Abstract
With the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with increased transmissibility and potential resistance, antibodies and vaccines with broadly inhibitory activity are needed. Here, we developed a panel of neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) that bound the receptor binding domain of the spike protein at distinct epitopes and blocked virus attachment to its host receptor, human angiotensin converting enzyme-2 (hACE2). Although several potently neutralizing mAbs protected K18-hACE2 transgenic mice against infection caused by ancestral SARS-CoV-2 strains, others induced escape variants in vivo or lost neutralizing activity against emerging strains. One mAb, SARS2-38, potently neutralized all tested SARS-CoV-2 variants of concern and protected mice against challenge by multiple SARS-CoV-2 strains. Structural analysis showed that SARS2-38 engaged a conserved epitope proximal to the receptor binding motif. Thus, treatment with or induction of neutralizing antibodies that bind conserved spike epitopes may limit the loss of potency of therapies or vaccines against emerging SARS-CoV-2 variants.
Keywords: neutralizing antibodies, SARS-CoV-2, variants of concern
Graphical abstract
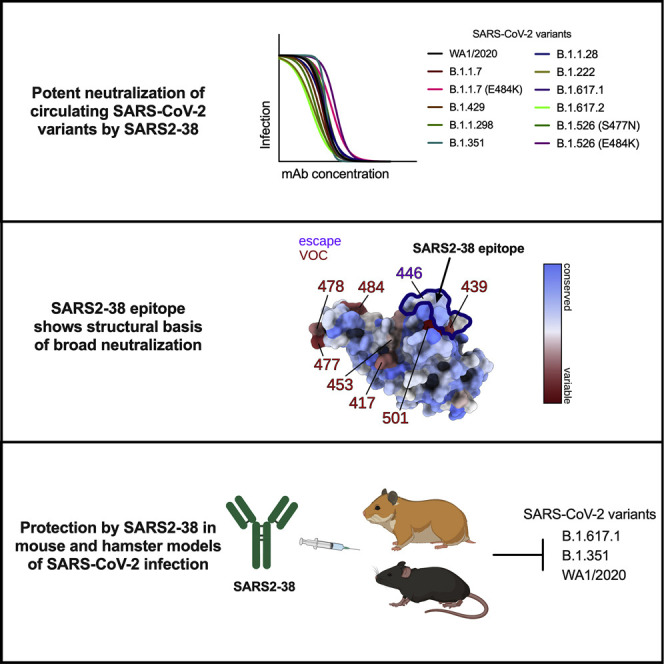
VanBlargan et al. describe a potently neutralizing mAb, SARS2-38, that recognizes a panel of SARS-CoV-2 variants and confers therapeutic protection in vivo. Structure analysis of SARS2-38 bound to the viral spike protein reveals the basis of its broadly neutralizing activity, highlighting an epitope target for antibody therapeutics and vaccine design.
Introduction
Severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2 belong to the Sarbecovirus subgenus of Betacoronaviruses (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has resulted in 200 million infections and 4 million deaths (https://covid19.who.int/). Multiple effective vaccines against SARS-CoV-2 that prevent COVID-19 have been developed and deployed rapidly (Baden et al., 2021; Polack et al., 2020; Sadoff et al., 2021; Voysey et al., 2021). Monoclonal antibodies (mAb) have also shown efficacy in animal models of SARS-CoV-2 infection (Alsoussi et al., 2020; Baum et al., 2020a; Fagre et al., 2020; Hansen et al., 2020; Hassan et al., 2020; Kreye et al., 2020; Rogers et al., 2020; Shi et al., 2020; Zost et al., 2020), and three mAb treatments are approved for use under Emergency Use Authorization (EUA) (Cathcart et al., 2021; Chen et al., 2021b; Weinreich et al., 2021). Therapy with mAbs may be beneficial for high-risk individuals following exposure to SARS-CoV-2 and can complement vaccines as a means of combating the COVID-19 pandemic (O’Brien et al., 2021).
The majority of characterized potently neutralizing and protective anti-SARS-CoV-2 mAbs bind the receptor binding domain (RBD) of the viral spike protein (Barnes et al., 2020; Baum et al., 2020a; Cao et al., 2020; Tortorici et al., 2020; Zost et al., 2020), but some inhibitory mAbs against the N-terminal domain (NTD) and S2 domains of spike have been described (Chi et al., 2020; Liu et al., 2020; Ng et al., 2020; Song et al., 2021; Suryadevara et al., 2021). Under immune selection pressure, SARS-CoV-2 can select for mutations that enable escape from antibody recognition and neutralization (Baum et al., 2020b; Greaney et al., 2021; Liu et al., 2021; Starr et al., 2021b; Suryadevara et al., 2021). Indeed, several emerging SARS-CoV-2 variants have mutations in the spike protein that confer resistance to mAbs or polyclonal antibodies (pAbs) elicited by vaccines or natural infection (Chen et al., 2021d; Hoffmann et al., 2021; Thomson et al., 2021; Weisblum et al., 2020; Wibmer et al., 2021). Therefore, additional mAbs or vaccines that retain efficacy against emerging SARS-CoV-2 variants may be needed to combat evolving strains.
In this study, we describe a panel of potently neutralizing murine mAbs against the RBD of SARS-CoV-2 that bind epitopes proximal to the receptor binding motif (RBM) of the RBD or at the base of the RBD. Although some neutralizing mAbs demonstrated limited ability to protect against infection by the historical SARS-CoV-2 WA1/2020 strain in a mouse disease model and selected rapidly for escape in vivo, others protected completely in the context of prophylactic or therapeutic administration. Two protective mAbs, SARS2-02 and SARS2-38, showed variable capacity to neutralize variants of concern (VOC) and variants of interest (VOI): SARS2-02 binds an epitope that includes residues E484 and L452 and shows reduced potency against strains (B.1.351 [Beta], B.1.617.2 [Delta], B.1.429 [Epsilon], B.1.1.28/P.1 [Gamma], B.1.526 [Iota], and B.1.617.1 [Kappa]) encoding these mutations. In contrast, SARS2-38 binds an epitope centered on residues K444 and G446 and neutralized all tested VOC and VOI. Analysis of a cryoelectron microscopy (cryo-EM) structure of SARS2-38 bound to spike reveals that this mAb binds a conserved epitope on the RBD that is also engaged, albeit through distinct geometries, by other neutralizing and protective human mAbs. Thus, treatment with mAbs or induction of pAbs targeting this conserved region of the RBD may confer protection against many emerging SARS-CoV-2 variants.
Results
Development and characterization of anti-SARS-CoV-2 mAbs
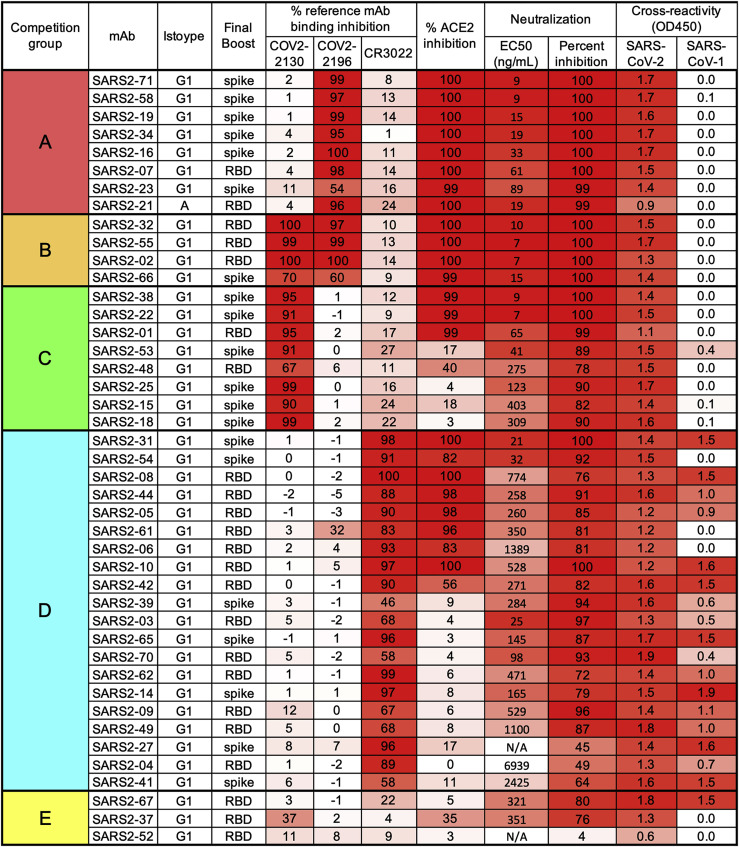
We generated a panel of anti-SARS-CoV-2 mAbs from BALB/c mice that were immunized with purified RBD and/or ectodomain of the spike protein mixed with AddaVax, a squalene-based adjuvant (Figure 1 ). After splenocyte-myeloma fusions, hybridoma supernatants were screened for antibody binding to recombinant spike protein and permeabilized, SARS-CoV-2-infected Vero cells by ELISA and flow cytometry, respectively. Sixty-four hybridomas producing anti-SARS-CoV-2 antibodies were cloned by limiting dilution. Forty-three of these mAbs bound to recombinant RBD and were selected for further study because prior experiments showed that this class includes potently inhibitory antibodies (Barnes et al., 2020; Baum et al., 2020a; Cao et al., 2020; Tortorici et al., 2020; Zost et al., 2020); the majority of these mAbs were of the immunoglobulin G1 (IgG1) subclass (Figure 1).
Figure 1.
Panel of anti-SARS-CoV-2 mAbs
Hybridoma supernatants were assayed for neutralization of SARS-CoV-2 by FRNT, cross-reactivity to SARS-CoV-1 spike protein, and ability to inhibit SARS-CoV-2 spike protein binding to hACE2 or a panel of reference human mAbs through competition ELISA. mAbs are grouped by reference mAb competition properties. Data represent the mean (or geometric mean for EC50 values) from two to four experiments. Hybridomas were produced from splenocytes of mice that received three immunizations (once with RBD and then twice with spike) prior to a final pre-fusion boost with RBD or spike, as indicated in the “Final Boost” column.
The mAbs were evaluated by competition binding analysis using three previously characterized human mAbs that recognize distinct antigenic sites on the RBD (COV2-2196, COV2-2130, and CR3022) (Yuan et al., 2020; Zost et al., 2020; Figure 1). Although the relatively large size of antibodies limits the precision of this mapping approach, competition binding analysis can allow classification of mAb interaction regions in a high-throughput manner. Eight mAbs competed for spike protein binding with the neutralizing mAb COV2-2196 only, eight mAbs competed with the neutralizing mAb COV2-2130 only, four mAbs competed with COV2-2196 and COV2-2130, and 20 mAbs competed with CR3022, a mAb that recognizes a more conserved, cryptic epitope on the SARS-CoV-2 spike protein distal from the receptor binding site. Three RBD-binding mAbs did not compete with COV2-2196, COV2-2130, or CR3022. Based on the binding analysis, mAbs were divided into five competition groups, A–E (Figure 1).
One potential mechanism of antibody-mediated neutralization of SARS-CoV-2 is through inhibition of viral spike protein binding to the human ACE2 receptor. The COV2-2196 epitope directly overlaps the ACE2 binding site on the RBD, whereas the COV2-2130 epitope lies proximal to residues in the RBM that interact with ACE2 (Dong et al., 2021); nonetheless, both mAbs can block spike binding to ACE2. In contrast, CR3022 engages the base of the RBD and does not block ACE2 binding to spike (Yuan et al., 2020). Of the 43 RBD-binding antibodies in our panel, all mAbs in groups A and B inhibited ACE2 binding to the spike protein, mAbs in groups C and D variably inhibited ACE2 binding, and mAbs in group E failed to inhibit ACE2 binding (Figure 1).
The mAbs were also tested for cross-reactive binding to the SARS-CoV-1 spike protein. The majority of mAbs in group D, which competed with the cross-reactive mAb CR3022 for spike binding, cross-reacted with SARS-CoV-1 spike protein, indicating that they bind conserved sarbecovirus epitopes. MAbs in groups A, B, and C did not bind to SARS-CoV-1 (Figure 1), and one group E mAb recognized SARS-CoV-1. Based on competition analysis, many anti-RBD mAbs in our panel bind within or proximal to the RBM and are type-specific for SARS-CoV-2, whereas those binding near the base of the RBD are more cross-reactive with SARS-CoV-1.
Anti-SARS-CoV-2 RBD mAbs neutralize SARS-CoV-2 with varying potency
We next determined the neutralizing activity of mAb hybridoma supernatants using a focus-reduction neutralization test (FRNT) and Vero E6 cells (Case et al., 2020) with the WA1/2020 SARS-CoV-2 strain. Antibody concentrations were quantified by ELISA and used to calculate half-maximal inhibitory concentrations (EC50 values). The most potently inhibitory mAbs (EC50, <10 ng/mL) belonged to groups A, B, and C and also blocked ACE2 binding (Figure 1). Some mAbs in groups C and D that did not block ACE2 binding still showed robust neutralizing activity (EC50, 20–100 ng/mL), although the majority were weakly inhibitory. Group E mAbs were weakly neutralizing and did not block ACE2 binding.
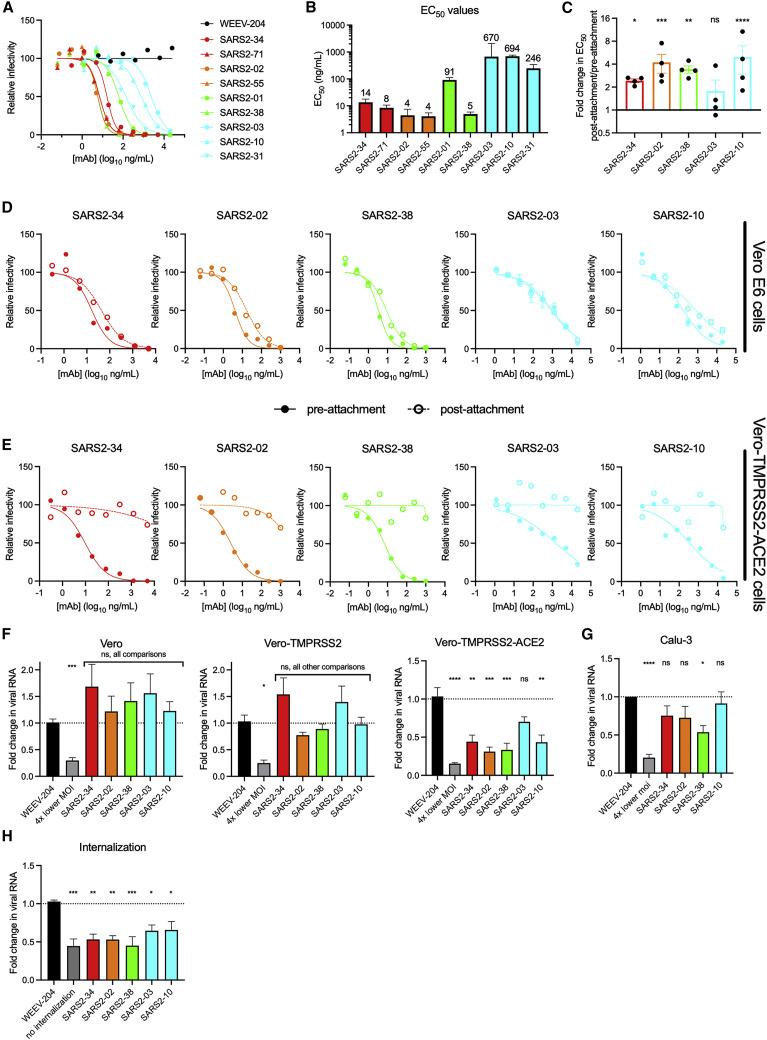
A subset of mAbs from groups A, B, C, and D were selected for detailed study. We chose two mAbs with the highest neutralization potency from each group; in cases where mAbs had high variable region sequence similarity, we selected only one of these mAbs for further study. We also selected SARS2-03, as it was one of the few neutralizing mAbs that did not block ACE2 binding. Nine mAbs were purified and retested for neutralization potency by FRNT using Vero cells and the WA1/2020 isolate (Figures 2 A and 2B). Again, the most potently neutralizing purified mAbs belonged to groups A, B, and C, with less inhibitory activity in those from group D. We also characterized these nine mAbs for competition binding with each other (Figure S1). The two group A mAbs (SARS2-34 and SARS2-71) competed for spike binding only with each other. In contrast, mAbs in groups B (SARS2-02 and SARS2-55) and C (SARS2-01 and SARS2-38) competed for spike binding across both groups. SARS2-03, a group D mAb, did not bind spike efficiently in the presence of group B or C mAbs and blocked binding of group C mAb SARS2-01. SARS2-10 and SARS2-31, the other two group D mAbs, however, competed only with each other. These results suggest that mAbs in group C may have overlapping epitopes with group B mAbs and group D mAb SARS2-03, whereas group A mAbs and the remaining group D mAbs likely engage physically distinct epitopes.
Figure 2.
Neutralization by anti-SARS-CoV-2 mAbs
(A and B) Anti-SARS-CoV-2 mAbs were assayed for neutralization by FRNT against SARS-CoV-2 using Vero E6 cells.
(A) Representative dose-response curves.
(B) Mean EC50 values; error bars represent standard error of the mean (SEM) from three to four experiments.
(C and D) Anti-SARS-CoV-2 mAbs were assayed for pre- or post-attachment neutralization of SARS-CoV-2 using Vero E6 cells.
(C) Fold change in EC50 values for post-attachment over pre-attachment neutralization. Error bars represent SEM from four experiments.
(D) Representative dose-response curves.
(E) Anti-SARS-CoV-2 mAbs were assayed for pre- or post-attachment neutralization on Vero-TMPRSS2-ACE2 cells. Dose response curves are shown. Data are representative of three experiments.
(F and G) Anti-SARS-CoV-2 mAbs were assayed for attachment inhibition of SARS-CoV-2 to Vero E6, Vero-TMPRSS2, or Vero-TMPRSS2-ACE2 (F) or Calu-3 (G) cells. Error bars represent SEM from three (F) to six (G) experiments.
(H) Anti-SARS-CoV-2 mAbs were assayed for inhibition of virus internalization in Vero E6 cells. Error bars represent SEM from four experiments.
In (C), we used ANOVA with Sidak’s post-test, comparing pre- versus post-attachment EC50 values for each mAb. In (F)–(H), we used one-way ANOVA with Dunnett’s post-test, comparing mAb treatment with isotype control mAb treatment. ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Mechanism of neutralization by anti-SARS-CoV-2 mAbs is cell-type dependent
We investigated whether the anti-SARS-CoV-2 mAbs inhibited infection at a pre- or post-attachment step of the entry process. For these experiments, we selected one representative mAb from groups A, B, and C (SARS2-34, SARS2-02, and SARS2-38, respectively) and two mAbs from group D (SARS2-10 and SARS2-03, which, respectively, blocks or does not block ACE2 binding). We compared the neutralization potency of mAbs when added before or after virus absorption to Vero E6 cells. All mAbs retained neutralizing activity when added post-attachment, although the potency of groups A, B, and C mAbs SARS2-02, SARS2-34, and SARS2-38 was reduced slightly (∼2- to 4-fold, p < 0.05) relative to pre-attachment neutralization titers (Figures 2C and 2D). SARS2-10, a group D mAb, also showed an ∼5-fold decrease (p < 0.0001) in neutralizing activity when added after attachment. In contrast, SARS2-03, another group D mAb and the only mAb in this smaller panel that did not block ACE2-spike interactions, had similar neutralization potencies (p = 0.79) when added before or after cell attachment. These data suggest that mAbs that inhibit spike protein binding to ACE2 neutralize SARS-CoV-2 slightly more efficiently when given at a pre-attachment step, although all of the tested mAbs retained the ability to inhibit infection when given after virus attachment to cells.
To determine the effect of entry factor expression on target cells on virus neutralization, we extended these findings to cells that ectopically express human ACE2 and TMPRSS2. In contrast to the relatively minor change in neutralization potency seen with all mAbs for pre- versus post-attachment observed using Vero E6 cells, mAbs no longer efficiently neutralized SARS-CoV-2 infection when added after attachment to Vero-TMPRSS2-ACE2 cells, although pre-attachment neutralization activity remained intact (Figure 2E). Thus, the ability of anti-SARS-CoV-2 mAbs to neutralize at a post-attachment step was modulated by expression levels of viral entry factors and the cell line.
We also tested the ability of the mAbs to directly block virus attachment to cells, including Vero E6, Vero-TMPRSS2, and Vero-TMPRSS2-ACE2 cells. None of the mAbs efficiently blocked SARS-CoV-2 attachment to Vero or Vero-TMPRSS2 cells (Figure 2F). However, with the exception of SARS2-03, all mAbs reduced virus attachment to Vero-TMPRSS2-ACE2 cells. To corroborate these findings with cells that endogenously express human ACE2, we repeated experiments with Calu-3 cells, a human lung epithelial cell line. We observed an intermediate phenotype with Calu-3 cells, with modest attachment inhibition by mAbs in groups A, B, and C; levels of attached virus were ∼25%–50% lower than the isotype mAb control, with inhibition by only SARS2-38 attaining statistical significance (Figure 2G). This result suggests that the anti-RBD mAbs can inhibit viral attachment to cells, but this activity depends on levels of human ACE2 expression. Because the mAbs did not efficiently inhibit attachment to Vero E6 cells lacking human ACE2 expression, we tested whether they block a later step in the entry process by using a virus internalization assay (Dejarnac et al., 2018; Earnest et al., 2021). In Vero E6 cells, pre-incubation with all of the anti-RBD mAbs tested resulted in reduced levels of internalized virus (Figure 2H).
Because we observed cell-type-dependent differences in the mechanism of neutralization, we tested the effect of cell substrate on the inhibitory potency of our anti-RBD mAbs by FRNT. Notably, the anti-RBD mAbs neutralized SARS-CoV-2 WA1/2020 equivalently in Vero E6, Vero-TMRPSS2, and Vero-TMPRSS2-ACE2 cells (Figure S2). Thus, although the mAbs block SARS-CoV-2 attachment variably on different cell types, the potency of infection inhibition was similar across Vero cell substrates. This result may be explained by the ability of anti-RBD mAbs to block a required ACE2-dependent entry interaction in all of the tested cell substrates even though the attachment step is affected variably.
Neutralization escape analysis identifies epitopes recognized by anti-SARS-CoV-2 mAbs
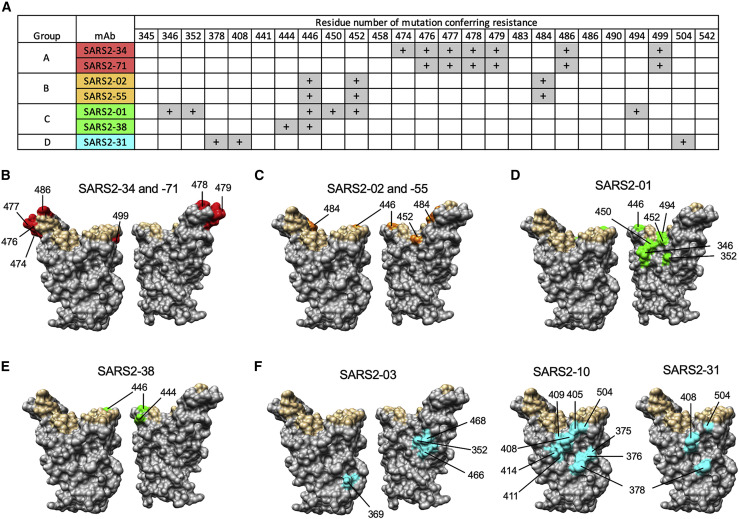
To determine spike residues important for recognition by anti-SARS-CoV-2 mAbs, we previously isolated neutralization escape mutants by passaging a vesicular stomatitis virus (VSV)-EGFP-SARS-CoV-2-S chimeric virus in the presence of neutralizing mAbs, including some of the antibodies described in this study (Liu et al., 2021). The above-described subset of nine mAbs from groups A–D were tested for neutralization against the panel of sequenced mutants, as well as against additional escape mutants generated against SARS2-03 and SARS2-10. Neutralizing activity was lost for the group A mAbs SARS2-34 and SARS2-71 with G476D, S477G/N/R, T478I/P, P479S, F486S/Y, and P499L substitutions; for the group B mAbs SARS2-02 and SARS2-55 with G446D/V, L452R, and E484K substitutions; for the group C mAb SARS2-01 with R346G, A352D, G446V, N450Y/K, L452R, and S494P substitutions; for the group C mAb SARS2-38 with K444E/N and G446D/V substitutions; for the group D mAb SARS2-03 with A352D, Y369H, R466I, and I468S substitutions; for the group D mAb SARS2-10 with S375P, T376I, K378E/Q/N, D405N/Y, R408G, Q409H, A411V, Q414K, and G504D substitutions; and for the group D mAb SARS2-31 with K378E/Q, R408K, and G504D substitutions (Figures 3 A–3F). These data are consistent with the competition binding analysis and suggest that group A, B, and C mAbs bind near the RBM and group D mAbs bind the base of the RBD.
Figure 3.
Epitopes recognized by anti-SARS-CoV-2 mAbs
mAbs were tested for inhibition of VSV-EGFP-SARS-CoV-2-S neutralization escape mutants.
(A) The “+” symbol indicates resistance to neutralization when a mutation at the indicated residue number is present.
(B–F) Residues from (A) are highlighted on the RBD structure (PDB: 6M0J) in red, orange, green, or cyan for mAbs from group A, B, C, or D, respectively, and indicated. Residues that engage hACE2 are highlighted in tan.
Anti-SARS-CoV-2 mAbs protect against virus challenge in vivo
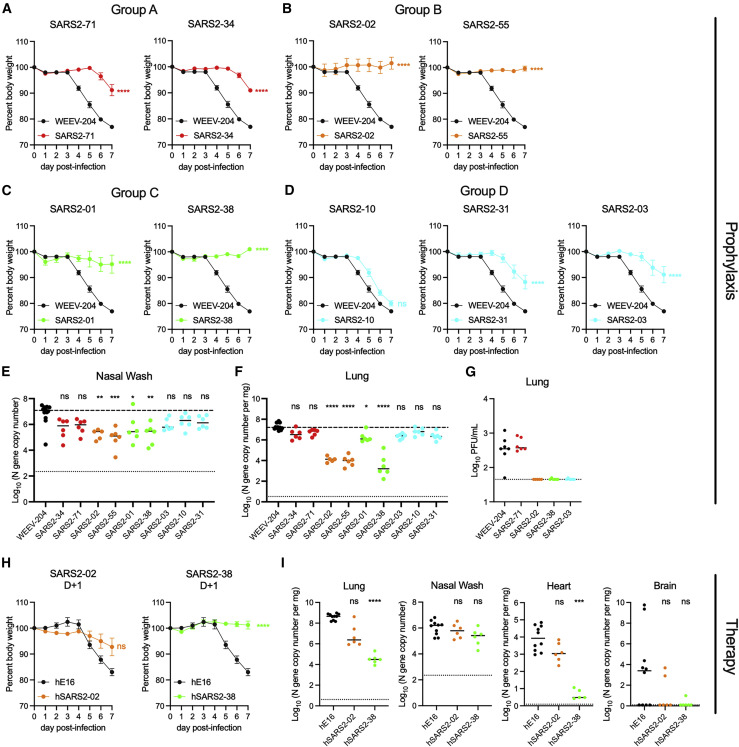
We tested the anti-SARS-CoV-2 mAbs for protection in vivo. Eight- to ten-week-old K18 human ACE2 (hACE2) transgenic mice were administered a single 100-μg dose (∼5 mg/kg) of anti-SARS-CoV-2 mAb 24 h prior to intranasal inoculation with 103 focus-forming units (FFU) of SARS-CoV-2 WA1/2020. Mice treated with the isotype control mAb lost up to 25% body weight by 7 days post-infection (dpi), the designated endpoint of the study (Figure 4 A). Mice treated with the group A mAbs SARS2-71 and SARS2-34 maintained body weight until 6–7 dpi, when we observed a 10% weight loss (Figure 4A). Mice treated with the group B mAbs SARS2-02 and SARS2-55 and group C mAbs SARS2-01 and SARS2-38 all maintained body weight throughout the experiment (Figures 4B and 4C). Animals treated with group D mAbs SARS2-10, SARS2-31, or SARS2-03 were generally less protected against virus-induced weight loss (Figure 4D).
Figure 4.
Anti-SARS-CoV-2 mAbs protect against SARS-CoV-2 infection in vivo
(A–G) K18-hACE2 mice were passively administered 100 μg (~5 mg/kg) of the indicated mAb by intraperitoneal injection 24 h prior to intranasal inoculation with 103 FFU of SARS-CoV-2 WA1/2020.
(A–D) Mice were monitored for weight change for 7 days following viral infection. Mean weight change is shown. Error bars represent SEM.
(E and F) At 7 dpi, nasal washes (E) and lungs (F) were collected, and viral RNA levels were determined. Median levels are shown. The top dotted line indicates the median viral load of control mAb-treated mice. The bottom dotted line represents the limit of detection (LOD) of the assay.
(H) A subset of the lungs from (F) were assessed for infectious viral burden by plaque assay. Median PFU/mL is shown. The dotted line indicates the LOD.
(H and I) K18-hACE2 mice were given 200 μg (~10 mg/kg) of the indicated mAb by intraperitoneal injection 24 h after intranasal inoculation with 102 FFU of SARS-CoV-2 WA1/2020. Data are from two or three experiments; WEEV-204 (isotype control): n = 10; SARS2-02 and SARS2-38: n = 6 per group.
(H) Mean weight change. Error bars represent SEM.
(I) At 7 dpi, lungs, nasal washes, heart, and brain were collected, and viral RNA levels were determined. Median levels are shown.
In (A)–(F), data for each mAb are from two experiments; WEEV-204 (isotype control): n = 12; all other mAbs: n = 5–6 per group. In (A)–(D) and (H), we used one-way ANOVA with Dunnett’s post-test of the area under the curve; ∗∗∗∗p < 0.0001. In (E), (F), and (I), we used a Kruskal-Wallis with Dunn’s post-test.; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To corroborate and extend these findings, we measured the effect of mAb treatment on viral burden in the nasal washes and lungs on 7 dpi. The greatest decreases in viral RNA levels (∼30- to 100-fold) in the nasal washes relative to isotype control mAb-treated mice were observed in animals treated with mAbs in groups B (SARS2-02 and SARS2-55) and C (SARS2-01 and SARS2-38) (Figure 4E). The largest reductions in viral RNA levels in the lung (∼100- to 1,000-fold) again were observed for mice treated with mAbs in groups B (SARS2-02 and SARS2-55) and C (SARS2-38) (Figure 4F). A smaller (∼10-fold) decrement of virus RNA levels in the lungs was observed for the group D mAb SARS2-03. We also measured effects on infectious viral load in the lungs by plaque assay for a subset of representative mAbs from each group. Although group A mAb SARS2-71 did not decrease the number of plaque-forming units (PFUs) in the lungs relative to isotype control mAb-treated mice, SARS2-02, SARS2-38, and SARS2-03 reduced infectious virus levels to the limit of detection of the assay (Figure 4G). The lack of protection conferred by SARS2-71 in vivo was unanticipated given its neutralizing activity in cell culture (EC50 of 8 ng/mL; Figure 2). Sequencing of viral RNA from the lungs of SARS2-71-treated mice at 7 dpi revealed an S477N mutation in all samples that was not present in the input WA1/2020 virus. Notably, S477N also emerged in vitro as an escape mutant under SARS2-71 selection pressure using the VSV-eGFP-SARS-CoV-2-S virus (Figure 3A). Thus, despite its potent inhibitory activity in vitro, SARS2-71 failed to protect in vivo likely because of rapid emergence of a fully pathogenic escape mutant.
To further evaluate the level of protection conferred by a subset of mAbs in our panel, we measured levels of cytokines and chemokines in lung tissues at 7 dpi, which are markers of the inflammatory and pathological outcomes in this mouse model (Golden et al., 2020; Oladunni et al., 2020; Winkler et al., 2020; Yinda et al., 2021). SARS2-38 and SARS2-02 treatment resulted in reduced cytokine and chemokine levels relative to isotype control mAb-treated mice, with levels equivalent to those seen in naive mice (Figure S3). In contrast, treatment with SARS2-71 and SARS2-03 did not result in these reductions.
To test for post-exposure therapeutic protection against SARS-CoV-2 challenge, we cloned the variable regions of group B mAb SARS2-02 and group C mAb SARS2-38 and inserted them into a human IgG1 backbone to make chimeric antibodies. We did this because chimeric, humanized, or fully human mAbs are more likely to be used in humans and because Fc effector functions contribute to the therapeutic activity of neutralizing SARS-CoV-2 mAbs in vivo (Winkler et al., 2021); the original murine IgG1 isotype of these mAbs binds poorly to activating murine FcγRI and FcγRIV, whereas human IgG1 binds these murine Fc receptors with higher affinity and, thus, could have enhanced effector function (Dekkers et al., 2017). We confirmed the neutralizing activity of the chimeric mAbs hSARS2-02 and hSARS2-38 relative to the original murine versions of the mAbs (Figure S4A). Next we inoculated K18-hACE2 mice with 102 FFU of SARS-CoV-2 WA1/2020. Twenty-four hours later, we administered a single 200-μg (∼10 mg/kg) dose of hSARS2-02, hSARS2-38 or an isotype control mAb. hSARS2-02 and hSARS2-38 protected against weight loss following infection (Figure 4H). At 7 dpi, hSARS2-38 reduced viral RNA levels in the lungs and heart by ∼10,000-fold, whereas hSARS2-02 reduced infection by only ∼10- to 100-fold in these tissues (Figure 4I). These data demonstrate that mAb neutralization potency in vitro does not directly predict protective efficacy in vivo.
SARS2-38 neutralizes in vitro and protects in vivo against SARS-CoV-2 variants
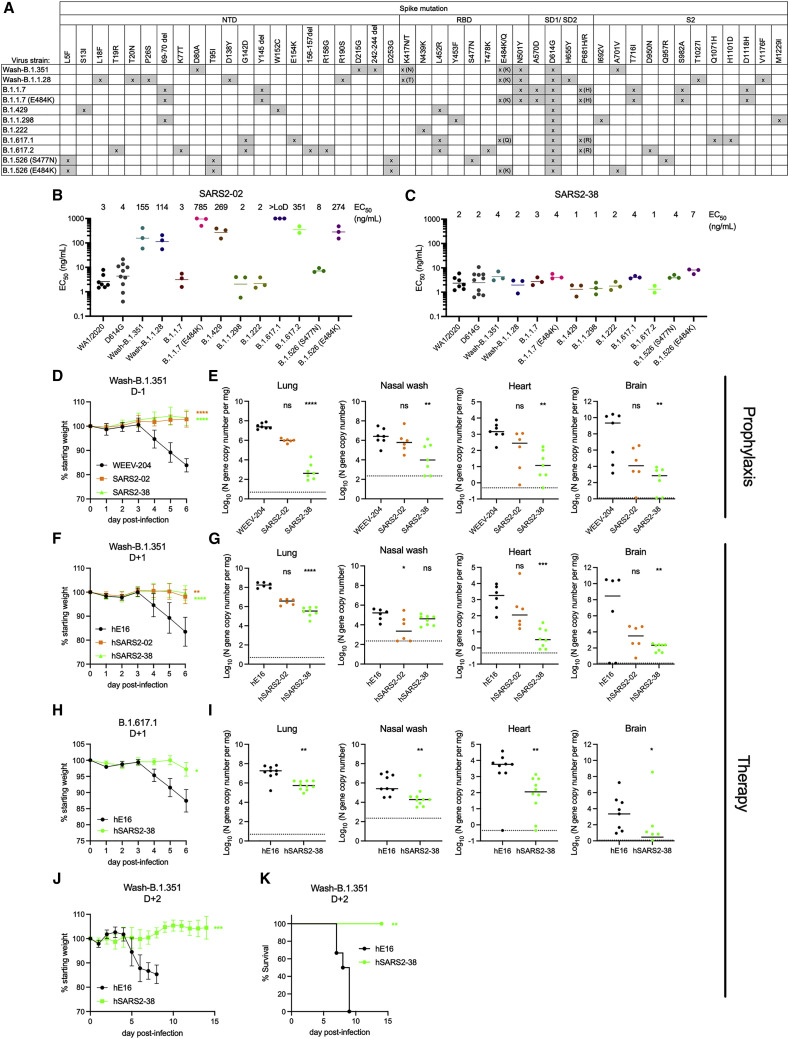
We tested the two mAbs (SARS2-02 and SARS2-38) that conferred the greatest protection against WA1/2020 in vivo for neutralization of viruses with spike proteins corresponding to circulating VOC and VOI. Recombinant chimeric WA1/2020 viruses encoding the spike protein from B.1.351 or B.1.1.28 (P.1 lineage) were utilized for these studies (Wash-B.1.351 and Wash-B.1.1.28), as well as WA1/2020 with an engineered D614G mutation. We also tested viral isolates B.1.1.7 (with and without E484K), B.1.429, B.1.1.298, B.1.222, B.1.617.1, and B.1.617.2 and two B.1.526 isolates (with E484K or S477N mutation). Several of these variants encode amino acid changes in spike that can affect mAb binding (Figure 5 A; Chen et al., 2021d; Shen et al., 2021; Wang et al., 2021), including changes we identified in our VSV-EGFP-SARS-CoV-2-S escape mutant panel: L452R and E484K showed reduced sensitivity to neutralization of VSV-EGFP-SARS-CoV-2-S by the group B mAbs SARS2-02 and SARS2-55. Indeed, SARS2-02 exhibited reduced (∼50- to 200-fold) neutralizing activity against SARS-CoV-2 strains with E484K (Wash-B.1.351, Wash-B.1.1.28, B.1.1.7+E484K, and B.1.526+E484K) or L452R (B.1.429 and B.1.617.2) substitutions and no inhibitory activity against B.1.617.1, which encodes E484Q and L452R mutations (Figures 5B and S4B). SARS2-38 did not lose potency against any of the variant viruses, with EC50 values ranging from 1–7 ng/mL across the tested panel (Figures 5C and S4B).
Figure 5.
Neutralization of SARS-CoV-2 variants by anti-SARS-CoV-2 mAbs
(A) SARS-CoV-2 variants and their mutations in spike.
(B and C) SARS2-2 (B) and SARS2-38 (C) were tested for neutralization of the indicated variants by FRNT. Mean EC50 values are shown; data are from 2–10 experiments. See Figures S4B and S4C for representative dose-response curves.
(D and E) K18-hACE2 mice were administered 100 μg (~5 mg/kg) of the indicated mAb by intraperitoneal injection 24 h prior to intranasal inoculation with 103 FFU of SARS-CoV-2 Wash-B.1.351.
(D) Mean weight change is shown. Error bars represent SEM.
(E) At 6 dpi, the indicated tissues were collected, and viral RNA levels were determined. Data are from two experiments; WEEV-204 (isotype control) and SARS2-38: n = 7; SARS2-02: n = 6.
(F and G) K18-hACE2 mice were inoculated with 103 FFU of SARS-CoV-2 Wash-B.1.351 and 24 h later were administered 200 μg (~10 mg/kg) of the indicated mAb.
(F) Mean weight change. Error bars represent SEM.
(G) At 6 dpi, tissues were collected, and viral RNA levels were determined. Data are from two experiments; hWNV-E16 and hSARS2-02: n = 6; hSARS2-38: n = 8.
(H and I) K18-hACE2 mice were inoculated with 5 × 103 FFU of SARS-CoV-2 B.1.617.1 and 24 h later were administered 200 μg (~10 mg/kg) of the indicated mAb.
(H) Mean weight change. Error bars represent SEM.
(I) At 7 dpi, tissues were collected, and viral RNA levels were determined. Data are from three experiments; hWNV-E16: n = 9; hSARS2-38: n = 10.
(J and K) K18-hACE2 mice were inoculated with 103 FFU of SARS-CoV-2 Wash-B.1.351 and 48 h later were administered 200 μg (~10 mg/kg) of the indicated mAb.
(J) Mean weight change. Error bars represent SEM.
(K) Survival with log-rank test.
In (D) and (F), we used one-way ANOVA with Dunnett’s post-test of the area under the curve. In (H) and (I), we used a t test of the area under the curve. In (E) and (G), we used a Kruskal-Wallis test with Dunn’s post-test. In (I), we used a Mann-Whitney test. For all statistical tests: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To expand this analysis, we tested the VSV-EGFP-SARS-CoV-2-S viruses that were resistant to SARS2-02 and SARS2-38 for neutralization using full dose-response curve analyses. SARS2-02 showed ∼20-fold reduced potency against E484K and ∼100-fold reduced potency against L452R and G446V and did not neutralize G446D at the highest concentration of mAb tested (Figure S4C). SARS2-38 showed virtually no neutralizing activity against K444E, K444N, G446D, or G446V mutants even at the highest concentration (1 μg/mL) of tested mAbs (Figure S4C). Despite these results with VSV-EGFP-SARS-CoV-2-S viruses, when we serially passaged authentic WA1/2020 D614G or Wash-B.1.351 SARS-CoV-2 in Vero-TMPRSS2-ACE2 cells in the presence of neutralizing mAbs, we readily isolated resistant viruses following SARS-02 but not SARS2-38 selection with both strains.
We tested SARS2-02 and SARS2-38 for protection against Wash-B.1.351 in K18-hACE2 mice as pre-exposure prophylaxis or post-exposure therapy. Animals treated with 100 μg of SARS2-02 or SARS2-38 24 h prior to infection were protected from weight loss (Figure 5D) despite the reduced neutralization potency of SARS2-02 against Wash-B.1.351. SARS2-38 treatment greatly reduced viral titers in the lungs, nasal washes, heart, and brain at 7 dpi compared with the isotype control-treated mice, whereas SARS2-02 had a less protective effect (Figure 5E). When hSARS2-02 and hSARS2-38 were administered to the K18-hACE2 transgenic mice as therapy 24 h after infection with Wash-B.1.351, a similar phenotype was observed; although they both protected mice against weight loss (Figure 5F), hSARS2-38 resulted in a greater reduction in viral titers at 7 dpi in the lungs, heart, and brain than hSARS2-02 (Figure 5G). We also tested hSARS2-38 for therapeutic protection against another variant, B.1.617.1, in K18-hACE2 mice. When administered 24 h after infection with B.1.617.1, hSARS2-38 protected mice against weight loss and viral infection in the lungs, heart, and brain (Figures 5H and 5I). To further evaluate its therapeutic window, we administered hSARS2-38 2 days after Wash-B.1.351 inoculation in K18-hACE2 mice. hSARS2-38 treatment fully protected mice against weight loss and lethality following Wash-B.1.351 infection (Figures 5J and 5K). Finally, hSARS2-38 also protected Syrian golden hamsters from Wash-B.1.351 when administered 24 h prior to infection, with hSARS2-38-treated hamsters showing no weight loss and decreased viral RNA and infectious virus levels in the lung (Figures S4D–S4F). Thus, SARS2-38 neutralizes a large panel of SARS-CoV-2 circulating variants in vitro and confers protection against multiple variants in vivo.
SARS2-38 targets the proximal RBM ridge with extensive light chain contact
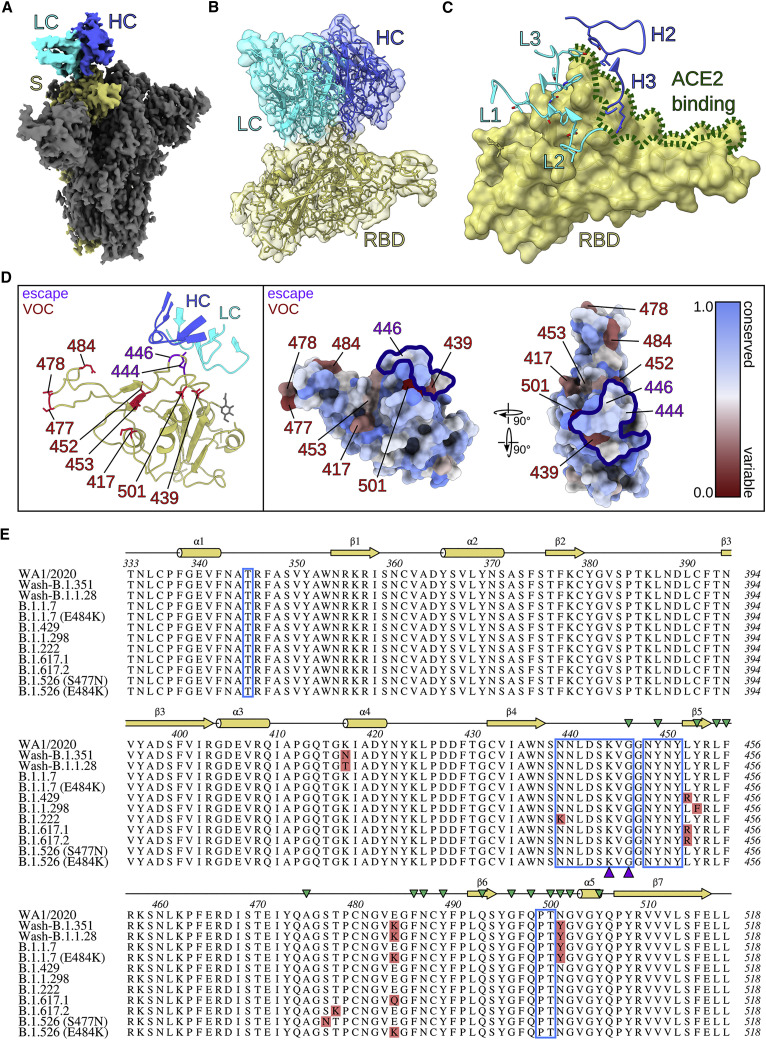
To further define the mechanistic basis for the broad and potent neutralization by SARS2-38, we first analyzed the interaction of antigen binding fragments (Fab) of SARS2-38 with SARS-CoV-2 spike using biolayer interferometry (BLI). SARS2-38 bound spike with high monovalent affinity (kinetically derived KD of 6.5 nM) and had a half-life of 4.8 min (Figure S5A). To understand the structural basis for this binding, we performed cryo-EM on complexes of SARS2-38 Fab and the SARS-CoV-2 spike protein (Figure S5B; Table S1). Using a large spherical mask and an ab initio spike density reference, we generated three-dimensional classes to sample the oligomeric states of the Fab/spike complex, and the class of highest resolution and clearest Fab density was refined further. This class consisted of trimeric spike with all RBDs in the down position (D/D/D) and one RBD bound by Fab (Figures 6 A and S6A). Using non-uniform refinement, we achieved an overall resolution of 3.20 Å, with local resolution ranging from ∼2.5 Å in the core of the spike to ∼5.5 Å in the constant region of the Fab, which was visible only at high contour (Figures S6B–S6D). Other binding configurations also were seen, although these classes aligned less clearly. The predominant class consisted of spike with one RBD up and two RBDs down (U/D/D), with only the up RBD bound by Fab (31.1%). Less frequently, all three RBDs were bound by Fab in the U/D/D conformation (22.0%; Figure S5B). Although SARS2-38 could bind SARS-CoV-2 spike with full occupancy, 61.1% of spike trimers were bound only by a single Fab molecule. Thus, despite binding spike with high affinity, it is likely that steric effects disfavor the interaction of multiple SARS2-38 Fabs with a single spike trimer.
Figure 6.
SARS2-38 targets a conserved portion of the RBM with extensive light chain contact
(A) Density map of SARS2-38 Fv bound to trimeric SARS-CoV-2 spike protein with all RBDs in the down position. The spike monomer bound by SARS2-38 is shown in yellow, with the rest of the trimer colored gray. The SARS2-38 heavy chain is shown in royal blue and the light chain in cyan.
(B) Focused density map of the Fv/RBD complex encompassing a refined atomic model. The RBD is shown in yellow. The SARS2-38 heavy and light chains are colored royal blue and cyan, respectively.
(C) Complementarity-determining regions (CDRs) of SARS2-38 overlay a surface rendering of the RBD. CDRs from the heavy and light chains are colored royal blue and cyan, respectively, with the RBD colored yellow. ACE2-binding residues of the receptor binding motif (RBM) are outlined in green.
(D) Left panel: a ribbon diagram of the RBD and SARS2-38 CDRs, with escape mutations and variants of concern (VOC) noted in purple and red, respectively. The RBD is otherwise colored yellow, with a gray glycan linked to N343. CDRs of the SARS2-38 heavy and light chains are colored royal blue and cyan, respectively. Right panel: surface renderings of RBD colored according to conservation of surface residues (blue = conserved, red = variable). Escape mutations and VOCs are noted in purple and red, respectively. The SARS2-38 epitope as determined by protein interfaces, surfaces, and assemblies (PISA) solvent exclusion analysis is outlined in navy.
(E) Multiple sequence alignment of RBD (residues 333–518) from WA1/2020 and SARS-CoV-2 VOC, with the PISA binding footprint of SARS2-38 boxed in blue. Mutations within SARS-CoV-2 VOC are highlighted in red, and SARS2-38 escape mutation contacts are marked with purple triangles. Secondary structure annotation is displayed above the alignment in yellow, with ACE2 contacts designated by green triangles (Lan et al., 2020).
To improve resolution at the Fab/RBD interface in the D/D/D reconstruction, we performed a focused refinement of the SARS2-38 variable domain (Fv) and RBD, excluding the rest of the spike and the constant region of the Fab. Optimal Fv density was generated using a highly constrained local search with starting orientations derived from the global refinement. This local refinement of the Fv/RBD complex achieved a nominal resolution of 3.16 Å, allowing placement of the protein backbone, secondary structures, and most side chains at the interface (Figures S5B, S6E, and S6F). The SARS2-38 Fv sits atop three loops protruding at the proximal end of the RBM between helix α1 and strand β1 (contact residue T345), strands β4 and β5 (N439–G446, N448–Y451), and strand β6 and helix α5 (P499–T500; Figures 6A and 6B); these results correspond well with our VSV-based escape mutant mapping (Figure 3). All three light-chain complementarity-determining regions (CDRs) contact loop β4-β5, with CDR2 and CDR3 forming additional contacts with loops α1-β1 and β6-α5, respectively (Figure 6C). In comparison, the heavy chain interacts in a more limited manner with loops β4-β5 and β6-α5 via CDR2 and CDR3 (Figure 6C). CDR1 of the heavy chain makes no contact at all with the RBD. The heavy chain does, however, engage ACE2 contact residues of the RBM (Figure 6C). This and other steric effects likely explain the inhibition of ACE2 binding by SARS2-38. These experiments provide a structural perspective for how SARS2-38 recognizes the RBD and blocks ACE2 interactions with spike protein.
The SARS2-38 epitope is conserved among circulating SARS-CoV-2 VOCs
SARS2-38 potently neutralized all tested VOC. To understand this broadly neutralizing activity, we mapped the SARS2-38 epitope alongside VOC mutations within the RBD (Figures 6D, left panel, and 6E). One mutation in the SARS2-38 footprint, N439K, is present in variant B.1.222 and resides at the periphery of the epitope. However, B.1.222 remained sensitive to neutralization by SARS2-38, and escape mutants at this residue were not generated in vitro, suggesting that N439 is not critical for SARS2-38 binding. N439 forms no close contacts (<3.9 Å) with SARS2-38 and accounts for only 1.4% of total buried surface area at the interface (Table S2). The SARS2-38 epitope includes no other residues corresponding to VOC mutations, which explains its performance against these variants. Notwithstanding this, we could select escape mutations in vitro in the context of VSV-EGFP-SARS-CoV-2-S chimeric virus, namely K444E/N and G446D/V substitutions, which reside on the β4-β5 loop central to the SARS2-38 epitope (Figures 6D and 6E). The substitutions generated at K444 result in loss of positive charge (K444N) or charge reversal (K444E), whereas mutations at G446 may distort the entire loop structure; in our model, G446 adopts a stereochemistry unique to the glycine residue (ϕ = 88.9° and ψ = 4.3°). Moreover, the distance between Cα of G446 and heavy chain I51 is only 4.6 Å, imposing space constraints on side chains at this position. This structural analysis likely explains the resistance conferred by these amino acid substitutions.
To understand the efficacy of SARS2-38 amidst the landscape of all circulating variants, we used the COVID-19 CoV Genetics Browser (https://covidcg.org) to probe RBD sequences in the global initiative on sharing avian influenza data (GISAID) database (786,273 isolates as of March 28, 2021; Chen et al., 2021a, Shu and McCauley, 2017). We then developed a log-scale conservation score for RBD residues 333–520. In this model, perfect conservation of the reference amino acid (from 2019n-CoV/WA1/2020) across all isolates corresponds to a score of 1, and complete loss of the reference amino acid results in a score of 0. Visualizing these scores on a color-coded RBD surface rendering (blue = 1, more conserved; red = 0, more variable) revealed that the RBM is generally more variable than the rest of the RBD, with VOC clearly seen as red patches (Figure 6D, right panel). This analysis also suggested that, in addition to not being affected by the VOC tested in this study, SARS2-38 targets a portion of the RBM that is conserved among circulating SARS-CoV-2 variants. The positions at which we identified escape mutants using VSV-EGFP-SARS-CoV-2-S chimeric viruses were substituted in only 0.02% (K444) and 0.04% (G446) of isolates, with the specific escape mutations (K444E/N and G446D/V) observed in only 0.007% and 0.03% of isolates, respectively. Overall, 99.96% of isolates lacked the escape mutations for SARS2-38 identified in our study. These analyses reveal the structural basis of the broadly neutralizing activity of SARS2-38 against VOC and VOI.
SARS2-38 targets a similar but distinct epitope than other broadly neutralizing SARS-CoV-2 mAbs
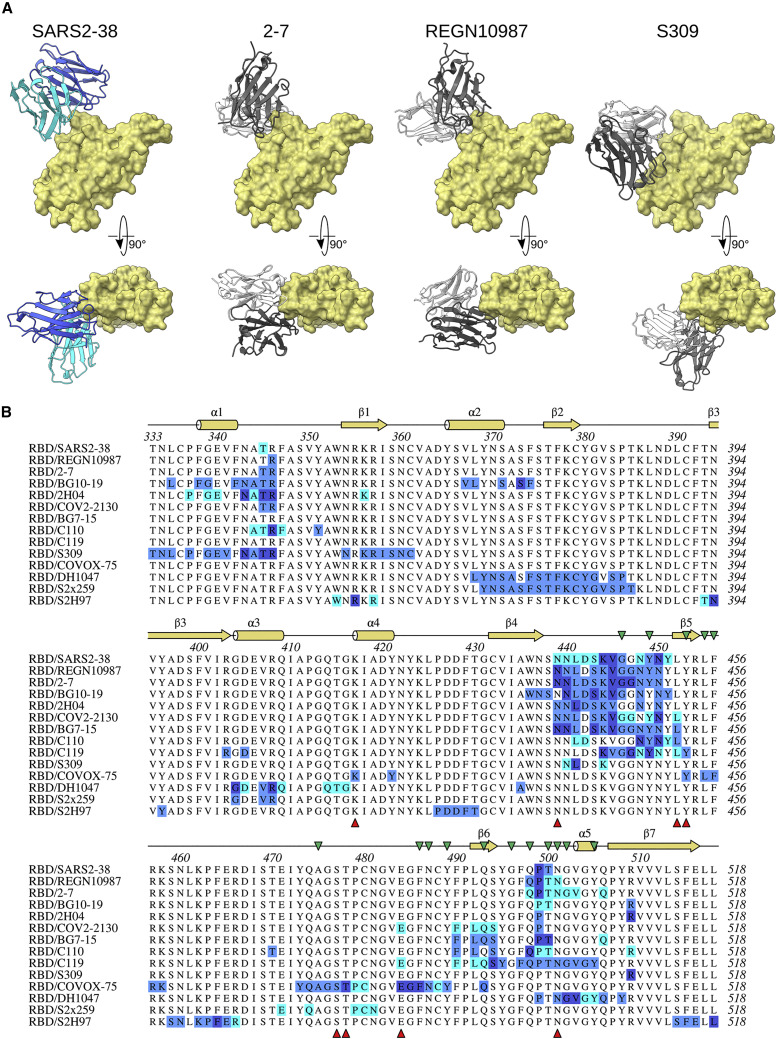
Relatively few mAbs targeting the SARS2-38 epitope have been described, and those characterized bind the RBD in distinct orientations with heavy chain predominance (Figure 7 A). These include murine mAb 2H04 as well as human mAbs REGN10987, COV2-2130, and, though less similar, S309 (Dong et al., 2021; Hansen et al., 2020; Liu et al., 2021; Pinto et al., 2020). SARS2-38 differs in two respects: (1) the baseline neutralizing activity of SARS2-38 against WA1/2020 in Vero cells (EC50, ∼5 ng/mL) is 30-fold, 20-fold, and 16-fold more potent than that of 2H04, COV2-2130, and S309, respectively (Alsoussi et al., 2020; Pinto et al., 2020; Zost et al., 2020); and (2) SARS2-38 retains strong neutralization potency against all VOC evaluated in this study, whereas the inhibitory activity of 2H04, COV2-2130, and S309 is somewhat reduced against B.1.1.7, B.1.429, and B.1.351, respectively (Chen et al., 2021c, 2021d; R.E.C. and M.S.D., unpublished data; Table S3). Similarly, REGN10987 exhibited a 10-fold reduction in neutralizing activity against B.1.429 compared with WA1/2020 (Chen et al., 2021c; Hansen et al., 2020; Wang et al., 2021). A structural examination of these other antibody footprints within the context of VOC mutations does not provide a clear explanation for some of the resistance (Figure 7B). Instead, allostery may play a role. While other broadly and potently neutralizing mAbs (including mAbs 2C08, COV2-2196, 58G6, 510A5, S2X259, S2H97, and DH1047) have been reported that bind RBD epitopes at residues G476, F486, and N487 or loops near residues 369–386, 404–411, 450–458, 460–466, and 499–508 (Dong et al., 2021; Li et al., 2021b; Martinez et al., 2021; Schmitz et al., 2021; Starr et al., 2021a; Tortorici et al., 2021), SARS2-38 targets a distinct epitope proximal to the RBM. Although other broadly neutralizing mAbs targeting a similar portion of the RBD have been described (e.g., mAbs 2-7, BG10-19, and BG7-15; Table S3), highlighting the conservation of this epitope (Cerutti et al., 2021; Dejnirattisai et al., 2021; Li et al., 2021a; Robbiani et al., 2020; Scheid et al., 2021; Schäfer et al., 2021), SARS2-38 interacts with different contact residues and employs a unique binding mode.
Figure 7.
Similarity of SARS2-38 epitope to other mAbs
(A) Structural comparison of SARS2-38 to selected mAbs targeting a similar region of the RBD. SARS2-38 heavy chain and light chain are colored royal blue and cyan, respectively. Antibodies 2-7 (PDB: 7LSS), REGN10987 (PDB: 6XDG), and S309 (PDB: 6WPS) are depicted with white light chains and dark gray heavy chains.
(B) Multiple sequence alignment of the SARS-CoV-2 RBD (residues 333–518) with mAb binding footprints as determined by PISA solvent exclusion analysis. The heavy chain, light chain, and shared contacts are shown in blue, cyan, and dark blue, respectively. Secondary structure annotation is displayed above the alignment in yellow, with ACE2 contacts designated by green triangles (Lan et al., 2020). VOC substitutions are designated below the alignment by red triangles and include residues from B.1.1.7, B.1.429, B.1.1.298, Wash-B.1.351, Wash-B.1.1.28, B.1.222, B.1.617.1, B.1.526+S477N, B.1.526+E484K, and B.1.617.2.
Discussion
In this study, we describe a panel of mAbs that bind the RBD of the SARS-CoV-2 spike protein. Several anti-RBD mAbs protected in vivo against SARS-CoV-2 infection in K18-hACE2 transgenic mice. Although the less potently neutralizing mAbs directed against epitopes on the base of the RBD (SARS2-10, SARS2-31, and SARS2-03) exhibited diminished protection compared with mAbs recognizing the RBM, neutralization potency was not the only predictor of in vivo efficacy. Indeed, SARS2-71 neutralized SARS-CoV-2 with a potency similar to that of protective mAbs SARS2-02 and SARS2-38 but failed to confer protection in mice. Notwithstanding this result, antibodies targeting proximal competing epitopes as SARS2-71, such as COV2-2196, have been shown to confer protection in vivo (Zost et al., 2020). The failure of SARS2-71 to protect in particular is likely due to the emergence of the escape variant S477N in vivo. This finding demonstrates that SARS-CoV-2 can escape rapidly from mAb inhibition in vivo and that mAb or mAb cocktails that limit rapid escape mutant generation will likely have greater therapeutic utility. Although currently authorized mAb treatments include cocktails, the emergence of VOC that are resistant to one or both component mAbs could compromise drug efficacy.
The most potently inhibitory mAbs in our panel bind epitopes within or proximal to the RBM and inhibit spike interaction with hACE2, as observed for other anti-SARS-CoV-2 mAbs (Zost et al., 2020). Several of these mAbs inhibited virus attachment to Calu-3 and Vero-TMPRSS2-ACE2 cells but not to Vero E6 cells or Vero-TMPRSS2 cells. Infection of Vero E6 cells by SARS-CoV-2 is dependent on endogenous levels of monkey ACE2 expression because pretreatment with anti-ACE2 mAbs inhibits infection (Hoffmann et al., 2020). However, other host factors, such as heparan sulfate, can also mediate virus attachment to cells (Chu et al., 2021; Clausen et al., 2020). If binding to other cell surface ligands occurs prior to the RBD-ACE2 interaction, then mAbs that block ACE2 binding may not efficiently inhibit SARS-CoV-2 attachment but, instead, block a downstream ACE2-dependent entry step. This idea is supported by our data showing that several neutralizing mAbs block viral internalization in Vero E6 cells. Moreover, anti-RBD mAbs have only moderate decreases in neutralization potency when added after virus absorption to Vero E6 cells. In contrast, when SARS-CoV-2 attaches to the cell surface via hACE2 interaction, such as in Vero-TMPRSS2-ACE2 cells, addition of anti-RBD mAbs after attachment failed to neutralize virus infection. A higher density of ACE2 or higher affinity of spike protein for hACE2 (relative to monkey ACE2) on the Vero-TMPRSS2-ACE2 cells may drive initial virus attachment through the RBD-ACE2 interaction and explain why mAbs can block this step in these cells. It is possible that multiple ACE2 interaction events occur during viral entry (during attachment or post-attachment steps), which ACE2-blocking mAbs can inhibit. These data suggest that the ability of anti-RBD mAbs to inhibit SARS-CoV-2 attachment depends on cellular ACE2 expression levels. Because these mechanistic differences did not markedly affect mAb potency on the different cellular substrates, we conclude that, in the cells we tested, there is a required entry interaction with ACE2 at attachment, post-attachment, or internalization steps.
Several mutations and deletions in emerging VOC and VOI occur in the NTD and RBD that allow them to evade antibody recognition, including RBD mutations K417N/T (B.1.351 and B.1.1.28), N439K (B.1.222), L452R (B.1.429, B.1.617.1, and B.1.617.2), Y453F (B.1.1.298), E484K (B.1.351, B.1.1.28, and some isolates in B.1.526 and B.1.1.7 lineages), E484Q (B.1.617.1), and N501Y (B.1.1.7, B.1.351, and B.1.1.28) (reviewed by Plante et al., 2021), highlighting the importance of developing mAbs against a variety of spatially distinct epitopes. In our panel, SARS2-38 potently neutralized viruses encoding any of the above mutations, did not readily select for escape mutations with authentic SARS-CoV-2 strains, and retained therapeutic activity in vivo against viruses containing VOC substitutions. Moreover, functional mapping and structural analysis of the binding footprint of SARS-CoV-2 defined a conserved RBD epitope that could be recognized by other potently neutralizing and protective human mAbs.
We characterized a panel of anti-SARS-CoV-2 mAbs, defined their cellular mechanism of action in different cells, tested in vitro neutralizing and in vivo protection capacity against historical and circulating variants, and determined the structure of the viral spike protein bound to SARS2-38, a potently and broadly neutralizing mAb that recognizes emerging VOC. The conserved epitope bound by SARS2-38 may be a potential target for antibodies with therapeutic potential or those induced by vaccines with more limited potential for resistance against VOC.
Limitations of the study
Although we demonstrated that hSARS2-38 confers protection against SARS-CoV-2 in rodent models of disease, corroboration in non-human primates and humans remains to be established. Furthermore, SARS-CoV-2 variants continue to emerge and will need to be tested for neutralization sensitivity to hSARS2-38 and other similar mAbs with therapeutic potential. Antibody cocktails that include multiple mAbs of different specificities against conserved epitopes may ensure broad protection against existing and future SARS-CoV-2 variants.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS2-01 | This paper | N/A |
| SARS2-02 | This paper | N/A |
| SARS2-03 | This paper | N/A |
| SARS2-10 | This paper | N/A |
| SARS2-16 | This paper | N/A |
| SARS2-31 | This paper | N/A |
| SARS2-34 | This paper | N/A |
| SARS2-38 | This paper | N/A |
| SARS2-55 | This paper | N/A |
| SARS2-71 | This paper | N/A |
| COV2-2130 | Zost et al., 2020 | N/A |
| COV2-2196 | Zost et al., 2020 | N/A |
| CR3022 | ter Meulen et al., 2006 | N/A |
| hSARS2-02 (chimeric human mAb) | This paper | N/A |
| hSARS2-38 (chimeric human mAb) | This paper | N/A |
| Anti-mouse IgG-HRP | Sigma | Cat # A8924, RRID: AB_258426 |
| goat anti-mouse IgA/G/M | SeraCare | Cat# 5210-0187 |
| Bacterial and virus strains | ||
| VSV-SARS-CoV-2 (WT and panel of escape mutants) | Liu et al., 2021 and this paper | N/A |
| SARS-CoV-2 WA1/2020 | CDC | N/A |
| SARS-CoV-2 D614G | Chen et al., 2021 | N/A |
| SARS-CoV-2 Wash-B.1.351 | Chen et al., 2021 | N/A |
| SARS-CoV-2 Wash-B.1.1.28 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.1.7 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.429 | C. Chiu and R. Andino labs (UCSF) | N/A |
| SARS-CoV-2 B.1.1.298 | BEI Resources | NR-53953 |
| SARS-CoV-2 B.1.222 | BEI Resources | NR-53945 |
| SARS-CoV-2 B.1.617.1 | M. Suthar laboratory (Emory University) | N/A |
| SARS-CoV-2 B.1.617.2 | R. Webby laboratory (St Jude Children Research Hospital) | N/A |
| SARS-CoV-2 B.1.526 (S477N) | M. Suthar laboratory (Emory University) | N/A |
| SARS-CoV-2 B.1.526 (E484K) | M. Suthar laboratory (Emory University) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 spike protein | Fremont laboratory | N/A |
| SARS-CoV-2 spike RBD | Fremont laboratory | N/A |
| Deposited data | ||
| SARS2-38/RBD structure (local) | This paper | PDB: 7MKM |
| SARS2-38/RBD EM density map (local) | This paper | EMD-23899 |
| SARS2-38/spike structure (three down conformation) | This paper | PDB: 7MKL |
| SARS2-38/spike EM density map (three down conformation) | This paper | EMD-23898SARS |
| SARS2-02 VH sequence | This paper | GenBank: MZ703121 |
| SARS2-02 VK sequence | This paper | GenBank: MZ703122 |
| SARS2-38 VH sequence | This paper | GenBank: MZ703123 |
| SARS2-38 VK sequence | This paper | GenBank: MZ703124 |
| Experimental models: Cell lines | ||
| Vero-E6 | ATCC | CRL-1586; RRID:CVCL_0574 |
| Vero+TMPRSS2 | Chen et al., 2021 | N/A |
| Vero+TMPRSS2+ACE2 | Chen et al., 2021 | N/A |
| MA104 | ATCC | CRL-2378.1 |
| Calu-3 | ATCC | HTB-55 |
| Experimental models: Organisms/strains | ||
| K18-hACE2 transgenic mice | Jackson laboratory | strain # 34860 |
| Golden Syrian Hamsters | Charles River | Strain 049 |
| Oligonucleotides | ||
| SARS-CoV-2 qPCR primer 5′- ATGCTGCAATCGTGCTACAA-3′ | Case et al., 2020 | N/A |
| SARS-CoV-2 qPCR primer 5′- GACTGCCGCCTCTGCTC-3′ | Case et al., 2020 | N/A |
| SARS-CoV-2 qPCR probe 5′-/56-FAM/ TCAAGGAAC/Zen/ AACATTGCCAA/3IABkFQ-3′ | Case et al., 2020 | N/A |
| GAPDH qPCR primer+probe | IDT PrimeTime Assay | Hs.PT.39a.22214836 |
| Software and algorithms | ||
| MotionCor2 v1.3.1 | Zheng et al., 2017 | N/A |
| GCTF v1.18 | Zhang, 2016 | N/A |
| CrYOLO v1.7.6 | Wagner et al., 2019 | N/A |
| Relion 3.1 | Scheres, 2012; Zivanov et al., 2018 | N/A |
| cryoSPARC v3.1.0 | Punjani et al., 2017 | N/A |
| DeepEMhancer | Sanchez-Garcia et al., 2021 | N/A |
| Coot v0.9.5 | Emsley et al., 2010 | N/A |
| Isolde v1.1.0 | Croll, 2018 | N/A |
| Phenix v1.19 | Adams et al., 2010 | N/A |
| qtPISA | Krissinel and Henrick, 2007 | N/A |
| MDTraj | McGibbon et al., 2015 | N/A |
| UCSF ChimeraX | Goddard et al., 2018 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Michael S. Diamond (diamond@wusm.wustl.edu).
Materials availability
All requests for resources and reagents should be directed to the Lead Contact author. This includes mice, antibodies, viruses, and proteins. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Viruses
The 2019n-CoV/USA_WA1/2020 (WA1/2020) isolate of SARS-CoV-2 was obtained from the US Centers for Disease Control (CDC). WA1/2020 stocks were propagated on Vero CCL81 cells and used at passage 6 and 7. Viral titer was determined by focus-forming assay (FFA) on Vero E6 cells as described (Case et al., 2020). The D614G virus was produced by introducing the mutation into an infectious clone of WA1/2020, and the B.1.351 and B.1.1.28 spike genes were cloned into the WA1/2020 infectious clone to produce Wash-B.1.351 and Wash-B.1.1.28 chimeric viruses, as described previously (Chen et al., 2021d). The B.1.1.7, B.1.429, B.1.1.298, B.1.222, B.1.617.1, B.1.617.2, B.1.526+S477N, and B.1.526+E484K isolates were isolated from infected individuals. Viruses were propagated on Vero-TMPRSS2 cells and subjected to deep sequencing to confirm the presence of the substitutions indicated in Figure 5A.
Cells
Cell lines were maintained at 37°C in the presence of 5% CO2. Vero E6 cells were passaged in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Omega Scientific) and 100 U/mL penicillin-streptomycin (P/S) (Invitrogen). Vero cells that overexpress TMPRSS2 or TMPRSS2-ACE2 were maintained as Vero CCL81 cells, with the addition of 5 μg/mL blasticidin (Vero-TMPRSS2) or 10 μg/mL puromycin (Vero-TMPRSS2-ACE2). Calu-3 cells were maintained in DMEM with 20% FBS and 100 U/mL P/S.
Proteins
Genes encoding SARS-CoV-2 spike protein (residues 1-1213, GenBank: MN908947.3) and RBD (residues 319-514) were cloned into a pCAGGS mammalian expression vector with a C-terminal hexahistidine tag. The spike protein was prefusion stabilized and expression optimized via six proline substitutions (F817P, A892P, A899P, A942P, K986P, V987P) (Hsieh et al., 2020), with a disrupted S1/S2 furin cleavage site and a C-terminal foldon trimerization motif (YIPEAPRDGQAYVRKDGEWVLLSTFL). Expi293F cells were transiently transfected, and proteins were recovered via cobalt-charged resin chromatography (G-Biosciences) as previously described (Alsoussi et al., 2020; Hassan et al., 2020). For ACE2 binding inhibition analysis, the SARS-CoV-2 spike protein was made by synthesizing a gene encoding the ectodomain of a prefusion conformation-stabilized SARS-CoV-2 spike (S6Pecto) protein (Hsieh et al., 2020) containing C-terminal Twin-Strep-tag. The spike gene was then cloned it into a DNA plasmid expression vector for mammalian cells. Protein was produced in FreeStyle 293-F cells (Thermo Fisher Scientific) and purified from culture supernatants using StrepTrap HP affinity column (Cytiva).
Mice
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
K18-hACE2 transgenic mice were purchased from Jackson Laboratories (#034860) and housed in a pathogen-free animal facility at Washington University in St. Louis. For passive transfer studies, mAbs were diluted in PBS and administered to mice via intraperitoneal injection in a 100 μL total volume. Viral infections were performed via intranasal inoculation with 103 FFU of virus. Mice were monitored daily for weight loss.
Hamsters
Five to six-week-old male Syrian hamsters were purchased from Charles River Laboratories and housed in microisolator units. All hamsters were allowed free access to food and water and cared for under United States Department of Agriculture (USDA) guidelines for laboratory animals. Hamsters were administered by intraperitoneal injection hSARS2-38 or isotype control (hE16; 10 mg/kg) antibody. One day later, hamsters were given 5 × 105 PFU of Wash-B.1.351 SARS-CoV-2 by the intranasal route in a final volume of 100 μL. All hamsters were monitored for body weight loss until humanely euthanized at 4 dpi. All procedures were approved by the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with 5% isoflurane. All efforts were made to minimize animal suffering.
Method details
mAb generation
BALB/c mice were immunized with 10 μg of SARS-CoV-2 RBD adjuvanted with 50% AddaVax (InvivoGen), via intramuscular route (i.m.), followed by i.m. immunization two and four weeks later with SARS-CoV-2 spike protein (5 μg and 10 μg, respectively) supplemented with AddaVax. Mice received a final, non-adjuvanted boost of 25 μg of SARS-CoV-2 spike or RBD (12.5 μg intravenously and 12.5 μg interperitoneally) 3 days prior to fusion of splenocytes with P3X63.Ag.6.5.3 myeloma cells. Hybridomas producing antibodies that bound to SARS-CoV-2-infected permeabilized Vero CCL81 cells by flow cytometry and to SARS-CoV-2 recombinant spike protein by direct ELISA were cloned by limiting dilution. All hybridomas were screened initially with a single-endpoint neutralization assay using hybridoma supernatant diluted 1:3 and incubated with SARS-CoV-2 for 1 h at 37°C prior to addition to Vero E6 cells. Following a 30-h incubation, cells were fixed, permeabilized, and stained for SARS-CoV-2 infection with CR3022 as described (Case et al., 2020). A subset of neutralizing hybridoma supernatants were purified commercially (Bio-X Cell) after adaptation for growth under serum-free conditions.
VSV-eGFP-SARS-CoV-2-S escape mutants
VSV-eGFP-SARS-CoV-2-S escape mutants were produced as described previously (Liu et al., 2021). Briefly, plaque assays were performed to isolate escape mutants on Vero-TMPRSS2 cells with neutralizing mAb in the overlay. Escape clones were plaque-purified on Vero-TMPRSS2 cells in the presence of mAb. Plaques in agarose plugs and viral stocks were amplified on MA104 cells at an MOI of 0.01 in Medium 199 containing 2% FBS and 20 mM HEPES pH 7.7 (Millipore Sigma) at 34°C. Viral supernatants were harvested upon extensive cytopathic effect and clarified of cell debris by centrifugation at 1,000 x g for 5 min.
Determination of mAb concentration in hybridoma supernatant
The mAb concentration in each hybridoma supernatant was quantified by ELISA. Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 1 μg/mL of goat anti-mouse IgG (Southern Biotech) in 50 μL of NaHCO3 (pH 9.6) coating buffer and incubated overnight at 4°C. Plates were washed three times with ELISA wash buffer (PBS containing 0.05% Tween-20), and then incubated with 200 μL of blocking buffer (PBS, 2% BSA, 0.05% Tween-20) for 1 h at room temperature. Plates were incubated with hybridoma supernatant diluted 1:500 or 1:2000 in blocking buffer, or serial dilutions of purified isotype control mAb as a standard, for 1 h at room temperature. Plates were washed three times with ELISA wash buffer, and incubated with 50 μL of anti-mouse IgG-HRP (Sigma) diluted 1:500 for 1 h at room temperature. Plates were washed three times with ELISA wash buffer and three times with PBS, before incubation with 100 μL of TMB substrate (Thermo Fisher Scientific) for 3 min at room temperature before quenching with the addition of 50 μL of 2 N H2SO4 and measuring OD 450 nm. Antibody concentrations in hybridoma supernatant were interpolated from a standard curve produced using an isotype control mAb.
Spike and RBD binding analysis
96-well Maxisorp plates were coated with 2 μg/mL of SARS-CoV-2 spike or RBD protein in 50 mM Na2CO3 (70 μL) overnight at 4 °C. Plates were washed three times with PBS + 0.05% Tween-20 and blocked with 200 μL of PBS + 0.05% Tween-20 + 1% BSA + 0.02% NaN3 for 2 h at room temperature. 75 μL of blocking buffer and 50 μL of hybridoma supernatant were combined, and 50 μL/well of diluted supernatants were added to the plates and incubated for 1 h at room temperature. Bound IgG was detected using HRP-conjugated goat anti-mouse IgG (at 1:2,000). Following a 1 h incubation, washed plates were developed with 50 μL of 1-Step Ultra TMB-ELISA, quenched with 2 N H2SO4, and the absorbance was read at 450 nm.
Competition binding analysis
The assay was performed as described previously (Zost et al., 2020). Briefly, for screening study wells of 384-well microtiter plates were coated with 1 μg/mL of purified SARS-CoV-2 S6Pecto protein at 4 °C overnight. Plates were blocked with 2% bovine serum albumin (BSA) in DPBS-T for 1 h. Mouse hybridoma culture supernatants were diluted five-fold in blocking buffer, added to the wells (20 μL per well) in duplicates for each tested reference mAb and incubated for 1 h at room temperature. Biotinylated reference human mAbs with known epitope specificity (COV2-2130, COV2-2196 [Zost et al., 2020] and CR3022 [ter Meulen et al., 2006]) were added to each of well with the respective hybridoma culture supernatant at 1.25 μg/mL in a volume of 5 μL per well (final concentration of biotinylated mAb, 0.25 μg/mL) without washing of the plates, and then incubated for 1 h at room temperature. Plates then were washed, and bound antibodies were detected using HRP-conjugated avidin (Sigma, A3151, 0.3 μg/mL final concentration) and a TMB substrate. The signal obtained for binding of the biotin-labeled reference antibody in the presence of the hybridoma culture supernatant was expressed as a percentage of the binding of the reference antibody alone after subtracting the background signal. Tested mAbs were considered competing if their presence reduced the reference antibody binding to less than 41% of its maximal binding and non-competing if the signal was greater than 71%. A level of 40%–70% was considered intermediate competition.
Human ACE2 binding inhibition analysis
The assay was performed as described previously (Zost et al., 2020). Briefly, for screening study wells of 384-well microtiter plates were coated with 1 μg/mL purified recombinant SARS-CoV-2 S6Pecto protein at 4 °C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS-T for 1 h. Mouse hybridoma culture supernatants were diluted five-fold in blocking buffer, added to the wells (20 μL per well) in quadruplicate, and incubated for 1 h at room temperature. Recombinant human ACE2 with a C-terminal Flag tag peptide was added to wells at 2 μg/mL in a 5 μL per well volume (final 0.4 μg/mL concentration of human ACE2) without washing of the plates, and then incubated for 40 min at room temperature. Plates were washed and bound human ACE2 was detected using HRP-conjugated anti-Flag antibody (Sigma-Aldrich, A8592, 1:5,000 dilution) and TMB substrate. ACE2 binding without antibody served as a control for maximal binding. Antibody COV2-2196 (RBD) served as a control for ACE2 binding inhibition. The signal obtained for binding of the human ACE2 in the presence of each dilution of tested culture supernatant was expressed as a percentage of the human ACE2 binding without antibody after subtracting the background signal.
Sequencing, cloning, and expression of chimeric IgG1
To generate chimeric human IgG1 from mouse hybridoma cell lines, cells were lysed in Trizol (Thermo) followed by RNA purification with Direct-Zol Micro kit (Zymo). 5′ RACE products were generated with Template Switching RT Enzyme Mix (New England Biolabs) using anchored poly(dT)23 and TSO (GCT AAT CAT TGC AAG CAG TGG TAT CAA CGC AGA GTA CAT rGrGrG) oligonucleotides according to the manufactures instructions. Heavy and light chain sequences were amplified with primers specific for the TSO handle-sequence and the respective constant region sequence with Q5 Polymerase (New England Biolabs). Following Sanger sequencing, full-length variable regions were synthesized as gene blocks (Integrated DNA Technologies) and cloned into hIgG1 and hKappa expression vectors by Gibson assembly. Recombinant antibodies were expressed in Expi293 cells following co-transfection of heavy and light chain plasmids (1:1 ratio) using Expifectamine 293 (Thermo Fisher Scientific). Supernatants were harvested after 5-6 days, purified by affinity chromatography (Protein A Sepharose, GE), and desalted with a PD-10 (Cytiva) column.
Binding analysis via biolayer interferometry
Biolayer interferometry (BLI) was used to quantify the binding capacity of SARS2-38 Fab fragments to trimerized SARS-CoV-2 spike. 10 μg/mL of biotinylated spike was immobilized onto streptavidin biosensors (ForteBio) for 3 min. After a 30 s wash, the pins were submerged in running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% P20 surfactant, and 1% BSA) containing SARS2-38 Fab ranging from 1 to 1,000 nM, followed by a dissociation step in running buffer alone. The BLI signal was recorded and analyzed using BIAevaluation Software (Biacore).
Cryo-EM sample preparation
Data were collected on lacey carbon grids with or without ultra-thin carbon film. For standard lacey carbon grids (Ted Pella #01895-F), SARS-CoV-2 spike was prepared at 1 mg/mL in TBS (30 mM Tris pH 8, 150 mM NaCl). For lacey carbon grids with ultra-thin carbon film (Ted Pella #01824G), SARS-CoV-2 spike was prepared at 0.2 mg/mL in TBS. Each sample was incubated for 15 min with 1 molar equivalent of SARS2-38 Fab fragments, applied to glow-discharged grids, then flash-frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific).
Cryo-EM data collection
Grids were loaded into a Cs-corrected FEI Titan Krios 300kV microscope equipped with a Falcon 4 direct electron detector. Images were collected at a nominal magnification of 59000x, resulting in a pixel size of 1.16Å. Each movie consisted of 50 frames at 260ms each with a dose of 1e-/Å2/frame, yielding a total dose of 50e-/Å2/movie.
Cryo-EM data processing
Movies were motion corrected using MotionCor2 v1.3.1 (Zheng et al., 2017), and contrast transfer function parameters were estimated using GCTF v1.18 (Zhang, 2016). Particles were picked using a general model in CrYOLO v1.7.6 (Wagner et al., 2019). 2D classification was performed in Relion 3.1 (Scheres, 2012; Zivanov et al., 2018), and particles in good classes from grids with or without ultra-thin carbon were combined for further processing. These particles were subjected to 3D classification with a large spherical mask and a low resolution spike reference generated ab initio. The class of highest resolution and clearest Fab density (consisting of all RBDs in the down position, with one bound by Fab) was selected for iterative Bayesian polishing and per-particle CTF refinement in Relion 3.1 (Zivanov et al., 2019). These particles were then used in non-uniform refinement in cryoSPARC v3.1.0 to generate a full-spike map (Punjani et al., 2017). To improve map quality at the Fab/spike interface, a mask was generated encompassing only the Fv and RBD, and particles were subjected to highly constrained, local non-uniform refinement with dynamic masking in cryoSPARC v3.1.0. Final maps were sharpened via deep learning employed through DeepEMhancer (Sanchez-Garcia et al., 2021).
Model building
The locally refined map was used to construct a model of the RBD bound by SARS2-38 Fv. An initial model for the RBD was adapted from a crystal structure of RBD bound to ACE2 (PDB: 6M0J). For initial modeling of SARS2-38 Fv, pBLAST was used to identify pre-existing Fab structures with high sequence similarity (PDB: 1KIQ for VH, and PDB: 5XJM for VL). These starting components were combined and docked into the map, then refined in Coot v0.9.5 (Emsley et al., 2010), Isolde v1.1.0 (Croll, 2018), and Phenix v1.19 (Adams et al., 2010). Epitope and paratope contacts were identified using Protein Interfaces, Surfaces and Assemblies (PISA) solvent exclusion analysis (Krissinel and Henrick, 2007); close contacts and buried surface area were determined using MDTraj (McGibbon et al., 2015); and structures were visualized using UCSF ChimeraX (Goddard et al., 2018).
The full-spike map was used to construct a model of the spike bound by one Fv with all RBDs in the down position. An initial model was generated by combining the locally refined Fv/RBD structure with a previously solved cryo-EM structure of trimeric SARS-CoV-2 spike in the proper RBD configuration (PDB: 6VXX). This model was docked into the full-spike map then refined using Coot v0.9.5, Isolde v1.1.0, and Phenix v1.19.
RBD conservation analysis
RBD sequence data (residues 333-520) were retrieved on March 28, 2021 from the COVID-19 CoV Genetics Browser (covidcg.org), enabled by data from GISAID (Chen et al., 2021a; Shu and McCauley, 2017). In total, 786,273 sequences were included in the analysis. Probability of conservation relative to the reference sequence (2019n-CoV/WA1/2020) was computed for each residue, and results were log-transformed and normalized to generate a per-residue conservation score (1 = complete conservation, 0 = zero conservation). Results were visualized using a color-coded surface rendering of the RBD in UCSF ChimeraX.
Neutralization assays
FRNTs were performed as described (Case et al., 2020). Briefly, serial dilutions of antibody were incubated with 2 × 102 FFU of SARS-CoV-2 for 1 h at 37°C. Immune complexes were added to cell monolayers (Vero E6 cells or other cell lines where indicated) and incubated for 1 h at 37°C prior to the addition of 1% (w/v) methylcellulose in MEM. Following incubation for 30 h at 37°C, cells were fixed with 4% paraformaldehyde (PFA), permeabilized and stained for infection foci with SARS2-16 (hybridoma supernatant diluted 1:6,000 to a final concentration of ∼20 ng/mL) when using SARS-CoV-2 isolate WA1/2020, or with a mixture of mAbs that bind various epitopes on the RBD and NTD of spike (SARS2-02, SARS2-11, SARS2-31, SARS2-38, SARS2-57, and SARS2-71; diluted to 1 μg/mL total mAb concentration) for the VOC. Antibody-dose response curves were analyzed using non-linear regression analysis (with a variable slope) (GraphPad Software). The antibody half-maximal inhibitory concentration (EC50) required to reduce infection was determined.
Pre- and post-attachment neutralization assays
For pre-attachment assays, serial dilutions of mAbs were prepared at 4°C in Dulbecco’s modified Eagle medium (DMEM) with 2% FBS and preincubated with 102 FFU of SARS-CoV-2 for 1 h at 4°C. mAb-virus complexes were added to a monolayer of Vero cells for 1 h at 4°C. Virus was allowed to internalize during a 37°C incubation for 30 min. Cells were overlaid with 1% (wt/vol) methylcellulose in MEM. For post-attachment assays, 2 × 102 FFU of SARS-CoV-2 was adsorbed onto a monolayer of Vero cells for 1 h at 4°C. After removal of unbound virus, cells were washed twice with cold DMEM, followed by the addition of serial dilutions of MAbs in cold DMEM. Virus-adsorbed cells were incubated with mAd dilutions for 1 h at 4°C. Virus then was allowed to internalize for 30 min at 37°C, and subsequently cells were overlaid with methylcellulose as described above. Thirty hours later, plates were fixed with 4% PFA and analyzed for antigen-specific foci as described above for FRNTs. Due to less efficient binding of virus to cells at 4°C, a 2-fold higher amount of input virus was used in the post-attachment assay; however, this is unlikely to affect mAb potency, as the final FFU count following removal of unbound virus in the post-attachment assay (which is prior to mAb incubation) is similar to that used in the pre-attachment assay.
Attachment inhibition assay
SARS-CoV-2 was incubated with mAbs at 10 μg/mL for 1 h at 4°C. The mixture then was added to pre-chilled Vero E6, Vero-TMPRSS2, Vero-TMPRSS2-ACE2, or Calu-3 cells at an MOI of 0.005 and incubated at 4°C for 1 h. Cells were washed six times with chilled PBS before addition of lysis buffer and extraction of RNA using MagMax viral RNA isolation kit (Thermo Fisher Scientific) and a Kingfisher Flex 96-well extraction machine (Thermo Fisher Scientific). SARS-CoV-2 RNA was quantified by qRT-PCR using the N-specific primer/probe set described below. GAPDH was measured using a predesigned primer/probe set (IDT PrimeTime Assay Hs.PT.39a.22214836). Viral RNA levels were normalized to GAPDH, and the fold change was compared with isotype control mAb. For each cell type, a control with a 4-fold lower MOI (0.00125) was included to demonstrate detection of decreased viral RNA levels.
Virus internalization assay
SARS-CoV-2 was incubated with mAbs at 10 μg/mL for 1 h at 4°C. The mixture was then added to pre-chilled Vero E6 cells at an MOI of 0.005 and incubated at 4°C for 1 h. Cells were washed twice with chilled PBS to remove unbound virus, and subsequently incubated in DMEM at 37°C for 30 min to allow virus internalization. Cells then were treated with proteinase K and RNaseA at 37°C for 10 min to removed uninternalized virus. Viral and cellular RNA were extracted and analyzed as described above for the attachment inhibition assay. A no internalization control was included, where proteinase K and RNase A treatments were performed directly after washing, without an internalization step.
Measurement of viral burden and cytokine and chemokine levels
On 7 dpi, mice were euthanized and organs were collected. Nasal washes were collected in 0.5 mL of PBS. Organs were weighed and homogenized using a MagNA Lyser (Roche). Viral RNA from homogenized organs or nasal wash was isolated using the MagMAX Viral RNA Isolation Kit (ThermoFisher) and measured by TaqMan one-step quantitative reverse-transcription PCR (RT-qPCR) on an ABI 7500 Fast Instrument. Viral burden is expressed on a log10 scale as viral RNA per mg for each organ or total nasal wash after comparison with a standard curve produced using serial 10-fold dilutions of viral RNA standard. Primers were 5′- ATGCTGCAATCGTGCTACAA-3′, 5′- GACTGCCGCCTCTGCTC-3′, and probe 5′-/56-FAM/ TCAAGGAAC/Zen/ AACATTGCCAA/3IABkFQ-3′ (Case et al., 2020). For the measurement of cytokine and chemokine levels in the lung, lung homogenates were treated with 1% Triton X-100 for 1 h at room temperature to inactivate virus. Cytokine and chemokine levels in the lung homogenate were then analyzed by multiplex array (Eve Technologies Corporation).
Quantification and statistical analysis
Statistical significance was assigned when p values were < 0.05 using Prism version 8 (GraphPad). Tests, number of animals (n), median values, and statistical comparison groups are indicated in the Figure legends.
Acknowledgments
This study was supported by contracts and grants from the NIH (75N93019C00062, HHSN272201700060C, 75N93019C00074, U01 AI151810, R01 AI118938, and R01 AI157155) and the Defense Advanced Research Project Agency (HR001117S0019). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. We acknowledge the originating and submitting laboratories who generated and shared genetic sequence data via the GISAID Initiative. We also thank Charles Chiu and Raul Andino for providing the B.1.429 isolate and Barney Graham for cell lines and experimental advice. The Graphical Abstract was created with BioRender.
Author contributions
Conceptualization and methodology, L.A.V., M.S.D., L.J.A., D.H.F., Z.L., and S.P.J.W; investigation, L.A.V., L.J.A., Z.L., R.E.C., P.G., S.R., B.K.S., J.B.C., B.M.W., L.D., I.D.A., T.L.B., and A.J.; formal analysis, L.J.A. and S.A.H.; key reagents, E.S.W., H.Z., P.-Y.S., A.P., and A.C.; supervision and funding, M.S.D., D.H.F., D.W., A.C.M.B., J.E.C., and S.P.J.W; writing – original draft, L.A.V., L.J.A., and M.S.D.; writing – review and editing, all authors.
Declaration of interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. Some of the mAbs described in this study have been licensed by Washington University to Bio X Cell. D.H.F is a founder of Courier Therapeutics and has received funding support in a sponsored research agreement from Emergent BioSolutions. J.E.C. has served as a consultant for Eli Lilly and Luna Biologics, is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines, and is the founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from AstraZeneca and IDBiologics. The Boon laboratory has received funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences, AbbVie, and Nano Targeting & Therapy Biopharma.
Published: August 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.08.016.
Supplemental information
Data and code availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. Structural datasets have been uploaded and are available at PDB (accession codes 7MKL and 7MKM). SARS2-02 and SARS2-38 sequences have been deposited to GenBank (Accession numbers are listed in the Key resources table).
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart A.L., Havenar-Daughton C., Lempp F.A., Ma D., Schmid M.A., Agostini M.L., Guarino B., Di iulio J., Rosen L.E., Tucker H., et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- Cerutti G., Rapp M., Guo Y., Bahna F., Bimela J., Reddem E.R., Yu J., Wang P., Liu L., Huang Y., et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29:655–663.e4. doi: 10.1016/j.str.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. eLife. 2021;10:e63409. doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y., Shuai H., Yang D., Hou Y., Zhang X., et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021;12:134. doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejarnac O., Hafirassou M.L., Chazal M., Versapuech M., Gaillard J., Perera-Lecoin M., Umana-Diaz C., Bonnet-Madin L., Carnec X., Tinevez J.Y., et al. TIM-1 Ubiquitination Mediates Dengue Virus Entry. Cell Rep. 2018;23:1779–1793. doi: 10.1016/j.celrep.2018.04.013. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers G., Bentlage A.E.H., Stegmann T.C., Howie H.L., Lissenberg-Thunnissen S., Zimring J., Rispens T., Vidarsson G. Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs. 2017;9:767–773. doi: 10.1080/19420862.2017.1323159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zost S.J., Greaney A.J., Starr T.N., Dingens A.S., Chen E.C., Chen R.E., Case J.B., Sutton R.E., Gilchuk P., et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Holmes A.C., Basore K., Mack M., Fremont D.H., Diamond M.S. The mechanistic basis of protection by non-neutralizing anti-alphavirus antibodies. Cell Rep. 2021;35:108962. doi: 10.1016/j.celrep.2021.108962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagre A.C., Manhard J., Adams R., Eckley M., Zhan S., Lewis J., Rocha S.M., Woods C., Kuo K., Liao W., et al. A Potent SARS-CoV-2 Neutralizing Human Monoclonal Antibody That Reduces Viral Burden and Disease Severity in Syrian Hamsters. Front. Immunol. 2020;11:614256. doi: 10.3389/fimmu.2020.614256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J., et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight. 2020;5:e142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. bioRxiv. 2021 [Google Scholar]
- Li T., Han X., Gu C., Guo H., Zhang H., Wang Y., Hu C., Wang K., Liu F., Luo F., et al. Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants. bioRxiv. 2021 doi: 10.1038/s41467-021-26539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schaefer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Manne K., Mansouri K., et al. A broadly neutralizing antibody protects against SARS-CoV, pre-emergent bat CoVs, and SARS-CoV-2 variants in mice. bioRxiv. 2021 [Google Scholar]
- McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernández C.X., Schwantes C.R., Wang L.P., Lane T.J., Pande V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M.P., Forleo-Neto E., Musser B.J., Isa F., Chan K.C., Sarkar N., Bar K.J., Barnabas R.V., Barouch D.H., Cohen M.S., et al. Covid-19 Phase 3 Prevention Trial Team Subcutaneous REGEN-COV Antibody Combination to Prevent Covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladunni F.S., Park J.G., Pino P.A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021;4:874. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Barnes C.O., Eraslan B., Hudak A., Keeffe J.R., Cosimi L.A., Brown E.M., Muecksch F., Weisblum Y., Zhang S., et al. B cell genomics behind cross-neutralization of SARS-CoV-2 variants and SARS-CoV. Cell. 2021;184:3205–3221.e24. doi: 10.1016/j.cell.2021.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A.J., Turner J.S., Liu Z., Aziati I.D., Chen R.E., Joshi A., Bricker T.L., Darling T.L., Adelsberg D.C., Alsoussi W.B., et al. A public vaccine-induced human antibody protects against SARS-CoV-2 and emerging variants. bioRxiv. 2021 doi: 10.1101/2021.03.24.436864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., Pajon R., Smith G., Glenn G.M., Shi W., Korber B., Montefiori D.C. Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. N. Engl. J. Med. 2021;384:2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021 doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Iulio J.D., Wang Z., et al. Structural basis for broad sarbecovirus neutralization by a human monoclonal antibody. bioRxiv. 2021 doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Port J.R., Bushmaker T., Offei Owusu I., Purushotham J.N., Avanzato V.A., Fischer R.J., Schulz J.E., Holbrook M.G., Hebner M.J., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021;17:e1009195. doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. Structural datasets have been uploaded and are available at PDB (accession codes 7MKL and 7MKM). SARS2-02 and SARS2-38 sequences have been deposited to GenBank (Accession numbers are listed in the Key resources table).