Abstract
Objective:
Vasomotor symptoms (VMS), hot flashes and night sweats, are cardinal symptoms of the menopausal transition. Little is known about genetic influences on VMS. This study evaluated whether previously-identified genetic factors predictive of VMS, age at menarche, and age at menopause were associated with VMS in a multi-racial/ethnic cohort.
Methods:
For 702 White, 306 Black, 126 Chinese, and 129 Japanese women from the Study of Women’s Health Across the Nation (SWAN) Genomic Substudy, we created polygenic risk scores (PRSs) from GWASs of VMS and ages at menarche and menopause. PRSs and single nucleotide polymorphisms (SNPs) from a previously-identified VMS locus (TACR3) were evaluated for associations with frequent VMS (VMS ≥6 days in past 2 weeks at any visit) and with VMS trajectories (persistently low, early onset, Final Menstrual Period (FMP) onset, persistently high).
Results:
The C-allele of rs74827081 in TACR3 was associated with reduced likelihood of frequent VMS in White women (OR=0.49 (95% CI, 0.29 to 0.83)). With higher menarche PRS (later menarche), Black women were less likely (OR=0.55 (95% CI, 0.38 to 0.78)) to report frequent VMS. With higher PRS for age at menarche, Black women were also less likely to have a persistently high VMS trajectory (OR=0.55 (95% CI, 0.34 to 0.91)) while White women (OR=0.75 (95% CI, 0.58 to 0.98)) were less likely to have an FMP onset trajectory (vs. persistently low). Chinese women with higher menopause PRS were more likely to have frequent VMS (OR=2.29 (95% CI, 1.39 to 3.78)). Associations were substantively similar after excluding rs74827081 C-allele carriers.
Conclusions:
Genetic factors predictive of reproductive aging are also associated with VMS, suggesting that VMS have a polygenic architecture. Further study in this area may help to identify new targets for novel VMS therapies.
Keywords: Menopause, Menarche, Vasomotor Symptoms, Hot Flashes, Genetic Variation, Polygenic Risk Score
INTRODUCTION
Vasomotor symptoms (VMS), or hot flashes and night sweats, are cardinal symptoms of the menopausal transition1-3 experienced by the majority of midlife women.1 Across women, VMS vary considerably in their frequency, timing, and persistence. There are pronounced racial/ethnic differences in the prevalence of VMS in the United States, with African American women having the greatest burden of VMS relative to White women, and Chinese and Japanese women being the least affected.1 In addition, recent research has indicated that VMS follow distinct trajectories over the menopause transition, with some women having their VMS relatively early in the transition, primarily when they are premenopausal, others later in the transition, and still others experiencing VMS persistently for well over a decade.4, 5 The determinants of VMS and their trajectories are not fully understood.
GWAS of symptoms characteristic of reproductive aging, such as VMS, are limited.6 Early candidate gene studies have suggested that VMS are associated with single nucleotide polymorphisms (SNPs) in sex steroid-metabolizing enzymes and receptors.7-11 To our knowledge, only one previous GWAS has focused on genetic variation in relation to VMS.6 That study analyzed data from 17,695 postmenopausal women aged 50 to 79 years participating in the Women’s Health Initiative study. After adjustment for covariates and population structure, meta-analysis of the multi-ethnic cohort revealed 14 single-nucleotide polymorphisms (SNPs) associated (p value <5x10−8) with ever experiencing VMS (vs. never having VMS). All 14 SNPs were located on chromosome 4 in the tachykinin receptor 3 (TACR3 locus). TACR is the receptor for neurokinin B. Previous animal and human studies have demonstrated an association between neurokinin B and VMS.12-15 Although only one genomic region reached genome-wide significance in this study, there may be other SNPs with smaller effects on VMS that did not attain the stringent genome-wide significance level. It remains to be determined if these sub-threshold SNPs also influence VMS.
GWAS studies have identifyed several candidate genes for reproductive timing, including age at menarche and age of menopause. 16-22 GWAS of age at menarche and menopause have been conducted among women of European ancestry with samples of over 370,000 and 70,000 women, respectively. In their GWAS meta analysis of age at menopause, Day and colleagues22 identified 54 independent SNPs in 44 genomic loci that explained approximately 6% of the variance in age at natural menopause, rising to 21% of the variance when using independent SNPs with at least nominal significance (p<0.05). These loci included genes related to DNA damage response, primary ovarian insufficiency, delayed puberty, and reproductive hormone synthesis. Cross-trait analysis suggested modest pleiotropy between age at natural menopause and adult body mass index (BMI). Age at menarche is highly polygenic, as a total of 389 independent genomic loci have been identified to date.23 These loci in aggregate explained about 7% of the trait variance in independent samples, with the set of identified genes most highly expressed in central nervous system tissues including the hypothalamus and pituitary gland. Cross-trait analysis suggested that approximately 10% of the identified genomic loci overlapped with loci for adult BMI. Body mass index in turn has been associated with frequency and severity of hot flashes, although the relationship is complex and dependent on stage of reproductive aging.2, 24
We have recently demonstrated in a multi-ethnic sample from the Study of Women’s Health Across the Nation (SWAN) that polygenic risk scores (PRSs) for menarche and menopause derived from these GWAS meta-analyses are associated with other characteristics of the menopausal transition including length of the reproductive lifespan, duration of the menopausal transition, and estradiol levels prior to and after the menopause in at least one race/ethnic group.25 Notably, lower estrogen levels have been associated with more prevalent and frequent VMS,26-29 a recent pooled analysis of six cohorts reported an association between early age at menarche and increased risk of frequent VMS,30 and some limited research suggests a potential relationship between the timing of menopause and VMS.31-34 Despite evidence of overlap in genes associated with menarche, menopause and risk factors for VMS, these two PRSs have not been investigated in relation to VMS.
Replication of previously-identified genetic loci associated with VMS is warranted as is consideration of whether VMS, menarche, or menopause PRSs are associated with risk of VMS. In this study in SWAN, we first replicated associations reported by Crandall et al. in the WHI study.6 Next we constructed standardized PRS using the genome-wide suggestive loci (p<1x10−4) for VMS,6 age at menarche,23 and age at natural menopause22 in SWAN and assessed the association between these PRSs and VMS.
MATERIALS AND METHODS
The Study of Women’s Health Across the Nation (SWAN).
SWAN, a multi-racial/ethnic cohort study of 3302 women, has prospectively observed the natural history of reproductive aging as women transitioned from pre-menopause through the post-menopause and ascertained whether and how often hot flashes and night sweats had been experienced at each study visit. SWAN enrolled 935 Black, 250 Chinese, 286 Hispanic, 281 Japanese, and 1,550 White women across seven sites in 1996/1997. Details of the recruitment and enrollment have been described elsewhere.35 Eligibility for the cohort included, being age 42-52 years of age, having an intact uterus, not being pregnant/lactating, having had a menstrual period and no use of reproductive hormones within the past 3 months, self-identification as White (all sites) or as Black (southeastern Michigan, Boston, Chicago, or Pittsburgh sites), Chinese (Northern California site), Japanese (Southern California site) or Hispanic (New Jersey site) and residence in the geographic area of the respective site. The study protocol was approved by the Institutional Review Boards at each study site and participants provided written, informed consent at each study visit. Women have been followed approximately annually for 15 follow-up visits and the study remains in contact with 75% of surviving participants.
At each visit women completed an interviewer-administered and self-administered questionnaires on a broad range of topics, including menstrual characteristics and current use of hormone therapy (HT) for menopausal symptoms, and symptom experience. Socio-demographic characteristics and health behaviors including education, smoking, alcohol intake, physical activity and history of contraceptive use were assessed at the baseline visit. Physical assessments at baseline included measurement of height and weight.
Vasomotor Symptoms
Women were asked at each study visit how frequently they had experienced VMS (hot flashes/flushes and/or night sweats) in the past 2 weeks with response categories of not at all, 1-5 days, 6-8 days, 9-13 days, or every day. Two binary variables were created for each annual visit indicating whether a woman reported any VMS (≥1 day versus not at all) and frequent VMS (≥6 days versus <6 days/not at all) in the past two weeks. SWAN routinely categorizes VMS in this manner as frequent VMS were judged to be clinically meaningful in prior analyses and frequent VMS have been associated with indicators of health risk.1, 5, 36-38 sing these data we created two summary variables indicating whether a woman ever reported any VMS (yes/no) and whether she ever reported frequent VMS (yes/no) at any study visit. Given that 93% of women reported ever having any VMS, we examine only frequent VMS in analyses.
In addition, women who had a natural menopause were categorized by their patterns of reporting VMS across the menopausal transition4, 39-41 using group-based trajectory models (SAS PROC TRAJ). Any VMS and frequent VMS were modeled separately as a function of time before and after the FMP using a logit distribution. To be included in trajectory analyses, women were required to contribute at least 2 observations. Observations when women reported using hormone therapy (HT) were excluded from the trajectory analysis. To select the number of groups, we used a combination of Bayesian Information Criteria (BIC), group size (≥ 5%), average posterior probability (AvePP) for each group (≥ 0.7) and odds of correct classification (OCC) (≥ 5) as well as clinical meaningfulness based on visual inspection of modeled trajectories. We evaluated models with 1 to 6 trajectory groups (all quartic trajectories). To determine the shape of each group trajectory, we eliminated highest-order terms one by one until all highest-order terms were significant (p<0.05). Finally, we selected the 4 trajectory groups model for both any VMS, including 3 quartic and 1 quadratic trajectory (AvePP range 0.81 to 0.90, OCC 12.19 to 23.90) (Figure 1a), and for frequent VMS, including 1 quartic, 2 cubic, and 1 quadratic trajectory (AvePP range 0.82 to 0.89, OCC 8.54 to 96.30) (Figure 1b). Women were assigned to one of the four trajectory groups for any VMS and for frequent VMS based on their highest posterior probability. The four distinct trajectories identified from modelling any VMS and from modelling frequent VMS were similar including three patterns with high probability of VMS and one of low probability of VMS, specifically: a) early onset of VMS, about one decade prior to the FMP, that declines soon after the FMP (Early onset); b) onset of VMS proximate to the FMP that declines more slowly after the FMP (FMP onset); c) early onset of VMS that does not decline (High); and d) persistently low frequency of VMS (Low).
Figure 1. The four trajectories based on any VMS (a) or high frequency VMS (b) in SWAN.
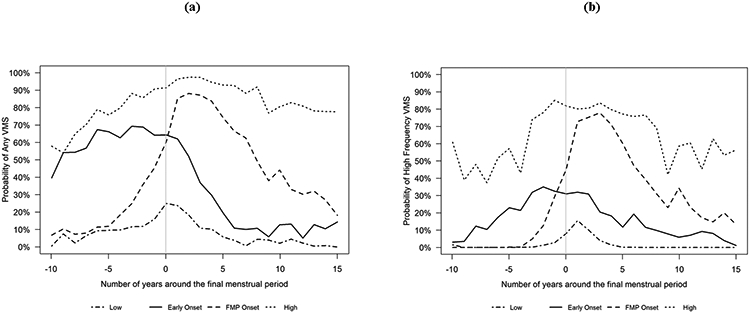
VMS, vasomotor symptoms; SWAN, Study of Women’s Health Across the Nation; FMP, final menstrual period.
Age at menarche and menopause
Age at menarche was based on self-reported age when the participant had their first menstrual period. Menopausal status was based on self-report of bleeding patterns, including the date of their last menstrual period, obtained at each annual study visit. Age at menopause was defined as age at FMP, with the FMP defined retrospectively after 12 months of amenorrhea. Surgical menopause was defined by report of either hysterectomy or bilateral oophorectomy prior the FMP. The FMP could not be determined in women using menopausal hormone therapy (HT) who had no subsequent untreated menstrual bleed. Women who had surgical menopause or whose FMP could not be determined because of hormone use were excluded from VMS trajectory analyses.
Covariates.
Covariates assessed at the baseline interview included smoking history (current vs. former/never), education (high school graduate/GED or less, more than high school, college and the above), and alcohol intake (none, <2 servings per week (moderate), or >2 servings per week (heavy)). Physical activity was assessed with a modified Kaiser Physical Activity Survey,42 adapted from the Baecke physical activity questionnaire,43 and consisting of subscales on sports and exercise, non-sports leisure, and household and childcare activity. The three subscales were summed to create a total physical activity score, ranging from 3 to 15. Body mass index (BMI) was calculated as weight in kilograms divided by the square of height in meters. Information on surgeries and hormone use was obtained at each study visit and we created indicator variables as ever used HT during study period (yes/no).
The SWAN Genomic Substudy.
A genomic substudy was undertaken at follow-up visits 5 and 6 at all sites except the New Jersey site. After women provided informed consent to participate in the SWAN Genomic Substudy, whole blood was collected, DNA was extracted and B-lymphocyte cells were immortalized. Of the 1,988 women completing a clinic visit at one of these two follow-up visits, 1,757 (88.3%) provided a whole blood sample. Immortalized cell lines were developed successfully for 1,588 (90.4%) with 1,536 (96.7%) processed into distributable diluted, extracted DNA.
Samples were genotyped on the Illumina (San Diego, CA) Multi-Ethnic Global Array (MEGA A1), and GenomeStudio (2011.1) was used to call the genotypes. Quality control procedures are described in Zhao et al.25 Briefly, samples with call rate <95% or unresolved identity issues were removed. SNPs with call rate <98%, Hardy Weinberg equilibrium p-value<10−4, more than 1 Mendelian error, or more than 0 discordant calls in duplicated samples were removed. The X chormosomes of two samples were removed due to chromosome anomalies. After QC, genotype data for 1,462 participants remained and were imputed to the 1000 Genome Phase 3 v5 reference panel using the Michigan Imputation server.44 SWAN genetic data are available from dbGaP (accession number: phs001470.v1.p1). The King-robust method45 coupled with principal component analysis (PCA) was used to select four sets of unrelated homogenous analysis samples: White (N=740), Black (N=368), Chinese (n=139) and Japanese (N=146). After accounting for missing phenotype data, the analytical sample included 702 White women, 306 Black women, 126 Chinese women, and 129 Japanese women.
VMS SNPs and Polygenic Risk Score (PRS).
The VMS GWAS using data from the WHI6 identified 14 SNPs (all from TARC3) from trans-ethnic meta-analysis and an additional 3 SNPs from African Americans alone. We obtained data for these SNPs in SWAN, and also contructed an “all SNPs” PRS for VMS using beta estimates from trans-ethnic meta-analysis. We used a similar procedure as described in Zhao et al.25 The PRS includes all independent SNPs with p-value<1x10−4. Clumping was used to select independent SNPs in each ethnic group using the 1000 Genomes Reference Panel (phase 3 v5a) (503 European (EUR) for Whites, 661 African (AFR) for Black, 504 East Asian (EAS) for Chinese and Japense women). We performed clumping in PLINK 1.9 using 0.5 as the LD r2 threshold and a distance of 250kb. 46, 47 After clumping, the total number of SNPs included in the “all SNPs” PRS for VMS was 493, 465, 537 and 537 for White, Black, Chinese and Japanese, respectively.
Polygenic Risk Scores (PRS) for ages at menarche and menopause.
We contructed PRSs for ages at menarche and menopause using effect estimates from the largest GWAS meta-analyses to date.22, 23 As described in Zhao et al.25, two versions of the PRSs were contructed for each trait. The “all SNPs” PRS included all independent SNPs with p-value<1x10−4 and the “top SNPs” PRS included all independent genome-wide significant SNPs reported by the authors. In general, the “all SNPs” and “top SNPs” versions of the corresponding PRSs were positively correlated with each other (r ranged from 0.52 to 0.69 for menarche PRSs and from 0.50 to 0.68 for menopause PRSs across race/ethnic groups). Menarche and menopause PRSs were not highly correlated with each other. Four outliers were identified in “all SNPs” menarche PRS and removed from analyses. Prior to the analyses, each PRS was standardized (mean=0, sd=1) within each race/ethnic group. We were primarily interested in the “all SNPs” PRS, but also examined the “top SNPs” PRS in secondary analyses.
Analysis.
Association between genetic risk factors and VMS.
For each genome-wide significant VMS risk SNP from the prior GWAS (6), VMS PRS, menarche PRS or menopause PRS, we constructed logistic regression models with each genetic factor as the predictor and frequent VMS (yes/no) as the outcome. Analyses were performed separately for each racial/ethnic group with the exception that the 3 SNPs identified only in the African ancestry population in the prior VMS GWAS were only tested in the Black participants. Covariates included the first five genetic principal components (PCs), and study site for analyses using White and Black women. We additionally adjusted for baseline age, BMI, smoking, education, physical activity score, alcohol intake (none, <2 servings per week, or >2 servings per week), ever taking HT during the study period, and bilateral oophorectomy before FMP. Because this is the first analysis exploring these hypotheses with traits that are recognized to be correlated, we chose 0.05 as our significance level (2-sided test) without any adjustment for multiple testing correction.
Association between genetic risk factors and VMS trajectories.
Given the small sample size of Chinese and Japanese women, association analyses between VMS trajectories and genetic risk factors were performed in the samples of White women and Black women only. For each SNP identified in the VMS GWAS6 that was associated with VMS in SWAN, we constructed multiple independent logistic regression models to assess the odds of being in each VMS trajectory group compared to the “low” VMS trajectory group. For the PRSs, we constructed multinomial logistic regression models with the PRS as the predictor and VMS trajectories as the outcome using the “low” VMS trajectory as the reference group. We adjusted for the same set of covariates listed above (except bilateral oophorectomy because trajectory analysis required an observed FMP). The primary analysis focused on any VMS trajectories and secondary analysis analyzed frequent VMS trajectories.
Sensitivity analysis.
As carrying one or more VMS-associated alleles identified in the prior GWAS6 might mask the effect of the PRSs on risk of VMS, we also assessed the associations between each PRS and both frequent VMS and VMS trajectories for the subgroup of participants who did not carry any copies of the rare protective alleles at the sentinel SNP identified in the prior VMS GWAS.
RESULTS
Table 1 describes characteristics of the study population. Median age at baseline across the race/ethnicity groups ranged from 46.2 to 47.0 years. Median BMI was 30.3, 25.6, 22.5 and 22.1 kg/m2 in Black, White, Japanese and Chinese women respectively. The proportion of women who ever used HT during follow-up ranged from 33.3% of Chinese women to 52.7% of White women, with 6.1% to 25.0% reporting HT use at any given visit.
Table 1.
Descriptive statistics of the study participants from the Study of Women’s Health Across the Nation (SWAN)
| Variable | White (N=702) | Black (N=306) | Chinese (N=126) | Japanese (N=129) | ||||
|---|---|---|---|---|---|---|---|---|
| N, Median (IQR) | N, Median (IQR) | N, Median (IQR) | N, Median (IQR) | |||||
| Age (year) | 702 | 46.30 (4.10) | 306 | 46.20 (4.20) | 126 | 46.35 (3.65) | 129 | 47.00 (3.50) |
| BMI (kg/m2) | 702 | 25.60 (8.10) | 306 | 30.30 (9.53) | 126 | 22.10 (4.15) | 129 | 22.50 (4.20) |
| Physical activity score | 702 | 8.20 (2.44) | 306 | 7.43 (2.30) | 126 | 7.35 (2.49) | 129 | 7.75 (1.85) |
| VMS polygenic risk score (“all SNPs”)a | 702 | 1.52 (0.01) | 306 | 1.47 (0.02) | 126 | 1.48 (0.01) | 129 | 1.48 (0.01) |
| Menarche polygenic risk score (“all SNPs”)b | 697 | 1.19 (0.01) | 306 | 1.05 (0.01) | 126 | 0.98 (0.01) | 129 | 0.97 (0.01) |
| Menopause polygenic risk score (“all SNPs”)c | 702 | 1.02 (0.06) | 306 | 1.03 (0.05) | 126 | 1.20 (0.03) | 129 | 1.21 (0.03) |
| Menarche polygenic risk score (“top SNPs”)d | 702 | 0.91 (0.04) | 306 | 0.91 (0.04) | 126 | 0.90 (0.04) | 129 | 0.89 (0.04) |
| Menopause polygenic risk score (“top SNPs”)e | 702 | 0.89 (0.11) | 306 | 0.88 (0.10) | 126 | 0.98 (0.09) | 129 | 1.00 (0.09) |
| N (%) | N (%) | N (%) | N (%) | |||||
| Ever reported hot flashes or night sweats occurring ≥6 days over past two weeks, N=1263 | ||||||||
| Yes | 496 (70.66) | 243 (79.41) | 65 (51.59) | 68 (52.71) | ||||
| No | 206 (29.34) | 63 (20.59) | 61 (48.41) | 61 (47.29) | ||||
| Trajectories for any hot flashes or night sweats, N=826 | ||||||||
| Low | 105 (24.31) | 32 (15.61) | 38 (39.18) | 36 (39.13) | ||||
| Early onset | 82 (18.98) | 25 (12.20) | 11 (11.34) | 16 (17.39) | ||||
| FMP onset | 153 (35.42) | 52 (25.37) | 33 (34.02) | 26 (28.26) | ||||
| High | 92 (21.30) | 96 (46.83) | 15 (15.46) | 14 (15.22) | ||||
| Trajectories for high frequency (≥6 days) hot flashes or night sweats, N=826 | ||||||||
| Low | 226 (52.31) | 81 (39.51) | 63 (64.95) | 68 (73.90) | ||||
| Early onset | 83 (19.21) | 46 (22.44) | 15 (15.46) | 6 (6.50) | ||||
| FMP onset | 95 (22.00) | 53 (25.85) | 19 (19.59) | 13 (14.10) | ||||
| High | 28 (6.48) | 25 (12.20) | 0 (0.00) | 5 (5.40) | ||||
| Alcohol intake daily | ||||||||
| None, N (%) | 273 (38.89) | 179 (58.50) | 98 (77.78) | 71 (55.04) | ||||
| Moderate, N (%) | 72 (10.26) | 20 (6.54) | 8 (6.35) | 13 (10.08) | ||||
| Heavy, N (%) | 357 (50.85) | 107 (34.97) | 20 (15.87) | 45 (34.88) | ||||
| Education | ||||||||
| High school degree or less, N (%) | 93 (13.25) | 93 (30.39) | 33 (26.19) | 18 (13.95) | ||||
| Some college, N (%) | 210 (29.91) | 123 (40.20) | 24 (19.05) | 48 (37.21) | ||||
| College degree or more, N (%) | 399 (56.84) | 90 (29.41) | 69 (54.76) | 63 (48.84) | ||||
| Ever used hormones during study, N (%) | 370 (52.71) | 120 (39.22) | 42 (33.33) | 59 (45.74) | ||||
| Ever had bilateral oophorectomy, N (%) | 37 (5.27) | 22 (7.19) | 4 (3.17) | 6 (4.65) | ||||
| Current smoker, N (%) | 87 (12.39) | 73 (23.86) | 2 (1.59) | 14 (10.85) | ||||
VMS “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (6). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menarche “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (23). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menopause “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (22). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menarche “top SNPs” PRS was constructed using the 389 independent genome-wide significant SNPs (339 SNPs available in SWAN) that were previously reported for age at menarche (23).
Menopause “top SNPs” PRS was constructed using the 54 independent genome-wide significant SNPs (51 SNPs available in SWAN) that were previously reported for age at menopause (22).
BMI, body mass index; VMS, vasomotor symptoms; FMP, final menstrual period; IQR, interquartile range.
Frequent VMS were ever reported by 70.7% of White, 79.4% of Black 51.6% of Chinese and 52.7% of Japanese participants. For the trajectories based on any VMS, White participants were categorized most often as “FMP onset” (35.4%) followed by “low” (24.3%), “high” (21.3%) and “early onset” (20%). Black women were most likely to be categorized as “high” (46.8%) and “FMP onset” (25.4%) and less likely to be categorized as “low” (15.6%) or “early onset” (12.2%). Chinese and Japanese women had similar distributions with larger proportions categorized as “low” and “FMP onset” trajectories and smaller proportions categorized as “early onset” or “high” VMS. The VMS trajectory distributions for frequent VMS were similar across racial/ethnic groups.
Associations between previously-identified VMS SNPs and ever reporting frequent VMS.
Nine out of the 14 SNPs in TARC3 identified in the VMS GWAS6 were significantly associated with frequent VMS in White women (Table 2), and are in high linkage disequilibrium with one another. None of these 14 SNPs nor the 3 SNPs unique to the African Ancestry population were significant in Black women. All of the 14 SNPs were either monomorphic or very rare in Chinese or Japanese women, thus associations could not be evaluated in these racial/ethnic groups.
Table 2.
Association between previously-identified VMS SNPs and ever reporting high frequency VMS (yes/no) in SWAN
| SNP | CHR:POS | GWAS P-valuea |
Gene | Non- effect |
Effect | White (N=702) |
Black (N=306) |
Chinese (N=129) |
Japanese (N=129) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | OR | Pvalue | EAF | OR | P value |
EAF | OR | Pvalue | EAF | OR | P value |
||||||
| rs74827081 | 4:104556732 | 4.77E-15 | TACR3 | G | C | 0.05 | 0.49 | 0.0086 | 0.01 | 21.29 | 0.3484 | 0 | NA | NA | 0 | NA | NA |
| rs74589515 | 4:104584258 | 7.11E-15 | TACR3 | T | G | 0.05 | 0.48 | 0.0079 | 0.01 | 42.45 | 0.3957 | 0 | NA | NA | 0 | NA | NA |
| rs79246187 | 4:104580809 | 2.64E-14 | TACR3 | C | T | 0.05 | 0.48 | 0.0075 | 0.02 | 2.26 | 0.4688 | 0.0003 | NA | NA | 0.0008 | NA | NA |
| rs112390256 | 4:104575473 | 2.81E-14 | TACR3 | G | A | 0.05 | 0.48 | 0.0072 | 0.04 | 1.58 | 0.4817 | 0 | NA | NA | 0 | NA | NA |
| rs75544266 | 4:104584997 | 3.15E-14 | TACR3 | C | T | 0.05 | 0.48 | 0.0083 | 0.02 | 2.16 | 0.4943 | 0.0002 | NA | NA | 0.0001 | NA | NA |
| rs78154848 | 4:104562840 | 5.65E-14 | TACR3 | T | C | 0.05 | 0.49 | 0.0076 | 0.02 | 2.29 | 0.4554 | 0 | NA | NA | 0 | NA | NA |
| rs76643670 | 4:104562842 | 5.65E-14 | TACR3 | T | A | 0.05 | 0.49 | 0.0076 | 0.02 | 2.29 | 0.4554 | 0 | NA | NA | 0 | NA | NA |
| rs78289784 | 4:104580155 | 6.55E-14 | TACR3 | C | A | 0.05 | 0.48 | 0.0076 | 0.02 | 2.25 | 0.4699 | 0 | NA | NA | 0 | NA | NA |
| rs77322567 | 4:104569676 | 2.17E-12 | TACR3 | C | A | 0.05 | 0.48 | 0.0072 | 0.04 | 1.68 | 0.4189 | 0 | NA | NA | 0 | NA | NA |
| rs78141901 | 4:104593977 | 7.27E-10 | TACR3 | C | A | 0.05 | 0.83 | 0.5060 | 0.01 | 0.66 | 0.7603 | 0 | NA | NA | 0 | NA | NA |
| rs78844131 | 4:104600029 | 3.34E-09 | TACR3 | T | G | 0.05 | 0.82 | 0.4772 | 0.01 | 0.63 | 0.7309 | 0 | NA | NA | 0 | NA | NA |
| rs79852843 | 4:104628587 | 4.38E-09 | TACR3 | T | C | 0.05 | 0.79 | 0.4081 | 0.01 | 0.61 | 0.7127 | 0 | NA | NA | 0 | NA | NA |
| rs80328778 | 4:104612447 | 4.97E-09 | TACR3 | C | T | 0.05 | 0.80 | 0.4336 | 0.01 | 0.62 | 0.7230 | 0 | NA | NA | 0 | NA | NA |
| rs112623956 | 4:104623714 | 6.19E-09 | TACR3 | A | G | 0.05 | 0.79 | 0.4126 | 0.01 | 0.61 | 0.7131 | 0 | NA | NA | 0 | NA | NA |
| rs75699757a | 11:56818268 | 1.60E-09 | OR5BP1P | A | G | 0.03 | 0.91 | 0.8923 | |||||||||
| rs11518608a | 11:56767184 | 3.47E-08 | NA | G | C | 0.03 | 0.82 | 0.7730 | |||||||||
| rs148680409a | 3:173330544 | 3.69E-08 | NLGN1 | G | A | 0.01 | 0.11 | 0.1446 | |||||||||
GWAS p values are from the African American SHARe GWAS (6).
VMS: vasomotor symptoms; SNP, single nucleotide polymorphism; SWAN: Study of Women’s Health Across the Nation; EAF: Effect allele frequency; OR, odds ratio; NA, not applicable.
Association between polygenic risk scores (PRSs) and ever reporting frequent VMS.
The associations between the VMS, menarche and menopause “all SNPs” PRSs and frequent VMS (yes/no) are presented in Table 3. The VMS PRS was not significantly associcated with frequent VMS in any race/ethnic group. For the menarche PRS, we observed a negative association between PRS and frequent VMS in Black women (OR=0.58, p=0.001). This finding suggests that Black women with genetic predisposition toward having a later age at menarche may be less likely to have frequent VMS. For the menopause PRSs, we observed a significant positive association with frequent VMS in Chinese women only (OR=2.29, p=0.001). This finding suggests that Chinese women with genetic predisposition toward having a later age at menopause were more likely to report frequent VMS. For the “top SNPs” PRSs, only the association between menopause PRS and frequent VMS in Chinese women was significant (Supplemental Digital Content 1).
Table 3.
Association between menarche/menopause “all SNPs” PRSs and ever reporting high frequency VMS (yes/no) in SWAN
| Race | PRS | N | OR | LL | UL | P value |
|---|---|---|---|---|---|---|
| White | VMS PRS (“all SNPs”)a | 702 | 1.15 | 0.97 | 1.35 | 0.114 |
| Menarche PRS (“all SNPs”)b | 697 | 0.97 | 0.82 | 1.15 | 0.727 | |
| Menopause PRS (“all SNPs”)c | 702 | 0.86 | 0.72 | 1.03 | 0.093 | |
| Black | VMS PRS (“all SNPs”)a | 306 | 0.95 | 0.70 | 1.31 | 0.772 |
| Menarche PRS (“all SNPs”)b | 306 | 0.55 | 0.38 | 0.78 | 0.001 | |
| Menopause PRS (“all SNPs”)c | 306 | 0.98 | 0.72 | 1.34 | 0.915 | |
| Chinese | VMS PRS (“all SNPs”)a | 126 | 1.37 | 0.88 | 2.14 | 0.163 |
| Menarche PRS (“all SNPs”)b | 126 | 1.41 | 0.92 | 2.17 | 0.117 | |
| Menopause PRS (“all SNPs”)c | 126 | 2.29 | 1.39 | 3.78 | 0.001 | |
| Japanese | VMS PRS (“all SNPs”)a | 129 | 0.86 | 0.56 | 1.31 | 0.470 |
| Menarche PRS (“all SNPs”)b | 129 | 0.85 | 0.56 | 1.29 | 0.445 | |
| Menopause PRS (“all SNPs”)c | 129 | 1.01 | 0.66 | 1.53 | 0.977 |
All PRSs were standardized (mean=0, SD =1) within each race/ethnic group prior to analyses
VMS “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (6). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menarche “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (23). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menopause “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (22). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
VMS, vasomotor symptoms; PRS, polygenic risk score; SWAN: Study of Women’s Health Across the Nation; SNP, single nucleotide polymorphism; OR, odds ratio; LL, lower limit of the 95% confidence interval; UL, upper limit of the 95% confidence interval.
Association between genetic risk factors and VMS trajectories.
The association between the TARC3 SNP rs74827081, the “all SNPs” PRSs, and trajectories of any VMS are presented in Table 4. Since all the TARC3 SNPs that were associated with frequent VMS in SWAN are in linkage disequilibrium and represent one signal, we only analyzed the most significant SNP from the VMS GWAS (rs74827081) in this analysis. Neither the VMS risk SNP nor the VMS PRS was significantly associated with VMS trajectories. White women who had higher PRS for age at menarche were less likely to be in the “FMP onset” trajectory group (OR=0.75, p=0.03) than the “low” trajectory group. Black women who had higher PRS for age at menarche were less likely to be in the “high” trajectory group (OR=0.55, p=0.02) relative to the “low” trajectory group. When we estimated trajectories using frequent VMS (Supplemental Digital Content 2), white women who had a higher PRS for age at menopause were less likely to be in the “FMP onset” VMS trajectory group (OR=0.69, p=0.01) relative to the “low” VMS group. Also this latter association was the only significant association in the trajectory analysis using the “top SNPs” PRS (Supplemental Digital Content 3).
Table 4.
Association between genetic risk factors and trajectories for any VMS in SWAN
| Outcome | Predictor | White | Black | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | OR | LL | UL | P value | N | OR | LL | UL | P value | ||
| Early onset vs. low | rs74827081a | 187 | 0.57 | 0.20 | 1.65 | 0.30 | 57 | NA | NA | NA | NA |
| FMP onset vs. low | 258 | 0.48 | 0.20 | 1.13 | 0.09 | 84 | NA | NA | NA | NA | |
| High vs. low | 197 | 0.55 | 0.20 | 1.51 | 0.25 | 128 | NA | NA | NA | NA | |
| Early onset vs. low | VMS PRS (“all SNPs”)b,c | 432 | 1.21 | 0.89 | 1.65 | 0.23 | 205 | 0.79 | 0.44 | 1.44 | 0.44 |
| FMP onset vs. low | 0.96 | 0.73 | 1.25 | 0.74 | 0.79 | 0.46 | 1.36 | 0.39 | |||
| High vs. low | 1.13 | 0.83 | 1.53 | 0.44 | 0.75 | 0.45 | 1.22 | 0.24 | |||
| Early onset vs. low | Menarche PRS (“all SNPs”)b,d | 428 | 0.83 | 0.60 | 1.14 | 0.24 | 205 | 1.10 | 0.59 | 2.06 | 0.75 |
| FMP onset vs. low | 0.75 | 0.58 | 0.98 | 0.03 | 0.74 | 0.43 | 1.30 | 0.30 | |||
| High vs. low | 0.77 | 0.57 | 1.05 | 0.10 | 0.55 | 0.34 | 0.91 | 0.02 | |||
| Early onset vs. low | Menopause PRS (“all SNPs”)b,e | 432 | 1.18 | 0.86 | 1.61 | 0.31 | 205 | 0.93 | 0.51 | 1.72 | 0.83 |
| FMP onset vs. low | 1.00 | 0.76 | 1.31 | 0.99 | 0.72 | 0.42 | 1.22 | 0.22 | |||
| High vs. low | 1.04 | 0.77 | 1.42 | 0.79 | 0.77 | 0.48 | 1.25 | 0.29 | |||
All PRSs were standardized (mean=0, SD =1) within each race/ethnic group prior to analyses
Multiple independent logistic regression models were constructed to assess the odds of being in each VMS trajectory group compared to the “low” VMS trajectory group. When a category had a minor allele count < 5, logistic regression was not conducted and “NA” was reported.
Multinomial logistic regression model was constructed with the PRS as the predictor and VMS trajectories as the outcome using the “low” VMS trajectory as the reference group.
VMS “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (6). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menarche “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (23). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
Menopause “all SNPs” PRS was constructed using all available independent SNPs that were significant at p<10−4 in GWAS meta-analysis (22). Independent SNPs were selected using a clumping approach (LD r2 threshold = 0.5, distance from index SNP = 250kb).
VMS, vasomotor symptoms; PRS, polygenic risk score; SWAN: Study of Women’s Health Across the Nation; SNP, single nucleotide polymorphism; FMP, final menstrual cycle; OR, odds ratio; LL, lower limit of the 95% confidence interval; UL, upper limit of the 95% confidence interval; NA, not applicable.
Sensitivity analysis.
When we excluded participants who carried 1 or 2 copies of the minor allele (C) at rs74827081, which is protective for reporting VMS, results were very similar to results of analyses that included these women with a negative association between the age at menarche PRS and frequent VMS in Black women and associations between the PRSs and VMS trajectories in White and Black women (Supplemental Digital Content 4 and 5), suggesting these findings were independent of TACR3.
DISCUSSION
This study focused on the association between VMS and genetic risk factors, singly and in PRSs, in a multi-racial/ethnic cohort of women studied across the menopause transition. To our knowledge, this is the first study to replicate the findings of the association between SNPs in TARC3 and VMS reported recently in Crandall et al. in the WHI.6 The association was replicated in 9 of 14 SNPs in the White women in SWAN. While none of the SNPs were replicated in the Black women, the sample size was not large (N=306) and the minor alleles were all low frequency (≤ 0.04).
One important difference between the Crandall et al. GWAS in the WHI and our study is the different definitions of VMS. Specifically, the Crandall et al. study employed retrospective report of VMS often many years after the menopause, whereas SWAN employed repeated prospective reporting of VMS. With this prospective approach in SWAN, over 90% of women reported ever having any VMS. In fact, the power to examine an ever vs. never VMS categorization in this sample was limited due to the fact that few women never reported having had VMS. Notably, the use of prospective measures of VMS to address study questions represents a significant advancement over the prior literature.
The “top SNPs” identified by Crandall et al.6 were not amenable to constructing a PRS, because 14 of the SNPs were in one gene and in high linkage disequilibrium with one another while the other three SNPs either were monomorphic in some race/ethnic groups or not statistically significantly associated with VMS after meta-analysis. As an alternative, we constructed an “all SNPs” PRS of VMS to capture all the sub-threshold SNPs with p<10e-4. Studies that utilize a PRS have advantages over use of a single SNP or just a small number of SNPs. A PRS summarizes the genetic effect among an ensemble of SNPs that do not individually have large effects on the trait of interest and provide a quantitive measure of the variation in genetic risk among individuals in a study. However, the fact that the VMS “all SNPs” PRS was not associated with frequent VMS or with VMS trajectories suggests that those sub-threshold SNPs probably do not convey any additional risk of VMS beyond the TARC3 signal alone.
We also constructed PRSs from GWAS for age at menarche and age at menopause. Earlier work in SWAN found that a PRS based on SNPs associated with age at menarche was associated with age at menarche in all the race/ethnic groups in SWAN with the exception of the Chinese, which is the smallest group in SWAN.25 Similarly, a PRS based on SNPs associated with age at menopause was associated with age at menopause in all race/ethnic groups in SWAN with the exception of Black women. The same study also found that the age at menarche and the age at menopause PRSs were associated with additional traits related to reproductive aging including length of the reproductive lifespan, duration of the menopausal transition, and estradiol levels prior to and after the menopause in at least one race/ethnic group in SWAN. Lower estrogen levels have been associated with the prevalence and frequency of VMS in several cohorts,26-29 and early age at menarche has recently been associated with increased risk of frequent VMS in a pooled analysis across multiple cohorts.30
New findings reported here include associations between the PRSs for age at menarche and age at menopause with VMS frequency and with VMS patterns in relation to time before and after the FMP. The results are racial/ethnic group-specific. Chinese women with genetic predisposition toward older age at menopause were at greater risk for having frequent VMS. In contrast, Black women with a genetic predisposition toward older age of menarche had less risk of having frequent VMS. No assocations between the PRSs and frequent VMS were found in White or Japanese women.
When the outcome was VMS pattern in relation to timing of the FMP based on any VMS, Black women with genetic predisposition toward older age at menarche were less likely to be in the “persistently high” trajectory group relative to the “persistently low” group, consistent with the association observed for these women being less likely to ever report frequent VMS. White women with genetic predisposition toward older age at menarche were less likely to be in the “FMP onset” trajectory group relative to the “persistently low” VMS group. When we considered the VMS trajectories based on frequent VMS, White women with genetic predisposition toward older age at menopause were less likely to be in the “FMP onset” trajectory group relative to the “low” VMS group.
Importantly, when women who carried the the sentinel SNP in TARC3 were excluded, the magnitude of associations were unchanged for the both frequent VMS analyses and the VMS trajectory analyses. This suggests that the sentinel SNP in TARC3 was neither masking nor driving results of the PRS analyses. TACR is the receptor for neurokinin B. Previous animal and human studies support an association between neurokinin B (NKB)and VMS. Evidence of association includes co-expression of NKB and estrogen receptor alpha messenger RNA in the human infundicular nucleus,12 the dense neurokinin B fiber plexuses located in the descending gonadotropin-releasing hormone tract that courses through the infundibular stalk to the neurohypophysis in humans.13 Neurokinin B gene expression in the human infundibular nucleus increases after menopause, and postmenopausal women have hypertrophy of neurokinins expressing neurokinin B.12, 14 A recent clinical trial has reported a reduction in hot flashes associated with administration of a neurokinin 3 receptor antagonist.48
This study has some limitations. VMS were self-reported via questionnaire, which may have been influenced by mood and memory relative to daily diary VMS measures or physiologic VMS measures,49, 50 however the duration of recall is short (in the past two weeks) lessening the potential for recall bias. Similarly, age at menopause is based on repeated self-reported date of the last menstrual period at each clinic visit, however as clinic visits occurred approximately annually recall bias is limited. Age at menarche is based on recall at the baseline visit and may be subject to digit preference. The PRS for menarche and for menopause were based on GWAS in women of European ancestry only. Also, the small sample size, particularly for the Chinese and Japanese women, limited study power. Finally, Black women were more likely than women of other race/ethnicities to have had a hysterectomy at the time of enrollment making them ineligible to participate in the SWAN cohort, which may have led to differential patterns of selection bias. Strengths of the study include the multi-racial/ethnic population and that the data on VMS was collected prospectively at multiple time points during the menopause transition, reducing recall biases and permitting characterization of the trajectories of VMS over the course of the menopause transition.
CONCLUSIONS
Genetic factors predictive of reproductive aging are also associated with VMS, suggesting that VMS have a polygenic architecture. We also found evidence that genetic influences may differ across racial/ethnic groups. If the results are confirmed in other study popuations, they could help to guide the development of new strategies to inform patients regarding risk for VMS. Validation of the causal effects of specific SNPs in vitro could also provide support for new targets for VMS alleviation, such as the use of NKB pathway agents.48, 51, 52
Supplementary Material
Supplemental Digital Content 1: Table S1. Association between “top SNPs” menarche/menopause PRS and ever reporting high frequency VMS (yes/no) in VMS_Supplemental Digital Content 1_CLEAN.DOCX
Supplemental Digital Content 2: Table S2. Association between genetic risk factors and trajectories for high frequency VMS (≥6 days) in VMS_Supplemental Digital Content CLEAN.DOCX
Supplemental Digital Content 3: Table S3. Association between “top SNPs” menarche/menopause PRS and VMS trajectories in VMSGeneticsSWAN_Supplemental Digital Content 3_CLEAN.DOCX
Supplemental Digital Content 4: Table S4. Association between PRS and ever reporting high frequency VMS (yes/no) in SWAN participants who do not carry any minor alleles at rs74827081. VMS_Supplemental Digital Content 4_CLEAN.DOCX
Supplemental Digital Content 5: Table S5. Association between PRS and VMS trajectories in SWAN participants who do not carry any minor alleles at rs74827081 in VMS_Supplemental Digital Content 5_CLEAN.DOCX
ACKNOWLEDGEMENTS
Clinical Centers:University of Michigan, Ann Arbor – Siobán Harlow, PI 2011 – present, MaryFran Sowers, PI 1994-2011; Massachusetts General Hospital, Boston, MA – Joel Finkelstein, PI 1999 – present; Robert Neer, PI 1994 – 1999; Rush University, Rush University Medical Center, Chicago, IL – Howard Kravitz, PI 2009 – present; Lynda Powell, PI 1994 – 2009; University of California, Davis/Kaiser – Ellen Gold, PI; University of California, Los Angeles – Gail Greendale, PI; Albert Einstein College of Medicine, Bronx, NY – Carol Derby, PI 2011 – present, Rachel Wildman, PI 2010 – 2011; Nanette Santoro, PI 2004 – 2010; University of Medicine and Dentistry – New Jersey Medical School, Newark – Gerson Weiss, PI 1994 – 2004; and the University of Pittsburgh, Pittsburgh, PA – Karen Matthews, PI. NIH Program Office: National Institute on Aging, Bethesda, MD – Chhanda Dutta 2016- present; Winifred Rossi 2012–2016; Sherry Sherman 1994 – 2012; Marcia Ory 1994 – 2001; National Institute of Nursing Research, Bethesda, MD – Program Officers. Central Laboratory: University of Michigan, Ann Arbor – Daniel McConnell (Central Ligand Assay Satellite Services). SWAN Repository: University of Michigan, Ann Arbor – Siobán Harlow 2013- present; Dan McConnell 2011 – 2013; MaryFran Sowers 2000 – 2011. Coordinating Center: University of Pittsburgh, Pittsburgh, PA – Maria Mori Brooks, PI 2012 - present; Kim Sutton-Tyrrell, PI 2001 – 2012; New England Research Institutes, Watertown, MA - Sonja McKinlay, PI 1995 – 2001. Steering Committee: Susan Johnson, Current Chair; Chris Gallagher, Former Chair
Sources of funding: The Study of Women's Health Across the Nation (SWAN) has grant support from the National Institutes of Health (NIH), DHHS, through the National Institute on Aging (NIA), the National Institute of Nursing Research (NINR) and the NIH Office of Research on Women’s Health (ORWH) (Grants U01NR004061; U01AG012505, U01AG012535, U01AG012531, U01AG012539, U01AG012546, U01AG012553, U01AG012554, U01AG012495) and for the SWAN Genomic Analyses an SWAN Legacy (U01AG017719). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIA, NINR, ORWH or the NIH. SDH gratefully acknowledge use of the services and facilities of the Population Studies Center at the University of Michigan, funded by NICHD Center Grant R24 HD041028.
Footnotes
Financial disclosures/conflicts of interest:RCT: Consulting/Advisory Board: Astellas, Pfizer, Procter & Gamble, Virtue Health. No disclosures for WZ, JAS, PP, CJC, ER, MU, MMH, SLRK, SDH, CJC.
We thank the study staff at each site and all the women who participated in SWAN.
Data Availability:
All data generated or analyzed during this study are included in data repositories. Genomic data are available through the NIH Database of Genotypes and Phenotypes (dbGaP, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001470.v1.p1 , accession number: phs001470.v1.p1). Phenotype data are available through the Aging Research Biobank (https://agingresearchbiobank.nia.nih.gov/studies/swan/?search_term=SWAN) )
REFERENCES
- 1.Gold EB, Colvin A, Avis N, et al. Longitudinal analysis of the association between vasomotor symptoms and race/ethnicity across the menopausal transition: Study of women's health across the nation. Am J Public Health. 2006;96(7):1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman EW, Sammel MD, Lin H, Liu ZY, Gracia CR. Duration of menopausal hot flushes and associated risk factors. Obstet Gynecol. 2011;117(5):1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods NF, Mitchell ES. Symptoms during the perimenopause: prevalence, severity, trajectory, and significance in women's lives. Am J Med. 2005;118 Suppl 12B:14–24. [DOI] [PubMed] [Google Scholar]
- 4.Tepper PG, Brooks MM, Randolph JF, et al. Characterizing the trajectories of vasomotor symptoms across the menopausal transition. Menopause. 2016;23(10):1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avis NE, Crawford SL, Greendale G, et al. Duration of menopausal vasomotor symptoms over the menopause transition. JAMA Intern Med. 2015;175(4):531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crandall CJ, Manson JE, Hohensee C, et al. Association of genetic variation in the tachykinin receptor 3 locus with hot flashes and night sweats in the Women's Health Initiative Study. Menopause. 2017;24(3):252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crandall CJ, Crawford SL, Gold EB. Vasomotor symptom prevalence is associated with polymorphisms in sex steroid-metabolizing enzymes and receptors. Am J Med. 2006;119(9):52–60. [DOI] [PubMed] [Google Scholar]
- 8.Kardia SR, Jian C, Sowers MR. Characterizing variation in sex steroid hormone pathway genes in women of 4 races/ethnicities: The Study of Women's Health Across the Nation (SWAN). Am J Med. 2006;119(9):3–15. [DOI] [PubMed] [Google Scholar]
- 9.Woods NF, Cray LA, Mitchell ES, Farrin F, Herting J. Polymorphisms in estrogen synthesis genes and symptom clusters during the menopausal transition and early postmenopause: observations from the Seattle Midlife Women's Health Study. Biol Res Nurs. 2018;20(2):153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebbeck TR, Su HI, Sammel MD, et al. Effect of hormone metabolism genotypes on steroid hormone levels and menopausal symptoms in a prospective population-based cohort of women experiencing the menopausal transition. Menopause. 2010;17(5):1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crandall CJ, Diamant AL, Maglione M, Thurston RC, Sinsheimer J. Genetic variation and hot flashes: A systematic review [published online ahead of print, 2020 Aug 14]. J Clin Endocrinol Metab. 2020;dgaa536. doi: 10.1210/clinem/dgaa536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rance NE, Krajewski SJ, Smith MA, Cholanian M, Dacks PA. Neurokinin B and the hypothalamic regulation of reproduction. Brain Res. 2010;1364:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borsay BA, Skrapits K, Herczeg L, et al. Hypophysiotropic Gonadotropin-Releasing Hormone Projections Are Exposed to Dense Plexuses of Kisspeptin, Neurokinin B and Substance P immunoreactive fibers in the human: a study on tissues from postmenopausal women. Neuroendocrinology. 2014;100(2–3):141–152. [DOI] [PubMed] [Google Scholar]
- 14.Rance NE, Bruce TR. Neurokinin-B gene-expression is increased in the arcuate nucleus of ovariectomized rats. Neuroendocrinology. 1994;60(4):337–345. [DOI] [PubMed] [Google Scholar]
- 15.Topaloglu AK, Reimann F, Guclu M, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41(3):354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruth KS, Campbell PJ, Chew S, et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur J Hum Genet. 2016;24(2):284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuring AN, Busch AS, Bogdanova N, Gromoll J, Tuttelmann F. Effects of the FSH-beta-subunit promoter polymorphism-211G -> T on the hypothalamic-pituitary-ovarian axis in normally cycling women indicate a gender-specific regulation of gonadotropin secretion. J Clin Endocrinol Metab. 2013;98(1):E82–E86. [DOI] [PubMed] [Google Scholar]
- 18.Prescott J, Thompson DJ, Kraft P, et al. Genome-Wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One. 2012;7(6):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigorova M, Punab M, Poolamets O, et al. Increased prevalance of the-211 T allele of follicle stimulating hormone (FSH) beta subunit promoter polymorphism and lower serum FSH in infertile men. J Clin Endocrinol Metab. 2010;95(1):100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigorova M, Punab M, Ausmees K, Laan M. FSHB promoter polymorphism within evolutionary conserved element is associated with serum FSH level in men. Hum Reprod. 2008;23(9):2160–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunning AM, Dowsett M, Healey CS,et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96(12):936–945. [DOI] [PubMed] [Google Scholar]
- 22.Day FR, Ruth KS, Thompson DJ, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day FR, Thompson DJ, Helgason H, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49(6):834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurston RC, Santoro N, Matthews KA. Adiposity and hot flashes in midlife women: a modifying role of age. J Clin Endocrinol Metab. 2011;96(10):E1588–E1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W, Smith JA, Bielak LF, et al. Polygenic risk scores for age at menarche and menopause are associated with reproductive timing and serum hormone levels. Manuscript submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell ES, Woods NF. Hot flush severity during the menopausal transition and early postmenopause: beyond hormones. Climacteric. 2015;18(4):536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods NF, Smith-Dijulio K, Percival DB, Tao EY, Taylor HJ, Mitchell ES. Symptoms during the menopausal transition and early postmenopause and their relation to endocrine levels over time: Observations from the Seattle Midlife Women's Health Study. Journal of Womens Health. 2007;16(5):667–677. [DOI] [PubMed] [Google Scholar]
- 28.Randolph JF, Sowers MF, Bondarenko I, et al. The relationship of longitudinal change in reproductive hormones and vasomotor symptoms during the menopausal transition. J Clin Endocrinol & Metabol. 2005;90(11):6106–6112. [DOI] [PubMed] [Google Scholar]
- 29.Freeman EW, Sammel MD, Lin H, et al. Symptoms associated with menopausal transition and reproductive hormones in midlife women. Obstet and Gynecol. 2007;110(2):230–240. [DOI] [PubMed] [Google Scholar]
- 30.Chung HF, Zhu D, Dobson AJ, et al. Age at menarche and risk of vasomotor menopausal symptoms: a pooled analysis of six studies. BJOG. 2020. 10.1111/1471-0528.16393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leidy LE. Symptoms of menopause in relation to the timing of reproductive events and past menstrual experience. Am J Hum Bio. 1996;8(6):761–769. [DOI] [PubMed] [Google Scholar]
- 32.Da Fonseca AM, Bagnoli VR, Souza MA, et al. Impact of age and body mass on the intensity of menopausal symptoms in 5968 Brazilian women. Gynecol Endocrinol. 2013;29(2):116–118. [DOI] [PubMed] [Google Scholar]
- 33.Nakano K, Pinnow E, Flaws JA, Sorkin JD, Gallicchio L. Reproductive history and hot flashes in perimenopausal women. J Womens Health. 2012;21(4):433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gjelsvik B, Rosvold EO, Straand J, Dalen I, Hunskaar S. Symptom prevalence during menopause and factors associated with symptoms and menopausal age. Results from the Norwegian Hordaland Women's Cohort study. Maturitas. 2011;70(4):383–390. [DOI] [PubMed] [Google Scholar]
- 35.Sowers MFR, Crawford SL, Sternfeld B, et al. SWAN: a multicenter, multiethnic, community-based cohort study of women and the menopausal transition. 2000. [Google Scholar]
- 36.Thurston RC, Aslanidou Vlachos HE, et al. Menopausal vasomotor symptoms and risk of incident cardiovascular disease events in SWAN. J Am Heart Assoc. 2021;10(3):e017416. Epub 2021 Jan 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurston RC, Sutton-Tyrrell K, Everson-Rose SA, Hess R, Powell LH, Matthews KA. Hot flashes and carotid intima media thickness among midlife women. Menopause. 2011;18(4):352–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crandall CJ, Tseng CH, Crawford SL, et al. Association of menopausal vasomotor symptoms with increased bone turnover during the menopausal transition. J Bone Miner Res. 2011;26(4):840–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagin DS. Group-Based Modeling of Development. Cambridge, MA: Harvard University Press; 2005. [Google Scholar]
- 40.Jones BL, Nagin DS, Roeder K. A SAS procedure based on mixture models for estimating developmental trajectories. Social Methods Res 2001;29:374–393. [Google Scholar]
- 41.Jones BL, Nagin DS. Advances in group-based trajectory modeling and an SAS procedure for estimating them. Social Methods Res 2007;35:542–571. [Google Scholar]
- 42.Sternfeld B, Ainsworth BE, Quesenberry CP. Physical activity patterns in a diverse population of women. Prev Med. 1999;28(3):313–323. [DOI] [PubMed] [Google Scholar]
- 43.Baecke JA, Burema J, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr. 1982;36(5):936–42. [DOI] [PubMed] [Google Scholar]
- 44.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. Published 2015 Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prague JK, Roberts RE, Comninos AN, et al. Neurokinin 3 receptor antagonism as a novel treatment for menopausal hot flushes: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet. 2017;389(10081):1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu P, Matthews KA, Thurston RC. How well do different measurement modalities estimate the number of vasomotor symptoms? Findings from the Study of Women's Health Across the Nation FLASHES Study. Menopause. 2014;21(2):124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thurston RC, Blumenthal JA, Babyak MA, Sherwood A. Emotional antecedents of hot flashes during daily life. Psychosom Med. 2005;67(1):137–146. [DOI] [PubMed] [Google Scholar]
- 51.Oakley AE, Steiner RA, Chavkin C, Clifton DK, Ferrara LK, Reed SD. Agonists as a novel therapy for menopausal hot flashes. Menopause. 2015;22(12):1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayasena CN, Comninos AN, Stefanopoulou E, et al. Neurokinin B administration induces hot flushes in women. Sci Rep. 2015;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content 1: Table S1. Association between “top SNPs” menarche/menopause PRS and ever reporting high frequency VMS (yes/no) in VMS_Supplemental Digital Content 1_CLEAN.DOCX
Supplemental Digital Content 2: Table S2. Association between genetic risk factors and trajectories for high frequency VMS (≥6 days) in VMS_Supplemental Digital Content CLEAN.DOCX
Supplemental Digital Content 3: Table S3. Association between “top SNPs” menarche/menopause PRS and VMS trajectories in VMSGeneticsSWAN_Supplemental Digital Content 3_CLEAN.DOCX
Supplemental Digital Content 4: Table S4. Association between PRS and ever reporting high frequency VMS (yes/no) in SWAN participants who do not carry any minor alleles at rs74827081. VMS_Supplemental Digital Content 4_CLEAN.DOCX
Supplemental Digital Content 5: Table S5. Association between PRS and VMS trajectories in SWAN participants who do not carry any minor alleles at rs74827081 in VMS_Supplemental Digital Content 5_CLEAN.DOCX
Data Availability Statement
All data generated or analyzed during this study are included in data repositories. Genomic data are available through the NIH Database of Genotypes and Phenotypes (dbGaP, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001470.v1.p1 , accession number: phs001470.v1.p1). Phenotype data are available through the Aging Research Biobank (https://agingresearchbiobank.nia.nih.gov/studies/swan/?search_term=SWAN) )


