Abstract
Schizophrenia is a complex brain disorder with genetic and environmental factors contributing to its etiology. Complement C4 genes are schizophrenia susceptibility loci and are activated in response to infections and gut microbiome imbalances. We hypothesize that C4 genetic susceptibility predisposes individuals to neuropathological effects from pathogen exposures or a microbiome in dysbiosis. In 214 individuals with schizophrenia and 123 non-psychiatric controls, we examined C4 gene copy number and haplotype groups for associations with schizophrenia and microbial plasma biomarkers. C4A copy number and haplotypes containing HERV-K insertions (C4A-long; C4AL-C4AL) conferred elevated odds ratios for schizophrenia diagnoses (OR 1.58–2.56, p<0.0001), while C4B-short (C4BS) haplogroups conferred decreased odds (OR 0.43, p<0.0001). Haplogroup-microbe combinations showed extensive associations with schizophrenia including C4AL with Candida albicans IgG (OR 2.16, p<0.0005), C4AL-C4BL with cytomegalovirus (CMV) IgG (OR 1.79, p<0.008), C4BS with lipopolysaccharide-binding protein (LBP) (OR 1.18, p<0.0001), and C4AL-C4AL with Toxoplasma gondii IgG (OR=17.67, p<0.0001). In controls, only one haplogroup-microbe combination was significant: C4BS with CMV IgG (OR 0.52, p<0.02). In schizophrenia only, LBP and CMV IgG levels were inversely correlated with C4A and C4S copy numbers, respectively (R2=0.13–0.16, p<0.0001). C4 haplogroups were associated with altered scores of cognitive functioning in both cases and controls and with psychiatric symptom scores in schizophrenia. Our findings link complement C4 genes with a susceptibility to infections and a dysbiotic microbiome in schizophrenia. These results support immune system mechanisms by which gene-environmental interactions may be operative in schizophrenia.
Keywords: complement system, microbiome, gut-brain axis, pathogens, gene-environment interactions, biomarkers
1. Introduction
Schizophrenia is a complex brain disorder with compelling evidence pointing to immune system dysfunctions as a place where genetic and environmental factors might converge and become neuropathological. Genetic studies show consistent associations of schizophrenia with polymorphisms in 6p21–6p22, a chromosomal region that contains the major histocompatibility complex (MHC) and that encodes hundreds of proteins integral to the immune system (Harrison, 2015). Environmental factors that interact with the immune system at elevated rates in schizophrenia include those involving specific pathogens such as bacteria, viruses, and parasites, as well as increased risks conferred by the infectious disease process (Benros et al., 2011; Kirch, 1993; Kohler et al., 2017; Miller et al., 2013; Severance and Yolken, 2020a; Torrey et al., 2012; Torrey and Peterson, 1976; Yolken and Torrey, 2008). In recent years, microbial associations with schizophrenia have taken a new direction, as research across many medical fields including psychiatry focuses on the gut microbiome as a potential source of novel disease mechanisms and treatments.
Residing in the MHC genomic region is complement C4, a gene that has long shown genetic and other biological associations with schizophrenia (Laskaris et al., 2019; Mayilyan et al., 2008b; Mondelli et al., 2020; Pouget, 2018; Prasad et al., 2018; Rey et al., 2020; Rudduck et al., 1985; Sekar et al., 2016; Woo et al., 2019). Complement C4 is an intriguing susceptibility gene candidate in schizophrenia because of its putative functional role in central nervous system synaptic pruning (Johnson and Stevens, 2018; Presumey et al., 2017; Sellgren et al., 2019). Prefrontal cortical and hippocampal dendritic spine deficiencies in schizophrenia may be a result of abnormally active synaptic pruning during adolescence (Bennett et al., 2013; Clarke et al., 2018). A C4 gene variant that is overexpressed or underexpressed during this sensitive time period would support a dysregulated pruning model mechanism for schizophrenia. Sekar et al (2016) demonstrated that increasing C4A copy number was associated with increased C4A expression in post-mortem brains of individuals with schizophrenia compared to controls. In that study, C4 protein localized to neuronal synapses, processes and cell bodies (Sekar et al., 2016).
Technological advances in basic research and bioinformatic methods have made accessible the many encoded microbial genes and inferred metabolic pathways of the gut microbiome and as such represent unique opportunities to discover novel disease mechanisms and treatments in fields not traditionally considered to have an infectious basis. In psychiatry, for example, there are accumulating reports of alterations in diversities and abundances of presumably commensal gut microbial taxa which distinguish the schizophrenia microbiome from that of controls (Castro-Nallar et al., 2015; Dickerson et al., 2017; Li et al., 2020; Nguyen et al., 2019; Shen et al., 2018; Xu et al., 2019; Yolken et al., 2015; Zheng et al., 2019). These findings have contributed a resurgence of interest in understanding the role of the gut brain-axis in psychiatric disorders and overall idea that properly functioning peripheral systems have important ramifications for brain health. Experiments in germ-free mice continue to shed light on the neurobiological mechanisms by which gut microbes enter the brain and control behaviors (Collins et al., 2012; Diaz Heijtz et al., 2011; Erny et al., 2015; Foster and McVey Neufeld, 2013; Hsiao et al., 2013; Luczynski et al., 2016; Sampson and Mazmanian, 2015; Stilling et al., 2014). As relevant for studies of gene-environmental interactions involving the immune system, the gut microbiome is also critical for immune system development and maturation (Chistiakov et al., 2014; Dinan and Cryan, 2015; Sandhya et al., 2016; Severance et al., 2018; Wekerle, 2017).
Evaluating C4 genotypes and functional changes in the microbiome have previously been experimentally cumbersome to manage. Complement C4A and C4B are extensively polymorphic loci with significant variation in copy number and structure, making it difficult to accurately genotype individuals. C4A and C4B genes are paralogous and each can contain a human endogenous retroviral sequence (HERV-K) that confers a long (L) or short (S) form of the gene (Figure 1). C4A and C4B can also vary in the number of copies with a typical diploid genome containing two to eight copies. The availability of algorithms to impute C4 genotypes from GWAS data has accelerated the progress of C4 gene analyses in schizophrenia (Sekar et al., 2016). Dysfunction in the microbiome can be assessed by direct nucleic acid sequencing of bacterial taxa in relevant mucosal biospecimens, or via indirect measures of host physiological changes that occur during gut dysbioses. For the latter, plasma biomarkers of gastrointestinal inflammation and microbial translocation are considered valuable surrogate measures of an unhealthy gut microbiome (Severance and Yolken, 2020b). Markers that specifically target the bacterial endotoxin, lipopolysaccharide (LPS), are important validators that systemic inflammation has a microbial source such as a disturbed microbiome. In this paper, we test the hypothesis that complement C4 associations with microbes contribute to gene-environmental interactions in schizophrenia by examining C4A and C4B gene associations with plasma biomarkers of pathogen exposures and a dysregulated gut microbiome. Plasma biomarkers examined here included C-Reactive Protein (CRP), LPS-binding Protein (LBP), soluble CD14 (sCD14), Candida albicans IgG, Saccharomyces cerevisiae IgG, Cytomegalovirus IgG, and Toxoplasma gondii IgG.
Figure 1. Structural complexity of the complement C4 gene.
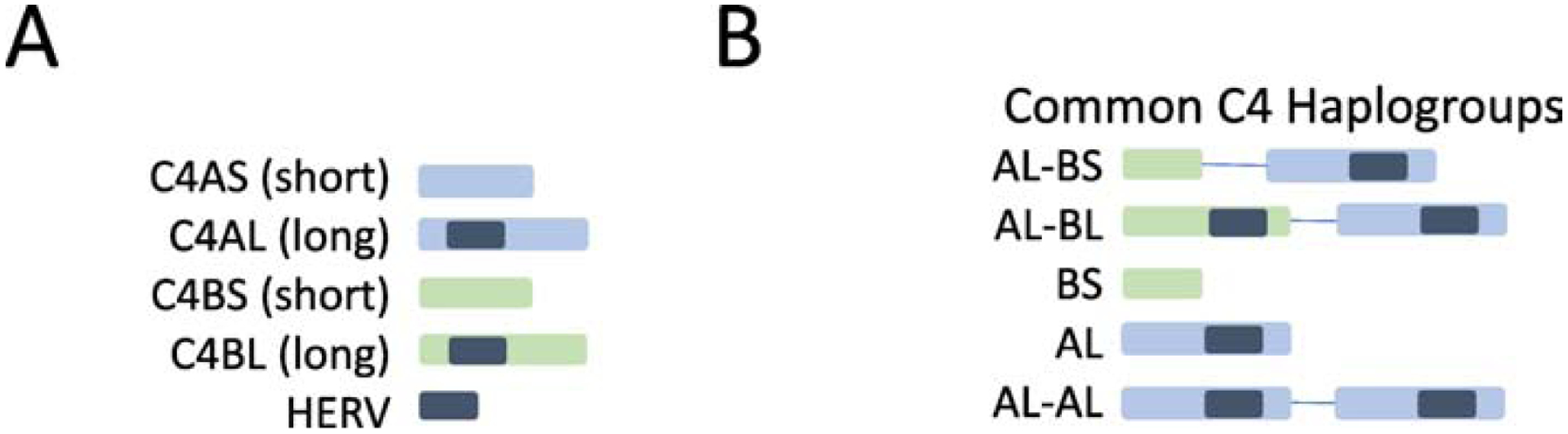
Panel A: C4 genes exist in four forms (1) C4A gene short (C4AS - no HERV insertion); (2) C4A gene long (C4AL – contains HERV insertion); (3) C4B gene short (C4BS no HERV insertion); (4) C4B gene long (C4BL – contains HERV insertion). Panel B: Five common C4 haplogroups that reflect copy number variation and presence/absence of the HERV insertion. This diagram is based on material from Sekar et al (2016)(Sekar et al., 2016).
2. Material and methods
2.1. Study population
A total of 337 individuals were recruited from Sheppard Pratt located in Baltimore, MD, U.S.A.: 123 were control individuals with no history of psychiatric disorder; 214 individuals were diagnosed with schizophrenia. Diagnoses were made in accordance with DSM-IV-TR (APA, 2000) and have been previously described (Dickerson et al., 2015; Dickerson et al., 2013). For the schizophrenia group, individuals received a DSM-IV-TR diagnosis of schizophrenia, schizophreniform disorder, or schizoaffective disorder and were between the ages of 18 and 65. Individuals without a history of psychiatric disorder were interviewed to rule out current or past psychiatric disorders with the Structured Clinical Interview for DSM-IV Axis I Disorders Non-Patient Edition (First, 1998). Controls were between the ages of 20 and 60, inclusive. Exclusion criteria for both groups included: mental retardation; clinically significant medical disorder that would affect cognitive performance; any history of intravenous substance abuse or a primary diagnosis of substance abuse or substance dependence. For controls, any active substance misuse was considered an exclusion criterion. Cognitive function was evaluated with the Repeatable Battery for the Assessment of Neuropsychological Status (RBANS) Form A (Randolph, 1998) and psychiatric symptoms rated according to the Positive and Negative Syndrome Scale (PANSS) (Kay et al., 1987). Mean total RBANS scores ± standard deviation (SD) were 85.10+11.71 for controls and 64.30±11.64 for schizophrenia. In individuals with schizophrenia, mean total PANSS scores ± SD were 77.74±13.82; mean PANSS positive symptom scores were 19.70±5.56; and mean PANSS negative symptom scores were 21.07±4.57. Basic demographic and other data (age, sex, race, body mass index (BMI)) for this study population are shown in Table 1. Age, sex, race and BMI were significantly different between diagnostic groups.
Table 1.
Demographics and other variables of the study populations.
| Population | n Individuals |
Age Mean years+SD1 |
Sex n (% Female) |
Race n (% Caucasian) |
BMI Mean score+SD |
|---|---|---|---|---|---|
| Controls | 123 | 32.34+11.202 | 79 (64.22)3 | 80 (65.04)4 | 26.76+6.335 |
| Schizophrenia | 214 | 37.40+12.43 | 73 (34.11) | 105 (49.06) | 30.67+8.07 |
SD refers to standard deviation;
T =−4.66, two-tailed p<0.001;
Chi-square = 28.61, p<0.001;
Chi-square = 8.05, p<0.005;
T =−4.65, two-tailed p<0.0001
These studies were approved by the Institutional Review Boards (IRB) of Sheppard Pratt and the Johns Hopkins Medical Institution following established guidelines. All participants provided written informed consent after study procedures were explained. This research was performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans.
2.2. Laboratory tests
Blood was drawn at the time of interview using Becton-Dickinson’s Cell Preparation Tubes containing sodium citrate. Plasma was separated and stored at −80°C. Peripheral blood mononuclear cells (PBMCs) were isolated and stored at −80oC. DNA was extracted from PBMCs using Qiagen’s DNAeasy kit and stored at −80oC.
2.3. Biomarkers of pathogen exposures and gut dysbioses
The following biomarkers were measured in plasma using commercially-available enzyme-linked immunosorbent assays (ELISAs): CRP, LBP, sCD14, Candida albicans IgG, Saccharomyces cerevisiae IgG, Cytomegalovirus IgG, and Toxoplasma gondii IgG. Methods and analyses reporting psychiatric case and control levels of these biomarkers were previously described (Dickerson et al., 2007a; Dickerson et al., 2007b; Leweke et al., 2004; Severance et al., 2012; Severance et al., 2016a; Severance et al., 2013).
2.4. Data analyses
Complement C4A, C4B, C4S and C4L haplotypes and copy numbers were imputed from GWAS SNPs data using previously described methods (Sekar et al 2016; https://github.com/freeseek/imputec4). T-tests were used to detect bivariate associations between continuous variables. Chi-square analyses were used to detect bivariate associations between categorical variables. Multiple linear regression models were used to examine correlations among continuous variables. Multivariate logistic regression models were used to assign odds ratios for copy numbers and haplotype group associations with diagnoses, plasma biomarker levels, RBANS scores, and PANSS scores. We focused on the most common haplogroups found in our population and as characterized by Sekar et al (2016). Haplogroup associations in multivariate models were further analyzed in relation to homozygous and heterozygous status compared to individuals in whom the haplogroup was absent. Multivariate regressions included the covariates: age, gender, race, BMI; an assay plate covariate was included with the ELISA data analyses to correct for plate-to-plate variation. These regression models applied robust standard error corrections to accommodate multiple samples per individual. We did not formally account for multiple comparisons in order to prevent the possibility that a too stringent threshold would lead to type B errors. Therefore, a p-value of less than 0.05 was considered significant. However, it is of note that for most of the analyses, our p-value fell well below 0.01.
3. Results
We found that the haplogroup containing two copies of C4A-long (C4AL-C4AL) was associated with an elevated odds of schizophrenia compared to controls (Table 2; OR=2.56, 95th%CI 1.05–6.27, p<0.0001). The haplogroup containing one copy of C4B-short (C4BS) was associated with a significantly decreased odds of a schizophrenia diagnosis (OR=0.43, 95th%CI 0.20–0.94, p<0.0001). None of the other haplogroups shown in Table 2 were associated with a significantly altered odds for schizophrenia. In comparisons of mean C4 copy number differences between cases and controls, we found significantly greater copy numbers of C4A in schizophrenia and greater numbers of C4B in controls (controls vs schizophrenia, mean levels ± standard error: C4A 1.94±0.04 vs 2.08±0.04, t=−2.53, p<0.01; C4B 1.91±0.03 vs 1.76±0.03, t=3.19, p<0.002). In multivariate logistic models, C4A copy numbers conferred increased odds of a schizophrenia diagnosis compared to controls (Table 2; OR=1.58, 95%CI 1.00–2.53, p<0.0001). There were no detectable differences in mean number of copies for C4B, C4S or C4L between diagnostic groups. C4A copy numbers were significantly inversely correlated with C4B copy numbers in controls and schizophrenia (controls: R2=0.16, coefficient=−0.44, 95th% CI=−0.62−−0.27, p<0.0001; schizophrenia: R2=0.46, coefficient=−0.85, 95th% CI=−0.97−−0.73, p<0.0001). Similarly, C4S copy numbers were significantly inversely correlated with C4L copy numbers in controls and schizophrenia (controls: R2=0.69, coefficient=−0.78, 95th% CI=−0.87−−0.69, p<0.0001; schizophrenia: R2=0.77, coefficient=−0.73, 95th% CI=−0.79−−0.67, p<0.0001).
Table 2.
C4 haplogroup & copy number odds ratios for a schizophrenia diagnosis as compared to controls
| OR | 95th%CI | p-value | |
|---|---|---|---|
| Haplogroup | |||
| AL-BS | 1.07 | 0.76–1.51 | NS |
| AL-BL | 0.94 | 0.66–1.34 | NS |
| BS | 0.43 | 0.20–0.94 | 0.0001 |
| AL | 0.73 | 0.28–1.90 | NS |
| AL-AL | 2.56 | 1.05–6.27 | 0.0001 |
| Copy Number | |||
| C4A | 1.58 | 1.00–2.53 | 0.0001 |
| C4B | 0.55 | 0.29–1.06 | NS |
| C4S | 0.85 | 0.61–1.18 | NS |
| C4L | 1.20 | 0.89–1.60 | NS |
Controls n=123, Schizophrenia n=214.
OR – odds ratio; CI – confidence interval; NS - not statistically significant at p<0.05.
Plasma biomarkers of pathogen exposure and gut dysbiosis were extensively associated with C4 haplogroups in schizophrenia but minimally in controls. In multiple linear regression models, cytomegalovirus IgG was inversely correlated with C4S copy numbers in schizophrenia but not controls (R2=0.16, regression coefficient=−0.30, 95th%CI=−0.56− −0.05, p<0.0001). Also, only in schizophrenia, LBP was inversely correlated with C4A copy numbers (R2=0.13, regression coefficient=−2.41, 95th%CI=−4.03− −0.79, p<0.0001). As shown in Table 3, C. albicans IgG, cytomegalovirus IgG, LBP and T. gondii IgG all showed significantly elevated odds ratios for associations with specific C4 haplogroups and significantly reduced odds ratios for other C4 haplogroups in schizophrenia. These associations varied according to homozygous and heterozygous states. For haplogroup-biomarker combinations associated with significantly elevated odds ratios, we further depicted these relationships for exploratory purposes by charting several representative biomarker levels according to homozygous and heterozygous status of the haplogroup, as shown in Figure 2. In controls, only one haplogroup-microbe combination was significant: C4BS with CMV IgG (OR 0.52, 95th%CI=0.29–0.95,−p<0.02). Overall, no significant associations with C4 were observed for S. cerevisiae IgG, CRP or sCD14.
Table 3.
Odds ratios for C4 haplogroup associations with biomarkers of pathogen exposures and gut dysbioses
| C. albicans IgG | CMVIgG | LBP | T. gondii IgG | |||||
|---|---|---|---|---|---|---|---|---|
| HTZ | HMZ | HTZ | HMZ | HTZ | HMZ | HTZ | HMZ | |
| Control | ||||||||
| AL-BS | NS | NS | NS | NS | NS | NS | NS | NS |
| AL-BL | NS | NS | NS | NS | NS | NS | NS | NS |
| BS | NS | NA | 0.52 (0.29–0.95) 0.02 |
NA | NS | NA | NS | NA |
| AL | NS | NA | NS | NA | NS | NA | NS | NA |
| AL-AL | NS | NA | NS | NA | NS | NA | NS | NA |
| Schizophrenia | ||||||||
| AL-BS | NS | NS | 0.62 (0.44–0.87) 0.001 |
0.62 (0.43–0.90) 0.001 |
NS | NS | NS | NS |
| AL-BL | NS | NS | NS | 1.79 (1.21–2.64) 0.008 |
0.97 (0.94–1.00) 0.002 |
NS | NS | NS |
| BS | 0.20 (0.05–0.82) 0.0001 |
NS | 1.55 (1.01–2.38) 0.0001 |
0.0003 (1E-6–0.10) 0.0001 |
NS | 1.18 (1.05–1.33) 0.0001 |
NS | 0.43 (0.26–0.70) 0.001 |
| AL | 2.16 (1.22–3.83) 0.0005 |
NS | 1.56 (1.09–2.24) 0.0001 |
0.20 (0.06–0.62) 0.0001 |
NS | 1.09 (1.02–1.16) 0.0001 |
NS | 0.33 (0.18–0.60) 0.0001 |
| AL-AL | NS | NS | NS | NS | 0.96 (0.92–0.99) 0.0001 |
NS | NS | 17.67 (0.98–319.19) 0.0001 |
OR – odds ratio; CI – confidence interval; p – p-value; HTZ – heterozygote; HMZ – homozygote; NS - not statistically significant at p<0.05; NA - not applicable (due to absence of homozygotes in the control group); CMV - cytomegalovirus
Comparison is HTZ and HMZ relative to absence of haplotype.
Figure 2. C4 haplogroup associations with microbial biomarkers in schizophrenia.
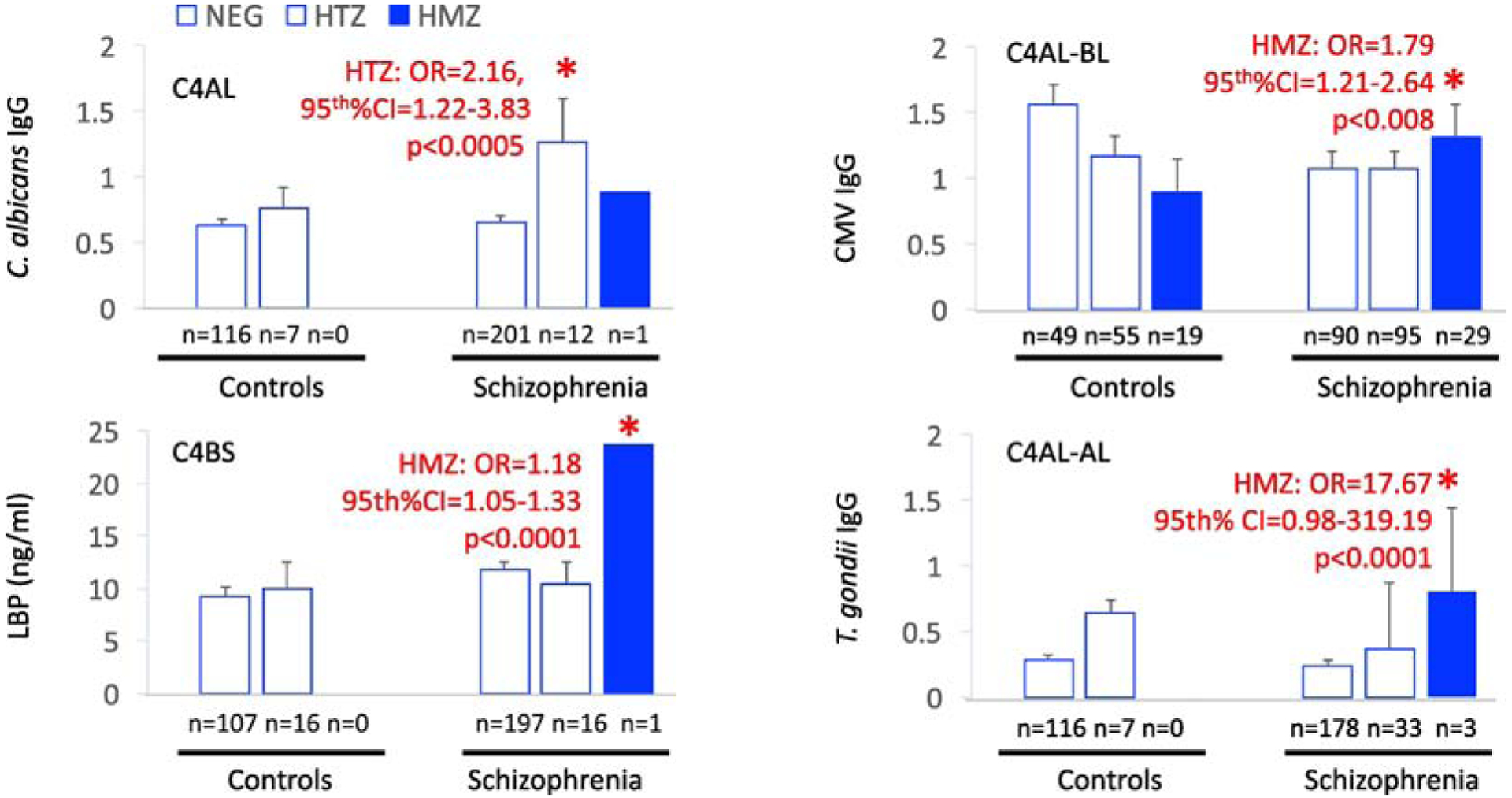
NEG refers to individuals not having the listed haplogroup; HTZ refers to heterozygous for the listed haplogroup; HMZ refers to homozygous. Plotted are mean levels of plasma biomarkers. Error bars indicate standard error of the mean. Asterisk designates significance at p<0.05 of multivariate models that included age, race, sex, BMI and assay plate as covariates.
In analyses of cognitive functioning, decreased RBANS scores were significantly associated with schizophrenia in individuals who were homozygous for the haplogroups C4AL and C4BS compared to individuals with schizophrenia who did not have these haplogroups (Table 4; C4AL: OR 0.68, 95th%CI 0.55–0.83, p<0.0001; C4BS: OR 0.82, 95th%CI 0.76–0.90, p<0.0001. Controls who were homozygous for the haplogroup C4AL-C4BS showed decreased RBANS scores compared to those who did not have this haplotype (OR 0.94, 95th%CI 0.89–1.00, p<0.03). Control individuals who were homozygous for the haplotype C4AL-C4BL showed elevated scores on RBANS compared to those who did not have this haplotype (OR 1.07, 95th%CI 1.00–1.14, p<0.02).
Table 4.
Odds ratios for C4 haplogroup associations with PANSS and RBANS scores
| PANSS positive | PANSS negative | RBANS | ||||
|---|---|---|---|---|---|---|
| HTZ | HMZ | HTZ | HMZ | HTZ | HMZ | |
| Control | ||||||
| AL-BS | NA | NA | NA | NA | NS | 0.94 (0.89–1.00) 0.03 |
| AL-BL | NA | NA | NA | NA | NS | 1.07 (1.00–1.14) 0.02 |
| BS | NA | NA | NA | NA | NS | NA |
| AL | NA | NA | NA | NA | NS | NA |
| AL-AL | NA | NA | NA | NA | NS | NA |
| Schizophrenia | ||||||
| AL-BS | NS | NS | NS | NS | NS | NS |
| AL-BL | NS | NS | NS | NS | NS | NS |
| BS | NS | 0.54 (0.33–0.88) 0.0001 |
NS | NS | NS | 0.82 0.76–0.90 0.0001 |
| AL | NS | NS | 1.24 (1.09–1.40) 0.0001 |
8.62 (1.48–50.34) 0.0001 |
NS | 0.68 (0.55–0.83) 0.0001 |
| AL-AL | 0.92 (0.85–1.00) 0.0001 |
NS | NS | NS | NS | NS |
OR – odds ratio; CI – confidence interval; p – p-value; HTZ – heterozygote; HMZ – homozygote; NS - not statistically significant at p<0.05; NA - not applicable (due to absence of homozygotes in the control group); PANSS positive refers to positive psychiatric symptoms; PANSS negative refers to negative psychiatric symptoms.
Comparison is HTZ and HMZ relative to absence of haplotype.
In multivariate models, homozygosity of the C4BS haplogroup and heterozygosity of the C4AL-AL haplogroup were associated with less severe positive symptoms (C4BS: OR 0.54, 95th%CI 0.33–0.88, p<0.0001; C4AL-AL: OR 0.92, 95th%CI 0.85–1.00, p<0.0001). Homozygotes and heterozygotes of the C4AL haplogroup showed increased severity of negative psychiatric symptoms compared to those who did not have this haplogroup (Heterozygotes: OR 1.24, 95th%CI 1.09–1.40, p<0.0001; Homozygote: OR 8.62, 95th%CI 1.48–50.33 p<0.0001).
4. Discussion
Complement C4 is an interesting susceptibility gene candidate for studies of gene-environmental interactions in schizophrenia because of its multifactorial neurobiological ties to the immune system. Not only is it located in the MHC, an immune gene region long associated with schizophrenia, but it is activated following environmental immune challenges such as infections that are known risk factors for schizophrenia. It also likely has a functional role in central nervous system synaptic pruning, a neurobiological mechanism relevant to schizophrenia (Nimgaonkar et al., 2017; Presumey et al., 2017; Sekar et al., 2016; Stevens et al., 2007). Here we demonstrated that in people with schizophrenia, complement C4A and C4B polymorphisms conferred a diversity of phenotypes related to microbial environmental variables such as pathogen exposures and gut dysbioses. These findings are especially relevant as examples of how gene and environmental variables may be interactive in schizophrenia via the immune system. We also observed that C4A and C4B copy numbers were inversely correlated with each other, likely reflecting the nonrandom linkage of C4A and C4B loci. Our study further detected significant associations of C4A and C4B haplotypes not only with a diagnosis of schizophrenia and environmental factors, but with psychiatric symptoms and cognitive functioning. These phenotypes in schizophrenia appeared most extreme when haplogroups were in the homozygous state.
Pathogen exposures have been studied as risk factors for the development of schizophrenia for a long time (Severance and Yolken, 2020a). More recently, interest in the microbiome and gut-brain axis has expanded the scope of the focus on infectious agents to the trillions of commensal microbes that inhabit the gastrointestinal tract and other mucosal surfaces. Alterations in the microbiome have been associated with gut dysbiosis, a condition that is increased in individuals with schizophrenia and other serious psychiatric disorders. This gut dysbiosis generally reflects an inflammatory state in the GI tract which leads to microbial translocation and a toxic cycle of systemic inflammation and leaky endothelial barriers at the blood-gut and blood-brain barriers (Dickerson et al., 2017; Severance et al., 2012; Severance et al., 2014; Severance et al., 2015; Severance et al., 2016c). Exposures to pathogens and a gut in dysbiosis represent immune-related environmental variables that are potentially impacted by a dysregulated complement system. In our study, only one of the measured biomarkers was associated with C4 haplogroups in controls, whereas in schizophrenia, there were widespread associations of these environmental immune factors with C4 haplogroups. For example, case-control differences in peripheral biomarkers in conjunction with C4 haplotypes were detected for IgG class antibodies to C. albicans, cytomegalovirus, and T. gondii.
The IgG-based markers can be generally categorized as biomarkers of exposure to pathogens; however, there are additional implications of these markers for GI-specific pathologies. C. albicans IgG, for example, has been shown in our previous studies to be associated with GI conditions in schizophrenia (Severance et al., 2016a; Severance et al., 2017; Severance et al., 2015; Severance et al., 2016b; Severance et al., 2016c). CMV is a virus that can infect the GI tract and has been implicated as a source of inflammation in inflammatory bowel diseases such as ulcerative colitis (Jentzer et al., 2020; Sager et al., 2015). Furthermore, studies of animal models implicate T. gondii in gut-related inflammation and dysbiosis, as this parasite primarily infects its host by the GI tract (Severance et al., 2016a; Severance et al., 2016b; Severance et al., 2016c). Here, the C4AL-AL haplogroup showed the strongest association with a schizophrenia diagnosis and with T. gondii IgG antibodies. T. gondii is historically one of the best-replicated pathogens that is associated with an increased risk of schizophrenia (Torrey et al., 2012; Yolken et al., 2009). The non-IgG based measure, LBP, on the other hand, specifically reflects the response to microbial translocation and the circulation of bacterial LPS, as would be apparent if a gut microbiome were in dysbiosis (Severance et al., 2013). In individuals with schizophrenia but not controls, LBP was inversely correlated with C4A copy numbers, a finding of interest since C4A copy numbers showed the strongest association with a schizophrenia diagnosis. While these markers are useful surrogate indices of microbial exposures, it will be important to compare marker patterns with direct measures of the microbiome in future studies.
Most of the common C4 haplogroups studied were associated with altered severity of psychiatric symptoms and measures of cognitive functioning, especially C4 haplogroups in a homozygous state. For example, the C4BS haplogroup was associated with decreased severity of positive psychiatric symptoms (OR 0.54, p<0.0001) and the C4AL haplogroup was associated with increased severity of negative psychiatric symptoms (OR 8.62, p<0.0001). C4 haplogroups were associated with scores of cognitive functioning in people with schizophrenia and in controls suggesting an interaction between C4 and cognition irrespective of its association with schizophrenia. Interestingly, in controls the C4AL-BS haplogroup was associated with worse performance on the RBANS while C4AL-BL conferred a beneficial effect (i.e high scores). The effects of C4 variants on cognition in schizophrenia and controls have previously been examined in various forms. For example, C4A RNA expression predicted from C4A structural variation was associated with poorer memory recall in individuals with schizophrenia compared to healthy controls. Healthy controls with higher predicted C4A expression, however, had reduced cortical activity during a visual processing task (Donohoe et al., 2018). Likewise, a strong association of complement pathway genes on cognitive function was observed independently of C4A structural variation and independently of an association with schizophrenia (Holland et al., 2019).
The association between C4A and an increased risk of schizophrenia has been previously demonstrated in several studies. However, the driving force behind the inverse relationship between C4B and schizophrenia risk may be more tenuous (Mayilyan et al., 2008a; Rudduck et al., 1985; Sekar et al., 2016; Woo et al., 2019). It is not clear if the decreased C4B copy number confers a protection against risk for the disorder or if the decrease reflects an associated pathological protein deficit. The finding of an inverse correlation between C4A and C4B and between C4L and C4S copy numbers more likely reflects that these loci may be in linkage disequilibrium with risk (C4A and C4L) vs protection (C4B and C4S) at either end of the spectrum. Based on studies of chemical-binding preferences, C4A has an affinity for amino groups and C4B for hydroxyl groups, suggesting that C4A may function more to bind immune complexes and antigenic proteins and C4B in binding to antigens with carbohydrate-rich domains such as microbes (Presumey et al., 2017). Because a multitude of studies demonstrate exposure to pathogens as a risk factor for schizophrenia (Severance and Yolken, 2020a), a deficiency of C4B protein in schizophrenia may reflect compromised microbe-binding ability. Conversely, it is also conceivable that low C4B copy number confers a degree of protection with respect to preserving the body’s microbiome from C4B-mediated over-harvesting of beneficial microbes. In a study of pediatric inflammatory bowel disease, a low C4B copy number was associated with less inflammation and greater diversity of gut microbes than individuals with higher C4B gene copy numbers (Nissila et al., 2017). As reports of the gut microbiome in schizophrenia begin to populate the literature, it will be important to examine C4 genotypes in conjunction with microbiome profiling. It will also be necessary to further interrogate these haplotype associations to rule out that variation in nearby HLA genes which segregate with specific C4 alleles via linkage disequilibrium are not driving the observed immune traits (Kamitaki et al., 2020).
The understanding of complement C4 genotypes and C4A/C4B dynamics is just beginning to be elucidated as improved genomic techniques illuminate previously inaccessible regions of the genome in larger and more diverse study populations. Our finding of several deleterious haplotype combinations and associations with pathogens, inflammatory gut processes, psychiatric symptom severity and cognitive functioning suggests that C4 gene screening might aid the identification of subsets of individuals with a potentially more severe disease course. Complement-based therapies with the aim of decreasing C4A activity and/or increasing C4B activity in different genetic backgrounds will require examination in a research setting.
Acknowledgements
This work was supported by a NIMH P50 Silvio O. Conte Center at Johns Hopkins (grant# MH-94268) and by the Stanley Medical Research Institute. We thank Giulio Genovese and Steven McCarroll for technical assistance related to C4 haplotype imputations and for constructive comments on the manuscript.
Role of funding source
The funding sources had no involvement in study design; collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Abbreviations:
- BMI
Body mass index
- C4AL
C4A-long
- C4AS
C4A-short
- C4BL
C4B-long
- C4BS
C4B-short
- CMV
Cytomegalovirus
- CRP
C-reactive protein
- ELISA
Enzyme-linked immunosorbent assay
- HERV
Human endogenous retrovirus
- HMZ
Homozygous
- HTZ
Heterozygous
- IgG
Immunoglobulin G
- LBP
Lipopolysaccharide binding protein
- MHC
Major histocompatibility complex
- OR
Odds ratio
- PBMCs
Peripheral blood mononuclear cells
- sCD14
Soluble CD14
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
Dr. Yolken is a member of the Stanley Medical Research Institute Board of Directors and Scientific Advisory Board. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. None of the other authors report any potential conflicts of interest.
7. References
- APA, 2000. Diagnostic and statistical manual of mental disorders : DSM-IV-TR, 4th ed.American Psychiatric Association, Washington, DC. [Google Scholar]
- Bennett MR, Farnell L, Gibson WG, 2013. Fiber pathway pathology, synapse loss and decline of cortical function in schizophrenia. PLoS One 8(4), e60518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benros ME, Nielsen PR, Nordentoft M, Eaton WW, Dalton SO, Mortensen PB, 2011. Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am J Psychiatry 168(12), 1303–1310. [DOI] [PubMed] [Google Scholar]
- Castro-Nallar E, Bendall ML, Perez-Losada M, Sabuncyan S, Severance EG, Dickerson FB, Schroeder JR, Yolken RH, Crandall KA, 2015. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ 3, e1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, Orekhov AN, 2014. Intestinal mucosal tolerance and impact of gut microbiota to mucosal tolerance. Front Microbiol 5, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ, Chohan TW, Kassem MS, Smith KL, Chesworth R, Karl T, Kuligowski MP, Fok SY, Bennett MR, Arnold JC, 2018. Neuregulin 1 Deficiency Modulates Adolescent Stress-Induced Dendritic Spine Loss in a Brain Region-Specific Manner and Increases Complement 4 Expression in the Hippocampus. Schizophr Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P, 2012. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10(11), 735–742. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S, 2011. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108(7), 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson F, Boronow J, Stallings C, Origoni A, Yolken R, 2007a. Toxoplasma gondii in individuals with schizophrenia: association with clinical and demographic factors and with mortality. Schizophr Bull 33(3), 737–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson F, Severance E, Yolken R, 2017. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun 62, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson F, Stallings C, Origoni A, Boronow J, Yolken R, 2007b. C-reactive protein is associated with the severity of cognitive impairment but not of psychiatric symptoms in individuals with schizophrenia. Schizophrenia Research 93(1–3), 261–265. [DOI] [PubMed] [Google Scholar]
- Dickerson F, Stallings C, Origoni A, Katsafanas E, Schweinfurth LA, Savage CL, Khushalani S, Yolken R, 2015. Pentraxin 3 is reduced in bipolar disorder. Bipolar disorders 17(4), 409–414. [DOI] [PubMed] [Google Scholar]
- Dickerson F, Stallings C, Origoni A, Vaughan C, Khushalani S, Yang S, Yolken R, 2013. C-reactive protein is elevated in schizophrenia. Schizophrenia Research 143(1), 198–202. [DOI] [PubMed] [Google Scholar]
- Dinan TG, Cryan JF, 2015. The impact of gut microbiota on brain and behaviour: implications for psychiatry. Curr Opin Clin Nutr Metab Care 18(6), 552–558. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Holland J, Mothersill D, McCarthy-Jones S, Cosgrove D, Harold D, Richards A, Mantripragada K, Owen MJ, O’Donovan MC, Wtccc, Gill M, Corvin A, Morris DW, 2018. Genetically predicted complement component 4A expression: effects on memory function and middle temporal lobe activation. Psychol Med 48(10), 1608–1615. [DOI] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermohlen O, Chun E, Garrett WS, McCoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, Prinz M, 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18(7), 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW, 1998. Structured Clinical Interview for DSM-IV Axis I Disorders - Non-patient Edition (SCID I/NP). Biometrics Research, New York State Psychiatric Institute, New York. [Google Scholar]
- Foster JA, McVey Neufeld KA, 2013. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 36(5), 305–312. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, 2015. Recent genetic findings in schizophrenia and their therapeutic relevance. J Psychopharmacol 29(2), 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JF, Cosgrove D, Whitton L, Harold D, Corvin A, Gill M, Mothersill DO, Morris DW, Donohoe G, 2019. Beyond C4: Analysis of the complement gene pathway shows enrichment for IQ in patients with psychotic disorders and healthy controls. Genes Brain Behav 18(8), e12602. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK, 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155(7), 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentzer A, Veyrard P, Roblin X, Saint-Sardos P, Rochereau N, Paul S, Bourlet T, Pozzetto B, Pillet S, 2020. Cytomegalovirus and Inflammatory Bowel Diseases (IBD) with a Special Focus on the Link with Ulcerative Colitis (UC). Microorganisms 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Stevens B, 2018. Pruning hypothesis comes of age. Nature 554(7693), 438–439. [DOI] [PubMed] [Google Scholar]
- Kamitaki N, Sekar A, Handsaker RE, de Rivera H, Tooley K, Morris DL, Taylor KE, Whelan CW, Tombleson P, Loohuis LMO, Boehnke M, Kimberly RP, Kaufman KM, Harley JB, Langefeld CD, Seidman CE, Pato MT, Pato CN, Ophoff RA, Graham RR, Criswell LA, Vyse TJ, McCarroll SA, 2020. Complement genes contribute sex-biased vulnerability in diverse disorders. Nature 582(7813), 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay SR, Fiszbein A, Opler LA, 1987. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophrenia bulletin 13(2), 261–276. [DOI] [PubMed] [Google Scholar]
- Kirch DG, 1993. Infection and autoimmunity as etiologic factors in schizophrenia: a review and reappraisal. Schizophr Bull 19(2), 355–370. [DOI] [PubMed] [Google Scholar]
- Kohler O, Petersen L, Mors O, Mortensen PB, Yolken RH, Gasse C, Benros ME, 2017. Infections and exposure to anti-infective agents and the risk of severe mental disorders: a nationwide study. Acta Psychiatr Scand 135(2), 97–105. [DOI] [PubMed] [Google Scholar]
- Laskaris L, Zalesky A, Weickert CS, Di Biase MA, Chana G, Baune BT, Bousman C, Nelson B, McGorry P, Everall I, Pantelis C, Cropley V, 2019. Investigation of peripheral complement factors across stages of psychosis. Schizophrenia Research 204, 30–37. [DOI] [PubMed] [Google Scholar]
- Leweke FM, Gerth CW, Koethe D, Klosterkotter J, Ruslanova I, Krivogorsky B, Torrey EF, Yolken RH, 2004. Antibodies to infectious agents in individuals with recent onset schizophrenia. Eur Arch Psychiatry Clin Neurosci 254(1), 4–8. [DOI] [PubMed] [Google Scholar]
- Li SJ, Zhuo M, Huang X, Huang YY, Zhou J, Xiong DS, Li JH, Liu Y, Pan ZL, Li HH, Chen J, Li XB, Xiang ZM, Wu FC, Wu K, 2020. Altered gut microbiota associated with symptom severity in schizophrenia. PeerJ 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczynski P, Whelan SO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, Cryan JF, 2016. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci 44(9), 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayilyan KR, Dodds AW, Boyajyan AS, Soghoyan AF, Sim RB, 2008a. Complement C4B protein in schizophrenia. World J Biol Psychiatry 9(3), 225–230. [DOI] [PubMed] [Google Scholar]
- Mayilyan KR, Weinberger DR, Sim RB, 2008b. The complement system in schizophrenia. Drug News Perspect 21(4), 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BJ, Graham KL, Bodenheimer CM, Culpepper NH, Waller JL, Buckley PF, 2013. A prevalence study of urinary tract infections in acute relapse of schizophrenia. J Clin Psychiatry 74(3), 271–277. [DOI] [PubMed] [Google Scholar]
- Mondelli V, Di Forti M, Morgan BP, Murray RM, Pariante CM, Dazzan P, 2020. Baseline high levels of complement component 4 predict worse clinical outcome at 1-year follow-up in first-episode psychosis. Brain Behavior and Immunity 88, 913–915. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Kosciolek T, Maldonado Y, Daly RE, Martin AS, McDonald D, Knight R, Jeste DV, 2019. Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophr Res 204, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimgaonkar VL, Prasad KM, Chowdari KV, Severance EG, Yolken RH, 2017. The complement system: a gateway to gene-environment interactions in schizophrenia pathogenesis. Mol Psychiatry 22(11), 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissila E, Korpela K, Lokki AI, Paakkanen R, Jokiranta S, de Vos WM, Lokki ML, Kolho KL, Meri S, 2017. C4B gene influences intestinal microbiota through complement activation in patients with paediatric-onset inflammatory bowel disease. Clin Exp Immunol 190(3), 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget JG, 2018. The Emerging Immunogenetic Architecture of Schizophrenia. Schizophr Bull 44(5), 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KM, Chowdari KV, D’Aiuto LA, Iyengar S, Stanley JA, Nimgaonkar VL, 2018. Neuropil contraction in relation to Complement C4 gene copy numbers in independent cohorts of adolescent-onset and young adult-onset schizophrenia patients-a pilot study. Transl Psychiatry 8(1), 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presumey J, Bialas AR, Carroll MC, 2017. Complement System in Neural Synapse Elimination in Development and Disease. Adv Immunol 135, 53–79. [DOI] [PubMed] [Google Scholar]
- Randolph C, 1998. RBANS Manual - Repeatable Battery for the Assessment of Neuropsychological Status. Psychological Corporation, San Antonio. [Google Scholar]
- Rey R, Suaud-Chagny MF, Bohec AL, Dorey JM, d’Amato T, Tamouza R, Leboyer M, 2020. Overexpression of complement component C4 in the dorsolateral prefrontal cortex, parietal cortex, superior temporal gyrus and associative striatum of patients with schizophrenia. Brain Behav Immun. [DOI] [PubMed] [Google Scholar]
- Rudduck C, Beckman L, Franzen G, Jacobsson L, Lindstrom L, 1985. Complement factor C4 in schizophrenia. Hum Hered 35(4), 223–226. [DOI] [PubMed] [Google Scholar]
- Sager K, Alam S, Bond A, Chinnappan L, Probert CS, 2015. Review article: cytomegalovirus and inflammatory bowel disease. Alimentary pharmacology & therapeutics 41(8), 725–733. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK, 2015. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17(5), 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya P, Danda D, Sharma D, Scaria V, 2016. Does the buck stop with the bugs?: an overview of microbial dysbiosis in rheumatoid arthritis. Int J Rheum Dis 19(1), 8–20. [DOI] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Schizophrenia Working Group of the Psychiatric Genomics, C., Daly MJ, Carroll MC, Stevens B, McCarroll SA, 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530(7589), 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, Fu T, Worringer K, Brown HE, Wang J, Kaykas A, Karmacharya R, Goold CP, Sheridan SD, Perlis RH, 2019. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci 22(3), 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Alaedini A, Yang S, Halling M, Gressitt KL, Stallings CR, Origoni AE, Vaughan C, Khushalani S, Leweke FM, Dickerson FB, Yolken RH, 2012. Gastrointestinal inflammation and associated immune activation in schizophrenia. Schizophr Res 138(1), 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Dickerson FB, Yolken RH, 2018. Autoimmune phenotypes in schizophrenia reveal novel treatment targets. Pharmacol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Stallings CR, Katsafanas E, Schweinfurth LA, Savage CL, Adamos MB, Sweeney KM, Origoni AE, Khushalani S, Leweke FM, Dickerson FB, Yolken RH, 2016a. Candida albicans exposures, sex specificity and cognitive deficits in schizophrenia and bipolar disorder. NPJ Schizophr 2, 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Stallings CR, Katsafanas E, Schweinfurth LA, Savage CLG, Adamos MB, Sweeney KM, Origoni AE, Khushalani S, Dickerson FB, Yolken RH, 2017. Probiotic normalization of Candida albicans in schizophrenia: A randomized, placebo-controlled, longitudinal pilot study. Brain Behav Immun 62, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Stallings CR, Origoni AE, Khushalani S, Leweke FM, Dickerson FB, Yolken RH, 2013. Discordant patterns of bacterial translocation markers and implications for innate immune imbalances in schizophrenia. Schizophr Res 148(1–3), 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Gressitt KL, Yang S, Stallings CR, Origoni AE, Vaughan C, Khushalani S, Alaedini A, Dickerson FB, Yolken RH, 2014. Seroreactive marker for inflammatory bowel disease and associations with antibodies to dietary proteins in bipolar disorder. Bipolar Disorders 16(3), 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Prandovszky E, Castiglione J, Yolken RH, 2015. Gastroenterology issues in schizophrenia: why the gut matters. Curr Psychiatry Rep 17(5), 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Xiao J, Jones-Brando L, Sabunciyan S, Li Y, Pletnikov M, Prandovszky E, Yolken R, 2016b. Toxoplasma gondii-A Gastrointestinal Pathogen Associated with Human Brain Diseases. Int Rev Neurobiol 131, 143–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Yolken RH, 2020a. From Infection to the Microbiome: An Evolving Role of Microbes in Schizophrenia. Curr Top Behav Neurosci 44, 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance EG, Yolken RH, 2020b. Tracking a dysregulated gut-brain axis with biomarkers of the microbiome. Biomarkers in Neuropsychiatry 2, 100009. [Google Scholar]
- Severance EG, Yolken RH, Eaton WW, 2016c. Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: more than a gut feeling. Schizophr Res 176(1), 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J, Zhang M, Hu S, Liang Y, 2018. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophr Res. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA, 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131(6), 1164–1178. [DOI] [PubMed] [Google Scholar]
- Stilling RM, Dinan TG, Cryan JF, 2014. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav 13(1), 69–86. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Bartko JJ, Yolken RH, 2012. Toxoplasma gondii and other risk factors for schizophrenia: an update. Schizophr Bull 38(3), 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF, Peterson MR, 1976. The viral hypothesis of schizophrenia. Schizophr Bull 2(1), 136–146. [DOI] [PubMed] [Google Scholar]
- Wekerle H, 2017. Brain Autoimmunity and Intestinal Microbiota: 100 Trillion Game Changers. Trends Immunol 38(7), 483–497. [DOI] [PubMed] [Google Scholar]
- Woo JJ, Pouget JG, Zai CC, Kennedy JL, 2019. The complement system in schizophrenia: where are we now and what’s next? Mol Psychiatry. [DOI] [PubMed] [Google Scholar]
- Xu R, Wu B, Liang J, He F, Gu W, Li K, Luo Y, Chen J, Gao Y, Wu Z, Wang Y, Zhou W, Wang M, 2019. Altered gut microbiota and mucosal immunity in patients with schizophrenia. Brain Behav Immun. [DOI] [PubMed] [Google Scholar]
- Yolken RH, Dickerson FB, Torrey EF, 2009. Toxoplasma and schizophrenia. Parasite Immunol 31(11), 706–715. [DOI] [PubMed] [Google Scholar]
- Yolken RH, Severance EG, Sabunciyan S, Gressitt KL, Chen O, Stallings C, Origoni A, Katsafanas E, Schweinfurth LA, Savage CL, Banis M, Khushalani S, Dickerson FB, 2015. Metagenomic Sequencing Indicates That the Oropharyngeal Phageome of Individuals With Schizophrenia Differs From That of Controls. Schizophr Bull 41(5), 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken RH, Torrey EF, 2008. Are some cases of psychosis caused by microbial agents? A review of the evidence. Mol Psychiatry 13(5), 470–479. [DOI] [PubMed] [Google Scholar]
- Zheng P, Zeng B, Liu M, Chen J, Pan J, Han Y, Liu Y, Cheng K, Zhou C, Wang H, Zhou X, Gui S, Perry SW, Wong ML, Licinio J, Wei H, Xie P, 2019. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv 5(2), eaau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]


