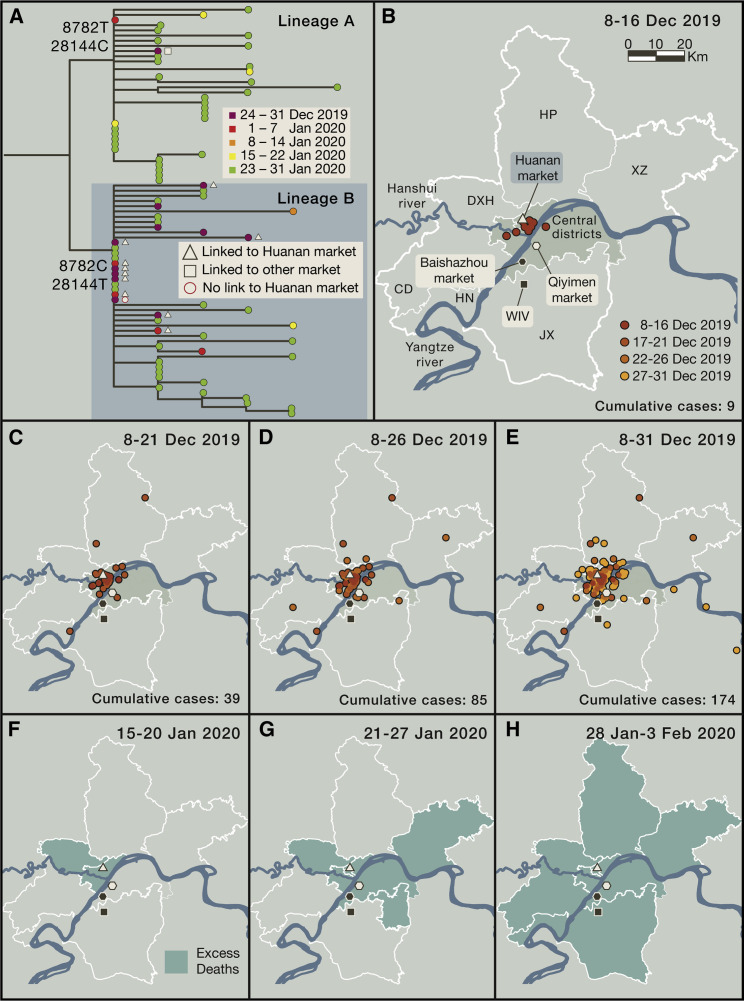
Based on epidemiological data, the Huanan market in Wuhan was an early and major epicenter of SARS-CoV-2 infection. Two of the three earliest documented coronavirus disease 2019 (COVID-19) cases were directly linked to this market selling wild animals, as were 28% of all cases reported in December 2019 (World Health Organization, 2021). Overall, 55% of cases during December 2019 had an exposure to either the Huanan or other markets in Wuhan, with these cases more prevalent in the first half of that month (World Health Organization, 2021). Examination of the locations of early cases shows that most cluster around the Huanan market, located north of the Yangtze river (Figures 1 B–1E), although case reporting may be subject to sampling biases reflecting the density and age structure of the population in central Wuhan, and the exact location of some early cases is uncertain. These districts were also the first to exhibit excess pneumonia deaths in January 2020 (Figures 1F–1H), a metric that is less susceptible to the potential biases associated with case reporting. There is no epidemiological link to any other locality in Wuhan, including the Wuhan Institute of Virology (WIV) located south of the Yangtze and the subject of considerable speculation. Although some early cases do not have a direct epidemiological link to a market (World Health Organization, 2021), this is expected given high rates of asymptomatic transmission and undocumented secondary transmission events and was similarly observed in early SARS-CoV cases in Foshan (Xu et al., 2004).
Figure 1.
Phylogenetic and epidemiological data on the early COVID-19 pandemic in Wuhan
(A) Phylogenetic tree of early SARS-CoV-2 genomes sampled from Wuhan during December 2019–January 2020. The split between lineages A and B is labeled with the coordinates and base of the two differentiating nucleotide mutations. Cases with a known association to the Huanan or other markets are denoted by symbols (reported in World Health Organization, 2021).
(B) Map of districts of Wuhan showing the location of markets, the Wuhan National Biosafety Laboratory at the Zhengdian Scientific Park of the Wuhan Institute of Virology (denoted WIV), where the coronavirus isolation and culture work of Dr. Shi Zhengli is performed, and the earliest known cases.
(C–E) Location of recorded COVID-19 cases in Wuhan from December 8 to December 31, 2019. Cases with a home address outside of Wuhan city are not shown.
(F–H) Map of districts of Wuhan indicating the first record of excess deaths due to pneumonia (shaded green) from January 15, 2020. Case and excess death data were extracted and redrawn from figures provided in World Health Organization (2021).
See also Document S1.
During 2019, markets in Wuhan—including the Huanan market—traded many thousands of live wild animals including high-risk species such as civets and raccoon dogs (Xiao et al., 2021). Following its closure, SARS-CoV-2 was detected in environmental samples at the Huanan market, primarily in the western section that traded in wildlife and domestic animal products, as well as in associated drainage areas (World Health Organization, 2021). Although animal carcasses retrospectively tested negative for SARS-CoV-2, these were unrepresentative of the live animal species sold and specifically did not include raccoon dogs and other animals known to be susceptible to SARS-CoV-2 (Xiao et al., 2021).
The earliest split in the SARS-CoV-2 phylogeny defines two lineages—denoted A and B (Rambaut et al., 2020)—that likely circulated contemporaneously (Figure 1A). Lineage B, which became dominant globally, was observed in early cases linked to the Huanan market and environmental samples taken there, whereas lineage A contains a case with exposure to other markets (Figures 1A and 1B) as well as with later cases in Wuhan and other parts of China (World Health Organization, 2021). This phylogenetic pattern is consistent with the emergence of SARS-CoV-2 involving one or more contacts with infected animals and/or traders, including multiple spill-over events, as potentially infected or susceptible animals were moved into or between Wuhan markets via shared supply chains and sold for human consumption (Xiao et al., 2021). The potential emergence of SARS-CoV-2 across multiple markets again mirrors SARS-CoV in which high levels of infection, seroprevalence, and genetic diversity in animals were documented at both the Dongmen market in Shenzhen (Yaqing, 2004; Guan et al., 2003) and the Xinyuan market in Guangzhou (Tu et al., 2004; Wang et al., 2005).
Viruses closely related to SARS-CoV-2 have been documented in bats and pangolins in multiple localities in South-East Asia, including in China, Thailand, Cambodia, and Japan (Lytras et al., 2021; Zhou et al., 2021), with serological evidence for viral infection in pangolins for more than a decade (Wacharapluesadee et al., 2021). However, a significant evolutionary gap exists between SARS-CoV-2 and the closest related animal viruses: for example, the bat virus RaTG13 collected by the WIV has a genetic distance of ∼4% (∼1,150 mutations) to the Wuhan-Hu-1 reference sequence of SARS-CoV-2, reflecting decades of evolutionary divergence (Boni et al., 2020). Widespread genomic recombination also complicates the assignment of which viruses are closest to SARS-CoV-2. Although RaTG13, sampled from a Rhinolophus affinis bat in Yunnan (Zhou et al., 2020b), has the highest average genetic similarity to SARS-CoV-2, a history of recombination means that three other bat viruses—RmYN02, RpYN06, and PrC31—are closer in most of the virus genome (particularly ORF1ab) and thus share a more recent common ancestor with SARS-CoV-2 (Li et al., 2021; Lytras et al., 2021; Zhou et al., 2021). None of these three closer viruses were collected by the WIV and all were sequenced after the pandemic had begun (Li et al., 2021; Zhou et al., 2020a, 2021). Collectively, these data demonstrate beyond reasonable doubt that RaTG13 is not the progenitor of SARS-CoV-2, with or without laboratory manipulation or experimental mutagenesis.
No bat reservoir or intermediate animal host for SARS-CoV-2 has been identified to date. This is presumably because the right animal species and/or populations have not yet been sampled and/or any progenitor virus may be at low prevalence. Initial cross-species transmission events are also very likely to go undetected. Most SARS-CoV-2 index case infections will not have resulted in sustained onward transmission (Pekar et al., 2021), and only a very small fraction of spillovers from animals to humans result in major outbreaks. Indeed, the animal origins of many well-known human pathogens, including Ebola virus, hepatitis C virus, poliovirus, and the coronaviruses HCoV-HKU1 and HCoV-NL63, are yet to be identified, while it took over a decade to discover bat viruses with >95% similarity to SARS-CoV and able to use hACE-2 as a receptor (Hu et al., 2017).
Could SARS-CoV-2 have escaped from a laboratory?
There are precedents for laboratory incidents leading to isolated infections and transient transmission chains, including SARS-CoV (Parry, 2004). However, with the exception of Marburg virus (Ristanović et al., 2020), all documented laboratory escapes have been of readily identifiable viruses capable of human infection and associated with sustained work in high titer cultures (Geddes, 2006; Lim et al., 2004; Senior, 2003). The 1977 A/H1N1 influenza pandemic, that most likely originated from a large-scale vaccine challenge trial (Rozo and Gronvall, 2015), is the only documented example of a human epidemic or pandemic resulting from research activity. No epidemic has been caused by the escape of a novel virus, and there is no data to suggest that the WIV—or any other laboratory—was working on SARS-CoV-2, or any virus close enough to be the progenitor, prior to the COVID-19 pandemic. Viral genomic sequencing without cell culture, which was routinely performed at the WIV, represents a negligible risk because viruses are inactivated during RNA extraction (Blow et al., 2004). No case of laboratory escape has been documented following the sequencing of viral samples.
Known laboratory outbreaks have been traced to both workplace and family contacts of index cases and to the laboratory of origin (Geddes, 2006; Lim et al., 2004; Ristanović et al., 2020; Senior, 2003). Despite extensive contact tracing of early cases during the COVID-19 pandemic, there have been no reported cases related to any laboratory staff at the WIV, and all staff in the laboratory of Dr. Shi Zhengli were said to be seronegative for SARS-CoV-2 when tested in March 2020 (World Health Organization, 2021), with the laboratory reportedly following the appropriate biosafety protocols during their coronavirus work (Cohen, 2020). During a period of high influenza transmission and other respiratory virus circulation (Liu et al., 2020a), reports of illnesses would need to be confirmed as caused by SARS-CoV-2 to be relevant. Epidemiological modeling suggests that the number of hypothetical cases needed to result in multiple hospitalized COVID-19 patients prior to December 2019 is incompatible with observed clinical, genomic, and epidemiological data (Pekar et al., 2021).
The WIV possesses an extensive catalog of samples derived from bats (Latinne et al., 2020) and has reportedly successfully cultured three SARSr-CoVs from bats—WIV1, WIV16, and Rs4874 (Ge et al., 2013; Hu et al., 2017; Yang et al., 2015). Importantly, all three viruses are more closely related to SARS-CoV than to SARS-CoV-2 (Ge et al., 2013; Hu et al., 2017; Yang et al., 2015). In contrast, bat virus RaTG13 from the WIV has reportedly never been isolated or cultured and only exists as a nucleotide sequence assembled from short sequencing reads (Cohen, 2020). The three cultured viruses were isolated from fecal samples through serial amplification in Vero E6 cells, a process that consistently results in the loss of the SARS-CoV-2 furin cleavage site (Davidson et al., 2020; Klimstra et al., 2020; Liu et al., 2020b; Ogando et al., 2020; Sasaki et al., 2021; Wong et al., 2021; Zhu et al., 2021b). It is therefore highly unlikely that these techniques would result in the isolation of a SARS-CoV-2 progenitor with an intact furin cleavage site. No published work indicates that other methods, including the generation of novel reverse genetics systems, were used at the WIV to propagate infectious SARSr-CoVs based on sequence data from bats. Gain-of-function research would be expected to utilize an established SARSr-CoV genomic backbone, or at a minimum a virus previously identified via sequencing. However, past experimental research using recombinant coronaviruses at the WIV has used a genetic backbone (WIV1) unrelated to SARS-CoV-2 (Hu et al., 2017), and SARS-CoV-2 carries no evidence of genetic markers one might expect from laboratory experiments (Andersen et al., 2020). There is no rational experimental reason why a new genetic system would be developed using an unknown and unpublished virus, with no evidence nor mention of a SARS-CoV-2-like virus in any prior publication or study from the WIV (Ge et al., 2012; Hu et al., 2017; Menachery et al., 2015), no evidence that the WIV sequenced a virus that is closer to SARS-CoV-2 than RaTG13, and no reason to hide research on a SARS-CoV-2-like virus prior to the COVID-19 pandemic. Under any laboratory escape scenario, SARS-CoV-2 would have to have been present in a laboratory prior to the pandemic, yet no evidence exists to support such a notion and no sequence has been identified that could have served as a precursor.
A specific laboratory escape scenario involves accidental infection in the course of serial passage of a SARSr-CoV in common laboratory animals such as mice. However, early SARS-CoV-2 isolates were unable to infect wild-type mice (Wan et al., 2020). Although murine models are useful for studying infection in vivo and testing vaccines, they often result in mild or atypical disease in hACE2 transgenic mice (Bao et al., 2020; Hassan et al., 2020; Israelow et al., 2020; Rathnasinghe et al., 2020; Sun et al., 2020b). These findings are inconsistent with a virus selected for increased pathogenicity and transmissibility through serial passage through susceptible rodents. Although SARS-CoV-2 has since been engineered (Dinnon et al., 2020) and mouse-adapted by serial passage (Gu et al., 2020; Leist et al., 2020; Sun et al., 2020a), specific mutations in the spike protein, including N501Y, are necessary for such adaptation in mice (Gu et al., 2020; Sun et al., 2020a). Notably, N501Y has arisen convergently in multiple SARS-CoV-2 variants of concern in the human population, presumably being selected to increase ACE2 binding affinity (Khan et al., 2021; Kuzmina et al., 2021; Liu et al., 2021; Starr et al., 2020). If SARS-CoV-2 resulted from attempts to adapt a SARSr-CoV for study in animal models, it would likely have acquired mutations like N501Y for efficient replication in that model, yet there is no evidence to suggest such mutations existed early in the pandemic. Both the low pathogenicity in commonly used laboratory animals and the absence of genomic markers associated with rodent adaptation indicate that SARS-CoV-2 is highly unlikely to have been acquired by laboratory workers in the course of viral pathogenesis or gain-of-function experiments.
Evidence from genomic structure and ongoing evolution of SARS-CoV-2
Considerable attention has been devoted to claims that SARS-CoV-2 was genetically engineered or adapted in cell culture or “humanized” animal models to promote human transmission (Zhan et al., 2020). Yet, since its emergence, SARS-CoV-2 has experienced repeated sweeps of mutations that have increased viral fitness (Deng et al., 2021; Otto et al., 2021; Simmonds, 2020). The first clear adaptive mutation, the D614G substitution in the spike protein, occurred early in the pandemic (Korber et al., 2020; Volz et al., 2021). Recurring mutations in the receptor binding domain of the spike protein, including N501Y, K417N/T, L452R, and E484K/Q—constituent mutations of the variants of concern—similarly enhance viral infectivity (Cai et al., 2021; Khan et al., 2021; Kuzmina et al., 2021) and ACE2 binding (Liu et al., 2021; Starr et al., 2020; Zhu et al., 2021a), refuting claims that the SARS-CoV-2 spike protein was optimized for binding to human ACE2 upon its emergence (Piplani et al., 2021). Further, some pangolin-derived coronaviruses have receptor binding domains that are near-identical to SARS-CoV-2 at the amino acid level (Andersen et al., 2020; Xiao et al., 2020) and bind to human ACE2 even more strongly than SARS-CoV-2, showing that there is capacity for further human adaptation (Dicken et al., 2021). SARS-CoV-2 is also notable for being a host generalist virus (Conceicao et al., 2020), capable of efficient transmission in multiple mammalian species (including mink, tigers, cats, gorillas, dogs, raccoon dogs, and ferrets), and large outbreaks have been documented in mink with spill-back to humans (Oude Munnink et al., 2021) and to other animals (van Aart et al., 2021). Combined, these findings show that no specific human “pre” adaptation was required for the emergence or early spread of SARS-CoV-2, and the claim that the virus was already highly adapted to the human host (Zhan et al., 2020), or somehow optimized for binding to human ACE2, is without validity.
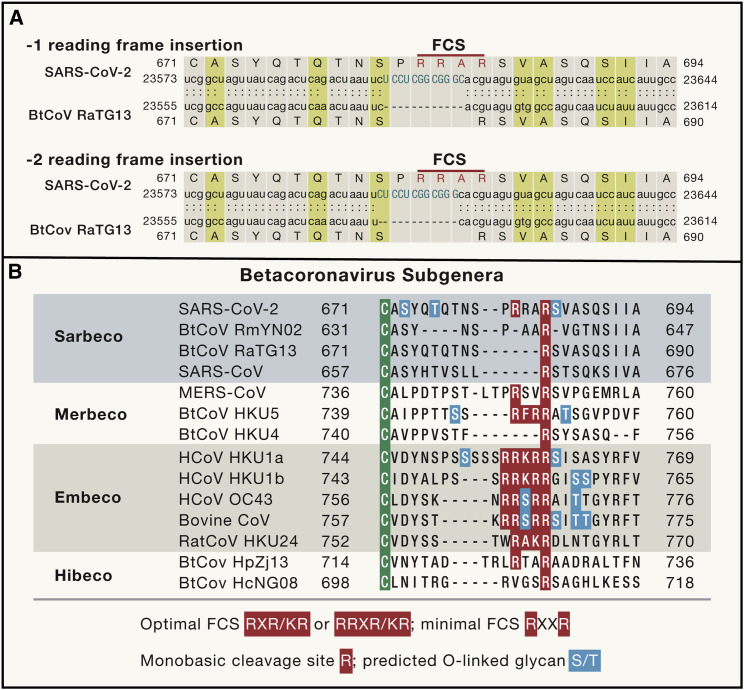
The genesis of the polybasic (furin) cleavage site in the spike protein of SARS-CoV-2 has been subject to recurrent speculation. Although the furin cleavage site is absent from the closest known relatives of SARS-CoV-2 (Andersen et al., 2020), this is unsurprising because the lineage leading to this virus is poorly sampled and the closest bat viruses have divergent spike proteins due to recombination (Boni et al., 2020; Lytras et al., 2021; Zhou et al., 2021). Furin cleavage sites are commonplace in other coronavirus spike proteins, including some feline alphacoronaviruses, MERS-CoV, most but not all strains of mouse hepatitis virus, as well as in endemic human betacoronaviruses such as HCoV-OC43 and HCoV-HKU1 (Gombold et al., 1993; de Haan et al., 2008; Kirchdoerfer et al., 2016). A near identical nucleotide sequence is found in the spike gene of the bat coronavirus HKU9-1 (Gallaher, 2020), and both SARS-CoV-2 and HKU9-1 contain short palindromic sequences immediately upstream of this sequence that are indicative of natural recombination break-points via template switching (Gallaher, 2020). Hence, simple evolutionary mechanisms can readily explain the evolution of an out-of-frame insertion of a furin cleavage site in SARS-CoV-2 (Figure 2 ).
Figure 2.
Evolution of the furin cleavage site in the spike protein of betacoronaviruses
(A) Sequence alignment of the region around the furin cleavage site (FCS) in SARS-CoV-2 (NCBI: MN908947) and bat coronavirus RaTG13 (NCBI: MN996532) showing that the former was the result of an out-of-frame nucleotide sequence insertion.
(B) Amino acid sequence alignment of the FCS region in representative members of the different subgenera of betacoronaviruses, highlighting the evolutionary volatility of this site and that the relevant amino acid motif (RRAR) in SARS-CoV-2 is functionally suboptimal. The residues predicted to be O-linked glycans are also marked.
See also Document S1.
The SARS-CoV-2 furin cleavage site (containing the amino acid motif RRAR) does not match its canonical form (R-X-R/K-R), is suboptimal compared to those of HCoV-HKU1 and HCoV-OC43, lacks either a P1 or P2 arginine (depending on the alignment), and was caused by an out-of-frame insertion (Figure 2). The RRAR and RRSR S1/S2 cleavage sites in feline coronaviruses (FCoV) and cell-culture adapted HCoV-OC43, respectively, are not cleaved by furin (de Haan et al., 2008). There is no logical reason why an engineered virus would utilize such a suboptimal furin cleavage site, which would entail such an unusual and needlessly complex feat of genetic engineering. The only previous studies of artificial insertion of a furin cleavage site at the S1/S2 boundary in the SARS-CoV spike protein utilized an optimal “RRSRR” sequence in pseudotype systems (Belouzard et al., 2009; Follis et al., 2006). Further, there is no evidence of prior research at the WIV involving the artificial insertion of complete furin cleavage sites into coronaviruses.
The recurring P681H/R substitution in the proline (P) residue preceding the SARS-CoV-2 furin cleavage site improves cleavage of the spike protein and is another signature of ongoing human adaptation of the virus (Peacock et al., 2021a). The SARS-CoV-2 furin site is also lost under standard cell culture conditions involving Vero E6 cells (Ogando et al., 2020; Peacock et al., 2021b), as is true of HCoV-OC43 (Follis et al., 2006). The presence of two adjacent CGG codons for arginine in the SARS-CoV-2 furin cleavage site is similarly not indicative of genetic engineering (Maxmen and Mallapaty, 2021). Although the CGG codon is rare in coronaviruses, it is observed in SARS-CoV, SARS-CoV-2, and other human coronaviruses at comparable frequencies (Maxmen and Mallapaty, 2021). Further, if low-fitness codons had been artificially inserted into the virus genome they would have been quickly selected against during SARS-CoV-2 evolution, yet both CGG codons are more than 99.8% conserved among the >2,300,000 near-complete SARS-CoV-2 genomes sequenced to date, indicative of strong functional constraints (supplemental information; Table S1).
Conclusions
As for the vast majority of human viruses, the most parsimonious explanation for the origin of SARS-CoV-2 is a zoonotic event. The documented epidemiological history of the virus is comparable to previous animal market-associated outbreaks of coronaviruses with a simple route for human exposure. The contact tracing of SARS-CoV-2 to markets in Wuhan exhibits striking similarities to the early spread of SARS-CoV to markets in Guangdong, where humans infected early in the epidemic lived near or worked in animal markets. Zoonotic spillover by definition selects for viruses able to infect humans. Although strong safeguards should be consistently employed to minimize the likelihood of laboratory accidents in virological research, those laboratory escapes documented to date have almost exclusively involved viruses brought into laboratories specifically because of their known human infectivity.
There is currently no evidence that SARS-CoV-2 has a laboratory origin. There is no evidence that any early cases had any connection to the WIV, in contrast to the clear epidemiological links to animal markets in Wuhan, nor evidence that the WIV possessed or worked on a progenitor of SARS-CoV-2 prior to the pandemic. The suspicion that SARS-CoV-2 might have a laboratory origin stems from the coincidence that it was first detected in a city that houses a major virological laboratory that studies coronaviruses. Wuhan is the largest city in central China with multiple animal markets and is a major hub for travel and commerce, well connected to other areas both within China and internationally. The link to Wuhan therefore more likely reflects the fact that pathogens often require heavily populated areas to become established (Pekar et al., 2021).
We contend that although the animal reservoir for SARS-CoV-2 has not been identified and the key species may not have been tested, in contrast to other scenarios there is substantial body of scientific evidence supporting a zoonotic origin. Although the possibility of a laboratory accident cannot be entirely dismissed, and may be near impossible to falsify, this conduit for emergence is highly unlikely relative to the numerous and repeated human-animal contacts that occur routinely in the wildlife trade. Failure to comprehensively investigate the zoonotic origin through collaborative and carefully coordinated studies would leave the world vulnerable to future pandemics arising from the same human activities that have repeatedly put us on a collision course with novel viruses.
Acknowledgments
We gratefully acknowledge the authors and the laboratories responsible for the genome sequence data shared via the GISAID Initiative, and we provide a complete acknowledgment table for the data used in Data S1. E.C.H. is supported by an ARC Australian Laureate Fellowship (FL170100022). S.A.G. is supported by the NIH (F32AI152341). J.O.W. acknowledges support from the NIH (AI135992). A.L.R. acknowledges that VIDO receives operational funding from the Canada Foundation for Innovation-Major Science Initiatives Fund and from the Government of Saskatchewan through Innovation Saskatchewan and the Ministry of Agriculture. D.L.R. acknowledges support of the Medical Research Council (MC_UU_12014/12) and the Wellcome Trust (220977/Z/20/Z). S.J.A. acknowledges funding from the NIH (R01AI149693). W.B. receives support from the Wellcome Trust (Z/205100 and Z/200187), BBSRC (BB/S008292), and MRC (MR/W005611/1). M.F.B. acknowledges funding from the Bill and Melinda Gates Foundation (INV-005517). J.L.G. is supported by a New Zealand Royal Society Rutherford Discovery Fellowship (RDF-20-UOO-007). J.L.L. is supported by the NIH (R01AI141607 and R21AI139738) and the NSF (2029949). S.J.N. is supported by a Wellcome Trust Senior Fellowship (WT098049AIA), the MRC, and the Huo Family Charitable Foundation 2. T.S. was funded by the Austrian Science Fund (FWF) (P 28183). S.R.W. is supported by the NIH (R01AI140442, R01AI104887, R21AI138564, and R21AI157147), as well as the Penn Center for Research on Coronaviruses and Other Emerging Pathogens. M.W. is supported by Bill and Melinda Gates Foundation (INV004212) and the Arizona Board of Regents. K.G.A. acknowledges support from the NIH (U19AI135995, U01AI151812, and UL1TR002550). R.F.G. is supported by the NIH (R01AI132223, R01AI132244, U19AI135995, U54HG007480, U19AI142790, and U01AI151812), the Coalition for Epidemic Preparedness Innovations (INTU1901 and ESEP1904), and the European and Developing Countries Clinical Trials Partnership (RIA2019LV-3053). A.R. acknowledges the support of the Wellcome Trust (Collaborators Award 206298/Z/17/Z-ARTIC network) and the European Research Council (725422-ReservoirDOCS).
Declaration of interests
E.C.H. is an honorary visiting professor at Fudan University (Shanghai Public Health Clinical Center), Shanghai, China and, from 2014–2020, was a guest professor at the Chinese Center for Disease Control and Prevention, Beijing, China. These affiliations are only used in papers co-authored with Prof. Yong-Zhen Zhang (Shanghai Public Health Clinical Center) and involve no formal appointment, no duties, and no remuneration nor research funding. J.O.W. receives funding from the U.S. Centers for Disease Control and Prevention (ongoing) via grants and contracts to his institution unrelated to this research. S.R.W. consults for Immunome and Ocugen. A.R., A.L.R., M.F.B., S.A.G., and K.G.A. have received consulting fees and compensation for expert testimony on SARS-CoV-2 and the COVID-19 pandemic. R.F.G. is co-founder of Zalgen Labs.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.08.017.
Supporting citations
The following references appear in the supplemental information: Chen et al. (2021); Li (2018); Minh et al. (2020); Ren et al. (2020); Wu et al. (2020).
Supplemental information
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.A., Dohm D.J., Negley D.L., Mores C.N. Virus inactivation by nucleic acid extraction reagents. J. Virol. Methods. 2004;119:195–198. doi: 10.1016/j.jviromet.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373:642–648. doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Prevalence of IgG antibody to SARS-associated coronavirus in animal traders--Guangdong Province, China, 2003. MMWR Morb. Mortal. Wkly. Rep. 2003;52:986–987. [PubMed] [Google Scholar]
- Chen L., Lu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan oubreak. Emerg Microbes Infect. 2021;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Wuhan coronavirus hunter Shi Zhengli speaks out. Science. 2020;369:487–488. doi: 10.1126/science.369.6503.487. [DOI] [PubMed] [Google Scholar]
- Conceicao C., Thakur N., Human S., Kelly J.T., Logan L., Bialy D., Bhat S., Stevenson-Leggett P., Zagrajek A.K., Hollinghurst P., et al. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020;18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Haijema B.J., Schellen P., Wichgers Schreur P., te Lintelo E., Vennema H., Rottier P.J.M. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008;82:6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Xing K., He X. Mutation signatures inform the natural host of SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.07.05.451089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicken S.J., Murray M.J., Thorne L.G., Reuschl A.-K., Forrest C., Ganeshalingham M., Muir L., Kalemera M.D., Palor M., McCoy L.E., et al. Characterisation of B.1.1.7 and Pangolin coronavirus spike provides insights on the evolutionary trajectory of SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.22.436468. [DOI] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling C.M., Breithaupt A., Müller T., Sehl J., Balkema-Buschmann A., Rissmann M., Klein A., Wylezich C., Höper D., Wernike K., et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg. Infect. Dis. 2020;26:2982–2985. doi: 10.3201/eid2612.203733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher W.R. A palindromic RNA sequence as a common breakpoint contributor to copy-choice recombination in SARS-COV-2. Arch. Virol. 2020;165:2341–2348. doi: 10.1007/s00705-020-04750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., Shi Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes A.M. The history of smallpox. Clin. Dermatol. 2006;24:152–157. doi: 10.1016/j.clindermatol.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Gombold J.L., Hingley S.T., Weiss S.R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J. Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.-P., Yang X.-L., Ge X.-Y., Zhang W., Li B., Xie J.-Z., Shen X.-R., Zhang Y.-Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. Published online March 23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528.e2. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lednicky J.A., Tagliamonte M.S., White S.K., Elbadry M.A., Alam M.M., Stephenson C.J., Bonny T.S., Loeb J.C., Telisma T., Chavannes S., et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv. 2021 doi: 10.1101/2021.03.19.21253391. [DOI] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-L., Wang J.-L., Ma X.-H., Sun X.-M., Li J.-S., Yang X.-F., Shi W.-F., Duan Z.-J. A novel SARS-CoV-2 related coronavirus with complex recombination isolated from bats in Yunnan province, China. Emerg. Micro. Infect. 2021;10:1683–1690. doi: 10.1080/22221751.2021.1964925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.L., Kurup A., Gopalakrishna G., Chan K.P., Wong C.W., Ng L.C., Se-Thoe S.Y., Oon L., Bai X., Stanton L.W., et al. Laboratory-acquired severe acute respiratory syndrome. N. Engl. J. Med. 2004;350:1740–1745. doi: 10.1056/NEJMoa032565. [DOI] [PubMed] [Google Scholar]
- Liu M., Deng L., Wang D., Jiang T. Influenza activity during the outbreak of coronavirus disease 2019 in Chinese mainland. Biosaf Health. 2020;2:206–209. doi: 10.1016/j.bsheal.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., et al. Identification of common deletions in the spike protein of Severe Acute Respiratory Syndrome coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00790-20. e00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv. 2021 doi: 10.1101/2021.03.08.434499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytras S., Hughes J., Martin D., de Klerk A., Lourens R., Kosakovsky Pond S.L., Xia W., Jiang X., Robertson D.L. Exploring the natural origins of SARS-CoV-2 in the light of recombination. bioRxiv. 2021 doi: 10.1101/2021.01.22.427830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxmen A., Mallapaty S. The COVID lab-leak hypothesis: what scientists do and don’t know. Nature. 2021;594:313–315. doi: 10.1038/d41586-021-01529-3. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.-Y., Donaldson E.F., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Van Domselaar G., Wu J., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry J. Breaches of safety regulations are probable cause of recent SARS outbreak, WHO says. BMJ. 2004;328:1222. doi: 10.1136/bmj.328.7450.1222-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Sheppard C.M., Brown J.C., Goonawardane N., Zhou J., Whiteley M., de Silva T.I., Barclay W.S., PHE Virology Consortium The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. bioRxiv. 2021 doi: 10.1101/2021.05.28.446163. [DOI] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Pekar J., Worobey M., Moshiri N., Scheffler K., Wertheim J.O. Timing the SARS-CoV-2 index case in Hubei province. Science. 2021;372:412–417. doi: 10.1126/science.abf8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piplani S., Singh P.K., Winkler D.A., Petrovsky N. In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Sci. Rep. 2021;11:13063. doi: 10.1038/s41598-021-92388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., Strohmeier S., Amanat F., Gillespie V.L., Krammer F., García-Sastre A., Coughlan L., Schotsaert M., Uccellini M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9:2433–2445. doi: 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.-Z., Xiong Y., Li Y.-J., Li X.-W., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl). 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristanović E.S., Kokoškov N.S., Crozier I., Kuhn J.H., Gligić A.S. A forgotten episode of Marburg virus disease: Belgrade, Yugoslavia, 1967. Microbiol. Mol. Biol. Rev. 2020;84 doi: 10.1128/MMBR.00095-19. e00095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo M., Gronvall G.K. The reemergent 1977 H1N1 strain and the gain-of-function debate. MBio. 2015;6:e01013–e01015. doi: 10.1128/mBio.01013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., Hall W.W., Orba Y., Sawa H. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17:e1009233. doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior K. Recent Singapore SARS case a laboratory accident. Lancet Infect. Dis. 2003;3:679. doi: 10.1016/S1473-3099(03)00815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. MSphere. 2020;5:e00408–e00420. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., Yang G., Li R.-T., Fan H., Ye Q., Deng Y.-Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.10.377333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., et al. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aart A.E., Velkers F.C., Fischer E.A.J., Broens E.M., Egberink H., Zhao S., Engelsma M., Hakze-van der Honing R.W., Harders F., de Rooij M.M.T., et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. 2021 doi: 10.1111/tbed.14173. Published online June 3, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Diaz A., Damtie D., Xiu L., Toh T.-H., Lee J.S.-Y., Saif L.J., Gray G.C. Novel canine coronavirus Isolated from a hospitalized pneumonia patient, east Malaysia. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab456. Published online May 20, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. COG-UK Consortium Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Tan C.W., Maneeorn P., Duengkae P., Zhu F., Joyjinda Y., Kaewpom T., Chia W.N., Ampoot W., Lim B.L., et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Jing H.-Q., Xu H.-F., Jiang X.-G., Kan B., Liu Q.-Y., Wan K.-L., Cui B.-Y., Zheng H., Cui Z.-G., et al. [Surveillance on severe acute respiratory syndrome associated coronavirus in animals at a live animal market of Guangzhou in 2004] Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:84–87. [PubMed] [Google Scholar]
- Wang N., Li S.-Y., Yang X.-L., Huang H.-M., Zhang Y.-J., Guo H., Luo C.-M., Miller M., Zhu G., Chmura A.A., et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.C., Lau S.Y., Wang To K.K., Mok B.W.Y., Li X., Wang P., Deng S., Woo K.F., Du Z., Li C., et al. Natural transmission of bat-like Severe Acute Respiratory Syndrome Coronavirus 2 without proline-arginine-arginine-alanine variants in Coronavirus Disease 2019 patients. Clin. Infect. Dis. 2021;73:e437–e444. doi: 10.1093/cid/ciaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2021. WHO-Convened Global Study of Origins of SARS-CoV-2: China Part. [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.-M., Chan K.-H., Tsoi H.-W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.-K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Xiao X., Newman C., Buesching C.D., Macdonald D.W., Zhou Z.-M. Animal sales from Wuhan wet markets immediately prior to the COVID-19 pandemic. Sci. Rep. 2021;11:11898. doi: 10.1038/s41598-021-91470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.-H., He J.-F., Evans M.R., Peng G.-W., Field H.E., Yu D.-W., Lee C.-K., Luo H.-M., Lin W.-S., Lin P., et al. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-L., Hu B., Wang B., Wang M.-N., Zhang Q., Zhang W., Wu L.-J., Ge X.-Y., Zhang Y.-Z., Daszak P., et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of Severe Acute Respiratory Syndrome coronavirus. J. Virol. 2015;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqing H.E. Surveillance of SARS coronavirus among wild animal sold in Dongmen market in Shenzhen city. Disease Surveillance. 2004;19:287–291. [Google Scholar]
- Zhan S.H., Deverman B.E., Chan Y.A. SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence? bioRxiv. 2020 2020.05.01.073262. [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Liu D., Yang J., Holmes E.C., Hughes A.C., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:1–8. doi: 10.1016/j.cub.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., Hu T., Song H., Zhao R., Chen Y., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184:4380–4391.e14. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Mannar D., Srivastava S.S., Berezuk A.M., Demers J.-P., Saville J.W., Leopold K., Li W., Dimitrov D.S., Tuttle K.S., et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19:e3001237. doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.