Abstract
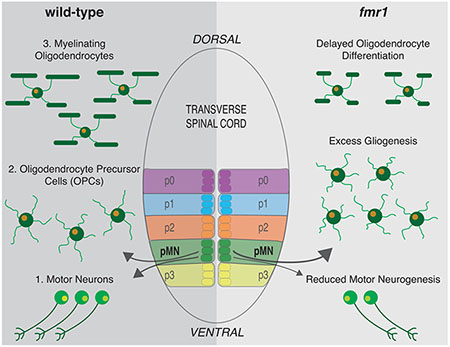
Neurodevelopment requires the precise integration of a wide variety of neuronal and glial cell types. During early embryonic development, motor neurons and then oligodendrocyte precursor cells (OPCs) are specified from neural progenitors residing in the periventricular pMN progenitor domain of the spinal cord. Following gliogenesis, OPCs can differentiate as oligodendrocytes (OLs)–the myelinating glial cells of the central nervous system–or remain as OPCs. To generate unique cell types capable of highly divergent functions, these specification and differentiation events require specialized gene expression programs. RNA binding proteins (RBPs) regulate mRNA localization and translation in the developing nervous system and are linked to many neurodevelopmental disorders. One example is Fragile X syndrome (FXS), caused by the loss of the RBP fragile X mental retardation protein (FMRP). Importantly, infants with FXS have reduced white matter and we previously showed that zebrafish Fmrp is autonomously required in OLs to promote myelin sheath growth. We now find that Fmrp regulates cell specification in pMN progenitor cells such that fmr1 mutant zebrafish generate fewer motor neurons and excess OPCs. Fmrp subsequently promotes differentiation of OPCs, leading to fewer differentiating OLs in the developing spinal cord of fmr1 larvae. Although the early patterning of spinal progenitor domains appears largely normal in fmr1 mutants during early embryogenesis, Shh signaling is greatly diminished. Taken together, these results suggest cell stage-specific requirements for Fmrp in the specification and differentiation of oligodendrocyte lineage cells.
Keywords: differentiation, FMRP, oligodendrocyte, OPC, specification
Graphical Abstract
1 |. INTRODUCTION
RNA binding proteins (RBPs), including fragile X mental retardation protein (FMRP), play essential roles in a wide variety of neurodevelopmental processes and are linked to autism spectrum disorders (ASDs; Barone et al., 2017; Lee et al., 2016; Verkerk et al., 1991). Fragile X syndrome (FXS) is caused by a trinucleotide expansion in the 5′ UTR of the FMR1 gene, leading to loss of FMRP expression (Verkerk et al., 1991). The RBP binds an array of mRNAs associated with ASDs (Ascano et al., 2012; Darnell et al., 2011) and can regulate the stability (Zalfa et al., 2007), transport and translation (Pilaz et al., 2016) of bound mRNAs. Though the bulk of FXS studies have centered on neuronal dysfunction (Dictenberg et al., 2008; Doll et al., 2017; Doll & Broadie, 2015; Pan et al., 2004), oligodendrocytes (OLs) are also implicated in the disease state, as white matter is reduced in infants with FXS (Swanson et al., 2018), and we recently showed that human FMRP can autonomously rescue the diminished myelin sheath growth seen in zebrafish fmr1 mutants (Doll et al., 2019). As myelin plays essential roles in neural circuit maturation and function (Mckenzie et al., 2014; Pan et al., 2020; Wang et al., 2020), this underscores the importance of FMRP in glial cell function.
Our current work highlights upstream roles for zebrafish Fmrp in the formation of oligodendrocyte precursor cells (OPCs) and subsequent differentiation into OLs, which could contribute to the diminished myelination seen in fmr1 mutants. We focus on the ventral spinal cord, where progenitor cells in the pMN domain sequentially generate spinal motor neurons and then OPCs. Importantly, Sonic hedgehog (Shh) signaling regulates both the transition from neurogenesis to gliogenesis in the pMN and the subsequent differentiation of OPCs to myelinating OLs (Dessaud et al., 2007; Ravanelli et al., 2018; Scott et al., 2020). However, not all OPCs differentiate into myelinating OLs and these precursors persist into adulthood (Bergles et al., 2000; Dawson et al., 2003; Tanaka et al., 2009).
As OPCs and OLs display widely divergent gene expression profiles (Perlman et al., 2020), RBPs must provide essential post transcriptional regulation of nascent mRNA in the OL lineage. Importantly, Fmr1 deletion leads to a fewer neurons and a surplus of neural progenitor cells (NPCs; Edens et al., 2019), thereby implicating the RBP in neuronal differentiation. Moreover, FXS models show disproportionate ratios of cell types throughout the nervous system, including excess glutamatergic neurons and astrocytes (Luo et al., 2010; Tervonen et al., 2009); fewer microglia and parvalbumin interneurons (Lee et al., 2019); and a reduction in white matter PDGFRα+/NG2+ OPCs at early postnatal stages (Pacey et al., 2013). It is therefore plausible that Fmrp could regulate both the balanced production of neuronal and glial cells from a common progenitor domain and the subsequent differentiation of glial precursors into mature OLs.
We find that fmr1 mutant zebrafish generate excess oligodendrocyte lineage cells (OLCs) throughout embryonic and larval development. Coupled with reduced motor neuron formation and Shh signaling, these data show a vital role for Fmrp in glial cell specification in embryonic development. In addition, we show that Fmrp promotes OL differentiation and limits the density of OPCs during subsequent larval stages. Taken together, Fmrp plays important roles in restricting glial precursor formation and driving myelin programming, two critical cellular programs in early neurodevelopment.
2 |. MATERIALS AND METHODS
2.1 |. Zebrafish lines and husbandry
The Institutional Animal Care and Use Committee at the University of Colorado School of Medicine approved all animal work, which is in compliance with US National Research Council’s Guide for the Care and Use of Laboratory Animals, the US Public Health Service’s Policy on Humane Care and Use of Laboratory Animals, and Guide for the Care and Use of Laboratory Animals. Larvae were raised at 28.5°C in embryo medium and staged as hours or days post fertilization (hpf/dpf) according to morphological criteria (Kimmel et al., 1995). Zebrafish lines used in this study included fmr1hu2787 (den Broeder et al., 2009), Tg(myrf:mScarlet)co66 (this paper), Tg(cspg4:mCherry)co28 (Ravanelli et al., 2018), and Tg(olig2:EGFP)vu12 (Shin et al., 2003). Genotyping for fmr1hu2787 was performed as previously described (Ng et al., 2013).
2.2 |. Imaging and analysis
We acquired images on a Zeiss LSM 880 or a Zeiss CellObserver SD 25 spinning disk confocal system (Carl Zeiss). Images were captured with Zen software (Carl Zeiss), then processed and analyzed using Fiji/ImageJ or Zen Blue (Carl Zeiss).
2.3 |. Immunohistochemistry
Larvae were fixed at indicated time points in 4% paraformaldehyde/1xPBS, rocking O/N at 4°C. Larvae were rinsed in 0.1%Triton/1xPBS (PBSTx), then embedded in 1.5% agar/30% sucrose and immersed in 30% sucrose O/N. Blocks were frozen on dry ice and 20 μm transverse sections were taken with a cryostat microtome and collected on polarized slides. Slides were mounted in Sequenza racks (Thermo Scientific), washed 3 × 5 min in PBSTx, blocked 1 h in 2% goat serum/2% bovine serum albumin/PBSTx and then placed in primary antibody (in block) O/N: rabbit α-Sox10 (1:500; Park et al., 2005), mouse α-Islet (1:500; Developmental Studies Hybridoma Bank, AB2314683), and mouse α-Elavl (1:250; Invitrogen, A-21271). Sections were washed 1.5 h in PBSTx, and then incubated 2 h at RT in secondary antibody (1:250; in block): AlexaFluor 488 goat α-rabbit (abcam, ab150077), AlexaFluor 488 goat α-mouse (abcam, ab150113), and AlexaFluor 568 goat α-mouse (abcam, ab175473). Sections were washed for 1 h in PBSTx, incubated with DAPI (1:2000 in PBSTx) for 5 min, washed 3 × 5 min in PBSTx, then mounted in Vectashield (Vector Laboratories, H-1000-10).
2.4 |. Fluorescent in situ RNA hybridization
Fluorescent in situ RNA hybridization (FISH) was performed with the RNAScope Multiplex Fluorescent V2 Assay Kit (Advanced Cell Diagnostics). Embryos and larvae at indicated timepoints were fixed in 4% paraformaldehyde/1xPBS, rocking O/N at 4°C. Samples were then embedded in 1.5% agar/30% sucrose and immersed in 30% sucrose O/N. Blocks were frozen on dry ice and 12 μm transverse sections were taken with a cryostat microtome and collected on polarized slides. FISH was performed as per the manufacturer’s protocol with the following modifications: slides were covered with Parafilm for all 40°C incubations to maintain moisture and disperse reagents across sections. Probes for zebrafish olig2-C1, nkx2.2-C2, ptch2-C3, sox10-C1, myrf-C2, cspg4-C3, neurog1-C2, and boc-C2 (1:50 dilution) were designed and synthesized by the manufacturer. Transcripts were labeled with the Opal 7 Kit (Perkin Elmer, NEL797001KT): Opal 520 (1:1500), Opal 570 (1:500), and Opal 650 (1:1500).
2.5 |. Quantification and statistical analysis
For all cell counts, the investigator was blind to genotype. For IHC (Figures 1 and 3), DAPI and Sox10/Islet channels were used to confirm cell number in a given section. For FISH, (Figure 4), DAPI sox10, myrf, and cspg4 were used to confirm cell number in acquired z-stacks. In experiments with transgenic larvae (Figure 5), all quantification includes transgenic larvae expressing myrf:mScarlet, though not all larvae co-expressed olig2:EGFP.
2.6 |. Fluorescent RNA in situ hybridization quantification
FISH puncta were quantified from z-projections collected at identical exposures with an adapted ImageJ custom script created by Karlie Fedder, University of Colorado, Department of Pediatrics, https://github.com/rebeccaorourke-cu/Prdm8-regulates-pMN-progenitor-specification. First, 10 z-intervals of 0.5 μm depth were maximum z-projected and background was subtracted with a two-rolling ball. Next, the image was thresholded by taking two standard deviations above the mean fluorescence intensity. A region of interest was drawn around the spinal cord and puncta were analyzed using the “Analyze Particles” feature with a size of 0.01-Infinity and circularity of 0.00–1.00. All thresholded puncta were inspected to ensure single molecules were selected. Puncta with an area of only 1 pixel were removed from the dataset. Data for each embryo was collected from a minimum of three consecutives trunk spinal cord sections and n represents the average number of puncta in a region of interest per section.
2.7 |. Statistics
All statistics were performed in Graphpad Prism (version 9). Normality was assessed with a D’Agostino and Pearson omnibus test. For two groups, unpaired comparisons were made using either unpaired two-tailed t tests (for normal distributions) or Mann–Whitney tests (abnormal distributions).
3 |. RESULTS
3.1 |. Fmrp restricts the production of OLCs
To examine the impact of Fmrp on gliogenesis in the developing spinal cord we used immunohistochemistry for Sox10—a canonical marker of the OL lineage—on transverse sections of wild-type and fmr1hu2787 mutant larvae at progressive embryonic and larval stages. Hereafter, we use OLC to denote Sox10-expressing cells in the central nervous system. Our results show surplus OLCs in the absence of Fmrp, beginning with a ~26% increase in fmr1 mutant embryos at 48 h postfertilization (hpf; Figure 1(a), (b), (i)). At 72 hpf, there was a ~29% increase in OLCs in fmr1 larvae (Figure 1(c), (d), (i)), and a comparable ~27% increase at 96 hpf (Figure 1(e), (f), (i)). Finally, at a late larval stage (7 days postfertilization; dpf) there was a less pronounced ~10% increase in fmr1 mutants compared to wild-type (Figure 1(g), (h), (i)). These results suggest that FMRP restricts the production of OLCs during early spinal cord development.
FIGURE 1.
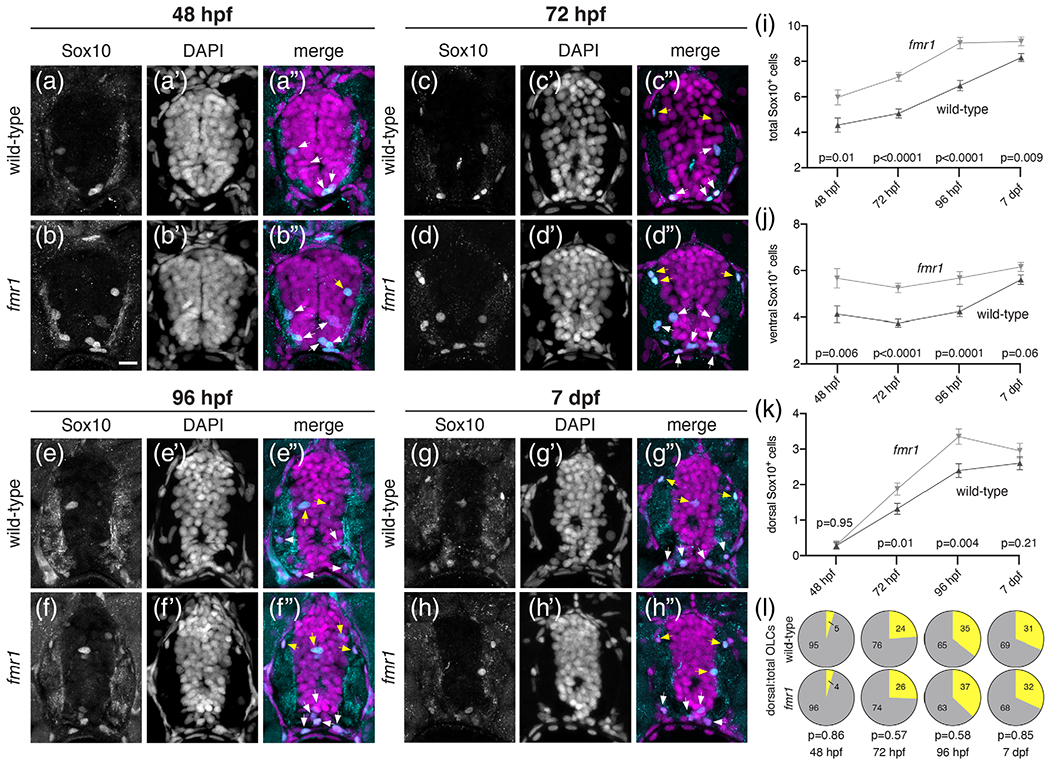
Fmrp restricts the production of OLCs. Representative images of trunk spinal cord transverse sections from wild-type and fmr1 mutant embryos and larvae processed to detect Sox10 antibody expression at progressive developmental stages. Wild-type (a) and fmr1 (b) embryos at 48 hpf (nWT = 11 embryos, 32 sections; nfmr1 = 10 embryos, 30 sections). Wild-type (c) and fmr1 (d) larvae at 72 hpf (nWT = 10 larvae, 46 sections; nfmr1 = 10 larvae, 47 sections). Wild-type (e) and fmr1 (f) larvae 96 hpf (nWT = 12 larvae, 38 sections; nfmr1 = 12 larve, 37 sections). Wild-type (g) and fmr1 (h) larvae at 7 dpf (nWT = 11 larvae, 43 sections; nfmr1 = 13 larvae, 42 sections). White arrowheads indicate Sox10+ OLCs in the ventral half of the cord. Yellow arrowheads indicate OLCs in the dorsal half of the cord. Graphs depict total OLCs (i), ventrally located OLCs (j), dorsally located OLCs (k), and the ratio of dorsal to total OLCs (L; dorsal OLCs in yellow; total OLCs in gray) at each developmental stage. Significance determined by unpaired t tests (total OLCs; 7 dpf dorsal OPCs; 72 hpf, 96 hpf, 7 dpf ratios) and Mann–Whitney tests (all others). Scale bars = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
Many OLCs generated from the pMN remain in the ventral cord, proximal to the prominent ventral axonal tracts. However, other OLCs migrate toward the myelinated tracts in the dorsal spinal cord (yellow arrowheads, Figure 1). We next quantified dorsal and ventral OLCs to examine whether Fmrp regulates the regional distribution of cells. At 48 hpf, fmr1 embryos had excess OLCs in the ventral cord compared to wild-type embryos (Figure 1(j)), before the onset of most dorsal migration (see Figure 2). By larval stages fmr1 mutants had more OLCs in both the ventral and dorsal halves of the cord (Figure 1(j), (k)), such that the regional distribution OLCs was actually quite similar to wildtype controls throughout our developmental timeline (Figure 1(l)). In short, these data indicate that Fmrp regulates the number of cells specified for OLC fate but not their proportional distribution between ventral and dorsal myelinated tracts of the spinal cord.
FIGURE 2.
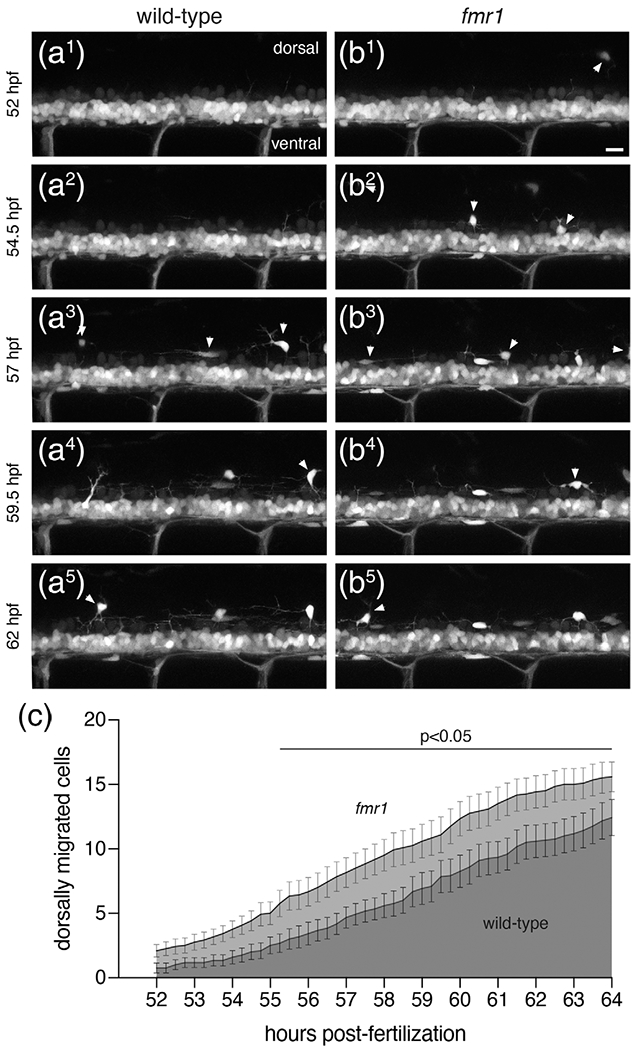
fmr1 larvae show premature dorsal migration of OLCs. Representative lateral trunk images of live transgenic wild-type (a1–a5) and fmr1 (b1–b5) embryos expressing olig2:EGFP at 2.5 h intervals, beginning at 52 hpf. White arrowheads indicate OPCs that have migrated from the ventral spinal cord. (c) Graph depicts the average number of OPCs that have migrated from the ventral cord in fmr1 (light gray) and wild-type (dark gray) at 15 min increments. Significance determined by unpaired t tests at each time point. n = 12 larvae for each genotype. Scale bar = 20 μm
We next used time-lapse microscopy to better understand the timeline of dorsal OLC migration in both wild-type and fmr1 mutants. We captured images of living transgenic larvae expressing olig2: EGFP—a marker of cells derived from the pMN domain, including OLCs (Shin et al., 2003)—at the onset of dorsal OLC migration, recording lateral three-dimensional z-stacks through the cord every 15 min from 52 to 64 hpf (Figure 2(a), (b); see Figures S1 and S2, e.g., videos). We then analyzed the timing of OLC emergence from the dense olig2: EGFP+ ventral domain. This developmental window was based on our Sox10 immunohistochemistry timeline, which showed that very few OLCs had migrated dorsally at 48 hpf (Figure 1). It is also important to note that migrating olig2+ cells are OLCs, as pMN-derived motor neurons do not leave the ventral cord (see Figure 3). At the onset of imaging at 52 hpf, there were very few cells in the dorsal spinal cord in both wild-type and fmr1 larvae, indicating that we captured the bulk of migration in most larvae. During much of the subsequent timeline, specifically 55–64 hpf, there was premature dorsal migration of OLCs in fmr1 larvae compared to wild-type controls (Figure 2(c)). Taken together with our previous data showing excess OLCs in fmr1 (Figure 1), these results raise the possibility that Fmrp restricts OLC formation and suggests that the precocious dorsal migration of cells in fmr1 larvae is likely due to increased cell density.
FIGURE 3.
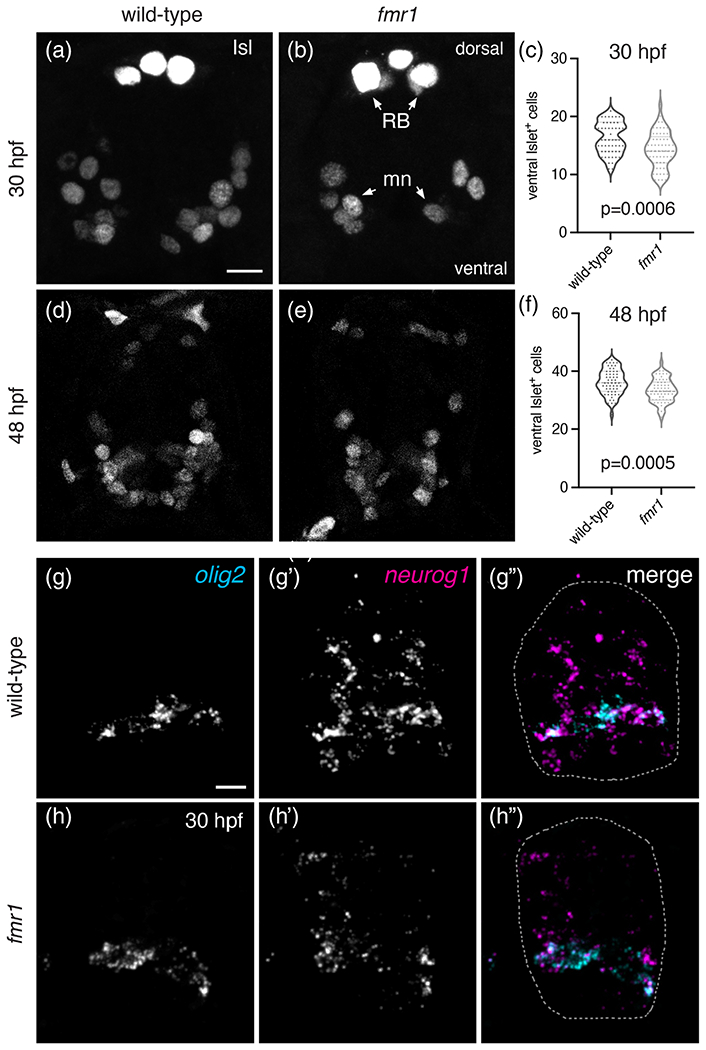
Fmrp promotes motor neuron formation. Representative images of trunk spinal cord transverse sections processed to detect expression of Islet protein (Isl) in wild-type (a) and fmr1 (b) embryos at 30 hpf. (c) Graph comparing the quantity of Isl+ motor neurons in wild-type and fmr1 embryos at 30 hpf (nWT = 15 embryos, 55 sections; nfmr1 = 15 embryos, 57 sections). Representative images of Isl+ motor neurons in wild-type (d) and fmr1 (e) embryos at 48 hpf. (c) Graph comparing the quantity of Isl+ motor neurons in wild-type and fmr1 larvae at 48 hpf (nWT = 15 embryos, 55 sections; nfmr1 = 15 embryos, 58 sections). RB = Rohon Beard neurons; mn = motoneurons. Significance determined by Mann–Whitney test (30 hpf) and unpaired t test (48 hpf). Representative images of transverse spinal cord sections processed to detect expression of olig2 and neurogenin1 (neurog1) mRNA in wild-type (g) and fmr1 mutant embryos (h) at 30 hpf (n = 5 embryos, 25 sections each genotype). Spinal cord in g″,h″ outlined in dashed line. Scale bars = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
3.2 |. Fmrp promotes motor neuron formation
The excess OLCs noted in fmr1 mutants led us to examine whether motor neuron formation was also affected by the loss of Fmrp, as pMN progenitor cells give rise to both motor neurons and OPCs during embryonic development (Park et al., 2002). We therefore quantified Islet+ (Isl) cells—an early marker of motor neurons (Ericson et al., 1992)—in transverse sections of wild-type and fmr1 mutant embryos (Figure 3). At 30 hpf, there was a ~13% reduction in Isl+ motor neurons in fmr1 embryos compared to wild-type control (Figure 3(a)–(c)). At 48 hpf, there was a less pronounced ~8% reduction in Isl+ cells in fmr1 embryos (Figure 3(d)–(f)). To examine whether Fmrp broadly regulates neurogenesis in the embryonic spinal cord, we next quantified cells expressing the pan-neuronal marker Elavl (Marusich et al., 1994) at 30 hpf, and found no difference in total Elavl+ cells in fmr1 mutants compared to control (Figure S3(a)–(c)). These results suggest that Fmrp promotes neurogenesis specifically in the pMN progenitor domain.
The excess OLCs and concomitant reduction in motor neurons in fmr1 mutants could indicate that Fmrp regulates cell formation in the pMN by promoting a pro-neurogenic gene expression program. To test this, we used fluorescent RNA in situ hybridization to examine proneuronal gene expression in wild-type and fmr1 embryos. At 30 hpf, neurogenin1 (neurog1; Blader et al., 1997) expression was evident throughout the spinal cord of wild-type embryos, with most prominent expression in the pMN domain, overlapping with olig2 (Figure 3(g)). In contrast, there was a sharp reduction in neurog1 expression in fmr1 mutants as compared to control embryos (Figure 3(h)). There were no obvious changes in olig2 expression in fmr1 embryos, which suggests that Fmrp is not required for progenitor cell formation. Taken together, these data indicate that FMRP promotes proneuronal gene expression and motor neurogenesis in the pMN domain, at a developmental stage that coincides with increased OLC formation in fmr1 mutants (Figure 1).
3.3 |. Fmrp promotes OL differentiation
The reduced white matter in infants with FXS (Swanson et al., 2018) and deficient myelination in FXS model organisms (Doll et al., 2019; Pacey et al., 2013) could reflect a requirement for Fmrp in the differentiation of OLCs into myelinating OLs. We examined this question in late embryonic and larval stages using three different methods. First, we used fluorescent RNA in situ hybridization to examine expression of three genes associated with progressive developmental stages of the OL lineage: olig2, a marker of pMN cells and OLCs; sox10 an OLC marker; and myelin regulatory factor (myrf), a gene expressed in differentiating OLs (Hornig et al., 2013). At 48 hpf, there was ~21% increase in total sox10+ OLCs in fmr1 mutants compared to wild-type (Figure 4(a)–(c)), a result comparable to our immunohistochemical results using Sox10 antibody (Figure 1). Although the quantity of sox10+ cells co-expressing myrf was not significantly different between genotypes (Figure 4(d), pink outlines in 4a‴, b‴), there was a ~33% increase in sox10+ cells that did not express myrf in fmr1 mutants (Figure 4(e), yellow outlines in 4a‴, b‴). When taken as a ratio of total sox10+ OLCs, 66% of wildtype cells expressed myrf, but only 59% of cells in fmr1 mutants co-expressed both genes (Figure 4(f)). These data suggest that proportionately fewer OLCs are undergoing differentiation in fmr1 mutants at this late embryonic stage, despite the preponderance of total OLCs.
FIGURE 4.
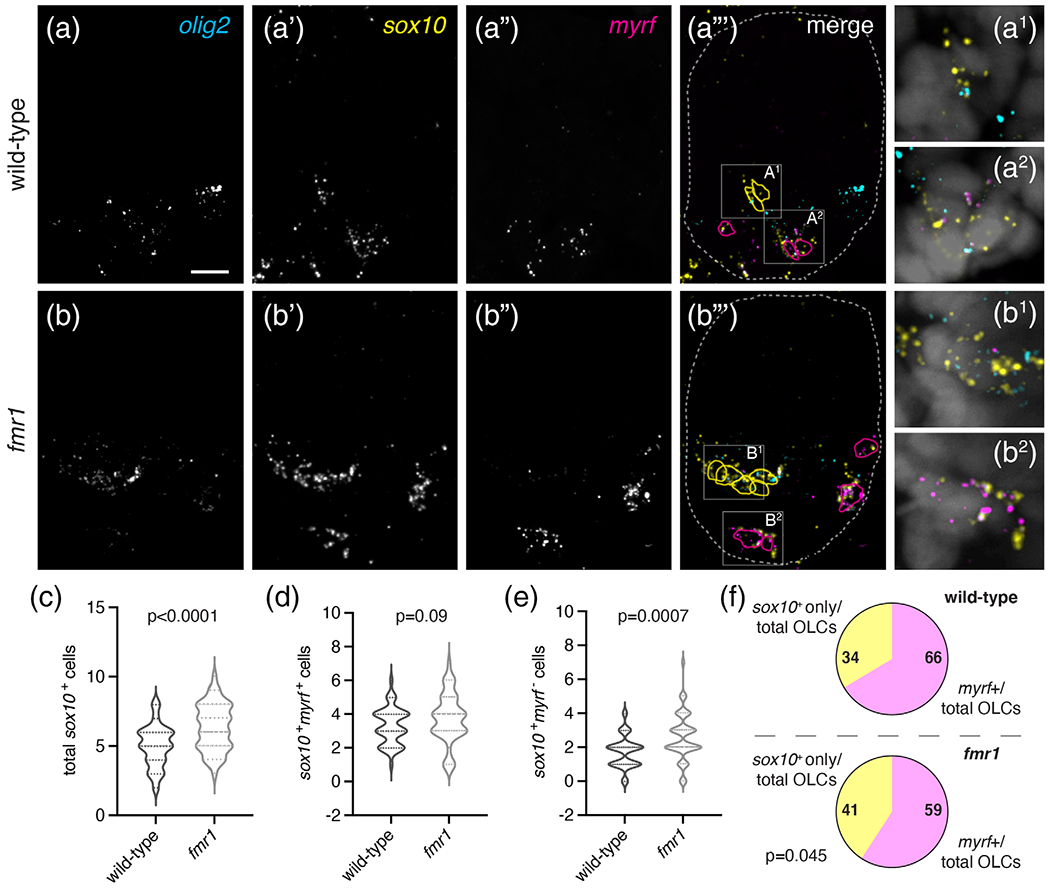
Fmrp promotes oligodendrocyte differentiation in the embryonic spinal cord. Representative images of trunk spinal cord sections from wild-type (a–a‴) and fmr1 mutant embryos (b–b‴) at 30 hpf, processed to detect expression of specific mRNAs using fluorescent in situ hybridization, including pMN descendants (olig2; a, b), oligodendrocyte lineage cells (sox10; a′, b′), and differentiating oligodendrocytes (myrf: a″, b″). cell outlines in a‴ and b‴ mark olig2+sox10+myrf-cells (yellow) and olig2+sox10+myrf+ cells (magenta), dashed lines represent the edges of the spinal cord. a1, a2 and b1, b2 show high magnification insets from a‴ and b‴, respectively, and include DAPI nuclear stain. Graphs comparing the average total OLCs (c), myrf+ OLCs (d), and myrf− OLCs (e) in wild-type (n = 9,45 sections) and fmr1 (n = 10,48 sections). (f) The relative ratio of myrf+ cells to total OLCs in wild-type and fmr1 mutants. Significance determined with unpaired t tests. Scale bar = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
As an independent method to assess the timing of OL differentiation in live animals, we next quantified differentiating OLs in subsequent larval stages using stable transgenic lines. We crossed Tg(olig2: EGFP)vu12, a reporter of pMN lineage cells (Figure 5(a)–(d)), with Tg (myrf:mScarlet)co66, a reporter of differentiating OLs (Figure 5(a′)–(d′)). At 72 hpf, there was a ~44% reduction in total myrf+ cells in fmr1 larvae compared to wild-type (Figure 5(e)), though it is important to note that this approach may be less time sensitive than FISH (Figure 4), as the maturation of mScarlet fluorescent protein takes up to 6 h (Balleza et al., 2018). At 96 hpf, fmr1 larvae had a less pronounced ~11% reduction in myrf+ cells compared to controls (Figure 5(e)), further indicating a delay in OL differentiation. Although fmr1 larvae had surplus OLCs in the dorsal spinal cord compared to wild-type controls (Figure 5(f))—consistent with our time-lapse imaging (Figure 2)—far fewer of these migrated cells expressed myrf (Figure 5(g)). Indeed, at these larval stages fmr1 mutants had a reduced ratio of differentiating myrf+ cells to total OLCs in the dorsal spinal cord compared to wild-type (Figure 5(h)). These results further validate the reduced OL differentiation seen in fmr1 mutants and suggest OL differentiation is delayed in the absence of Fmrp.
FIGURE 5.
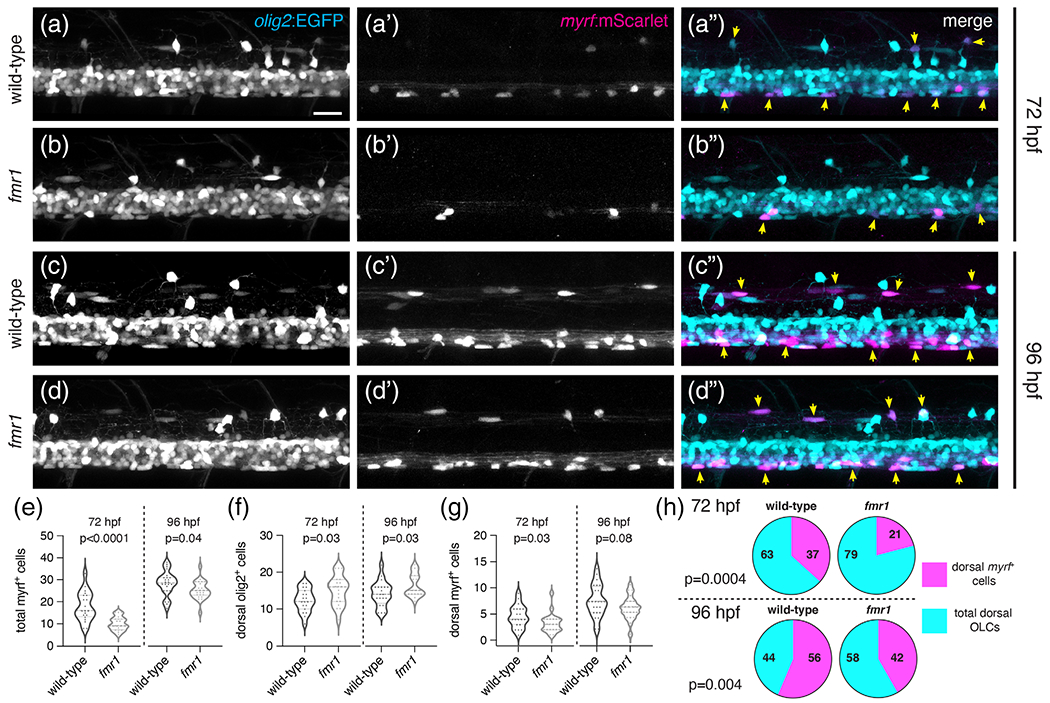
Fmrp regulates the timing of OL differentiation. Representative lateral images of live transgenic wild-type and fmr1 mutant larvae at 72 hpf (a, b) and 96 hpf (c, d) stably expressing olig2:EGFP (pMNderived cells) and myrf:mScarlet (differentiating oligodendrocytes). Yellow arrowheads indicate olig2+myrf+ cells. Graphs comparing the quantity of total myrf+ cells (e; n72hpf = 20 larvae each, n96hpf = 24WT,23fmr1), dorsal OLCs (f; n72hpf = 19 each, n96hpf = 20WT,23fmr1), dorsal myrf+ cells (G; n72hpf = 20 each, n 96hpf = 24WT,23fmr1), and the ratio of differentiating dorsal OLCs (h; n72hpf = 19 each, n96hpf = 20WT,22fmr1). Significance determined by a Mann–Whitney tests (g, 72 hpf) and unpaired t tests (all others). Scale bar = 20 μm [Color figure can be viewed at wileyonlinelibrary.com]
The above data suggest that a smaller proportion of OPCs undergo differentiation into myelinating OLs in fmr1 mutant larvae than in wild-type and thus OPCs may retain a longer-term precursor identity in the absence of Fmrp. To test this possibility directly, we again used fluorescent RNA in situ hybridization at a late larval stage to detect expression of chondroitin sulfate proteoglycan 4 (cspg4), which marks OPCs in larval and adult stages, alongside sox10 and myrf (Figure 6). At 6 dpf, fmr1 larvae had ~40% more cspg4+ cells than wild-type controls (Figure 6(a)–(c)) and ~42% of total OLCs in fmr1 larvae expressed cspg4, compared to just ~34% of OLCs in wild-type (Figure 6(f)), indicating that more OLCs retain a precursor identity in the absence of Fmrp. In addition, there was no change in the quantity of myrf+ differentiating OLCs in fmr1 larvae compared to wild-type at this late larval stage (Figure 6(a′), (b′), (d′)), which further suggests that Fmrp regulates the timing of differentiation, as there were fewer differentiating OLs in fmr1 mutants at earlier larval stages (Figure 5). fmr1 larvae also had ~23% more total sox10+ OLCs than wildtype at 6 dpf (Figure 6(a″), (b″), (e)), which was in line with Sox10 immunohistochemistry at late larval stages (Figure 1). Finally, to complement cspg4 RNA in situ hybridization data, we also quantified cspg4:mCherry+ cells in transgenic larvae at 5 dpf and found that fmr1 larvae had ~14% more total mCherry+ cells than wild-type and a ~46% increase in mCherry+ cells in the ventral spinal cord (Figure S4(a)–(d)). Taken together, these data suggest that Fmrp regulates both the timing of OL differentiation and the conversion of OPCs to OLs, such that fmr1 mutants have more OPCs at late larval stages.
FIGURE 6.
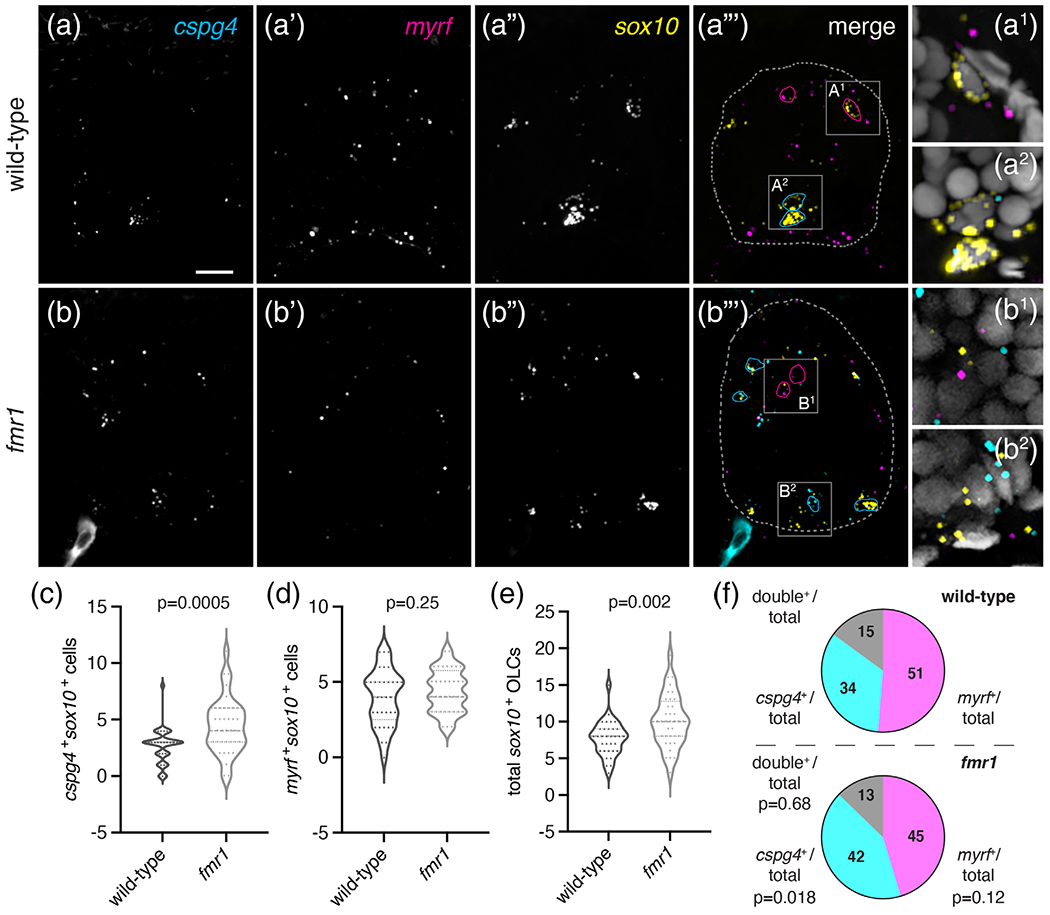
fmr1 mutant larvae have excess OPCs. Representative images of transverse trunk spinal cord sections of wild-type (a–a‴) and fmr1 mutant (b–b‴) larvae processed to detect expression of cspg4, myrf, and sox10 at 6 dpf via fluorescent in situ RNA hybridization. (a‴, b‴) sox10+myrf+cspg4− cells are outlined in magenta; cspg4+ cells not expressing myrf in cyan; spinal cord boundaries in dashed lines. (a1, a2) and (b1, b2) show high magnification insets from a‴ and b‴, respectively, and include DAPI nuclear stain. Graphs comparing myrf+ OLCs (c), cspg4+ OLCs (d), and total sox10+ cells in wild-type (n = 8 larvae, 33 sections) and fmr1 mutants (n = 8 larvae, 36 sections). (f) Relative ratios of OLCs expressing myrf, cspg4, or coexpressing both transcripts. Significance determined by unpaired t tests (total sox10+, myrf+ comparisons) and Mann–Whitney tests (double+ ratio, cspg4+ comparisons). Scale bar = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
3.4 |. fmr1 mutants have abnormally low levels of Shh signaling in the developing spinal cord
The Sonic hedgehog (Shh) signaling pathway is essential for the dorsoventral patterning of the spinal cord (Briscoe et al., 2000; Dessaud et al., 2007) and a transient increase in Shh signaling is necessary for OPC formation (Al Oustah et al., 2014; Scott et al., 2020). Moreover, OL formation appears to require slightly higher levels of Shh signaling than OPCs, as pharmacological inhibition of Shh leads to more OPCs and fewer OLs and overexpression of Shh shifts the balance toward OL fate (Ravanelli et al., 2018). To test whether altered populations of OLCs and motor neurons in fmr1 mutants are associated with changes in Shh signaling, we used fluorescent RNA in situ hybridization at early embryonic stages to examine the expression of genes marking ventral progenitor domains and Shh signaling: olig2, a marker of the pMN progenitor domain; nkx2.2, a marker of the p3 progenitor domain; ptch2, a positive transcriptional target of Shh (Concordet et al., 1996); and boc, a Shh co-receptor and negative transcriptional target of Shh (Kearns et al., 2021; Tenzen et al., 2006). fmr1 mutant embryos had no obvious changes relative to wild-type in the expression of olig2 at 24 or 30 hpf (Figure 7(a), (b), (e), (f)) or nkx2.2 at 30 hpf (Figure 7(e′), (f′)), which again suggests a general maintenance of spinal cord progenitor boundaries in fmr1 mutants. However, fmr1 embryos had a clear reduction in ptch2 expression at both 24 hpf (Figure 7(a″), (b″)) and 30 hpf (Figure 7(e″), (f″)), and boc expression was increased at 24 hpf compared with wild-type embryos (Figure 7(a′), (b′)). We quantified the total number of boc+ puncta at 24 hpf and ptch2+ puncta at 30 hpf in individual transverse trunk spinal cord sections (Figure 7(c), (g)) and normalized this data to the relative size of the spinal cord. At 24 hpf, fmr1 mutants had ~19% more boc+ puncta than wild-type (Figure 7(d)). In addition, fmr1 mutants had a ~26% decrease in ptch2+ puncta compared to wild-type at 30 hpf (Figure 7(h)). Taken together, these results raise the possibility that Fmrp regulates OLC formation, at least in part, by shaping or maintaining the Shh signaling gradient.
FIGURE 7.
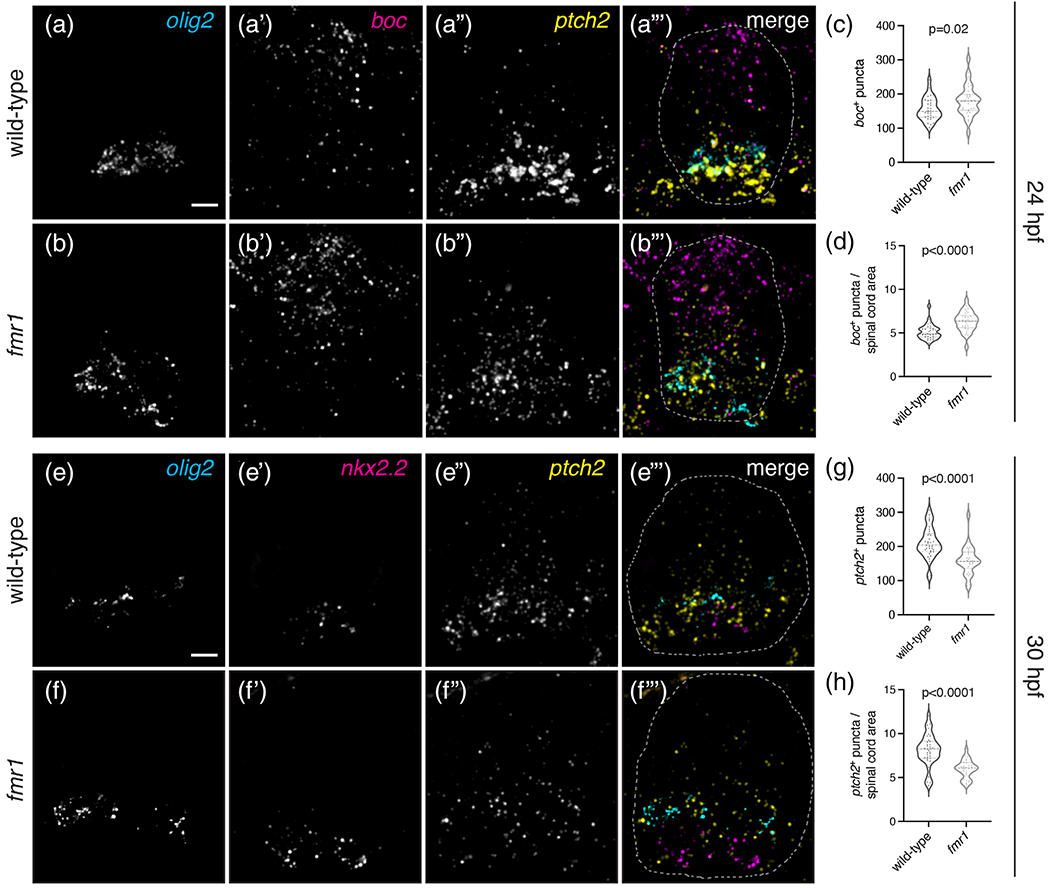
fmr1 mutants have abnormally low levels of Shh signaling in the developing spinal cord. Representative images of transverse trunk spinal cord sections from 24 hpf wild-type (a–a‴) and fmr1mutants (b–b‴) processed to detect expression of olig2, boc, and ptch2 using fluorescent in situ RNA hybridization. Graphs comparing the average number of boc+ puncta per section (c; unpaired t test), and average number of boc+ puncta per unit area of spinal cord (d; Mann–Whitney test) in wild-type (n = 6 embryos, 30 sections) and fmr1 mutants (n = 6 embryos, 30 sections). Representative images of transverse trunk spinal cord sections from 30 hpf wild-type (e–e‴) and fmr1 mutants (f–f‴) processed to detect expression of olig2, nkx2.2, and ptch2 using fluorescent in situ RNA hybridization. Graphs comparing the average number of ptch2+ puncta per section (g; Mann–Whitney test), and average number of ptch+ puncta per unit area (h; unpaired t test) in wild-type (n = 7 embryos, 35 sections) and fmr1 mutants (n = 6 embryos, 26 sections). Scale bars = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
4 |. DISCUSSION
The proportionate balance and integration of a wide variety of cell types is crucial for neural circuit formation and function. For example, altered quantities of excitatory and inhibitory neurons in the absence of FMRP (Lee et al., 2019; Tervonen et al., 2009) could directly contribute to the hyperexcitation theory of FXS (Contractor et al., 2015; Gibson et al., 2008; Paluszkiewicz et al., 2011). Imbalanced excitation (E) and inhibition (I) may contribute to debilitating symptoms in ASDs at large, such as hypersensitivity to sensory stimuli (Sapey-Triomphe et al., 2019). Importantly, infants with FXS have diminished white matter profiles (Swanson et al., 2018), and OLs autonomously require Fmrp for myelin sheath growth (Doll et al., 2019), which implicate glia in the disease state. In another interesting parallel, social isolation during an early critical period, leads to dramatic decreases in myelination (Makinodan et al., 2012). OLCs dramatically influence neural circuits, as myelin plasticity is required for many learning and memory paradigms (Mckenzie et al., 2014; Pan et al., 2020; Wang et al., 2020), and OPCs can also directly modulate synaptic transmission (Sakry et al., 2014). Finally, the fact that OLs ensheathe both inhibitory and excitatory axons with myelin (Micheva et al., 2016) further indicates that oligodendroglia can directly influence the balance of excitation and inhibition in the nervous system.
Many questions remain in regard to how Fmrp regulates gliogenesis and the potential functions of excess OPCs in fmr1 mutants. During late larval development, we found that OPC density appeared to reach an upper limit in fmr1 mutants between 4 and 7 dpf (Figure 1(i)), whereas wild-type larvae continued to produce OPCs during this window. This could be related to a progressive decline in Fmrp expression, which is very high in early embryonic stages but is reduced at maturity (Lu et al., 2004; Singh et al., 2007; Tessier & Broadie, 2008). Indeed, FXS phenotypes are often less severe or absent after early developmental defects, when FMRP expression fades (Doll et al., 2017; Doll & Broadie, 2016). It is also possible that other RBPs may play compensatory roles in late larval stages. However, neural circuits form during precise critical periods of development (Hensch, 2004) and it is likely that neurodevelopmental disorders such as ASDs are rooted in time sensitive changes within these critical periods (Meredith, 2015). Importantly, OPCs are not simply a reserve population for the myelinating OL lineage, as they are directly responsive to neuronal activity (Bergles et al., 2000) and can actively modulate synapses (Sakry et al., 2014).
The overabundance of OPCs in fmr1 embryos was also associated with a deficit of motor neurons, which suggests that Fmrp may be involved in regulating the transition from neurogenesis to gliogenesis in the pMN domain (Figure 3). In addition, reduced expression of the pro-neurogeneic gene neurog1 in fmr1 mutants further suggests that Fmrp drives neurogenesis in the pMN (Figure 3). Interestingly, Fmrp did not appear to broadly regulate spinal neurogenesis during early embryogenesis (Figure S3), which may indicate a unique mechanism for Fmrp in pMN progenitors. However, it is also possible that Fmrp regulates the specification or relative proportionality of other neuronal subtypes, as the pan-neuronal labeling of the HuC/D antibody does not allow insight into this question. We plan to assess Fmrp requirements in additional neuronal subtypes in our future work.
Are Fmrp-dependent changes in glial specification and differentiation somehow linked? Does excess OPC production in fmr1 mutants reflect a feedback mechanism linking diminished OL differentiation with prolonged gliogenesis? Interestingly, there is actually reduced NG2 expression and fewer deep white matter OPCs in a mouse model of FXS at early postnatal stages (P7), but NG2 expression was increased in Fmr1 mutants at P15 (Pacey et al., 2013). Therefore, Pacey et al. also show developmental requirements for FMRP in the production of OPCs, which highlights the necessity of developmental stage-specific analyses. Moreover, the fact that both mouse and zebrafish FXS models also show deficient myelination (Doll et al., 2019; Pacey et al., 2013), may indicate region-specific roles for the Fmrp in gliogenesis but common requirements in OL differentiation.
Our current study shows that Fmrp promotes OL differentiation, which could help explain the poor myelin sheath growth in the OLs that do manage to differentiate in fmr1 mutant zebrafish. This is perhaps not surprising given the catalog of mRNA targets of FMRP associated with differentiation (Liu et al., 2018). The reduced OL differentiation seen in fmr1 mutants could be linked to diminished Shh in early embryonic development (Figure 7), as our lab previously showed that pharmacological inhibition of Shh impedes OLC differentiation, resulting in excess OPCs and fewer mature OLs (Ravanelli et al., 2018). It is unclear how or why the absence of Fmrp is associated with such dramatic reductions in Shh signaling (Figure 7), but unfortunately we currently lack the tools to precisely manipulate Shh in a progenitor-specific and time-dependent manner. Interestingly, Fmr1-knockout NPCs are also differentiation-impaired, leading to surplus progenitors and fewer mature neurons (Edens et al., 2019). In this context, FMRP acts as an m6A reader driving nuclear export of mRNAs, including genes encoding Shh pathway components (Edens et al., 2019). In a fascinating tangent, m6A methylation also promotes OL differentiation and myelination (Xu et al., 2020). These studies therefore represent striking parallels to this study and our previous work (Doll et al., 2019). In addition, overexpression of the NG2 proteoglycan intracellular domain in OPCs leads to reduced FMRP and increased translation (Nayak et al., 2018), which suggests that Fmrp may repress translation of mRNAs maintaining OPC fate. Taken together, our current results (Figures 4–6) provide ample evidence that Fmrp promotes differentiation in the OL lineage.
Our work also links increased OPC density in fmr1 mutants with precocious dorsal migration in the developing spinal cord (Figures 1 and 2). OPC migration is heavily influenced by neighboring OPCs, which maintain spacing through contact-mediated repulsion (Hughes et al., 2013; Kirby et al., 2006). We speculate that the premature dorsal migration of OPCs in fmr1 mutants may stem from a similar repulsive mechanism, as we also noted clusters of Sox10+ OPCs in the ventral spinal cord during late embryogenesis (48 hpf), just prior to the onset of dorsal migration (Figure 1(b)). Although we cannot completely rule out a role for Fmrp in migration, the regional distribution of OLCs in the spinal cord was remarkably consistent between wild-type and fmr1 mutants (Figure 1(l)), which suggests that OPC migration is largely dependent on cell density.
FMRP is capable of regulating nearly every stage of the mRNA life-cycle, including nuclear shuttling (Edens et al., 2019), localization (Dictenberg et al., 2008; Pilaz et al., 2016), translation (Todd et al., 2003), and stability (Zalfa et al., 2007). Although we cannot yet determine the precise mRNA targets regulating gliogenesis and differentiation, our future work will identify the FMRP-bound mRNAs in defined stages of the OL lineage. We predict that FMRP requirements are stage specific, such that the catalog of mRNAs regulated in progenitor cells is largely distinct from the highly motile precursors, and also different from the mRNAs localized in distal OL processes driving myelin sheath growth (Doll et al., 2019). Here we show stage specific defects in the OL lineage of fmr1 mutants: FMRP appears to restrict the formation of persistent OPCs, while promoting differentiation of mature OLs. Taken together, our work unveils a crucial role for FMRP in the OL lineage with the potential for widespread consequences on neural circuit formation and function.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the members of the lab for insightful discussion. This work was supported by US National Institute of Health (NIH) grant R21 NS117886 to Caleb A. Doll, R21 NS110213 to Caleb A. Doll and Bruce Appel, and R01 NS095679 and a gift from the Gates Frontiers Fund to Bruce Appel. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility was supported by NIH grant P30 NS048154. The anti-Isl antibody, developed by T.M. Jessell and S. Brenner-Morton, was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
Funding information
National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: R01 NS095679, R21 NS110213, R21 NS117886, P30 NS048154
Footnotes
CONFLICT OF INTEREST
The authors have no financial or other interests to disclose.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Al Oustah A, Danesin C, Khouri-Farah N, Farreny MA, Escalas N, Cochard P, Glise B, & Soula C (2014). Dynamics of sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring sulfatase 1. Development, 141, 1392–1403. [DOI] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, & Tuschl T (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature, 492, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Kim JM, & Cluzel P (2018). Systematic characterization of maturation time of fluorescent proteins in living cells. Nature Methods, 15,47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone R, Fichera M, De Grandi M, Battaglia M, Lo Faro V, Mattina T, & Rizzo R (2017). Familial 18q12.2 deletion supports the role of RNA-binding protein CELF4 in autism spectrum disorders. American Journal of Medical Genetics Part A, 173, 1649–1655. [DOI] [PubMed] [Google Scholar]
- Bergles DE, Roberts JDB, Somogyi P, & Jahr CE (2000). Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature, 405, 187–191. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, & Strähle U (1997). The activity of Neurogenin1 is controlled by local cues in the zebrafish embryo. Development, 124, 4557–4569. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, & Ericson J (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell, 101, 435–445. [DOI] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, & Ingham PW (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase a in neural tube and somite patterning. Development, 122, 2835–2846. [DOI] [PubMed] [Google Scholar]
- Contractor A, Klyachko VA, & Portera-Cailliau C (2015). Altered neuronal and circuit excitability in fragile X syndrome. Neuron, 87, 699–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, & Darnell RB (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MRL, Polito A, Levine JM, & Reynolds R (2003). NG2-expressing glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Molecular and Cellular Neurosciences, 24(2), 476–488. [DOI] [PubMed] [Google Scholar]
- den Broeder MJ, van der Linde H, Brouwer JR, Oostra BA, Willemsen R, & Ketting RF (2009). Generation and characterization of Fmr1 knockout zebrafish. PLoS One, 4, 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, & Briscoe J (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature, 450, 717–720. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, & Bassell GJ (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental Cell, 14, 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, & Broadie K (2015). Activity-dependent FMRP requirements in development of the neural circuitry of learning and memory. Development, 142, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, & Broadie K (2016). Neuron class-specific requirements for fragile X mental retardation protein in critical period development of calcium signaling in learning and memory circuitry. Neurobiology of Disease, 89, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, Vita DJ, & Broadie K (2017). Fragile X mental retardation protein requirements in activity-dependent critical period neural circuit refinement. Current Biology, 27, 2318–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, Yergert KM, & Appel BH (2019). The RNA binding protein fragile X mental retardation protein promotes myelin sheath growth. Glia, 68, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, Miller N, Rojas Ringeling F, Ming G, He C, Song H, & Ma YC (2019). FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep, 28, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell T, & Yamada T (1992). Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science (80-), 256, 1555–1560. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Hays SA, & Huber KM (2008). Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. Journal of Neurophysiology, 100, 2615–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensch TK (2004). Critical period regulation. Annual Review of Neuroscience, 27, 549–579. [DOI] [PubMed] [Google Scholar]
- Hornig J, Fröb F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, & Wegner M (2013). The Transcription Factors Sox10 and Myrf Define an Essential Regulatory Network Module in Differentiating Oligodendrocytes. PLoS Genetics, 9(10), e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EG, Kang SH, Fukaya M, & Bergles DE (2013). Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nature Neuroscience, 16, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns CA, Walker M, Ravanelli AM, Scott K, Arzbecker MR, & Appel B (2021). Zebrafish spinal cord oligodendrocyte formation requires boc function. bioRxiv, pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995). Stages of embryonic development of the zebrafish. Developmental Dynamics, 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, & Appel B (2006). In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nature Neuroscience, 9, 1506–1511. [DOI] [PubMed] [Google Scholar]
- Lee FHF, Lai TKY, Su P, & Liu F (2019). Altered cortical cytoarchitecture in the Fmr1 knockout mouse. Molecular Brain, 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, & Martin KC (2016). Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron, 89, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li Y, Stackpole EE, Novak A, Gao Y, Zhao Y, Zhao X, & Richter JD (2018). Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America, 115, E11397–E11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, & Feng Y (2004). The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proceedings of the National Academy of Sciences of the United States of America, 101, 15201–15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, Li W, Liu C, Jin P, & Zhao X (2010). Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genetics, 6, e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan M, Rosen KM, Ito S, & Corfas G (2012). A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science (80-), 337, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusich MF, Furneaux HM, Henion PD, & Weston JA (1994). Hu neuronal proteins are expressed in proliferating neurogenic cells. Journal of Neurobiology, 25, 143–155. [DOI] [PubMed] [Google Scholar]
- Mckenzie IA, Ohayon D, Li H, De Faria JP, Emery B, Tohyama K, & Richardson WD (2014). Motor skill learning requires active central myelination. Science (80-), 346, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RM (2015). Sensitive and critical periods during neurotypical and aberrant neurodevelopment: A framework for neurodevelopmental disorders. Neuroscience and Biobehavioral Reviews, 50, 180–188. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Wolman D, Mensh BD, Pax E, Buchanan J, Smith SJ, & Bock DD (2016). A large fraction of neocortical myelin ensheathes axons of local inhibitory neurons. eLife, 5, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Trotter J, & Sakry D (2018). The intracellular cleavage product of the NG2 proteoglycan modulates translation and cell-cycle kinetics via effects on mTORC1/FMRP signaling. Frontiers in Cellular Neuroscience, 12, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MC, Yang YL, & Lu KT (2013). Behavioral and synaptic circuit features in a zebrafish model of fragile X syndrome. PLoS One, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey LKK, Xuan ICY, Guan S, Sussman D, Henkelman RM, Chen Y, Thomsen C, & Hampson DR (2013). Delayed myelination in a mouse model of fragile X syndrome. Human Molecular Genetics, 22, 3920–3930. [DOI] [PubMed] [Google Scholar]
- Paluszkiewicz SM, Olmos-Serrano JL, Corbin JG, & Huntsman MM (2011). Impaired inhibitory control of cortical synchronization in fragile X syndrome. Journal of Neurophysiology, 106, 2264–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, & Broadie K (2004). The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Current Biology, 14, 1863–1870. [DOI] [PubMed] [Google Scholar]
- Pan S, Mayoral SR, Choi HS, Chan JR, & Kheirbek MA (2020). Preservation of a remote fear memory requires new myelin formation. Nature Neuroscience, 23, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-C, Boyce J, Shin J, & Appel B (2005). Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. The Journal of Neuroscience, 25, 6836–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, & Appel B (2002). Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Developmental Biology, 248, 356–368. [DOI] [PubMed] [Google Scholar]
- Perlman K, Couturier CP, Yaqubi M, Tanti A, Cui QL, Pernin F, Stratton JA, Ragoussis J, Healy L, Petrecca K, Dudley R, Srour M, Hall JA, Kennedy TE, Mechawar N, & Antel JP (2020). Developmental trajectory of oligodendrocyte progenitor cells in the human brain revealed by single cell RNA sequencing. Glia, 68, 1291–1303. [DOI] [PubMed] [Google Scholar]
- Pilaz L-J, Lennox AL, Rouanet JP, Silver Correspondence DL, & Silver DL (2016). Dynamic mRNA transport and local translation in radial glial progenitors of the developing brain. Current Biology, 26, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli AM, Kearns CA, Powers RK, Wang Y, Hines JH, Donaldson MJ, & Appel B (2018). Sequential specification of oligodendrocyte lineage cells by distinct levels of hedgehog and notch signaling. Developmental Biology, 444, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakry D, Neitz A, Singh J, Frischknecht R, Marongiu D, Binamé F, Perera SS, Endres K, Lutz B, Radyushkin K, Trotter J, & Mittmann T (2014). Oligodendrocyte precursor cells modulate the neuronal network by activity-dependent Ectodomain cleavage of glial NG2. PLoS Biology, 12, e1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapey-Triomphe L-A, Lamberton F, Sonié S, Mattout J, Schmitz C (2019). Tactile hypersensitivity and GABA concentration in the sensorimotor cortex of adults with autism. Autism Research, 12(4), 562–575. [DOI] [PubMed] [Google Scholar]
- Scott K, O’Rourke R, Gillen A, & Appel B (2020). Prdm8 regulates pMN progenitor specification for motor neuron and oligodendrocyte fates by modulating the Shh signaling response. Development, 147, 8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Madwsley DJ, & Appel B (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods in Cell Science, 25, 7–14. [DOI] [PubMed] [Google Scholar]
- Singh K, Gaur P, Prasad S (2007). Fragile x mental retardation (Fmr-1) gene expression is down regulated in brain of mice during aging. Molecular Biology Reports, 34(3), 173–181. [DOI] [PubMed] [Google Scholar]
- Swanson MR, Wolff JJ, Shen MD, Styner M, Estes A, Gerig G, McKinstry RC, Botteron KN, Piven J, & Hazlett HC (2018). Development of white matter circuitry in infants with fragile x syndrome. JAMA Psychiatry, 75, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tozuka Y, Takata T, Shimazu N, Matsumura N, Ohta A, & Hisatsune T (2009). Excitatory GABAergic activation of cortical dividing glial cells. Cerebral Cortex, 19, 2181–2195. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, & McMahon AP (2006). The cell surface membrane proteins Cdo and Boc are components and targets of the hedgehog signaling pathway and feedback network in mice. Developmental Cell, 10, 647–656. [DOI] [PubMed] [Google Scholar]
- Tervonen TA, Louhivuori V, Sun X, Hokkanen M-E, Kratochwil CF, Zebryk P, Castrén E, & Castrén ML (2009). Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiology of Disease, 33, 250–259. [DOI] [PubMed] [Google Scholar]
- Tessier CR, & Broadie K (2008). Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development, 135, 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, & Malter JS (2003). The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proceedings of the National Academy of Sciences, 100, 14374–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell, 65, 905–914. [DOI] [PubMed] [Google Scholar]
- Wang F, Ren SY, Chen JF, Liu K, Li RX, Li ZF, Hu B, Niu JQ, Xiao L, Chan JR, & Mei F (2020). Myelin degeneration and diminished myelin renewal contribute to age-related deficits in memory. Nature Neuroscience, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng YL, Elbaz B, Fei Q, Jones JS, Li YI, Zhuang X, Ming G, He C, & Popko B (2020). m6A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron, 105, 293–309.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, Di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SGN, & Bagni C (2007). A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nature Neuroscience, 10, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


