Abstract
Background:
At least three syphilis typing systems are proposed. Recent work suggests that multilocus sequence typing (MLST) may be superior to enhanced CDC typing (ECDCT) by yielding a higher discriminatory power. The goal of this study was to compare the two systems and identify associations between neurosyphilis and strain types.
Methods:
MLST for tp0136, tp0548 and tp0705 was determined for DNA from 78 Treponema pallidum subspecies pallidum isolates propagated in rabbits, 10 oral and 10 genital or non-genital lesion swabs, and 10 blood samples from patients with syphilis. These samples were chosen because they were completely typeable by ECDCT. Using both systems, association between strain types and neurosyphilis, defined as a reactive cerebrospinal fluid Venereal Disease Research Laboratory test, was determined. Partial and complete ECDCT types were also determined for samples from different anatomical sites in 35 patients, and from blood and blood isolates (rabbit propagated) from 13 patients.
Results:
MLST type could be fully determined for 100 (92.6%) of 108 samples. While MLST subdivided three common ECDCT types, it failed to distinguish among others. Neurosyphilis was more common in individuals infected with type 1.1.2 and tp0705 type 2 using MLST, and tp0548 type f using ECDCT. ECDCT was stable among anatomical sites and between patient-derived and rabbit propagated organisms
Conclusions:
Compared to ECDCT, MLST was not uniformly more discriminating. Both typing systems demonstrate that specific types may be more neurotropic than others.
SUMMARY
Two Treponema pallidum subspecies pallidum strain typing systems differed in discriminatory ability. Individual strain types in both systems were more common in individuals with neurosyphilis.
INTRODUCTION
Syphilis, a sexually transmitted disease caused by the spirochete Treponema pallidum subspecies pallidum (T. pallidum), is a serious public health problem that has been increasing in incidence worldwide in recent years (1). In the United States, the reported number of individuals with primary and secondary syphilis increased 71% from 2013 to 2018, while the 2018 reported case counts of all stages of syphilis reached the highest number since 1993 (2). Similarly, according to Public Health – Seattle and King County, between 2014 and 2018, the number of cases of syphilis in King County, Washington, which includes Seattle, more than doubled (https://www.kingcounty.gov/depts/health/communicable-diseases/hiv-std/patients/epidemiology/std-data-reports.aspx).
Molecular strain typing has been an important tool for understanding the epidemiology of infectious diseases by identifying and tracing populations at high risk. For example, molecular typing of Neisseria gonorrhoeae has contributed to understanding the spread of antibiotic resistance within populations (3). Molecular genotyping of T. pallidum, in particular, has led to the discovery of types associated with an increased risk of neurosyphilis, and it has identified widespread macrolide resistance (4, 5).
Pillay et al. (6) published a molecular typing method for T. pallidum in 1998 that was developed at the US Centers for Disease Control and Prevention (CDC). It distinguished among types by the number of 60-base pair (bp) repeats in the acidic repeat protein (arp) gene, and restriction fragment length polymorphism (RFLP) analysis of sequence differences in the T. pallidum repeat (tpr) subfamily II genes (tprE, tprG and tprJ). In 2010, we published an enhanced CDC typing (ECDCT) system that added sequence analysis of a short variable region of tp0548, which was more discriminating than the CDC system (5). Using the ECDCT system, neurosyphilis was more common in individuals infected with type 14d/f compared to those infected with other types (5). In 2012, a study by Mikalová et al. (7) raised concerns about the genetic stability of the first two gene targets of CDC (or the ECDCT) system, reporting different subtypes among anatomical sites in 11 of 18 patients. An alternative system using multilocus sequence typing (MLST) was introduced in 2018 by Grillová et al. (8) from the same research group. The MLST is based on sequence analysis of variable regions in tp0136, tp0548 and tp0705. Using specimens collected in Seattle, WA from different patients with untreated syphilis, the goal of this study was to compare the discriminatory ability of MLST to that of the ECDCT, to identify whether an individual MLST type was more common in individuals with neurosyphilis, and to address the stability of the ECDCT within individuals with syphilis.
METHODS
Sample Sources
MLST analysis was performed on samples that were fully typeable using the ECDCT system and are representative of the variety of types seen in Seattle between 1999 and 2017. These included DNA from T. pallidum isolates from blood (n=51) or cerebrospinal fluid (CSF) (n=27) from 75 unique individuals with syphilis after one or two rounds of rabbit propagation. Both CSF and blood isolates were tested for three individuals. In addition, 10 swabs of oral mucosa, 10 swabs from genital or non-genital lesions, and 10 blood samples were tested from 30 individuals with untreated syphilis. An additional 87 DNA samples from at least two anatomic sites from the same individual (35 individuals; blood [n=34], CSF [n=2], lesion swabs [n=25] or oral swabs [n=26]) or from blood and blood isolate (13 individuals) were tested by the ECDCT system. Oral swabs, lesion swabs and blood were collected in 2015–2017 as part of a public health outbreak response, which did not require human subjects approval. Blood and CSF were collected in 1999–2015 from individuals enrolled in institutional review board approved studies after written informed consent was obtained.
Sample Collection and DNA Extraction
Bacterial isolates were stored in 200 μl 1X lysis buffer (10 mM Tris-HCl, 0.1 M anticoagulant ethylene-diamine-tetra-acetic acid [EDTA], 0.5% SDS). Blood was collected in EDTA tubes. 0.5 ml of blood was mixed with 0.5 ml of 2X lysis buffer (20 mM Tris-HCl, 0.2 M EDTA, 1% SDS). Swabs were stored in 1 ml 1X lysis buffer; 1 ml of uncentrifuged CSF was preserved without additives. All samples were stored at −80°C until DNA extraction.
T. pallidum DNA was extracted from isolates (1.3 × 106 to 3.8 × 107 organisms), swabs and CSF using a QIAamp DNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. T. pallidum DNA was extracted from blood using a QIAamp DNA Blood Midi kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Extracted DNA from blood and CSF were precipitated overnight at −20°C with 2.5 volumes of 100% ethanol, 0.1 volume of 3 M sodium acetate, and 1 μl of 20 mg/ml glycogen and resuspended after two washes with 75% ethanol in a final volume of 60 μl molecular grade water.
Molecular Methods
ECDCT typing was performed as previously described (5). MLST was performed as previously described (8) except that PCR products were purified using ExoSAP-IT PCR Product Cleanup Reagent (ThermoFisher Scientific, Waltham, MA) according to manufacturer’s instructions instead of a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). In addition, extra sequencing primers were used for tp0316 if the entire amplicon could not be sequenced (Supplementary Table 1). If a sequence was unable to be determined for any of the three gene targets, the PCRs were repeated once. The sequence types of the three MLST gene targets were compared to the reference sequences submitted to the PubMLST BIGSdb database of T. pallidum (9).
Statistical Methods
Neurosyphilis was defined as a reactive CSF Venereal Disease Research Laboratory test (CSF-VDRL). Association between strain type and neurosyphilis was determined in 74 patients using Chi-Square test or Fisher’s exact test. Comparisons of proportions used a two sample test of proportions calculator in Stata version 11.2. We restricted the analysis to strain types that were seen in more than 5 participants. Two sided p-values <0.05 were considered to be statistically significant.
RESULTS
Out of 108 completely typeable specimens using the ECDCT, complete MLST could be determined for 100 (92.6%) samples. Success was greater for bacterial isolates (77 [98.7%] of 78) than for other samples (23 [76.7%] of 30; p<0.001): 8 (80.0%) of 10 bloods, 9 (90.0%) of 10 lesion swabs, and 6 (60.0%) of 10 oral swabs). All remaining samples were at least partially typeable (Supplementary Table 2), meaning that at least one typing locus could be sequenced, except for two oral swabs in which no loci amplified for either tp0136, tp0548 or tp0705 on two separate occasions. Three new sequences were identified for tp0548 (Figure 1) that were not previously identified by the PubMLST BIGSdb database of T. pallidum (9) as of 21 January, 2020.
Figure 1. New MSLT variants identified in tp0548.
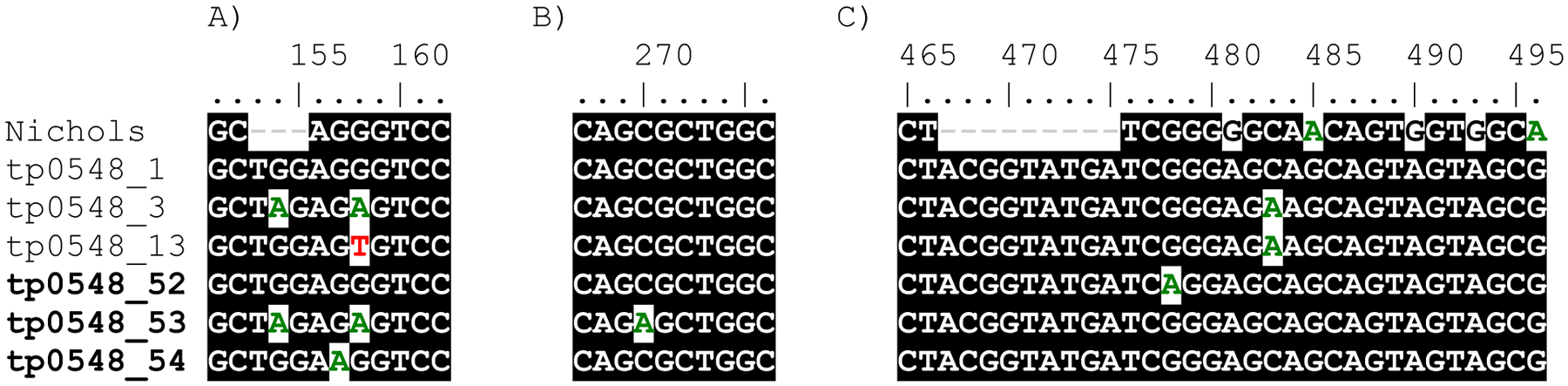
Three new tp0548 nucleotide variants (types 52–54, in bold) not previously identified in the tp0548 database (9) in the three different typing regions. Number coordinates correspond to nucleotide positions of TPA SS14 (CP004011.1) as listed in Grillová et al. (8).
MLST, multilocus sequence typing
Among the 100 specimens that were completely typeable using both systems, 11 types were identified using the ECDCT system, and 15 using MLST. While the MLST system identified additional genotypes among three ECDCT types (14d/f, 14d/g and 15d/f), it was unable to discriminate between several ECDCT types. Specifically, the MLST system assigned the same genotype (1.1.1) to three ECDCT types (14a/f, 14d/f and 14e/f) (Figure 2). In addition, the MLST assigned types 1.1.2 and 1.1.3 to ECDCT types 14d/f and 15d/f, type 1.1.9 to ECDCT types 14b/f and 14d/f, and type 1.3.1 to ECDCT types 8d/g, 14d/g and 14e/g (Figure 2).
Figure 2. ECDCT and MLST types.
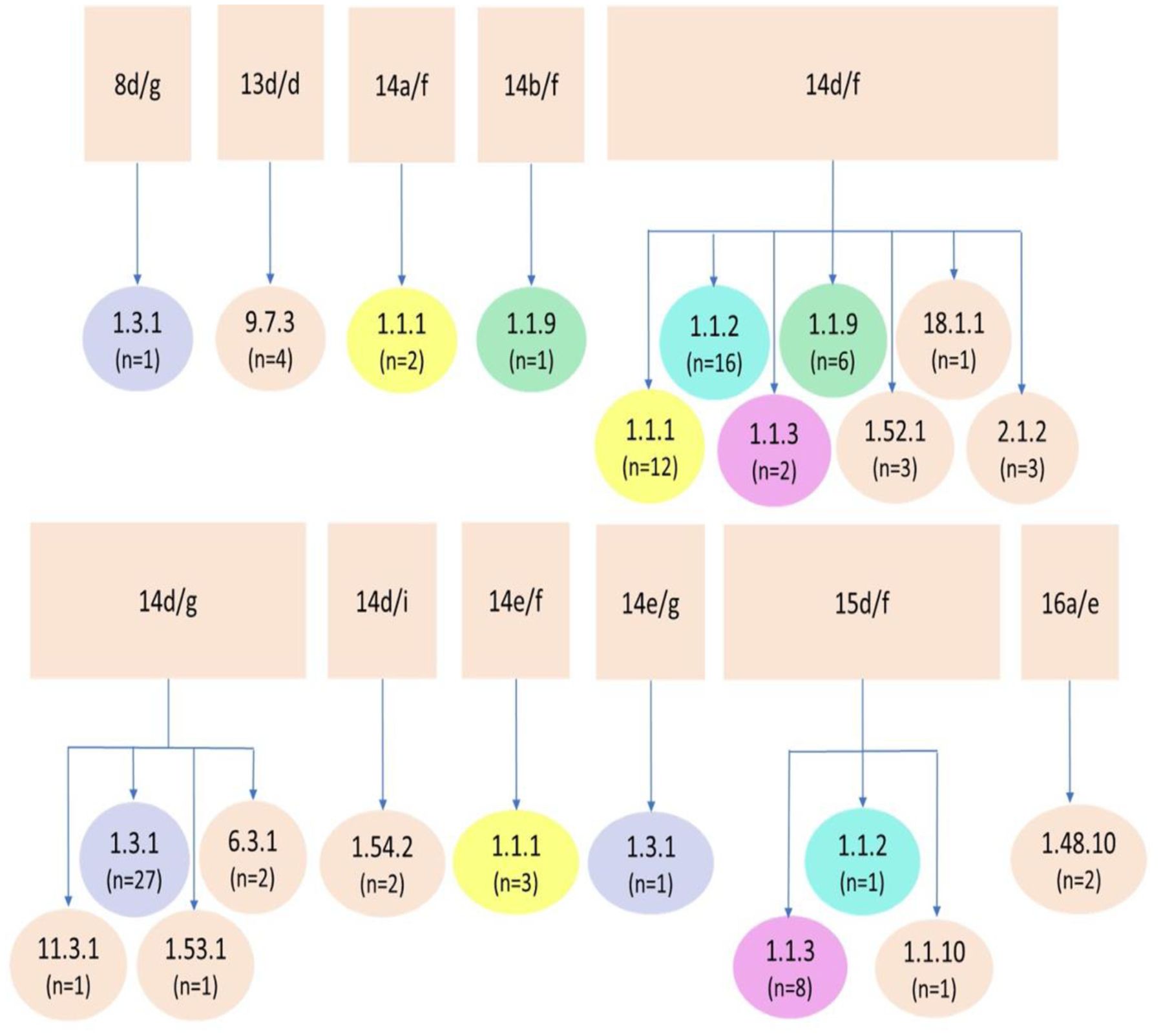
ECDCT types of samples from Seattle, in squares, separated into their respective complete types determined by MLST, in circles. Types shown in color indicate MLST genotypes found within more than one ECDCT type. MLST types found within the common 14d/f and 14d/g types are also found within unique ECDCT types, such as 8d/g, 14a/f, 14b/f, 14e/f, 14e/g and 15d/f.
ECDCT, enhanced CDC typing; MLST, multilocus sequence typing
Eighteen (24.3%) of 74 individuals with syphilis had a reactive CSF-VDRL. A reactive CSF-VDRL was found in 8 (50.0%) of 16 patients with MLST type 1.1.2 compared to 7 (17.9%) of 39 patients without type 1.1.2 (Figure 3). No patients out of 17 with MLST type 1.3.1 had a reactive CSF-VDRL, while 15 (39.5%) of 38 of those without type 1.3.1 were reactive (Figure 3). Likewise, only 1 (5.0%) out of 20 patients with 14d/g had a reactive CSF-VDRL compared to 16 (34.8%) out of 46 not infected with 14d/g. Nine (42.9%) of 21 patients with tp0705 type 2, irrespective of the first two target types, had a reactive CSF-VDRL compared to 9 (11.3%) of 53 without tp0705 type 2 (Figure 3). Similarly, CSF-VDRL was reactive in sixteen (34.8%) of 46 patients with tp0548 type f, compared to 2 (7.1%) of 28 without tp0548 type f. The sensitivity of tp0548 type f for neurosyphilis diagnosis was higher than for tp0705 type 2, and the specificity of tp0705 type 2 for neurosyphilis diagnosis was higher than for tp0548 type f (Table 1).
Figure 3. Association between reactive CSF-VDRL and strain type.
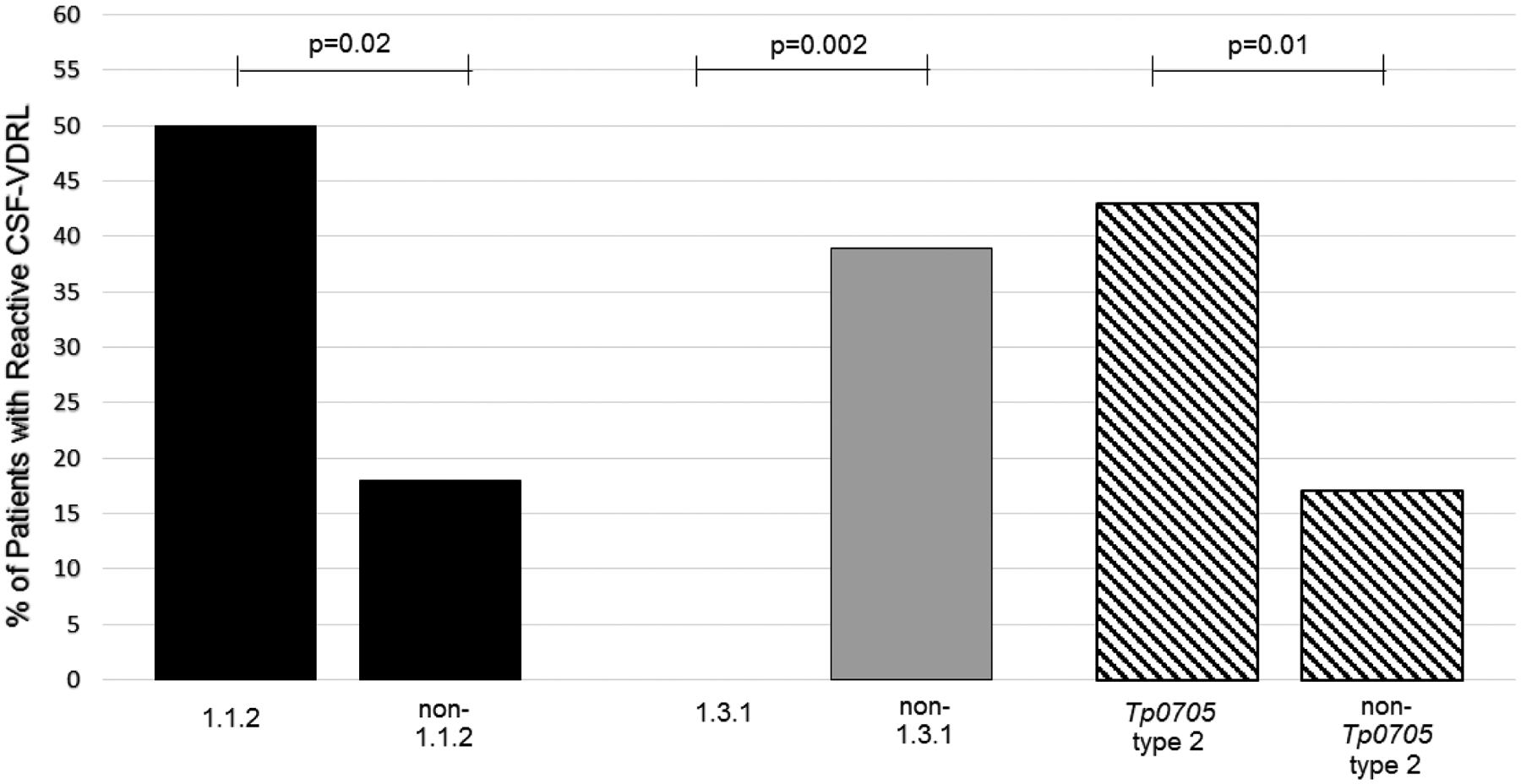
Percentage of patients with reactive CSF-VDRL based on MLST strain types. A reactive CSF-VDRL was more common in types 1.1.2, in patients without type 1.3.1, and in patients with tp0705 type 2.
CSF-VDRL, cerebrospinal fluid Venereal Disease Research Laboratory test; MLST, multilocus sequence typing
Table 1.
Sensitivity and specificity of individual gene types to diagnose neurosyphilis defined as a reactive cerebrospinal fluid Venereal Disease Research Laboratory test
| Test | Sensitivity % (95% CI) |
Specificity % (95% CI) |
|---|---|---|
| tp0548 type f | 88.9 (74.4 to 100.0) |
46.4 (33.4 to 59.5) |
| tp0705 type 2 | 50.0 (26.9 to 73.1) |
78.6 (67.9 to 89.3) |
The stability of the ECDCT system among different anatomical sites and between blood and blood isolates is shown in Table 2. Specifically, arp subtype was determined for parallel samples from 29 individuals, tprE, G and J RFLP subtype was determined for parallel samples from 19, and tp0548 subtype was determined in at least two parallel samples from all 48 individuals. No discrepancies in type were found within any gene target from the same individual. Complete ECDCT type was identical in 5 parallel lesion and oral swabs, 2 parallel blood and oral swabs, and 1 parallel blood, lesion and oral swabs. In addition, complete ECDCT type was identical in 8 parallel blood and blood isolates.
Table 2.
ECDCT types from multiple sites in the same individual or from blood and blood isolate from the same individual
| Participant Number | Type from Blood | Type from Lesion Swab | Type from Oral Swab | Type from CSF | Type from Isolate |
|---|---|---|---|---|---|
| 1 | 13d/d | 13d/d | |||
| 2 | 14X/f | XX/X | 14X/f | ||
| 3 | 14X/f | 14X/f | |||
| 4 | 14X/f | 14d/f | |||
| 5 | 14X/f | 14d/f | |||
| 6 | 14X/f | 14d/f | |||
| 7 | 14X/f | 14d/f | |||
| 8 | 14d/f | 14d/f | |||
| 9 | 14d/f | 14d/f | |||
| 10 | 14d/f | 14d/f | |||
| 11 | 14X/g | XX/X | Xd/g | ||
| 12 | 14X/g | 14b/g | |||
| 13 | 14X/g | 14d/g | |||
| 14 | 14X/g | 14d/g | XX/X | ||
| 15 | 14X/g | 14X/g | |||
| 16 | 14X/g | 14d/g | |||
| 17 | 14d/g | 14d/g | |||
| 18 | 14d/g | 14d/g | |||
| 19 | 14d/g | 14d/g | |||
| 20 | 14d/g | XX/g | XX/X | ||
| 21 | 14d/g | 14d/g | 14d/g | ||
| 22 | 14d/g | 14d/g | |||
| 23 | 14d/g | 14d/g | |||
| 24 | 14d/g | 14X/g | |||
| 25 | 14e/f | XX/X | 14X/f | ||
| 26 | 15d/f | 15d/f | |||
| 27 | Xa/f | 14a/f | |||
| 28 | Xe/e | 14e/e | |||
| 29 | Xd/f | 15d/f | XX/X | ||
| 30 | Xd/g | 14d/g | |||
| 31 | XX/e | 14e/e | XX/X | ||
| 32 | XX/f | 15d/f | |||
| 33 | XX/f | XX/X | 14a/f | ||
| 34 | XX/f | 14e/f | |||
| 35 | XX/f | Xd/f | |||
| 36 | XX/g | 14d/g | XX/X | ||
| 37 | XX/g | 14d/g | XX/X | ||
| 38 | XX/g | 14d/g | |||
| 39 | XX/g | 14e/g | |||
| 40 | XX/g | XX/g | |||
| 41 | XX/g | 13X/g | |||
| 42 | XX/X | 14a/f | 14a/f | ||
| 43 | XX/X | 14d/f | XX/f | ||
| 44 | XX/X | 14d/g | 14d/g | ||
| 45 | XX/X | 14d/g | 14d/g | ||
| 46 | XX/X | 14e/f | 14e/f | ||
| 47 | XX/X | 14X/f | XX/f | ||
| 48 | 14e/f | 14X/f |
ECDCT, enhanced Centers for Disease Control and Prevention typing; arp, acidic repeat protein gene; RFLP, restriction fragment length polymorphism (RFLP); tprE, G and J, T. pallidum repeat (tpr) subfamily II genes
DISCUSSION
Previous work has shown that the molecular typing efficiency, or success in obtaining complete types, of MLST to be comparable to ECDCT (8, 10–12).Our study showed that it was lower using DNA isolated from oral and lesion swabs, and from blood. The success of typing systems can vary for several reasons, but is likely directly related to the number of organisms in the sample tested. Other factors, including treatment history, specimen collection and storage, and DNA degradation may play a role. The low success rate for determining T. pallidum strain type from blood has been observed previously (13, 14) and is attributed to PCR inhibitors (15) and a lower yield of T. pallidum (13). Conversely, many studies using the CDC and ECDCT systems demonstrate high typing efficiency using lesion exudate (13, 16–18). Similarly, most studies using MLST (8–11) have used lesion exudate. Nevertheless, it is important to be able to genotype from specimens other than lesion exudate, such as oral fluid or blood, because swabs of lesions are not always easily acquired or available.
Worldwide, ECDCT type 14d/f has been most common (5, 19, 20) until more recently, when type 14d/g has gained prevalence (16, 21–23). MLST type 1.3.1 is designated to most ECDCT 14d/g types, and it is the most common type in Switzerland, France, Cuba, and in the Czech Republic. (8, 10, 11, 14). In our study, MLST discriminated among some ECDCT types, but failed to discriminate among others.
Individuals with syphilis who were infected with MLST type 1.1.2, the type corresponding to most patients with ECDCT type 14d/f, were more likely to have neurosyphilis (defined as reactive CDF-VDRL). Patients infected with MLST type 1.3.1, corresponding to type 14d/g, were less likely to have neurosyphilis, suggesting a less neuroinvasive strain is currently circulating. Our finding that neurosyphilis was more common in persons infected with organisms harboring tp0548 type f or tp0705 type 2 has potential clinical relevance. For example, if these two targets could be easily and quickly determined from blood, the results could be used to identify individuals with syphilis who might benefit the most from lumbar puncture.
Unlike in a previous study (7) where arp and tprE, G, and J RFLP subtypes differed by anatomic site in 11 out of 18 patients, we found that these subtypes were stable among anatomical sites and between blood and blood derived isolates in all 48 instances. Previous studies have demonstrated that ECDCT strain type is stable with repeated rabbit passages (5, 6). These findings are reassuring that the three gene targets of ECDCT are genetically stable. However, determination of arp and tprE, G, and J subtype are observer dependent, which is a drawback not shared by MLST systems.
Limitations of our study should be considered in interpreting our findings. Our samples were chosen because they were completely typeable by ECDCT and because they were representative of the types seen in Seattle over one and a half decades. While we cannot comment on typing efficiency of ECDCT, we can say that the MLST was less efficient for samples other than propagated bacterial isolates. In addition, our samples were from a single geographic area, and our conclusions regarding the discriminatory ability of the two systems may not apply to other regions.
Unlike in the past, in vitro culture of T. pallidum is becoming more feasible (24). This may make it possible to obtain full genomes of T. pallidum for routine sequencing, which may make all current typing systems irrelevant. However, cost will likely be an issue. Indeed, the cost of the MLST system was double the cost of ECDCT system in our laboratory (data not shown). For the foreseeable future, a robust T. pallidum typing system that can be applied to all clinical samples and that is independent of user interpretation is sorely needed, and we are not there yet. Based on discriminatory ability, stability and cost, the T. pallidum ECDCT system continues to have an important role in studies of syphilis epidemiology and clinical manifestations.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01 NS34235 to C.M.M. and by a gift from Liliana Eagan to the University of Washington Neuro-infectious Diseases Research Fund.
Footnotes
The authors have no conflicts of interest relevant to this work.
REFERENCES
- 1.Willeford WG, Bachmann LH. Syphilis ascendant: a brief history and modern trends. Trop Dis Travel Med Vaccines. 2016;2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Sexually Transmitted Disease Surveillance 2018. Atlanta: U.S.Department of Health and Human Services;2019. [Google Scholar]
- 3.Chisholm SA, Alexander S, Desouza-Thomas L, et al. Emergence of a Neisseria gonorrhoeae clone showing decreased susceptibility to cefixime in England and Wales. J Antimicrob Chemother. 2011;66(11):2509–12. [DOI] [PubMed] [Google Scholar]
- 4.Grimes M, Sahi SK, Godornes BC, et al. Two Mutations Associated With Macrolide Resistance in Treponema pallidum: Increasing Prevalence and Correlation With Molecular Strain Type in Seattle, Washington. Sexually transmitted diseases. 2012;39(12):954–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marra CM, Sahi SK, Tantalo LC, et al. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. The Journal of infectious diseases. 2010;202(9):1380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillay A, Liu H, Chen CY, et al. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex Transm Dis. 1998;25(8):408–14. [DOI] [PubMed] [Google Scholar]
- 7.Mikalova L, Pospisilova P, Woznicova V, Kuklova I, Zakoucka H, Smajs D. Comparison of CDC and sequence-based molecular typing of syphilis treponemes: tpr and arp loci are variable in multiple samples from the same patient. BMC Microbiol. 2013;13:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grillova L, Bawa T, Mikalova L, et al. Molecular characterization of Treponema pallidum subsp. pallidum in Switzerland and France with a new multilocus sequence typing scheme. PLoS One. 2018;13(7):e0200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grillova L, Jolley K, Smajs D, Picardeau M. A public database for the new MLST scheme for Treponema pallidum subsp. pallidum: surveillance and epidemiology of the causative agent of syphilis. PeerJ. 2019;6:e6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pospisilova P, Grange PA, Grillova L, et al. Multi-locus sequence typing of Treponema pallidum subsp. pallidum present in clinical samples from France: Infecting treponemes are genetically diverse and belong to 18 allelic profiles. PLoS One. 2018;13(7):e0201068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grillova L, Noda AA, Lienhard R, Blanco O, Rodriguez I, Smajs D. Multilocus Sequence Typing of Treponema pallidum subsp. pallidum in Cuba From 2012 to 2017. J Infect Dis. 2019;219(7):1138–45. [DOI] [PubMed] [Google Scholar]
- 12.Fu B, Li H, Zhao Y, et al. A comparison of genotyping tool in Treponema pallidum: Review and meta-analysis. Infect Genet Evol. 2020;78:104049. [DOI] [PubMed] [Google Scholar]
- 13.Peng RR, Wang AL, Li J, Tucker JD, Yin YP, Chen XS. Molecular typing of Treponema pallidum: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2011;5(11):e1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrbova E, Grillova L, Mikalova L, et al. MLST typing of Treponema pallidum subsp. pallidum in the Czech Republic during 2004–2017: Clinical isolates belonged to 25 allelic profiles and harbored 8 novel allelic variants. PLoS One. 2019;14(5):e0217611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39(2):485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grange PA, Allix-Beguec C, Chanal J, et al. Molecular subtyping of Treponema pallidum in Paris, France. Sex Transm Dis. 2013;40(8):641–4. [DOI] [PubMed] [Google Scholar]
- 17.Martin IE, Tsang RS, Sutherland K, et al. Molecular characterization of syphilis in patients in Canada: azithromycin resistance and detection of Treponema pallidum DNA in whole-blood samples versus ulcerative swabs. J Clin Microbiol. 2009;47(6):1668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton MY, Liu H, Steiner B, et al. Molecular subtyping of Treponema pallidum in an Arizona County with increasing syphilis morbidity: use of specimens from ulcers and blood. J Infect Dis. 2001;183(11):1601–6. [DOI] [PubMed] [Google Scholar]
- 19.Dai T, Li K, Lu H, Gu X, Wang Q, Zhou P. Molecular typing of Treponema pallidum: a 5-year surveillance in Shanghai, China. J Clin Microbiol. 2012;50(11):3674–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khairullin R, Vorobyev D, Obukhov A, et al. Syphilis epidemiology in 1994–2013, molecular epidemiological strain typing and determination of macrolide resistance in Treponema pallidum in 2013–2014 in Tuva Republic, Russia. APMIS. 2016;124(7):595–602. [DOI] [PubMed] [Google Scholar]
- 21.Giacani L, Ciccarese G, Puga-Salazar C, et al. Enhanced Molecular Typing of Treponema pallidum subspecies pallidum Strains From 4 Italian Hospitals Shows Geographical Differences in Strain Type Heterogeneity, Widespread Resistance to Macrolides, and Lack of Mutations Associated With Doxycycline Resistance. Sex Transm Dis. 2018;45(4):237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver S, Sahi SK, Tantalo LC, et al. Molecular Typing of Treponema pallidum in Ocular Syphilis. Sex Transm Dis. 2016;43(8):524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salado-Rasmussen K, Cowan S, Gerstoft J, et al. Molecular Typing of Treponema pallidum in Denmark: A Nationwide Study of Syphilis. Acta Derm Venereol. 2016;96(2):202–6. [DOI] [PubMed] [Google Scholar]
- 24.Edmondson DG, Hu B, Norris SJ. Long-Term In Vitro Culture of the Syphilis Spirochete Treponema pallidum subsp. pallidum. MBio 2018;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


