Abstract
Expansion of α-smooth muscle actin(α-SMA)-expressing airway smooth muscle of the large airways in asthma is well-studied. However, the contribution of α-SMA-expressing cells in the more distal alveolated parenchyma, including pericytes and myofibroblasts within the alveolar septum, to asthma pathophysiology remains relatively unexplored. The objective of this study was to evaluate α-SMA expression in the alveolated parenchyma of individuals with severe asthma, compared to healthy controls or individuals with chronic obstructive pulmonary disease (COPD). Using quantitative digital image analysis and video-assisted thoracoscopic surgery (VATS) lung biopsies, we show that alveolated parenchyma α-SMA expression is markedly reduced in severe asthma in comparison to healthy controls (mean %positive pixels: 12% vs. 23%, p=0.005). COPD cases showed a similar, but trending, decrease in α-SMA positivity compared to controls (mean %positivity: 17% vs. 23%, p=0.107), which may suggest loss of α SMA expression is a commonality of obstructive lung diseases. The severe asthma group had similar staining for ERG, a specific endothelial marker, comparatively to controls (mean %positive nuclei: 34% vs. 42%, p=0.218), which suggests intact capillary endothelium and likely intact capillary-associated, α-SMA-positive pericytes. These findings suggest that the loss of α-SMA expression in severe asthma may be due to changes in myofibroblast α-SMA expression or cell number. Further study is necessary to fully evaluate possible mechanisms and consequences of this phenomenon.
Keywords: Digital image analysis, severe asthma, smooth muscle actin, alveolated lung parenchyma, positive pixel count
Introduction
Asthma is a common chronic airway disease characterized by bronchial hyperreactivity, airway obstruction, and inflammation1. It is a highly heterogenous disease, with a wide range of clinical manifestations and severity. Severe asthma, a highly variable grouping itself, is defined by the ERS/ATS guidelines as asthma requiring high-dose inhaled corticosteroids and a second controller or asthma that remains uncontrolled despite these therapies2. While representing a relatively small portion of patients with asthma, severe asthma is responsible for a significant and disproportionate percentage of asthma-related costs and mortality3,4.
A number of variably consistent histologic changes have been described in asthma, such as reticular basement membrane thickening5, respiratory epithelium loss6, and eosinophil infiltration7, but one of the best described and functionally relevant changes is an increase in α-SMA-positive airway smooth muscle (ASM) mass8. Of note, the degree of ASM hypertrophy is strongly correlated with asthma severity and can discriminate severe asthma from milder forms9. Distal to the ASM of the small airways,, there also exist contractile, α-SMA-positive non-smooth muscle cell populations, including myofibroblasts, found in alveolar ducts and alveolar septa, and capillary-associated pericytes10. These cells are involved in a diverse array of processes. Pericytes have been ascribed roles in angiogenesis11, maintenance of capillary integrity and tissue perfusion12, and inflammation and leukocyte recruitment13,14. Myofibroblasts, on the other hand, have been identified as critical effector cells in fibrotic processes15 and may have a role in modulating lung elasticity16. Given the importance of these functions, these cells, particularly myofibroblasts, have been implicated and extensively studied in a variety of lung diseases, including idiopathic pulmonary fibrosis and COPD.
In contrast, there is a paucity of studies examining these cells in asthma. The histologic study of α-SMA-positive contractile cells in asthma has generally been limited to the ASM of larger airways, due to a reliance on endobronchial or transbronchial biopsies. These biopsies obtain only minute portions of small airway and alveolated lung parenchyma and cannot access distal, peripheral lung parenchyma17,18. As such, the potential pathophysiological roles and histological characteristics of distal lung α-SMA-positive cells in severe asthma remain relatively unexplored. Of the few studies investigating small airway and alveolated parenchyma pathology in asthma, including one examining distal lung α-SMA expression19, many have relied on decades-old fatal asthma autopsy tissue, which may affect tissue quality and is not necessarily synonymous with severe asthma during life. Video-assisted thoracoscopic surgery (VATS) biopsy, commonly used diagnostically for lung nodules or in interstitial lung disease, has very rarely been utilized in patients with asthma, even when severe. However, our group has recently contended that VATS obtained biopsies can provide useful diagnostic information in severe asthma20. Importantly, they also provide ample, high-quality small-airway and alveolar tissue for histologic analysis. In this study, we utilized a unique collection of severe asthma VATS lung biopsies, permitting novel evaluation of alveolated parenchymal α-SMA expression in living patients.
In the past few decades, use of whole slide imaging and quantitative digital image analysis has grown rapidly. It has now found utility in a wide variety of research and clinical applications. As an example, readily available commercial image analysis software and algorithms have been successfully used to quantify periprostatic fat adipokine expression in prostate cancer21, CD3+ cell number in renal allograft biopsies22, and markers of kidney damage in rat kidney failure models23. Importantly, a number of studies have demonstrated quantification with commercial image analysis software and algorithms shows strong agreement with traditional visual inspection in a number of measurements, including positive cell counts22,24, stained area24, and HER2 scoring25. While visual histological analysis has traditionally been a qualitative or quasi-quantitative evaluation, this technology provides the opportunity for true quantitative analysis26. This opens the possibility for study of details not easily assessable by traditional, purely manual assessments.
In this study, our objective was to utilize quantitative digital image analysis and our unique cohort of VATS biopsies to determine whether the expression of α-SMA in alveolated parenchyma differed in severe asthma, as compared to healthy controls and COPD.
Materials and Methods
Study cohorts
Video-assisted thoracoscopic surgery lung biopsies were obtained from our archives and severe asthma patients referred to the University of Pittsburgh Asthma Institute between 2007 and 2015 (n=9). All severe asthma patients met the ERS/ATS criteria for severe asthma2 and had a negative smoking history. Biopsies from severe asthma patients were included only after exclusion of immunodeficiency and other lung diseases, including infection, hypersensitivity pneumonitis, sarcoidosis, and aspiration pneumonia. For control cohorts, lung tissue from lifetime never-smoker, non-asthmatic control patients (n=7, all wedge resections for spontaneous pneumothorax) and COPD patients (n=4, all explanted lungs) were retrieved from the tissue archives of the University of Pittsburgh Department of Pathology considering the same time interval. Relevant demographic and clinical characteristics were obtained from the medical record. All patients gave informed consent for the use of their tissue and medical information for research, and this research received University of Pittsburgh Institutional Review Board approval (IRB protocol number: PRO10100527).
Immunohistochemistry
Freshly cut 4-μm thick formalin-fixed paraffin-embedded tissue sections were subjected to α-SMA (clone 1A4; Roche Diagnostics, Basel, Switzerland) or ERG (clone EPR3864; Roche Diagnostics, Basel, Switzerland) immunostaining on a BenchMark XT autostainer (Roche Diagnostics, Basel, Switzerland). Both antibodies were pre-diluted and used according to manufacturer protocol. The ultraView Universal DAB Detection Kit (Roche Diagnostics, Basel, Switzerland) was used for chromogenic detection.
Slide digitalization and image analysis
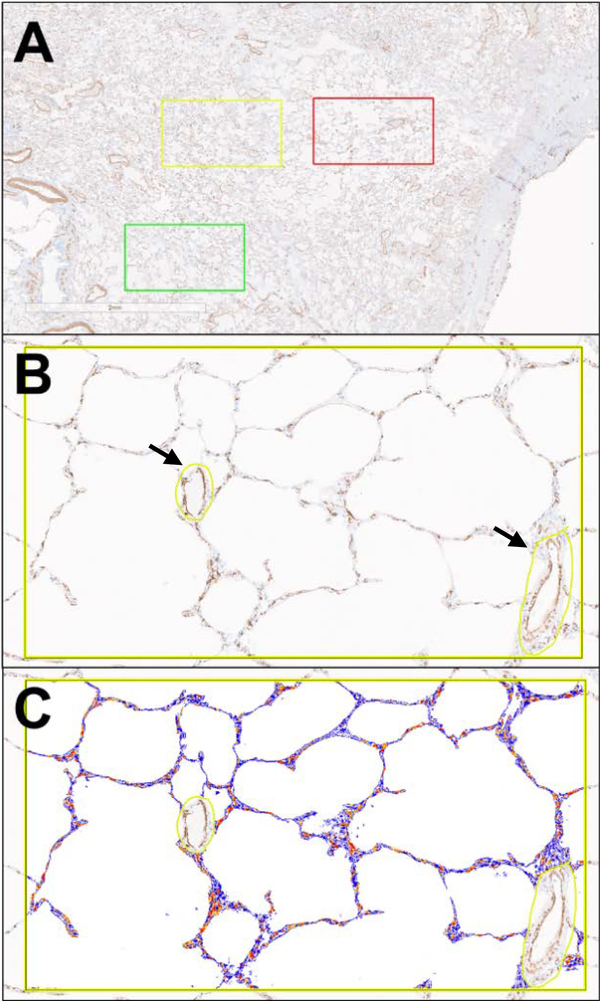
Slides were digitized using an Aperio AT2 whole slide scanner (Leica Biosystems, Wetzlar, Germany) at 20x magnification (0.25 micron/pixel resolution). After digitalization, slides were analyzed using Aperio ImageScope software (Leica Biosystems, Wetzlar, Germany). In an attempt to avoid selection bias, five selections per slide were made at digital 2X magnification, each the area of a digital 10X field. Bronchovascular bundles and collapsed tissue were avoided. Each selection was then evaluated as a digital 10X field and small airways and vessels were manually excluded (Figure 1), leaving only the alveoli and alveolar ducts included in the analysis. The software-embedded Aperio Positive Pixel Count algorithm (version 9) was used to quantify α-SMA expression and reported as %positive pixels (# positive pixels/total pixels). ERG expression was quantified using the Aperio Nuclear algorithm (version 9) and reported as %positive nuclei (# positive nuclei/total nuclei). Analyses were performed blinded to each sample’s group identity and individual characteristics.
Figure 1.
Digital image analysis process. The selection of 10X digital fields at 2X magnification (A), the visualization of a 10X field and manual exclusion of small airways and vessels, marked with arrows, (B), and the mark-up generated by the positive pixel software algorithm (C).
Statistics
When comparing two groups, p-values were calculated using an unpaired, two-tailed t-test. When comparing three or more groups, p-values were calculated using a one-way ANOVA, utilizing the Šídák correction for multiple comparisons. Linear regression was used to assess correlation between age and α-SMA %positive pixels; r and p values are reported. In all tests, a value of p < 0.05 was considered significant. All statistical analysis was performed using GraphPad Prism version 7 (GraphPad Software Inc., San Diego, CA).
Results
Patient characteristics
Patient characteristics for each case group are shown in Table 1. Of note, the control group’s mean age is younger and the age range is wider than the severe asthma or COPD group. However, age was not significantly correlated with α-SMA expression in any of the groups (Controls: r=−0.560, p=0.191; severe asthma: r=−0.060, p=0.879; COPD: r=−0.208, p=0.791). All nine severe asthma patients were currently undergoing oral corticosteroid treatment at the time of biopsy. In an attempt to account for this, COPD samples were included as controls, representing obstructive lung disease not treated with systemic corticosteroids. All control and severe asthma samples were obtained from never smokers, while all four COPD patients were current or previous smokers.
Table 1.
Basic patient characteristics for control, severe asthma (SA), and COPD groups.
| Controls (n=7) | SA (n=9) | COPD (n=4) | |
|---|---|---|---|
| Age: Mean (Range) | 28.6 (19–66) | 57 (42–65) | 66 (64–70) |
| Sex: M/F | 4/3 | 6/3 | 2/2 |
| Oral corticosteroid use: n (%) | 0 (0%) | 9 (100%) | 0 (0%) |
| Smoking history: n (%) | 0 (0%) | 0 (0%) | 4 (100%) |
| Biopsy location (n) | Right upper lobe (4), Left upper lobe (3) | Right upper lobe (3), Right middle lobe (4), Right lower lobe (2) | Right upper lobe (1), Left upper lobe (3) |
α-SMA expression
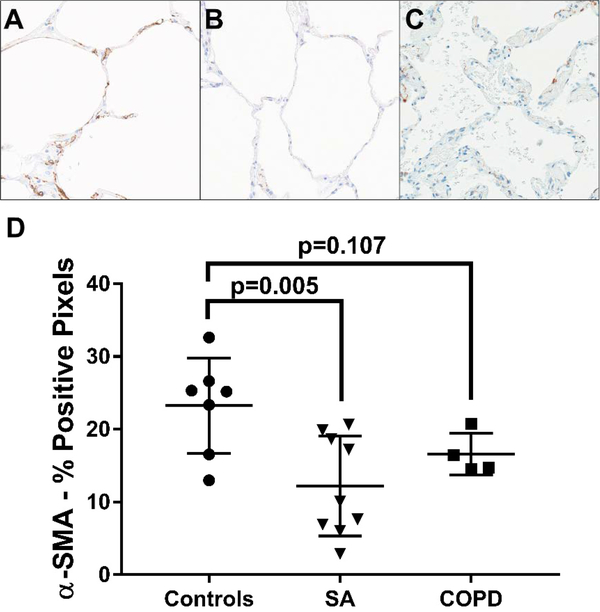
In all groups, there was α-SMA expression present around bronchi, bronchioles, arterioles, and venules and in alveolar duct tips and alveolar septa. As expected, all COPD cases showed notable alveolar septal thickening and abundant smoker’s macrophages. After manually excluding areas of airway and vascular smooth muscle in the regions of interest, leaving only the alveolated parenchymal α-SMA, the severe asthma group displayed significantly less α-SMA expression than the control group (mean %positive pixels: 12% vs. 23%, p=0.005) (Figure 2). Although not statistically significant, the COPD cases also demonstrated lesser alveolated parenchymal α-SMA expression compared to the controls (mean %positivity: 17% vs. 23%, p=0.107) (Figure 2). Interestingly, the alveolar α-SMA expression appeared to be reduced in both the alveolar septa and alveolar duct tips in both severe asthma and COPD.
Figure 2.
Alveolar parenchymal α-SMA positivity is significantly reduced in severe asthma (SA) compared to controls, with COPD tending to show similar reductions. Representative α-SMA immunostained images (20X) from a control sample (A), a severe asthma sample (B), and a COPD sample (C). The average percent α-SMA positive pixels in each sample, quantified using the Aperio ImageScope-embedded algorithm. Data expressed as mean ± SD (D).
Within the SA group, there appear to be two discrete groupings of α-SMA expression level. Given the clear separation in α-SMA expression, we were interested in whether α-SMA expression level correlated with lung function. However, α-SMA expression level did not significantly correlate with pre-VATS FEV1 % (r=0.176, p=0.650) or FEV1/FVC (r=0.055, p=0.887).
ERG expression
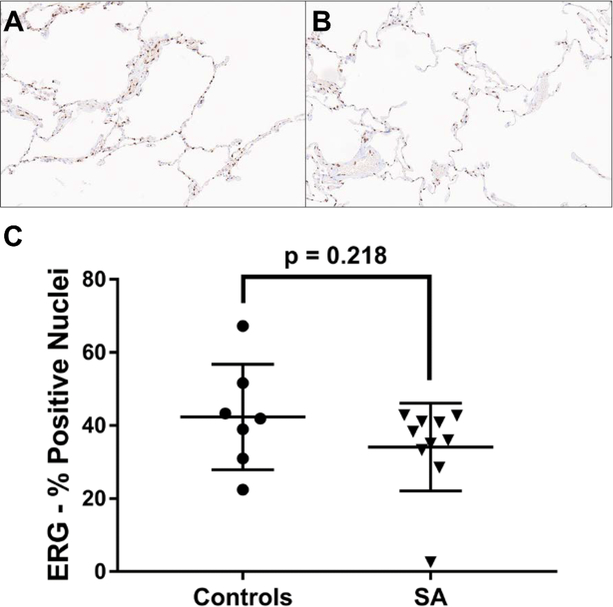
α-SMA expression alone is not sufficient to distinguish between parenchymal myofibroblasts and capillary-associated pericytes, which can both express α-SMA. To determine if the observed loss of α-SMA expression in severe asthma was due to loss of capillaries and the capillary-associated pericytes within the alveolar septa, the control and severe asthma cases were subjected to ERG immunostaining, a nuclear stain specific for endothelial cells27. After manual exclusion of any arteries, arterioles, or venules from the selections, levels of ERG expression were not significantly different between severe asthma cases and controls (mean %positive nuclei: 34% vs. 42%, p=0.218) and the staining pattern appeared similar in both groups (Figure 3).
Figure 3.
Expression of ERG, a specific endothelial cell marker, in the alveolated lung parenchyma is not significantly different in severe asthma (SA). Representative ERG immunostained images (20X) from a control sample (A) and a severe asthma sample (B). The average percent ERG positive nuclei in each sample, quantified using the Aperio ImageScope-embedded algorithm. Data expressed as mean ± SD (D).
Discussion
In asthma, much attention has been directed towards the hypertrophy and hyperplasia of α-SMA-expressing ASM within the large airways 8,28–30. However, studies examining the pathology of distal alveolated lung parenchyma in asthma are rare, due to the general inaccessibility of this tissue. Accordingly, the study of contractile, non-smooth muscle α-SMA-expressing cells, including myofibroblasts and pericytes, in this space has been limited. Of note, the few studies examining non-smooth muscle α-SMA-expression in the alveolated parenchyma have relied on either transbronchial biopsies31, which provide proximal alveolated parenchyma adjacent to large airways, or decades old post-mortem tissue from fatal asthmatics19. This study had the advantage of using entirely large tissue sections, which contain truly distal, peripheral alveolated parenchyma and provide ample tissue for analysis. To our knowledge, our study is the first to utilize VATS biopsies from living severe asthma patients to assess alpha-SMA expression in the distal lung.
As technology advances and analysis algorithms become more sophisticated, quantitative digital image analysis are becoming more popular in both research and clinical applications. Many studies have noted the utility of these commercially available analysis platforms and their strong agreement with standard visual assessments22,24,25. In this study, we used digital image analysis to quantify α-SMA expression in the distal alveolated lung parenchyma of control, severe asthma, or COPD patients. However, this methodology could be applied for any number of disease states or molecules of interest. Additionally, it allows for true quantification of immunohistochemical staining, as opposed to more traditional qualitative of quasi-quantitative methods.
Utilizing quantitative digital image analysis and this unique cohort of VATS biopsies, our study shows α-SMA expression is markedly reduced in the distal alveolated parenchyma of severe asthma patients compared to healthy controls. This decrease in α-SMA expression was unexpected, as increases in α-SMA expression or α-SMA-expressing cell number have been demonstrated in many areas in asthma, including the ASM of large airways8,28,30, bronchial submucosal glands32, and the vascular smooth muscle of bronchial arteries33.
Interestingly, the distribution of α-SMA expression in the severe asthma group appears to be bimodal. Asthma is a very heterogeneous disease and there may be currently undefined differences between these two groups of severe asthma patients that differentially impact alveolated parenchyma α-SMA expression. Given the inter-group differences in α-SMA expression, we explored whether α-SMA expression levels correlated with measures of lung function in the SA patients. However, α-SMA expression level did not significantly correlate with pre-VATS FEV1 % or FEV1/FVC. Although there was no significant correlation, it is still possible that alveolar α-SMA levels could correlate with other measures of asthma severity, such as symptom frequency, nighttime awakenings, or functional impairment, not accounted for in a single, static set of pulmonary function tests.
Within the alveolar parenchyma, there exists a population of non-smooth muscle, contractile α-SMA-expressing cells, namely myofibroblasts and pericytes. However, distinguishing these cells from alveolar septal myofibroblasts is difficult by histochemistry alone, as both cell types express α-SMA and neither expresses a truly cell-specific marker12. Pericytes play an important role in supporting the capillaries they are associated with34. As such, we postulated that if the observed loss of alveolar α-SMA in severe asthma was due to loss of pericytes, there would also be loss of the associated capillaries. To determine this, we performed ERG immunostaining on our control and severe asthma cases. Within the lung, ERG is a specific endothelial cell marker27. After manual exclusion of arteries, arterioles, and venules, ERG staining was not significantly different between the control and severe asthma groups, suggesting intact capillaries and capillary-associated pericytes. Considering this, it is likely myofibroblasts are the source of the loss of α-SMA expression observed in severe asthma, due to either reduced α-SMA expression or a decrease in myofibroblast number.
It is possible the loss of contractile, α-SMA-expressing myofibroblasts in severe asthma could be detrimental by decreasing alveolar wall tension and promoting expiratory airway collapse, similar to the mechanism seen in COPD. In fact, a number of studies have described loss of elastic recoil and small airway obstruction35–37 in asthma, processes thought to be further exacerbated in severe asthma38. Additionally, a study by Mauad et al. showed that alveolar elastic fibers, which myofibroblasts can produce, are diminished in fatal asthma, possibly promoting airway collapse39. It is also possible that the loss of α-SMA-expressing myofibroblasts represent an impaired repair response secondary to chronic alveolar hyperinflation and axial tension from occluded proximal airways, similar to the process of emphysema development described in COPD40. Additionally, studies have shown that gene expression profiles suggestive of inadequate epithelial repair and growth are associated with asthma severity and predict severity better than gene profiles suggestive of type-2 inflammation41,42. Inflammation is closely tied to the processes of airway remodeling in asthma, promoting structural and cellular changes43. Inflammatory mediators have complex and wide-ranging effects on the differentiation, contractility, and survival of myofibroblasts44 and, plausibly, could contribute to observed loss of α-SMA. However, within the alveolated parenchyma of the severe asthma selections, there were no obvious instances of inflammatory infiltrate, although this was not formally assessed. Ultimately, further work is needed to determine if α-SMA loss in severe asthma is associated with specific clinical or physiologic factors.
We also included a small sample of COPD cases as obstructive lung disease controls free from oral corticosteroid treatment, which all severe asthma patients in the study were receiving. Like the severe asthma samples, the COPD group showed a decrease in aveolated parenchyma α-SMA expression compared to the control group. This is in line with a study by Karvonen et al. which also showed decreased α-SMA expression in the alveolated parenchyma of COPD patients45. A comparable α-SMA expression reduction in both severe asthma and COPD, compared to controls, may suggest this decreased expression is a common factor of obstructive lung diseases and not merely an effect of oral corticosteroid use. Of note, the decrease in α-SMA expression in the COPD group failed to reach statistical significance; however, the true level of α-SMA expression may be overestimated, as the positive pixel algorithm counted the darkly pigmented smoker’s macrophages as falsely positive staining and there was no practical way to exclude this false positivity during the imaging analysis due to their large number and proximity to the alveolar septa. In addition, the very small sample size (n=4) limits statistical power.
To our knowledge, there are only two other studies that have investigated α-SMA expression in the alveolated parenchyma of patients with asthma. Interestingly, both studies showed results contradictory to ours, with increases in alveolar α-SMA expression and myofibroblast number in asthma19,31. However, there are many significant differences between our study and the two previous studies. Notably, the prior studies utilized tissue acquisition methods significantly different from our study. In the study by Boser et al., post-mortem tissue initially collected in the early-mid 1990s was used for analysis. The study from Weitoft et al. utilized tissue obtained by transbronchial biopsy. Importantly, both studies also targeted asthma populations different from the severe asthma population defined in our study. In the study by Boser et al., they examined a population of young patients who died of asthma, which is not synonymous with severe asthma. In the study from Weitoft et al., a population of patients with uncontrolled asthma defined by the GINA guidelines was studied. Uncontrolled asthma, unlike severe asthma as defined by the ERS/ATS criteria2, may include patients with residual symptoms due to modifiable factors like poor medication adherence, incorrect inhaler use technique, or comorbidities46. As such, a population of uncontrolled asthma patients likely represents less severe disease on average than a population made up solely of severe asthma patients. In addition, these studies did not utilize digital pathology techniques, and the study from Weitoft et al. utilized immunofluorescence microscopy and other markers, in addition to α-SMA, as surrogate markers of myofibroblasts numbers.
This study has a number of limitations. First, the mean age of the control group is lower than the severe asthma or COPD groups. As such, we cannot be certain age does not play a role in alveolar α-SMA expression. However, by linear regression, age was not significantly associated with α-SMA expression levels in the control, severe asthma, or COPD groups. Second, the sample size in all groups was small, limiting statistical power. Finally, while COPD cases were included as non-oral corticosteroid treated obstructive lung disease controls, we cannot completely rule out the possibility that regular oral corticosteroid use could affect alveolar α-SMA expression.
In summary, our study shows α-SMA expression, quantified by digital image analysis, is significantly reduced in the alveolated parenchyma of patients with severe asthma, compared to healthy controls. The COPD group shows a similar, although only trending, decrease in α-SMA expression, which may suggest the observed loss of α-SMA expression is a shared phenomenon of obstructive lung diseases. Lastly, the presence of intact capillary endothelial cells in severe asthma suggests the supporting pericytes are likely maintained as well. Given this, it may suggest myofibroblasts are the source of the decrease in α-SMA in severe asthma. Further work is needed to determine the possible physiologic and clinical ramifications of these findings.
Acknowledgments
This work was supported by the following sources of funding: NIH P01 AI106684-01A1 (Sally E. Wenzel) and the Dellenback Research Fund (Sally E. Wenzel).
References
- 1.National Asthma Education and Prevention Program, Third Expert Panel on the Diagnosis and Management of Asthma. Definition, Pathophysiology and Pathogenesis of Asthma, and Natural History of Asthma. In: Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. Bethesda, MD: National Heart, Lung, and Blood Institute; 2007:12. [Google Scholar]
- 2.Chung KF, Wenzel SE, Brozek JL, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343–373. doi: 10.1183/09031936.00202013 [DOI] [PubMed] [Google Scholar]
- 3.Braman SS. The global burden of asthma. Chest. 2006;130(1 Suppl):4S–12S. doi: 10.1378/chest.130.1_suppl.4S [DOI] [PubMed] [Google Scholar]
- 4.McFadden ER. Observations on asthma mortality. Ann Intern Med. 1997;127(2):142. doi: 10.7326/0003-4819-127-2-199707150-00009 [DOI] [PubMed] [Google Scholar]
- 5.Djukanović R, Roche WR, Wilson JW, et al. Mucosal inflammation in asthma. Am Rev Respir Dis. 1990;142(2):434–457. doi: 10.1164/ajrccm/142.2.434 [DOI] [PubMed] [Google Scholar]
- 6.Dunnill MS. The pathology of asthma, with special reference to changes in the bronchial mucosa. J Clin Pathol. 1960;13:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possa SS, Leick EA, Prado CM, Martins MA, Tibério IFLC. Eosinophilic inflammation in allergic asthma. Front Pharmacol. 2013;4:46. doi: 10.3389/fphar.2013.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James AL, Elliot JG, Jones RL, et al. Airway smooth muscle hypertrophy and hyperplasia in asthma. Am J Respir Crit Care Med. 2012;185(10):1058–1064. doi: 10.1164/rccm.201110-1849OC [DOI] [PubMed] [Google Scholar]
- 9.Benayoun L, Druilhe A, Dombret M-C, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167(10):1360–1368. doi: 10.1164/rccm.200209-1030OC [DOI] [PubMed] [Google Scholar]
- 10.Barron L, Gharib SA, Duffield JS. Lung pericytes and resident fibroblasts: busy multitaskers. Am J Pathol. 2016;186(10):2519–2531. doi: 10.1016/j.ajpath.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaengel K, Genové G, Armulik A, Betsholtz C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29(5):630–638. doi: 10.1161/ATVBAHA.107.161521 [DOI] [PubMed] [Google Scholar]
- 12.Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21(2):193–215. doi: 10.1016/j.devcel.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 13.Stark K, Eckart A, Haidari S, et al. Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and “instruct” them with pattern-recognition and motility programs. Nat Immunol. 2013;14(1):41–51. doi: 10.1038/ni.2477 [DOI] [PubMed] [Google Scholar]
- 14.Ayres-Sander CE, Lauridsen H, Maier CL, Sava P, Pober JS, Gonzalez AL. Transendothelial migration enables subsequent transmigration of neutrophils through underlying pericytes. PLoS One. 2013;8(3):e60025. doi: 10.1371/journal.pone.0060025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinz B, Phan SH, Thannickal VJ, et al. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180(4):1340–1355. doi: 10.1016/j.ajpath.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suki B, Stamenović D, Hubmayr R. Lung parenchymal mechanics. Compr Physiol. 2011;1(3):1317–1351. doi: 10.1002/cphy.c100033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balzar S, Wenzel SE, Chu HW. Transbronchial biopsy as a tool to evaluate small airways in asthma. Eur Respir J. 2002;20(2):254–259. doi: 10.1183/09031936.02.00261102 [DOI] [PubMed] [Google Scholar]
- 18.Wenzel SE, Szefler SJ, Leung DY, Sloan SI, Rex MD, Martin RJ. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156(3 Pt 1):737–743. doi: 10.1164/ajrccm.156.3.9610046 [DOI] [PubMed] [Google Scholar]
- 19.Boser SR, Mauad T, Araújo-Paulino BB de, et al. Myofibroblasts are increased in the lung parenchyma in asthma. PLoS One. 2017;12(8):e0182378. doi: 10.1371/journal.pone.0182378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doberer D, Trejo Bittar HE, Wenzel SE. Should lung biopsies be performed in patients with severe asthma? Eur Respir Rev. 2015;24(137):525–539. doi: 10.1183/16000617.0045-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahran N, Szewczyk-Bieda M, Vinnicombe S, Fleming S, Nabi G. Periprostatic fat adipokine expression is correlated with prostate cancer aggressiveness in men undergoing radical prostatectomy for clinically localized disease. BJU Int. 2019;123(6):985–994. doi: 10.1111/bju.14469 [DOI] [PubMed] [Google Scholar]
- 22.Moon A, Smith GH, Kong J, Rogers TE, Ellis CL, Farris ABB. Development of CD3 cell quantitation algorithms for renal allograft biopsy rejection assessment utilizing open source image analysis software. Virchows Arch. 2018;472(2):259–269. doi: 10.1007/s00428-017-2260-6 [DOI] [PubMed] [Google Scholar]
- 23.Klapczynski M, Gagne GD, Morgan SJ, et al. Computer-assisted imaging algorithms facilitate histomorphometric quantification of kidney damage in rodent renal failure models. J Pathol Inform. 2012;3:20. doi: 10.4103/2153-3539.95456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouzin C, Saini ML, Khaing K-K, et al. Digital pathology: elementary, rapid and reliable automated image analysis. Histopathology. 2016;68(6):888–896. doi: 10.1111/his.12867 [DOI] [PubMed] [Google Scholar]
- 25.Jakobsen MR, Teerapakpinyo C, Shuangshoti S, Keelawat S. Comparison between digital image analysis and visual assessment of immunohistochemical HER2 expression in breast cancer. Pathol Res Pract. 2018;214(12):2087–2092. doi: 10.1016/j.prp.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 26.Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. doi: 10.4103/jpi.jpi_82_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miettinen M, Wang Z-F, Paetau A, et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35(3):432–441. doi: 10.1097/PAS.0b013e318206b67b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentley JK, Hershenson MB. Airway smooth muscle growth in asthma: proliferation, hypertrophy, and migration. Proc Am Thorac Soc. 2008;5(1):89–96. doi: 10.1513/pats.200705-063VS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebina M, Takahashi T, Chiba T, Motomiya M. Cellular hypertrophy and hyperplasia of airway smooth muscles underlying bronchial asthma. A 3-D morphometric study. Am Rev Respir Dis. 1993;148(3):720–726. doi: 10.1164/ajrccm/148.3.720 [DOI] [PubMed] [Google Scholar]
- 30.Johnson PR, Roth M, Tamm M, et al. Airway smooth muscle cell proliferation is increased in asthma. Am J Respir Crit Care Med. 2001;164(3):474–477. doi: 10.1164/ajrccm.164.3.2010109 [DOI] [PubMed] [Google Scholar]
- 31.Weitoft M, Andersson C, Andersson-Sjöland A, et al. Controlled and uncontrolled asthma display distinct alveolar tissue matrix compositions. Respir Res. 2014;15:67. doi: 10.1186/1465-9921-15-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green FHY, Williams DJ, James A, McPhee LJ, Mitchell I, Mauad T. Increased myoepithelial cells of bronchial submucosal glands in fatal asthma. Thorax. 2010;65(1):32–38. doi: 10.1136/thx.2008.111435 [DOI] [PubMed] [Google Scholar]
- 33.Green FHY, Butt JC, James AL, Carroll NG. Abnormalities of the bronchial arteries in asthma. Chest. 2006;130(4):1025–1033. doi: 10.1378/chest.130.4.1025 [DOI] [PubMed] [Google Scholar]
- 34.Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97(6):512–523. doi: 10.1161/01.RES.0000182903.16652.d7 [DOI] [PubMed] [Google Scholar]
- 35.Woolcock AJ, Read J. The static elastic properties of the lungs in asthma. Am Rev Respir Dis. 1968;98(5):788–794. doi: 10.1164/arrd.1968.98.5.788 [DOI] [PubMed] [Google Scholar]
- 36.Gelb AF, Licuanan J, Shinar CM, Zamel N. Unsuspected loss of lung elastic recoil in chronic persistent asthma. Chest. 2002;121(3):715–721. doi: 10.1378/chest.121.3.715 [DOI] [PubMed] [Google Scholar]
- 37.McCarthy DS, Sigurdson M. Lung elastic recoil and reduced airflow in clinically stable asthma. Thorax. 1980;35(4):298–302. doi: 10.1136/thx.35.4.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carr TF, Altisheh R, Zitt M. Small airways disease and severe asthma. World Allergy Organiz J. 2017;10(1):20. doi: 10.1186/s40413-017-0153-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauad T, Silva LFF, Santos MA, et al. Abnormal alveolar attachments with decreased elastic fiber content in distal lung in fatal asthma. Am J Respir Crit Care Med. 2004;170(8):857–862. doi: 10.1164/rccm.200403-305OC [DOI] [PubMed] [Google Scholar]
- 40.Mitzner W Emphysema--a disease of small airways or lung parenchyma? N Engl J Med. 2011;365(17):1637–1639. doi: 10.1056/NEJMe1110635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modena BD, Tedrow JR, Milosevic J, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190(12):1363–1372. doi: 10.1164/rccm.201406-1099OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Modena BD, Bleecker ER, Busse WW, et al. Gene Expression Correlated with Severe Asthma Characteristics Reveals Heterogeneous Mechanisms of Severe Disease. Am J Respir Crit Care Med. 2017;195(11):1449–1463. doi: 10.1164/rccm.201607-1407OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergeron C, Al-Ramli W, Hamid Q. Remodeling in asthma. Proc Am Thorac Soc. 2009;6(3):301–305. doi: 10.1513/pats.200808-089RM [DOI] [PubMed] [Google Scholar]
- 44.van Caam A, Vonk M, van den Hoogen F, van Lent P, van der Kraan P. Unraveling ssc pathophysiology; the myofibroblast. Front Immunol. 2018;9:2452. doi: 10.3389/fimmu.2018.02452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karvonen HM, Lehtonen ST, Harju T, et al. Myofibroblast expression in airways and alveoli is affected by smoking and COPD. Respir Res. 2013;14:84. doi: 10.1186/1465-9921-14-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Global Initiative for Asthma. Diagnosis and Management of Difficult-to-treat and Severe Asthma. https://ginasthma.org/severeasthma/. Accessed October 20, 2019.