Abstract
Major histocompatibility complex class I (MHC-I) molecules play a critical role in both innate and adaptive immune responses. The heterodimeric complex of a polymorphic MHC-I heavy chain and a conserved light chain binds to a diverse set of peptides which are presented at the cell surface. Peptide-free (empty) versions of MHC-I molecules are typically retained intracellularly due to their low stability and bound by endoplasmic reticulum chaperones and assembly factors. However, emerging evidence suggests that at least some MHC-I allotypes are relatively stable and detectable at the cell-surface as peptide-deficient conformers, under some conditions. Such MHC-I conformers interact with multiple immune receptors to mediate various immunological functions. Furthermore, conformational sensing of MHC-I molecules by intracellular assembly factors and endoplasmic reticulum chaperones influences the peptide repertoire, with profound consequences for immunity. In this review, we discuss recent advances relating to MHC-I conformational variations and their pathophysiological implications.
Keywords: Major histocompatibility complex class I (MHC-I), Peptide-free MHC-I, Empty MHC-I, Endoplasmic reticulum (ER), Peptide repertoire
Introduction
Cell surface major histocompatibility complex class I (MHC-I) molecules comprise a highly polymorphic heavy chain (HC), a conserved light chain β2-microglobulin (β2m), and a diverse set of short peptides with sequences complementary to residues within the peptide-binding site in the HC. Peptides that bind MHC-I molecules are frequently derived from the cytosol of cells, where they undergo cleavage by the proteasomes. The resulting peptides are translocated to the endoplasmic reticulum (ER) lumen by the transporter associated with antigen processing (TAP). Before peptide binding, MHC-I molecules are localized in the ER as heterodimers of HC and β2m. Such peptide-free (empty) heterodimers are generally unstable and in complex with several ER chaperones and other factors that comprise what is called the peptide-loading complex (PLC). The PLC includes TAP, tapasin, calreticulin and ERp57 [1,2]. In the ER lumen, peptides are assembled with MHC-I molecules with the assistance of the PLC components, which facilitate the selection of high affinity peptides. After such peptide loading, MHC-I molecules are released from the PLC and transported to the cell surface via the Golgi [1]. Polymorphisms of MHC-I molecules influence their assembly pathways. Many MHC-I allotypes can be successfully assembled in the absence of specific PLC components [3,4].
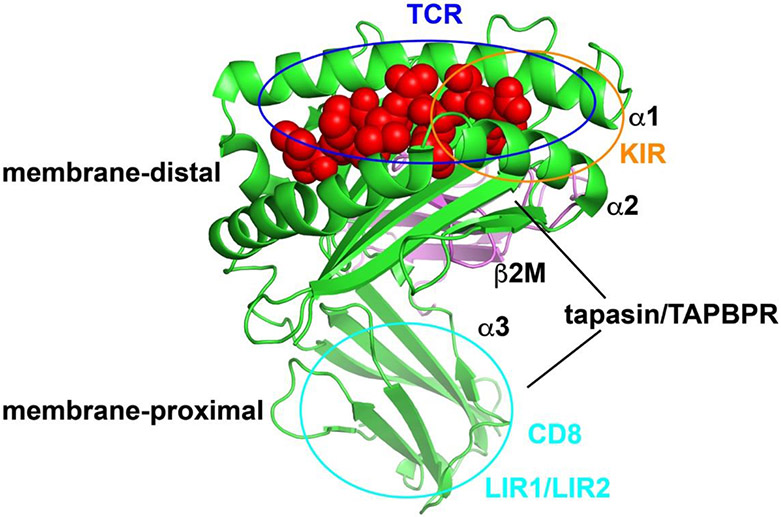
The α1 and α2 domains of MHC-I molecules, which include the peptide-binding site, constitute the recognition site for T cell receptors (TCRs) (Figure 1). This site partly overlaps with binding sites for other immune receptors for MHC-I, such as specific killer cell immunoglobulin-like receptors (KIRs) (Figure 1), expressed on natural killer (NK) cells and other immune cells [5]. In addition, the membrane-proximal α3 domain of MHC-I modulates immune responses. The major binding site for CD8, a co-receptor for TCRs [6] and some KIRs [7], is contained within the α3 domain (Figure 1). The α3 domain is also the binding site for leukocyte immunoglobulin-like receptors (LIR) 1 and LIR2 [8] (Figure 1), which carry immunoreceptor tyrosine-based inhibitory motifs in their cytosolic domains, and deliver inhibitory signals [9]. Although the membrane-proximal and membrane-distal receptor recognition sites correspond to distinct MHC-I surfaces, accumulating evidence suggests the domains are dynamically coupled [10-13]. Recent studies have revealed important functions of peptide-free MHC-I conformers. Here we review recent advances relating to MHC-I-specific immune receptors and assembly factors, which modulate immune responses through sensing MHC-I conformations.
Figure 1.
Structure and interaction sites of MHC-I. MHC-I is composed of a heavy chain (green) that has three domains (α1, α2 and α3), a light chain β2m (violet), and a peptide (red). The recognition sites of TCR, KIR, CD8, LIR1/LIR2 and tapasin/TAPBPR are indicated. There are two major tapasin/TAPBPR binding regions on MHC-I. The crystal structure of HLA-B*5701 in complex with peptide LSSPVTKSF (pdb: 2rfx) is used and the figure was generated with PyMOL.
Polymorphisms of Human MHC-I Molecules and Expression of Sub-Optimally Loaded and Empty Conformers
The human MHC-I HC genes (human leukocyte antigens (HLAs); HLA-A, HLA-B and HLA-C) are the most polymorphic genes in humans with thousands of variants in the population [14]. The structures of the variants are broadly conserved. The vast majority of the polymorphic residues are located within the peptide-binding cleft (Figure 1) conferring variants with different peptide-binding specificities. TCRs have fine specificity for individual cell-surface peptide-MHC-I (pMHC-I) complexes, whereas KIR receptor recognition has broad specificity [5,15], although some peptides are known to interfere with KIR binding [16].
MHC-I molecules typically exist as peptide-free forms in the ER during their folding and assembly and in peptide-loaded forms on the cell surface. However, peptide-free versions or free HCs are also observed on the cell surface under certain conditions, such as upon lymphocyte activation [17,18], viral infection [18], or TAP deficiency [19-21]. The exact mechanism of their expression is not elucidated under some of these conditions. One factor that can influence the cell surface expression of empty conformers is the peptide loading efficiency. When optimal peptide loading is compromised, MHC-I allotypes become loaded with suboptimal peptides, which are more likely to be released after trafficking to the cell surface, thus generating empty conformers of MHC-I [22]. The intrinsic peptide loading efficiency is dictated by the conformational stability of the empty form [23], and for many allotypes, stabilization by the PLC is critical for effective peptide loading and cell surface expression. MHC-I molecules on the surface of cells deficient in TAP [24,25], tapasin [24,26], calreticulin [27], or ERp57 [28], have been shown to be loaded with suboptimal peptides and are thus more peptide-receptive at the cell surface. Given the importance of peptide loading in the antigen presentation process, the PLC is widely targeted by tumor cells and viruses to escape MHC-I mediated immune surveillance. Therefore, under these pathological conditions, suboptimally loaded or empty conformers of MHC-I are more easily produced. Due to varying conformational stabilities of suboptimally loaded forms of different MHC-I allotypes, varying cell surface expression levels are measured in the presence of viral inhibitors of the PLC [21,29,30].
Although the interaction between the highly polymorphic MHC-I molecules and PLC is largely conserved, the functions of the PLC are not fully optimized for the assembly of all MHC-I allotypes [3]. For example, TAP is the major known source of peptides for MHC-I assembly, but in fact, some allotypes are mismatched with TAP in their peptide-binding specificity [31]. Human MHC-I (HLA-I) molecules from the HLA-B7 supertype [32], including the frequent HLA*B35:01 and HLA-B*07:02 allotypes, have specificities for peptides with a proline at the second position, while these peptides are highly disfavored by TAP [33]. For such allotypes, peptide precursors could be transported and further trimmed to optimal sequence by the ER aminopeptidase ERAP, that functions to optimize MHC-I assembly [34]. Nonetheless, the mismatch with TAP specificity results in lower expression and stability of members of the HLA-B7 supertype on the surface of lymphocytes [31]. These studies suggest that even under normal conditions, certain MHC-I allotypes have tendencies to be expressed as suboptimally-loaded or empty conformers.
Suboptimally loaded or empty MHC-I molecules are generally quickly internalized and degraded, and their presence on the cell surface depends on their stabilities [35]. Increasing the stability of MHC-I by lowering the cell culture temperature is known to enhance the cell surface expression level of peptide-receptive conformers of MHC-I in TAP-deficient cells [19]. Cell surface expression of empty or suboptimally loaded MHC-I in TAP-deficient cells is also elevated by introducing a disulfide bond at the C-terminal end of the peptide binding pocket, which increases the stability of MHC-I molecules but does not influence the peptide binding specificity [36]. The polymorphisms of HLA-I molecules influence not only the peptide binding specificities, but also the intrinsic stabilities in the absence of peptides. MD simulations showed that empty conformers of MHC-I variants have different conformational dynamics, particularly near the peptide binding region [11]. In a recent study, we compared the thermostabilities of purified empty conformers of several HLA-B allotypes (HC-β2m heterodimer) and found a strong hierarchy, with HLA-B*35:01 and HLA-B*18:01 displaying high stability compared with HLA-B*44:02 and HLA-B*51:01 [12]. Correspondingly, HLA-B*35:01 and HLA-B*18:01 are expressed at higher levels than HLA-B*44:02 and HLA-B*51:01 in TAP-deficient cells, in peptide-receptive forms [21]. TAP is widely targeted by tumor cells or virus to escape MHC-I mediated immune surveillance. Higher expression levels of certain HLA-I allotypes in TAP-deficient cells suggest such allotypes could be more efficient at antigen presentation under these conditions [21].
The most famous MHC-I molecules known to be expressed in multiple conformations are HLA-B*27 allotypes, some of which have strong genetic associations with ankylosing spondylitis (AS) , an inflammatory disease [37]. HLA-B*27 is known to be expressed on the cell surface as either free HCs or disulfide bond-linked homodimers, following recycling of endocytosed cell surface pMHC-I from endosomes, where some molecules lose their peptides [38]. HLA-B*27 allotypes have a unique free cysteine at position 67, which impairs the folding, and confers the ability to form homodimers [39]. However, the free thiol per se is not enough to explain the cell surface expression of HLA-B*27 as aberrant conformers. Since peptides are more easily released from suboptimally loaded MHC-I molecules, cell surface expression of suboptimally loaded HLA*B27 might also be a prerequisite for expression of aberrant conformers. In fact, expression of aberrant HLA-B*27 conformers is correlated with abnormal ERAP1 activity, via the presence of specific ERAP1 polymorphic variants, which reduce the level of optimal peptides in the ER [40,41]. Indeed, ERAP1 polymorphisms are linked to AS, based on genetic studies [42]. These findings are consistent with the view that compromised peptide loading efficiency is important for cell surface expression of aberrant conformers. Aberrant conformers of HLA-B*27 are shown to be ligands for multiple immune receptors [37], but the causal association of such interactions with AS remains to be established.
Tapasin and TAP-Binding Protein Related (TAPBPR) as Intracellular Sensors of Empty and Peptide-Loaded MHC-I Conformations
Tapasin, an ER chaperone specialized for MHC-I peptide loading, binds and stabilizes HC-β2m heterodimers within the PLC [1]. Tapasin preferentially binds peptide-free class I [43]. HLA-I variants are known to vary widely in their requirements for tapasin [23,44], which is correlated with the intrinsic stabilities of empty HLA-I [23] and their conformational plasticity during their folding in the ER [11,45,46] (reviewed in [13]). As a component of its functions, tapasin edits the MHC-I peptide repertoire, facilitating loading of HLA-I with high affinity peptide, replacing low affinity peptides [47-49]. An open question in the field relates to whether HLA-I allotypes that assemble in a tapasin-independent manner acquire a broader or more suboptimal peptide repertoire compared to tapasin-dependent allotypes that strictly assemble within the PLC and may thus be subject to more stringent quality control. In a recent study [44], tapasin dependencies of 97 HLA-I were determined by analyzing their expression levels on the surface of tapasin-deficient cells in comparison with the corresponding tapasin-sufficient cells. The tapasin dependency scores were negatively correlated with the breadth of HIV peptides that elicited responses from CD8+ T cells of HIV-infected individuals, and higher tapasin dependency was linked to higher viral load and more rapid progression to AIDS. An implication of these associations is that tapasin-independent allotypes in general display greater peptide repertoire diversities. Based on these studies, further assessments of these key differences between tapasin-dependent and the relatively tapasin-independent allotypes are important.
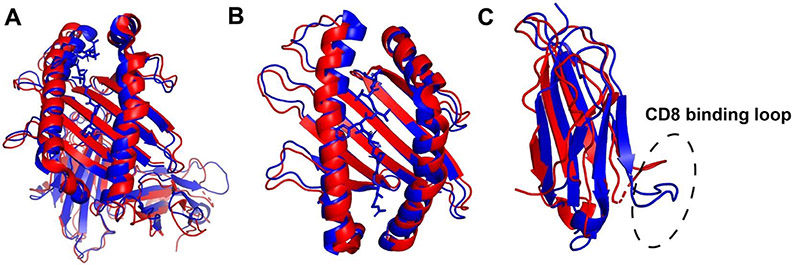
TAPBPR is a recently identified protein homologous to tapasin. Different from tapasin, TAPBPR is not a component of PLC [50]. It is localized in multiple organelles along the secretory pathway [50]. TAPBPR and tapasin share similar overall structures and interaction mode with HLA-I [2,51,52]. High resolution crystal structures (Figure 2A) and NMR studies of TAPBPR in complex with MHC-I have uncovered mechanisms underlying the modulation of MHC-I peptide loading and selection by TAPBPR and tapasin. TAPBPR and tapasin bind MHC-I at two major regions. While the N-terminal domain of TAPBPR interacts with the α2-1 helix and the bottom side of the β-sheet in the peptide-binding groove, the C-terminal domain interacts with both α3 domain and β2m [2,51,52]. Peptide binding stabilizes a relatively closed conformation of MHC-I molecules, and whereas the TAPBPR-bound conformation is more open (Figure 2B). In addition, peptide and TAPBPR binding induce different conformations of the α3 domain, including the CD8 binding loop region (Figure 2C). There is negative allosteric coupling between the peptide binding site and the TAPBPR binding site [53]. Only high affinity peptides can effectively stabilize the closed conformation, and thus induce MHC-I release from TAPBPR. Thus, like tapasin, TAPBPR preferentially binds MHC-I molecules lacking peptide or those bound to specific peptides, constricting the peptide repertoire, and the effects of TAPBPR are allele-specific [54-56]. TAPBPR is suggested to recognize discrete conformational states of peptide-bound MHC-I allotypes to achieve its peptide editing function [56]. Overall, interactions with both tapasin and TAPBPR alter the MHC-I peptidome. As noted above, recent studies indicate that enhanced peptide repertoire diversity is a functionally significant feature of tapasin-independent allotypes [44].
Figure 2.
Structural alignments of TAPBPR-associated (red) and peptide-filled (blue) MHC-I molecules show significant movements of both the α1-α2 and α3 domains. (A) The structure of H2-Dd-TAPBPR complex (5wer) was superimposed onto H2-Dd with a 10mer peptide (5weu). TAPBPR was omitted to compare the conformations of H2-Dd in peptide and TAPBPR-bound forms. (B) α1 and α2 domain of the structures shown in A. (C) α3 domain of the structures shown in A, with the CD8 binding loop indicated. The figure was generated with PyMOL.
CD8 Stabilizes Diverse MHC-I-Receptor Interactions and Senses the Conformation of Cell Surface MHC-I
CD8 is a well-known co-receptor for MHC-I molecules. It plays a crucial role in CD8+ T cell development and activation. The interaction between a TCR and pMHC-I complex determines the specificity of each CD8+ T cell. The binding affinity between TCR and MHC-I is generally weak, typically around 1~100 μM. The interactions are characterized by slow association rates and fast dissociation rates [57]. Such low binding affinity could reduce the likelihood of autoimmune responses. Fast dissociation rates are also good for broad immune surveillance. However, the disadvantages of fast dissociation rates are also obvious. CD8+ T cells can get readily released from antigen presenting cells. A major function of CD8 is to enhance the dwelling time of TCR-pMHC-I complexes, and thus allowing a given pMHC-I sufficient interaction time to activate CD8+ T cells [58]. Another function of CD8 is to synergize TCR/CD3 phosphorylation by recruiting Lck [59,60]. The combination of interactions mediated by TCR and CD8 allow CD8+ T cells to precisely and quickly identify target cells. Previous studies have shown that CD8+ T cells can lyse cells carrying extremely low numbers of HLA-I loaded with cognate peptides [61]. The exact mechanism underlying the synergistic effect of CD8 is still under debate.
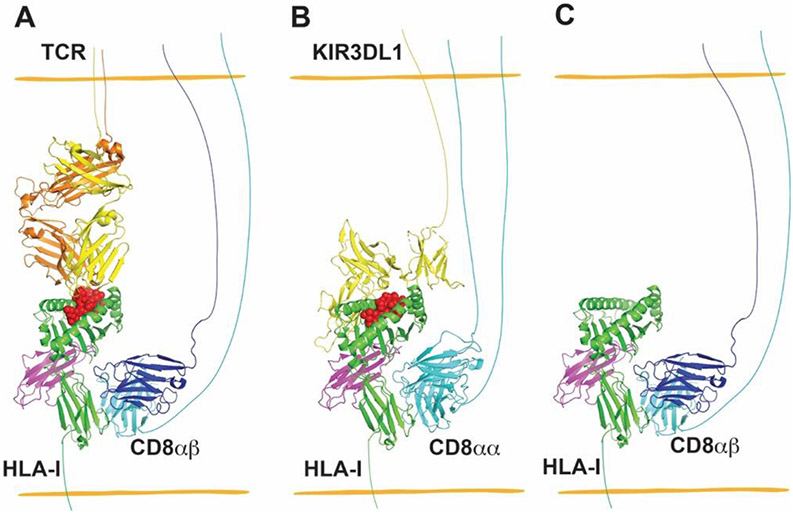
The diversity of TCRs is generated by somatic recombination during T cell development. While TCRs recognize the highly polymorphic α1 and α2 domains, CD8 binds MHC-I molecules at the highly conserved α3 domain, with loop residues 222-232 constituting the major binding site [6] (Figure 3A). CD8 has two isoforms, the CD8αβ heterodimer and the CD8αα homodimer. Cytotoxic T lymphocytes (CTLs) express both CD8αβ and CD8αα. However, it was proposed that only CD8αβ synergizes CTL activation. CD8αα, on the other hand, is thought to be inhibitory in CTL activation [62]. While the CD8αβ heterodimer is exclusively expressed in CTLs, the CD8αα homodimer is widely expressed by subsets of other immune cells, such as NK cells, γδ T cells and NKT cells. The expression levels of CD8 in these non-CTL immune cells are significantly lower than in CTL. The function of CD8 in such cells is less clear. Earlier studies have shown that CD8+ dendritic cells are more efficient at cross-presentation, and CD8+ NK cells are more cytotoxic, findings which imply that CD8 might also play a role in the function of these cells. CD8αα+ γδ T cells were recently shown to be activated by Qa-1b, a strong CD8 binder, via CD8αα binding [63]. In a recent study, we investigated the function of CD8 upon NK cell activation and education. We found that CD8αα functions as a co-receptor for the NK cell inhibitory receptor KIR3DL1 (Figure 3B), facilitating binding of its HLA-B ligands (those which are the Bw4 serotype), and enhancing the inhibitory signal delivered by HLA-Bw4 via KIR3DL1 [7]. Inhibitory signaling is known to be important for NK cell education. Consistent with the co-receptor function of CD8αα, in NK cells, our studies showed that CD8+ NK cells were more cytotoxic than CD8− NK cells only in HLA-Bw4+ donors [7]. These results suggest that CD8 might be a universal co-receptor for MHC-I, and that its function depends on the property of the receptor with which it cooperates.
Figure 3.
Interactions between MHC-I and CD8 provide co-receptor-based synergy for receptors of CD8+ T cells and NK cells (A and B), and enhanced binding between CD8 and peptide-free versions of some MHC-I allotypes contributes to increased adhesion between CD8-expressing cells and MHC-I-expressing target cells (C). (A) CD8αβ heterodimers are co-receptors for TCR recognizing cognate pHLA-I. The structure of TCR–HLA-A*02:01 complex (PDB: 5c0a) was superimposed with H2-Dd- CD8αβ (PDB: 3dmm) by aligning HLA-A*02:01 and H2-Dd, and H2-Dd was then deleted to generate a model for the TCR–HLA-A*02:01-CD8αβ complex. (B) A model for the complex between CD8αα and KIR3DL1 of NK cells with HLA-I of target cells, wherein CD8αα has co-receptor function in NK cells, similar to that for CD8αβ in T cells. The structure of KIR3DL1–HLA-B*57:01 complex (PDB: 3vh8) was superimposed onto HLA-A*02:01-CD8αα (PDB: 1akj) by aligning HLA-B*57:01 and HLA-A*02:01, followed by deletion of HLA-A*02:01 to generate a model for the KIR3DL1-HLA-B*57:01-CD8αα complex. (C) CD8 has been shown to preferentially bind to the empty version of HLA-B*35:01 relative to specific peptide-filled versions. To depict this type of interaction, peptide was deleted from the structure of H2-Dd-CD8αβ (PDB: 3dmm) to model a peptide-free MHC-I allotype binding to CD8αβ of CD8+ T cells. PyMOL was used to visualize the structures of HLA-I and its receptors.
MHC-I molecules bound to specific pathogen-derived or mutant peptides are typically recognized by a small subset of peripheral CD8+ T cells in a TCR dependent manner. Different from such interactions, our recent study also found that empty conformers of HLA-B*35:01 stain bulk CD8+ T cells in a TCR-independent but CD8-dependent manner [12]. In vitro binding assays indicated that empty conformers of HLA-B*35:01 bind CD8 with higher affinity than HLA-B*35:01 bound to specific peptides (Figure 3C). Further studies are needed to determine whether CD8 binding preferences for empty conformers are conserved across all HLA-I allotypes, and whether CD8 binding is also influenced by the sequence of the HLA-I-bound peptides. Although the CD8-binding site is distal from the peptide binding site, the two binding sites have correlated motions. MD simulations [11] and the structures of TAPBPR-MHC-I complexes [51,52] showed that opening of the F pocket induces an allosteric movement of the α3 domain. A comparison of the peptide-filled and TAPBPR-associated empty MHC-I also indicates a significant conformational change of the CD8-binding loop [51,52] (Figure 2C). Similar conformational changes between empty HLA-B*35:01 and specific peptide-filled versions could explain why empty HLA-B*35:01 was preferentially bound by CD8 [12]. Importantly, enhanced binding between empty HLA-I and CD8 did not bypass the TCR to activate CD8+ T cells directly. Rather, empty conformers of HLA-B*35:01 augmented pHLA-I-mediated CD8+ T cell activation. Empty conformers of MHC-I were found to be enriched within the immunological synapse, which enhanced the adhesion between target cells and CTLs. Enrichment of CD8 by empty conformers of HLA-B*35:01 might further activate CD3 through recruiting more Lck, although this was not examined in the study [12]. As described above, empty conformers of HLA-I are expressed under certain pathological conditions. The enhanced interaction between empty conformers of HLA-I and CD8 might be a mechanism to augment the ability of CD8+ T cells to eliminate target cells under pathogenic conditions that target HLA-I expression or assembly, which could result in low levels of HLA-I loaded with antigenic peptides.
Other Immune Receptors Recognize Empty MHC-I Conformers
LIR1 and LIR2 are two inhibitory innate immune receptors for HLA-I molecules. They share similar HLA-I binding sites as CD8 and compete with CD8 for HLA-I interactions [8]. LIR1 prefers peptide-filled HLA-I molecules, while LIR2 binds both conformations [64]. Thus, as seen with CD8-HLA-I interactions [12], peptide binding is linked to α3 domain conformational changes to regulate the functional interactions of HLA-I with other immune receptors. Engagement of LIR1 and LIR2 by MHC-I regulates the differentiation [65] and antigen-presenting properties of DCs [66] and phagocytosis by macrophages [67], correlating with HIV infection outcomes [66] and inhibition of tumor elimination by macrophages [67]. As noted previously, empty conformers of HLA-I were observed on the surface of certain viral infected cells and tumor cells. How these empty conformers affect the functions of DCs and phagocytosis by macrophages through LIR-1 and LIR-2 needs further investigation.
Concluding Remarks
It is becoming clear that MHC-I molecules exist in different conformations and that conformational variants can also represent functional variants. Specific conformers are recognized by intracellular chaperones to modulate the peptide repertoires and by cell surface immune receptors to modulate the immune cell activation or inhibition. MHC-I allotypes have different tendencies to be expressed as suboptimally loaded or empty conformers, or with variable peptide repertoire diversities because of their peptide specificities, assembly pathways and due to differential influences of pathophysiological conditions. These types of variations have key functional consequences for the immune response.
Highlights.
MHC-I allotypes have different tendencies to be expressed as empty or sub-optimally loaded conformers.
Tapasin and TAPBPR sense intracellular MHC-I conformations and modulate the peptide repertoire.
CD8 preferentially recognizes specific MHC-I conformations.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health Grants (R01AI044115 and RO1 AI123957 to MR), and the University of Michigan Fast Forward Protein Folding Diseases Initiative. The authors are grateful to the many contributors to the field, including our co-authors and collaborators.
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Blum JS, Wearsch PA, Cresswell P: Pathways of antigen processing. Annu Rev Immunol 2013, 31:443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampe R: Structure of the human MHC-I peptide-loading complex. Nature 2017, 551:525–528. [DOI] [PubMed] [Google Scholar]
- 3.Olson E, Geng J, Raghavan M: Polymorphisms of HLA-B: influences on assembly and immunity. Curr Opin Immunol 2020, 64:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaitouna AJ, Kaur A, Raghavan M: Variations in MHC class I antigen presentation and immunopeptidome selection pathways. F1000Res 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djaoud Z, Parham P: HLAs, TCRs, and KIRs, a Triumvirate of Human Cell-Mediated Immunity. Annu Rev Biochem 2020, 89:717–739. [DOI] [PubMed] [Google Scholar]
- 6.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, Jakobsen BK: Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature 1997, 387:630–634. [DOI] [PubMed] [Google Scholar]
- *7. Geng J, Raghavan M: CD8alphaalpha homodimers function as a coreceptor for KIR3DL1. Proc Natl Acad Sci U S A 2019, 116:17951–17956. In this study, CD8αα was shown to act as co-receptor for the NK cell inhibitory receptor KIR3DL1, revealing a new function of CD8. CD8αα engagement by MHC-I enhances interaction between MHC-I and KIR3DL1 and thus elevates inhibition of NK cell activity. Moreover, since inhibitory receptors are crucial for NK cell licensing, the enhanced interaction between MHC-I and KIR3DL1 confers CD8αα+ NK cells better education, which could explain why CD8αα+ NK cells are generally more cytotoxic.
- 8.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, et al. : Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A 2003, 100:8856–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson LE, Allen RL: Leukocyte Ig-Like Receptors - A Model for MHC Class I Disease Associations. Front Immunol 2016, 7:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hateren A, Carter R, Bailey A, Kontouli N, Williams AP, Kaufman J, Elliott T: A mechanistic basis for the co-evolution of chicken tapasin and major histocompatibility complex class I (MHC I) proteins. J Biol Chem 2013, 288:32797–32808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Hateren A, Bailey A, Werner JM, Elliott T: Plasticity of empty major histocompatibility complex class I molecules determines peptide-selector function. Mol Immunol 2015, 68:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12. Geng J, Altman JD, Krishnakumar S, Raghavan M: Empty conformers of HLA-B preferentially bind CD8 and regulate CD8+ T cell function. Elife 2018, 7. In this paper the authors prepared empty or peptide-filled conformers of HLA-B*35:01 and found that empty conformers have stonger CD8 binding affinity than those loaded with specific peptides, based on tetramer staining assays. Although CD8 engagement alone is insufficient to activate CD8+ T cells, the engagement between HLA-B*35:01 empty conformers and CD8 enhanced antigen-induced CD8+ T cell activation. In addition, open conformers were shown to be enriched in the immunological synapse during CD8+ T cell activation.
- 13.Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, Noe F, Freund C: Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol 2017, 8:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE: IPD-IMGT/HLA Database. Nucleic Acids Res 2020, 48:D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Natarajan K, Margulies DH: MHC Molecules, T cell Receptors, Natural Killer Cell Receptors, and Viral Immunoevasins-Key Elements of Adaptive and Innate Immunity. Adv Exp Med Biol 2019, 1172:21–62. [DOI] [PubMed] [Google Scholar]
- 16.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, et al. : Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol 2007, 178:33–37. [DOI] [PubMed] [Google Scholar]
- 17.Schnabl E, Stockinger H, Majdic O, Gaugitsch H, Lindley IJ, Maurer D, Hajek-Rosenmayr A, Knapp W: Activated human T lymphocytes express MHC class I heavy chains not associated with beta 2-microglobulin. J Exp Med 1990, 171:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madrigal JA, Belich MP, Benjamin RJ, Little AM, Hildebrand WH, Mann DL, Parham P: Molecular definition of a polymorphic antigen (LA45) of free HLA-A and -B heavy chains found on the surfaces of activated B and T cells. J Exp Med 1991, 174:1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TN, Townsend A, Karre K, et al. : Empty MHC class I molecules come out in the cold. Nature 1990, 346:476–480. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz-Navarrete V, Hammerling GJ: Surface appearance and instability of empty H-2 class I molecules under physiological conditions. Proc Natl Acad Sci U S A 1991, 88:3594–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng J, Zaitouna AJ, Raghavan M: Selected HLA-B allotypes are resistant to inhibition or deficiency of the transporter associated with antigen processing (TAP). PLoS Pathog 2018, 14:e1007171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montealegre S, Venugopalan V, Fritzsche S, Kulicke C, Hein Z, Springer S: Dissociation of beta2-microglobulin determines the surface quality control of major histocompatibility complex class I molecules. FASEB J 2015, 29:2780–2788. [DOI] [PubMed] [Google Scholar]
- 23.Rizvi SM, Salam N, Geng J, Qi Y, Bream JH, Duggal P, Hussain SK, Martinson J, Wolinsky SM, Carrington M, et al. : Distinct assembly profiles of HLA-B molecules. J Immunol 2014, 192:4967–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chefalo PJ, Grandea AG 3rd, Van Kaer L, Harding CV: Tapasin−/− and TAP1−/− macrophages are deficient in vacuolar alternate class I MHC (MHC-I) processing due to decreased MHC-I stability at phagolysosomal pH. J Immunol 2003, 170:5825–5833. [DOI] [PubMed] [Google Scholar]
- 25.Martien van Santen H, Woolsey A, Rickardt PG, Van Kaer L, Baas EJ, Berns A, Tonegawa S, Ploegh HL: Increase in positive selection of CD8+ T cells in TAP1-mutant mice by human beta 2-microglobulin transgene. J Exp Med 1995, 181:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandea AG 3rd, Golovina TN, Hamilton SE, Sriram V, Spies T, Brutkiewicz RR, Harty JT, Eisenlohr LC, Van Kaer L: Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity 2000, 13:213–222. [DOI] [PubMed] [Google Scholar]
- 27.Gao B, Adhikari R, Howarth M, Nakamura K, Gold MC, Hill AB, Knee R, Michalak M, Elliott T: Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity 2002, 16:99–109. [DOI] [PubMed] [Google Scholar]
- 28.Garbi N, Tanaka S, Momburg F, Hammerling GJ: Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol 2006, 7:93–102. [DOI] [PubMed] [Google Scholar]
- 29.Park B, Kim Y, Shin J, Lee S, Cho K, Fruh K, Lee S, Ahn K: Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 2004, 20:71–85. [DOI] [PubMed] [Google Scholar]
- 30.Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, et al. : Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med 2004, 200:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarzabek B, Zaitouna AJ, Olson E, Silva GN, Geng J, Geretz A, Thomas R, Krishnakumar S, Ramon DS, Raghavan M: Variations in HLA-B cell surface expression, half-life and extracellular antigen receptivity. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidney J, Peters B, Frahm N, Brander C, Sette A: HLA class I supertypes: a revised and updated classification. BMC Immunol 2008, 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uebel S, Kraas W, Kienle S, Wiesmuller KH, Jung G, Tampe R: Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc Natl Acad Sci U S A 1997, 94:8976–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang SC, Momburg F, Bhutani N, Goldberg AL: The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a "molecular ruler" mechanism. Proc Natl Acad Sci U S A 2005, 102:17107–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montealegre S, van Endert PM: Endocytic Recycling of MHC Class I Molecules in Non-professional Antigen Presenting and Dendritic Cells. Front Immunol 2018, 9:3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hein Z, Uchtenhagen H, Abualrous ET, Saini SK, Janssen L, Van Hateren A, Wiek C, Hanenberg H, Momburg F, Achour A, et al. : Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J Cell Sci 2014, 127:2885–2897. [DOI] [PubMed] [Google Scholar]
- 37.Bowness P: Hla-B27. Annu Rev Immunol 2015, 33:29–48. [DOI] [PubMed] [Google Scholar]
- 38.Bird LA, Peh CA, Kollnberger S, Elliott T, McMichael AJ, Bowness P: Lymphoblastoid cells express HLA-B27 homodimers both intracellularly and at the cell surface following endosomal recycling. Eur J Immunol 2003, 33:748–759. [DOI] [PubMed] [Google Scholar]
- 39.Dangoria NS, DeLay ML, Kingsbury DJ, Mear JP, Uchanska-Ziegler B, Ziegler A, Colbert RA: HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J Biol Chem 2002, 277:23459–23468. [DOI] [PubMed] [Google Scholar]
- 40.Haroon N, Tsui FW, Uchanska-Ziegler B, Ziegler A, Inman RD: Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann Rheum Dis 2012, 71:589–595. [DOI] [PubMed] [Google Scholar]
- 41.Reeves E, Colebatch-Bourn A, Elliott T, Edwards CJ, James E: Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc Natl Acad Sci U S A 2014, 111:17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et al. : Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet 2011, 43:761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizvi SM, Raghavan M: Direct peptide-regulatable interactions between MHC class I molecules and tapasin. Proc Natl Acad Sci U S A 2006, 103:18220–18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44. Bashirova AA, Viard M, Naranbhai V, Grifoni A, Garcia-Beltran W, Akdag M, Yuki Y, Gao X, O’HUigin C, Raghavan M, et al. : HLA tapasin independence: broader peptide repertoire and HIV control. Proc Natl Acad Sci U S A 2020, 117:28232–28238. The authors in this study quantified the tapasin dependencies of MHC-I allotypes common in European and African Americans and then investigated the relationship between tapasin dependencies of MHC-I and their abilities to mediated HIV-specific CTL responses. The study measured responses of CD8+ T cells derived from HIV-infected patients towards antigen presenting cells pulsed with a panel of 410 overlapping synthetic HIV-derived peptides. They found an inverse correlation between tapasin dependencies of HLA-I and the number of peptides that elicited CD8+ T cell responses, findings which suggest that tapasin-independent allotypes, in general, have broader peptide repertoires.
- 45.Sieker F, Straatsma TP, Springer S, Zacharias M: Differential tapasin dependence of MHC class I molecules correlates with conformational changes upon peptide dissociation: a molecular dynamics simulation study. Mol Immunol 2008, 45:3714–3722. [DOI] [PubMed] [Google Scholar]
- 46.Abualrous ET, Fritzsche S, Hein Z, Al-Balushi MS, Reinink P, Boyle LH, Wellbrock U, Antoniou AN, Springer S: F pocket flexibility influences the tapasin dependence of two differentially disease-associated MHC Class I proteins. Eur J Immunol 2015, 45:1248–1257. [DOI] [PubMed] [Google Scholar]
- 47.Praveen PV, Yaneva R, Kalbacher H, Springer S: Tapasin edits peptides on MHC class I molecules by accelerating peptide exchange. Eur J Immunol 2010, 40:214–224. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Bouvier M: Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J 2007, 26:1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wearsch PA, Cresswell P: Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol 2007, 8:873–881. [DOI] [PubMed] [Google Scholar]
- 50.Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, Duncan LM, Harbour ME, Rhodes DA, Skjodt K, et al. : Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci U S A 2013, 110:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang J, Natarajan K, Boyd LF, Morozov GI, Mage MG, Margulies DH: Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science 2017, 358:1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas C, Tampe R: Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science 2017, 358:1060–1064. [DOI] [PubMed] [Google Scholar]
- **53. McShan AC, Natarajan K, Kumirov VK, Flores-Solis D, Jiang J, Badstubner M, Toor JS, Bagshaw CR, Kovrigin EL, Margulies DH, et al. : Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat Chem Biol 2018, 14:811–820. In this paper, the authors investigated the conformational dynamics of MHC-I using solution NMR. The study suggests that TAPBPR modulates peptide binding by sensing the global conformational change induced by peptide binding. The TAPBPR and peptide binding sites are mutually exclusive yet negatively coupled. Binding of high affinity peptides to the empty groove results in a prolonged interaction between the peptide and the groove, and closure of the α2-l helix, a switch controlling the release of TAPBPR.
- 54.Morozov GI, Zhao H, Mage MG, Boyd LF, Jiang J, Dolan MA, Venna R, Norcross MA, McMurtrey CP, Hildebrand W, et al. : Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci U S A 2016, 113:E1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ilca FT, Drexhage LZ, Brewin G, Peacock S, Boyle LH: Distinct Polymorphisms in HLA Class I Molecules Govern Their Susceptibility to Peptide Editing by TAPBPR. Cell Rep 2019, 29:1621–1632 el623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56. McShan AC, Devlin CA, Overall SA, Park J, Toor JS, Moschidi D, Flores-Solis D, Choi H, Tripathi S, Procko E, et al. : Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection. Proc Natl Acad Sci U S A 2019, 116:25602–25613. By using bimolecular fluorescence complementation assays, the authors found that TAPBPR associates with a broad range of peptide-deficient MHC-I allotypes inside the cell. Using solution NMR, the authors showed that peptide filled-MHC-I molecules display allotype-specific conformational dynamics. Only certain allotypes exhibit a minor conformational state in the presence of peptide, which is recognized by TAPBPR, findings that explain the narrower specificity of TAPBPR toward peptide-filled MHC-I allotypes. Based on these results, the authors proposed that TAPBPR has the capacity to stabilize a broad range of peptide-deficient MHC-I inside the cell, but edit the peptide repertoires of a narrower range of allotypes.
- 57.Stone JD, Chervin AS, Kranz DM: T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology 2009, 126:165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hebeisen M, Schmidt J, Guillaume P, Baumgaertner P, Speiser DE, Luescher I, Rufer N: Identification of Rare High-Avidity, Tumor-Reactive CD8+ T Cells by Monomeric TCR-Ligand Off-Rates Measurements on Living Cells. Cancer Res 2015, 75:1983–1991. [DOI] [PubMed] [Google Scholar]
- 59.Horkova V, Drobek A, Mueller D, Gubser C, Niederlova V, Wyss L, King CG, Zehn D, Stepanek O: Dynamics of the Coreceptor-LCK Interactions during T Cell Development Shape the Self-Reactivity of Peripheral CD4 and CD8 T Cells. Cell Rep 2020, 30:1504–1514 e1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK: CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci U S A 2010, 107:16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM: T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol 2004, 5:524–530. [DOI] [PubMed] [Google Scholar]
- 62.Gangadharan D, Cheroutre H: The CD8 isoform CD8alphaalpha is not a functional homologue of the TCR co-receptor CD8alphabeta. Curr Opin Immunol 2004, 16:264–270. [DOI] [PubMed] [Google Scholar]
- *63. Goodall KJ, Nguyen A, McKenzie C, Eckle SBG, Sullivan LC, Andrews DM: The murine CD94/NKG2 ligand, Qa-1(b), is a high-affinity, functional ligand for the CD8alphaalpha homodimer. J Biol Chem 2020, 295:3239–3246. In this study, the murine non-classical MHC-I Qa-1b was shown to strongly bind CD8αα by tetramer staining and surface plasmon resonance assays, although this interaction was not conserved in the human counterpart of Qa-1b, HLA-E. The engagement between Qa-1b and CD8αα directly activated γδ T cells from small intestine.
- 64.Jones DC, Kosmoliaptsis V, Apps R, Lapaque N, Smith I, Kono A, Chang C, Boyle LH, Taylor CJ, Trowsdale J, et al. : HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol 2011, 186:2990–2997. [DOI] [PubMed] [Google Scholar]
- 65.Young NT, Waller EC, Patel R, Roghanian A, Austyn JM, Trowsdale J: The inhibitory receptor LILRB1 modulates the differentiation and regulatory potential of human dendritic cells. Blood 2008, 111:3090–3096. [DOI] [PubMed] [Google Scholar]
- 66.Bashirova AA, Martin-Gayo E, Jones DC, Qi Y, Apps R, Gao X, Burke PS, Taylor CJ, Rogich J, Wolinsky S, et al. : LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLoS Genet 2014, 10:e1004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY, George BM, Markovic M, Ring NG, Tsai JM, et al. : Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol 2018, 19:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]