Abstract
Epigenetic clocks are calculated by combining DNA methylation states across select CpG sites to estimate biologic age, and have been noted as the most successful markers of biologic aging to date. Yet, limited research has considered epigenetic clocks calculated in brain tissue. We used DNA methylation states in dorsolateral prefrontal cortex specimens from 721 older participants of the Religious Orders Study and Rush Memory and Aging Project, to calculate DNA methylation age using four established epigenetic clocks: Hannum, Horvath, PhenoAge, GrimAge, and a new Cortical clock. The four established clocks were trained in blood samples (Hannum, PhenoAge, GrimAge) or using 51 human tissue and cell types (Horvath); the recent Cortical clock is the first trained in postmortem cortical tissue. Participants were recruited beginning in 1994 (Religious Orders Study) and 1997 (Memory and Aging Project), and followed annually with questionnaires and clinical evaluations; brain specimens were obtained for 80–90% of participants. Mean age at death was 88.0 (SD 6.7) years. We used linear regression, logistic regression, and linear mixed models, to examine relations of epigenetic clock ages to neuropathologic and clinical aging phenotypes, controlling for chronologic age, sex, education, and depressive symptomatology.
Hannum, Horvath, PhenoAge and Cortical clock ages were related to pathologic diagnosis of Alzheimer’s disease (AD), as well as to Aβ load (a hallmark pathology of Alzheimer’s disease). However, associations were substantially stronger for the Cortical than other clocks; for example, each standard deviation (SD) increase in Hannum, Horvath, and PhenoAge clock age was related to approximately 30% greater likelihood of pathologic AD (all p<0.05), while each SD increase in Cortical age was related to 90% greater likelihood of pathologic AD (odds ratio=1.91, 95% confidence interval 1.38, 2.62). Moreover, Cortical age was significantly related to other AD pathology (eg, mean tau tangle density, p=0.003), and to odds of neocortical Lewy body pathology (for each SD increase in Cortical age, odds ratio=2.00, 95% confidence 1.27, 3.17), although no clocks were related to cerebrovascular neuropathology. Cortical age was the only epigenetic clock significantly associated with the clinical phenotypes examined, from dementia to cognitive decline (5 specific cognitive systems, and a global cognitive measure averaging 17 tasks) to Parkinsonian signs. Overall, our findings provide evidence of the critical necessity for bespoke clocks of brain aging for advancing research to understand, and eventually prevent, neurodegenerative diseases of aging.
Keywords: epigenetics, aging, neuropathology, epidemiology, dementia
INTRODUCTION
Epigenetic modifications, such as DNA methylation (DNAm), are a hallmark of aging.1 Over the last decade, several DNAm-based biomarkers of aging have been identified; these “epigenetic clocks” are calculated by combining DNAm states across select sites to estimate age.2–5 Interestingly, growing evidence indicates the epigenome may both provide a signature of accumulated biologic aging as well as influence subsequent aging processes.6 Thus, epigenetic clocks could not only distinguish those with accelerated (or decelerated) biologic aging, but also monitor interventions to delay aging.7 In research to date, a variety of epigenetic clocks have been published. The primary existing clocks were developed in blood samples (eg, the Hannum,2 PhenoAge4 and GrimAge5 clocks) or across many tissues (eg, the Horvath clock3); most were trained to predict chronologic age, although recent “second generation” clocks have been trained to predict aging-related phenotypes (PhenoAge4, GrimAge5), such as mortality. Epigenetic clocks have been noted as the most successful aging biomarkers yet.1
Although the prevalence of neurodegenerative diseases of aging, such as Alzheimer’s dementia or Parkinson’s Disease, is increasing,9,10 limited research has examined epigenetic clocks in brain tissue. In previous reports,11,12 calculating several different epigenetic clocks designed in peripheral blood samples or across many tissues simultaneously, there were reasonable correlations of epigenetic age with chronologic age even in specimens from up to 6 different brain regions – suggesting these clocks may have applications in brain tissue. Indeed, in initial research in the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP), the Horvath and PhenoAge clocks were calculated in post-mortem dorsolateral pre-frontal cortex (DLPFC); after controlling for chronologic age, clock age was related to amyloid beta (Aβ) load and to pathologic Alzheimer’s disease (AD) diagnosis, although there was little association with several clinical dementia-related outcomes.4,13
Recently, an epigenetic clock trained in human cortex tissue was developed,14 although little is known regarding how this “Cortical” clock may predict pathologic or clinical neurodegenerative phenotypes, and whether it may perform better for neurodegenerative outcomes than clocks developed in blood samples or across many tissues. To fully understand how blood-based, multi-tissue-based, and brain-based epigenetic clocks function as biomarkers of brain aging, further examination of clocks in brain tissue is crucial.
Thus, we used post-mortem DLPFC from 721 older participants of ROS and MAP, to calculate four established epigenetic clocks (Hannum, Horvath, PhenoAge, GrimAge); for comparison, we also calculated the recent Cortical clock, which was developed in cortical tissue and trained to predict chronologic age. We examined associations of each of these clocks to a variety of age-related neuropathologies, and to key age-related clinical phenotypes.
MATERIALS AND METHODS
Study Populations
The Religious Orders Study was initiated in 1994,15 and includes older priests, nuns and brothers from across the US, free of known dementia at the time of enrollment. Participants agreed to annual neurological exams, neuropsychological testing, and blood draw, and signed an informed consent and Anatomic Gift Act to donate their brains at death. Over 1,468 participants completed a baseline evaluation. The follow-up rate and autopsies exceed 90%. The Rush Memory and Aging Project was established in 1997,15 with virtually identical design and data collection, and includes older men and women from across the Chicago metropolitan area, without known dementia at enrollment; over 2,170 participants completed a baseline evaluation to date. The follow-up rate exceeds 90% and the autopsy rate exceeds 80%. Both studies were approved by an Institutional Review Board of Rush University Medical Center. All participants signed an informed consent and Anatomical Gift Act for organ donation. For the work described here, we leveraged DNA methylation profiling previously completed in frozen DLPFC tissue from over 700 decedents, representing all available specimens at the time profiling was completed.
Assessment of DNA Methylation States and Epigenetic Clocks
In the brain specimens, DNA methylation was measured in DLPFC tissue. Briefly, 100mg frozen sections were thawed on ice, with the gray matter dissected from the white matter, as previously described in detail.16 The Qiagen QIAamp DNA mini protocol was used for DNA isolation. DNA methylation profiles were generated using the Illumina Infinium HumanMethylation450 platform. In recent work with these specimens, processing methods were updated compared to our previous publications.16 The raw signal intensities were imported into the R statistical environment with functions from the methylumi package and further processed with the wateRmelon package. Initial quality control assessment was performed using functions in the methylumi package to exclude samples with inefficient bisulfite conversion (< 90%) as well as outliers. Further preprocessing was conducted using the wateRmelon package17 by applying a p-filter. Probes having more than 1% of samples with a detection p-value greater than 0.05 and a beadcount lower than 3 in more than 5% of samples were excluded. Finally, the filtered data were normalized with ‘dasen’. Non-CpG SNP (single nucleotide polymorphism) probes, probes that had been reported to contain common (MAF >5 %) SNPs in the CG or single base extension position, or probes that were non-specific or mismapped, were flagged and disregarded in the evaluation of our results. The resulting dataset for analysis here consisted of 730 samples with 423,841 probes each. At each probe, DNAm level was represented as a beta value, that is, the ratio of the methylated probe intensity to the sum of methylated and unmethylated probe intensities. The values ranged from 0 to 1, where a larger value indicates higher methylation.
We used open source software at https://dnamage.genetics.ucla.edu/home to calculate DNA methylation age from four commonly used epigenetic clocks in the DLPFC: Hannum clock, Horvath clock, PhenoAge clock and GrimAge clock. The Cortical clock was calculated using publically-available code provided by the authors (https://github.com/gemmashireby/CorticalClock). Briefly, the Hannum clock is based on 71 CpG sites,2 selected agnostically by regressing DNAm levels against chronologic age, in peripheral blood. The Horvath clock incorporates 353 CpG sites, also selected by regressing DNAm levels against chronologic age, but using 51 different tissue types.3 The newer PhenoAge clock is calculated from 513 CpGs,4 representing 9 plasma biomarkers that predict mortality (rather than chronologic age), and the GrimAge clock incorporates 1030 CpG sites, representing 12 plasma biomarkers that predict lifespan, plus cigarette smoking.5 Finally, the Cortical clock was designed in postmortem cortical specimens,14 and, similar to the Horvath and Hannum clocks, was trained to predict chronologic age; the Cortical clock includes 347 CpG sites.
Assessment of Neuropathologic Outcomes
In the brain specimens, we considered the primary outcomes of AD pathology, other neurodegenerative neuropathology, and cerebrovascular pathology. In collecting the brain donations, the mean post-mortem interval was 9.1 (SD=7.9) hours. Details of the autopsy procedures are described elsewhere.18 In brief, the brain was removed, one hemisphere was frozen, and the other hemisphere was fixed in 4% paraformaldehyde for at least three days. The fixed hemisphere was cut into 1 cm slabs, and tissue blocks were obtained from pre-specified brain regions for neuropathological evaluation.
Alzheimer Disease (AD) Pathology:
For assessing AD pathology, we calculated a global score representing overall AD pathologic burden combining neuritic plaques, diffuse plaques, and neurofibrillary tangles. For this score, we used a modified Bielschowsky silver stain to visualize neuritic plaques, diffuse plaques, and neurofibrillary tangles in five cortical areas. Because the regional counts had wide means, standard deviations, and ranges, we first converted the raw counts to a standard distribution by dividing each person’s count by the SD for that particular count; next, we formed summary measures of neuritic plaques, diffuse plaques, and neurofibrillary tangles by averaging the standardized scores across all brain regions. Finally, we constructed the global AD pathology score by averaging the summary measures of the three AD pathologic indices, such that higher scores represent more pathology.
We also separately examined the percent area occupied by Aβ and tau tangle density, using image analyses (Aβ) and stereology (tau tangles) of immunohistochemically-stained brain sections. For these two measures, we used tissue from 8 cortical regions and then averaged findings across the regions to create two separate summary measures of the brain’s load of Aβ and density of tau-labeled tangles; again, higher scores represent greater pathology.
A pathologic diagnosis of AD was also rendered according to the National Institute on Aging (NIA)-Reagan criteria.19
Other Neurodegenerative Neuropathology:
Immunohistochemistry with α-synuclein immunostain (1:20,000, Wako Chem. USA, Richmond, VA) was used for assessment of Lewy bodies in the substantia nigra, 2 limbic sites (entorhinal cortex, anterior cingulate cortex), and 3 neocortical sites (midfrontal cortex, superior or middle temporal cortex, inferior parietal cortex). We classified Lewy body disease as nigral, limbic, or neocortical using a modified version of the staging criteria of McKeith et al.20,21 Neocortical disease required Lewy bodies in frontal, temporal, or parietal cortex and was accompanied by nigral and/or limbic Lewy bodies. A dichotomous variable indicating presence of neocortical Lewy bodies was used in this analysis.
To identify limbic-predominant age-related TDP-43 encephalopathy neuropathological change (LATE-NC), we assessed transactive response DNA binding protein 43 kDa (TDP-43) pathology in 8 brain regions (amygdala, entorhinal cortex, hippocampus CA1 and subiculum, dentate gyrus, anterior temporal pole, interior frontal, middle temporal cortex, and midfrontal cortex) using monoclonal antibodies (pS409/410;1:100), which stain pathologically phosphorylated TDP-43 proteins but not normal nuclear TDP-43. We classified those with stage 0 (no presence of TDP-43) or stage 1 (presence of TDP-43 localized to the amygdala) as not having LATE-NC, and those with stage 2 (extension to the hippocampus and/or entorhinal cortex) or stage 3 (extension to the neocortex) as having LATE-NC.
Cerebrovascular Pathology:
To identify vessel disease, atherosclerosis severity grade was assessed by evaluation of circle of Willis vessels at the base of the brain. Vertebral, basilar, posterior cerebral, middle cerebral, and anterior cerebral arteries were evaluated along with their proximal branches. Severity of atherosclerosis was scaled semi-quantitatively (none or possible, mild, moderate, severe) on the basis of involvement of each artery and number of arteries.
Additionally, severity of arteriolosclerosis was graded based on concentric hyaline thickening of vessel walls, specifically small arterioles less than approximately 50 μm in diameter. The scale ranged from none, to mild if the arterial wall was minimally thickened, moderate if the arteriolar wall thickness was up to twice the normal thickness, and severe, if the arteriolar wall thickness was more than twice the normal thickness or completely occluded.
Cerebral amyloid angiopathy (CAA) was examined using Aβ immunostaining of meningeal and parenchymal vessels in four regions (midfrontal, midtemporal, angular, and calcarine cortices). Three monoclonal antibodies against Aβ were used in immunostaining of sections: 4G8 (1:9000; Covance Labs, Madison, WI), 6F/3D (1:50; Dako North America Inc., Carpinteria, CA), and 10D5 (1:600; Elan Pharmaceuticals, San Francisco, CA). In this analysis, we used a semi-quantitative scoring system as none, mild, moderate, and severe.
Uniform examination of cerebral infarcts was conducted to identify chronic infarcts; we identified all brain infarcts visible to the naked eye. Microscopic examination also provided identification of chronic microinfarcts. A minimum of 9 regions in 1 hemisphere were examined for microinfarcts on 6-μm paraffin-embedded sections stained with H&E. We created dichotomous summaries (presence vs. absence) for each of the two infarct variables.
Assessment of Clinical Neurologic and Other Outcomes
In addition to pathology, we considered several age-related clinical phenotypes, especially neurologic phenotypes. Participants underwent an annual, uniform structured clinical evaluation which included detailed cognitive testing. Scores from 17 cognitive tests were summarized in a global cognitive score, and 5 cognitive systems were evaluated, including episodic memory, semantic memory, working memory, perceptual speed, and visuospatial ability. For the global score and for each cognitive system, all test scores were transformed to z-scores, using the baseline means and standard deviations from the cohort, and then z-scores for each relevant test were averaged to create the composite scores.22 Further, all participants were evaluated annually by an experienced clinician who used cognitive and clinical data to diagnose any dementia, as well as Alzheimer’s dementia.23 As per research conventions regarding terminology,24 we use “Alzheimer’s dementia” to refer to clinical diagnosis prior to death (and “Alzheimer’s disease” [AD] to refer to post-mortem pathologic diagnosis).
In addition, we assessed Parkinsonian signs, using a composite measure averaging 4 separate signs based on a 26-item modified version of the motor portion of the United Parkinson’s Disease Rating Scale, which assessed bradykinesia, parkinsonian gait, rigidity, and tremor. Briefly, the score for each sign was calculated by adding the ratings for the individual items from each sign, dividing by the maximum possible score for the sign, then multiplying by 100.25 A summary score was created, ranging from 0 to 100, and averaging the four scores from each sign. A higher summary score reflects expression of more severe Parkinsonian signs.
We also calculated a summary score of motor function, based on: Purdue pegboard test (no. of pegs), finger-tapping test (taps/10 seconds), time to cover a distance of 8 feet (seconds), number of steps required to cover 8 feet (steps), 360 degree turn time (seconds), number of steps to complete a 360 degree turn (steps), leg stand (seconds), toe stand (seconds), grip strength (kilograms), and pinch strength (kilograms).24 The composite measure was constructed by converting each measure to a score using the mean from all participants at baseline and averaging all the motor tests together. Higher score represents better motor function.
Finally, since apolipoprotein E (APOE) genotype is strongly related to neuropathology and neurologic disease, we also examined APOE genotype;27 we excluded those with e2e4 genotype, since it is more challenging to interpret this group.
Population for Analysis
For examining epigenetic clocks in DLPFC, we used 730 specimens that were part of previous research on DNAm and neurodegeneration, representing all available specimens at the time DNAm was profiled.27 We excluded 9 participants who did not have full covariate data available. For analyses of specific neuropathologic and neurologic outcomes, some participants were missing data, thus analytic samples varied somewhat across analyses. Finally, since the original training set for the Cortical clock included 88 of the ROSMAP DLPFC specimens,14 for all analyses of the Cortical clock, we also excluded those 88 samples.
Statistical Analysis
To enhance interpretability of results across the epigenetic clocks, we standardized each clock (ie, to mean=0, SD=1), and used this variable as the independent variable in models.
We utilized different statistical models across the neuropathologic and neurologic variables. Because we were only interested in the ability of the clocks to predict phenotypes above and beyond the effects of chronologic age, all statistical models include chronologic age as a covariate. For the dependent variables which were continuous measures (i.e., global AD pathologic score, amyloid load, and tau tangle density), we used linear regression models adjusted for chronologic age at death (continuous), education (years), sex, and depressive symptoms at baseline (CES-D score). We used a square root transformation of these outcomes in the regression models, due to the positively skewed distributions of data. To examine associations of the epigenetic clocks with categorical outcomes (AD pathologic diagnosis, cerebrovascular and other neurodegenerative neuropathologies, clinical diagnosis of Alzheimer’s dementia or any dementia, APOE genotype), we used either logistic regression or ordinal logistic regression models, controlling for chronologic age, education, sex, and depressive symptoms at baseline. While genotype would standardly be considered as an independent rather than dependent variable, we chose to maintain clock age as our independent variable in all models to facilitate comparison of findings across variables of interest here. For analyses of cognitive decline, and for change over time in the scores for Parkinsonian signs and motor function, we used linear mixed models, controlled for age at death, sex, education and depressive symptoms at baseline. A term for the interaction of follow-up time by epigenetic clock age was used to estimate the relation of clock age to annual rates of decline.
We also conducted several sensitivity analyses. In one set of analyses, instead of epigenetic age, we examined epigenetic age acceleration, calculated as the residuals from regressing epigenetic age on chronologic age. Second, we were concerned about confounding from cell type proportions. Since there is no ideal method for calculating neuron proportion, we conducted two analyses using different approaches. In one, we used the most common approach of estimating neuron proportion using the CETS algorithm,28 which utilizes DNAm states from the Illumina array, and was thus available in all participants. In a second, we estimated neuron proportion using a deconvolution approach (digital sorting algorithm, DSA) based on cell type-specific genes derived from published reference data, using available transcriptomics from RNAseq in the DLPFC.29 We had adequate information available in 528 participants to calculate neuron proportion estimated from DSA. We then conducted two sets of analyses, creating models adding each of these variables to test for confounding by neuron proportion.
For all analyses, we calculated two-sided p-values, and 95% confidence intervals (CI); p-values <0.05 were considered statistically significant.
Data Availability
ROSMAP data can be requested at https://www.radc.rush.edu.
RESULTS
Among participants (Table 1), 36% were male and mean chronologic age at death was 88.0 (SD 6.7) years. Participants were highly educated, with nearly three-quarters having at least 15 years of education. Absolute epigenetic ages in the DLPFC samples ranged from a mean of 1.6 (SD 5.8) years for the PhenoAge clock to 87.3 (SD 5.6) years for the Cortical clock. Pathologic AD diagnosis was identified in over half of participants, while other neurodegenerative disease pathologies were much less common; LATE-NC was found in approximately one-third of participants, and neocortical Lewy bodies were identified in less than 10%. Approximately 40–45% of participants had moderate or severe levels of cerebral vessel pathology (i.e., atherosclerosis, arteriosclerosis, or cerebral amyloid angiopathy). Chronic microinfarcts were identified in about one-quarter and gross macroinfarcts in about one-third of the brain specimens. For clinical phenotypes, diagnosis of dementia, including Alzheimer’s dementia, was identified in nearly half of participants, as of death. Finally, one-quarter of participants had an APOE e4 allele (e4e4 or e3e4).
Table 1.
Characteristics of Participants1
| Mean (SD) or % (n) | |
|---|---|
| DEMOGRAPHIC FACTORS | |
| Male | 36% (260) |
| Mean age at death | 88.0 (6.7) |
| EPIGENETIC CLOCK AGE IN DORSOLATERAL PREFRONTAL CORTEX | |
| Mean Hannum clock age | 57.0 (3.2) |
| Mean Horvath clock age | 79.7 (6.3) |
| Mean PhenoAge clock age | 1.6 (5.8) |
| Mean GrimAge clock age | 82.6 (4.7) |
| Mean Cortical clock age | 87.3 (5.6) |
| ALZHEIMER’S DISEASE PATHOLOGY | |
| Mean global Alzheimer’s Disease pathology | 0.7 (0.4) |
| Mean amyloid | 3.1 (3.5) |
| Mean tau tangle density | 3.7 (6.6) |
| Pathologic Alzheimer’s Disease diagnosis | 60% (433) |
| NEURODEGENERATIVE PATHOLOGY | |
| LATE-NC2 | 30% (200) |
| Presence of Lewy bodies | 10% (72) |
| CEREBRAL VESSEL/CEREBROVASCULAR DISEASE PATHOLOGY | |
| Cerebral atherosclerosis | 46% (332) moderate or severe |
| Arteriolosclerosis | 41% (294) moderate or severe |
| Cerebral amyloid angiopathy | 35% (245) moderate or severe |
| Chronic microinfarcts, at least 1 | 26% (189) |
| Gross macroinfarcts , at least 1 | 35% (256) |
| COGNITIVE FUNCTION | |
| Mean baseline global cognition | −0.21 (0.69) |
| Mean baseline episodic memory | −0.23 (0.86) |
| Mean baseline semantic memory | −0.24 (0.91) |
| Mean baseline perceptual speed | −0.27 (0.91) |
| Mean baseline working memory | −0.12 (0.79) |
| Mean baseline visuospatial ability | −0.11 (0.85) |
| Any dementia diagnosis as of death | 44% (320) |
| Alzheimer’s dementia diagnosis as of death | 43% (301) |
| OTHER AGING/NEUROLOGIC-RELATED FACTORS | |
| Mean baseline Parkinsonian signs | 11.1 (8.5) |
| Mean baseline motor function | 0.9 (0.2) |
| ApolipoproteinE e4 genotype (e3e4, e4e4) | 25% (171) |
Sample size n=721, except n=674 for LATE-NC; n=716 for cerebral atherosclerosis; n=700 for cerebral amyloid angiopathy; n=714 for arteriolosclerosis; n=702 for clinical dementia; n=680 for global cognition, episodic memory, semantic memory, working memory; n=661 for perceptual speed, visuospatial ability; n=677 for Parkinsonian signs; n=635 for motor function; n=697 for apoliproteinE genotype. For the Cortical clock, sample size n=633 after excluding 88 specimens which had been part of the original training set for the Cortical clock.
LATE-NC, limbic-predominant age-related TDP-43 encephalopathy – neurological change
Epigenetic Clocks and Neuropathology
We first examined the relations of epigenetic clock ages to AD pathology (Figure 1). We found that each SD increase in Hannum, Horvath, and PhenoAge clock age was related to approximately 30% greater likelihood of pathologic AD, which was significant across these three clocks, although there was no relation with GrimAge. Findings were stronger for the Cortical clock, where each SD increase in clock age was related to 90% greater likelihood of pathologic AD (OR=1.91, 95% CI 1.38, 2.62). Similarly, for levels of global AD pathology as a continuous outcome (Figure 2), we found significant associations with all clocks except the GrimAge clock, such that each standard deviation increase in clock age was associated with greater mean levels of global AD pathology (all p<0.05 for Hannum, Horvath, PhenoAge, Cortical). Among the three blood- and multi-tissue based clocks, strongest relations were observed for the PhenoAge clock, where each SD increase in age was associated with an increment of 0.048 units of global AD pathology (p=0.005), after controlling for chronologic age at death, sex, educational attainment, and depressive symptoms at baseline. Nonetheless, there was a substantially greater association of Cortical age with global AD pathology; each SD increment in Cortical age was related to an increment of 0.12 units of global AD pathology (p<0.00001). Further, when considering molecularly specific Aβ load and tau tangle density (Figure 2), relations of Hannum, Horvath and PhenoAge clocks to pathology were primarily observed for Aβ; results were significant or borderline significant for the Hannum, Horvath and PhenoAge clocks (adjusted mean difference in Aβ for each SD increase in clock age=0.130, p=0.01 for Hannum; mean difference-0.10, p=0.08 for Horvath; ; and mean difference=0.119, p=0.009 for PhenoAge). Again, findings were most pronounced for the Cortical clock (mean difference=0.280, p=0.0003). In addition, Cortical age was strongly related to tau tangle density (mean difference for each SD increase in clock age=0.26, p=0.003), while among the other clocks, we found only a borderline significant association of older Hannum clock age to greater mean tangle density (adjusted mean difference for each SD increase in clock age=0.100, p=0.06).
Figure 1.
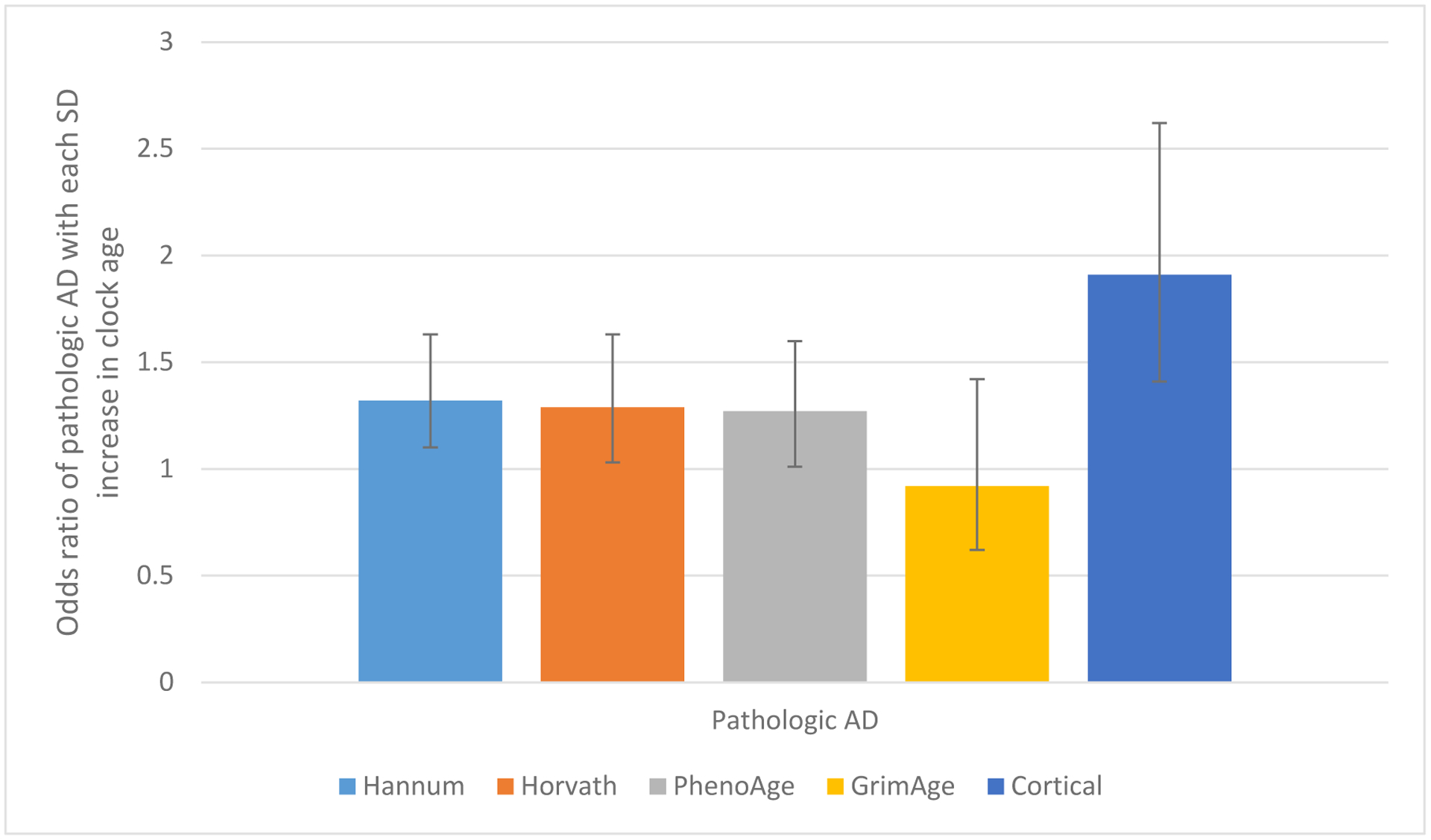
Relations of Epigenetic Clocks in Dorsolateral Prefrontal Cortex to Diagnosis of Pathologic Alzheimer’s Disease1
1 Odds ratios adjusted for age at death, sex, education, depressive symptoms at baseline. N=721 for Hannum, Horvath, PhenoAge, and GrimAge clocks; for Cortical clock, we excluded 88 specimens which had been in the original training set for this clock. Error bars represent 95% confidence intervals.
Figure 2.
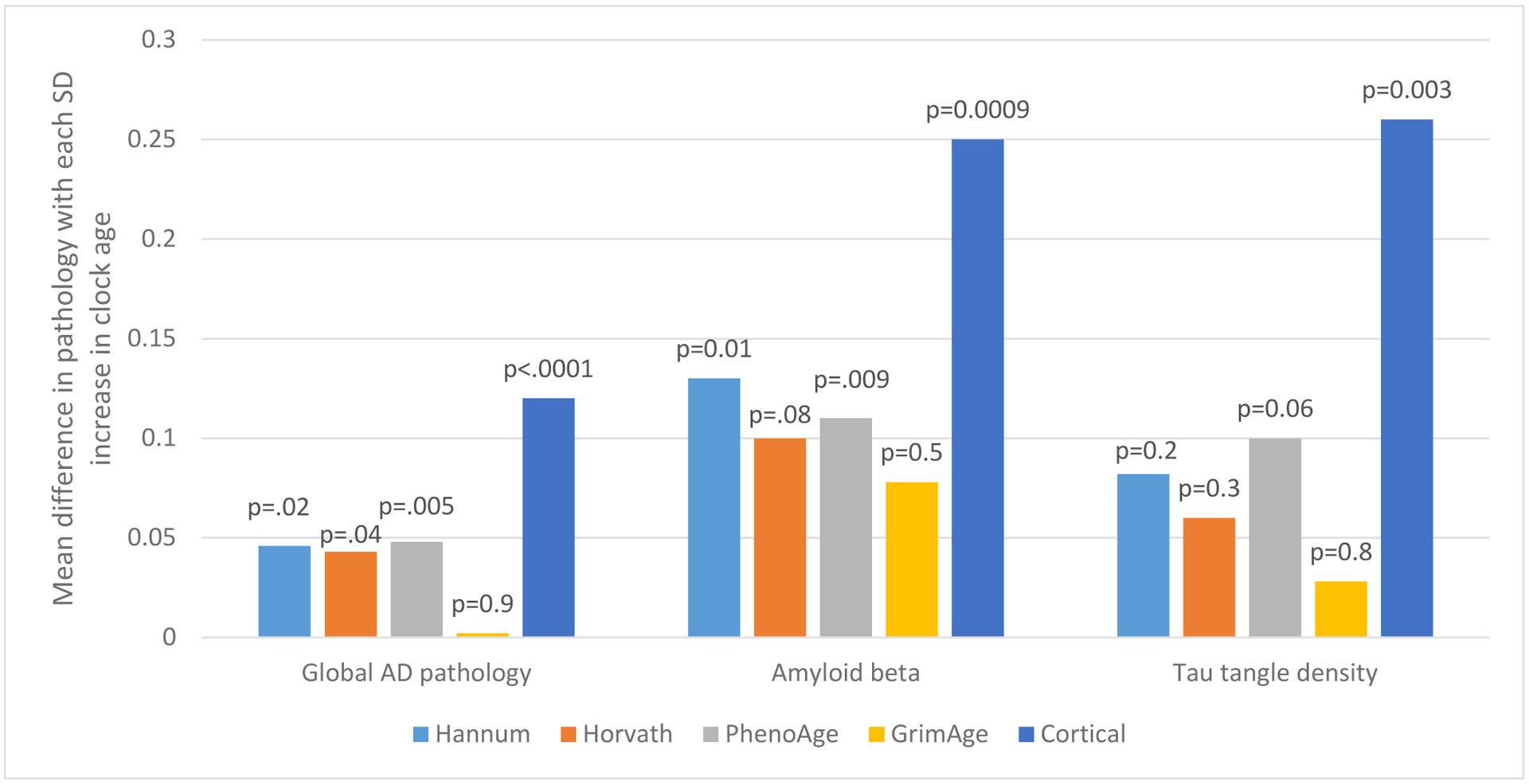
Relations of Epigenetic Clocks in Dorsolateral Prefrontal Cortex to Global Alzheimer’s Disease Pathologies, and Amyloid and Tau Tangle Density1
1 Mean differences adjusted for age at death, sex, education, depressive symptoms at baseline. N=721 for Hannum, Horvath, PhenoAge, GrimAge clocks; for Cortical clock, we excluded 88 specimens which had been in the original training set for this clock.
We next investigated the relation of epigenetic clock ages to other neurodegenerative disease pathologies and to cerebrovascular disease pathologies (Figure 3), to evaluate the breadth of age-related pathologies which may be associated with epigenetic age. Across the Hannum, Horvath, PhenoAge and GrimAge clocks, we observed no association of clock ages to LATE-NC or to neocortical Lewy body pathology, with odds ratios mostly ranging from approximately 1.0 to 1.2. However, we found a highly significant, 2-fold greater odds of Lewy body pathology with each SD increment in Cortical age (OR=2.00, 95% CI 1.27, 3.17).
Figure 3.
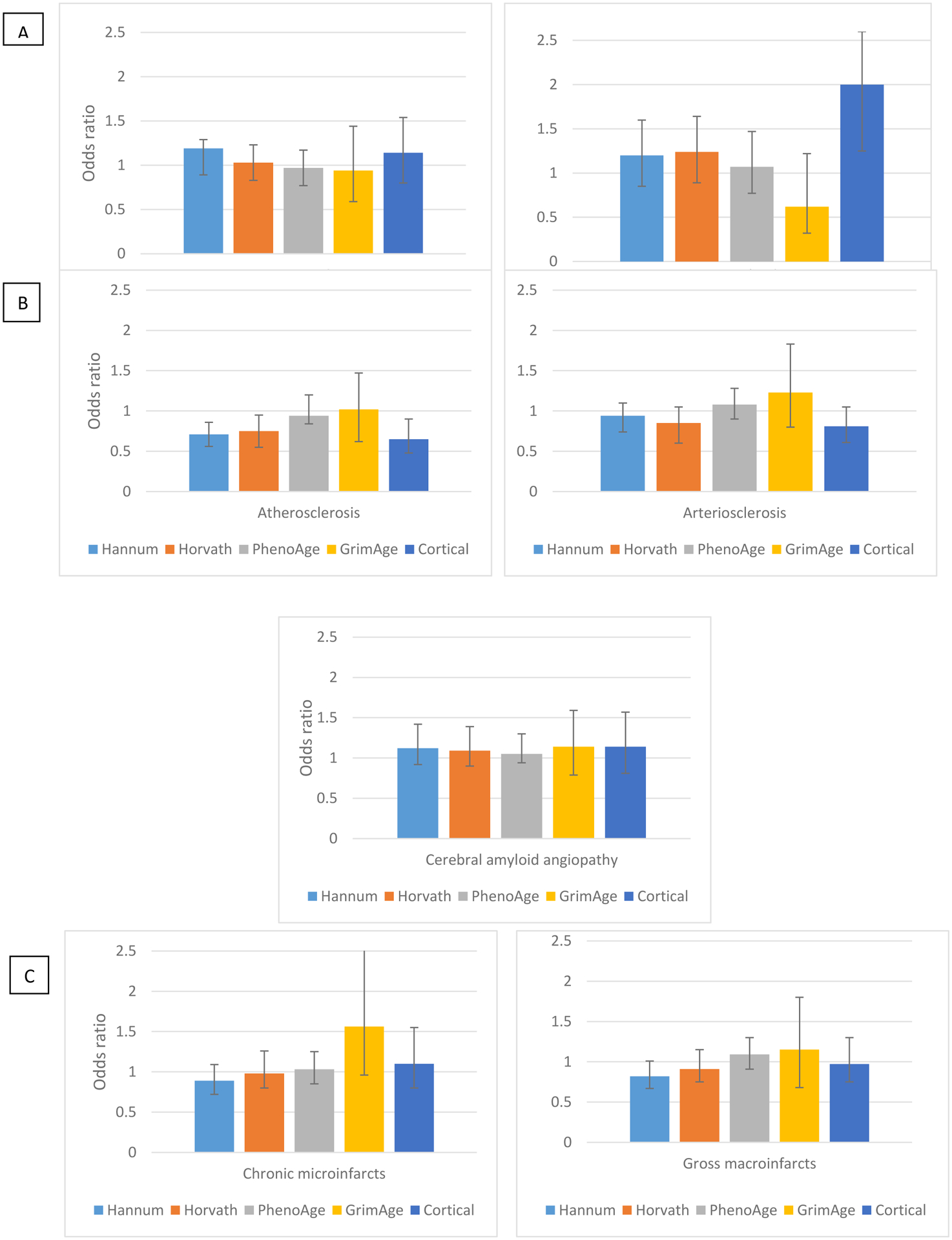
Odds Ratios of Neurodegenerative (Panel A), Cerebral Vessel (Panel B), and Cerebrovascular Pathology (Panel C) per Standard Deviation Increment in Epigenetic Clock Age in the Dorsolateral Prefrontal Cortex1
1Odds ratios adjusted for age at death, sex, education, depressive symptoms at baseline. LATE-NC, limbic-predominant age-related TDP-43 encephalopathy – neurologic change. N=721, except n=674 for LATE-NC, n=716 for cerebral atherosclerosis, n=714 for arteriolosclerosis, n=700 for cerebral amyloid angiopathy. For Cortical clock we further excluded 88 specimens which had been in the original training set for this clock.
For cerebrovascular disease pathologies (Figure 3), we first considered measures of cerebral vessel disease. We did not find that older clock age was related to higher levels of atherosclerosis, arteriolosclerosis, or CAA. Across the five clocks and the three measures of vessel diseases, most ORs were either less than 1.0, or near the null value of 1.0 (Figure 3). Unexpectedly, we found that for cerebral atherosclerosis, older Hannum, Horvath, and Cortical clock ages were related to less pathology, with statistically significant ORs of approximately 0.7. We found no relations of epigenetic clock ages to presence of infarcts; odds ratios of chronic microinfarcts with each SD increase in clock age were 0.89, 0.98, 1.03, 1.56, and 1.10, for the Hannum, Horvath, PhenoAge, and Cortical clocks, respectively, and none were significant. Similarly, odds ratios for the clocks ranged from 0.82–1.15 for gross macroinfarcts; none of these ORs were significant.
Secondary Analyses: Epigenetic Clocks and Clinical Aging Phenotypes
We also considered clinical aging-related phenotypes (Table 2): dementia, cognitive decline, Parkinsonian signs, motor decline, and APOE genotype. Specifically, for the Hannum, Horvath, and PhenoAge clocks, we found that older epigenetic ages were not related to greater odds of clinical Alzheimer’s dementia, or to worse rates of global cognitive decline over time, or to decline in any of five specific cognitive systems; we found unexpected inverse relations for older GrimAge with lower odds of dementia and significantly better rates of cognitive decline over time for global cognition, semantic memory and working memory. Across the four established blood-based and multi-tissue clocks, we observed no associations of epigenetic clock ages in DLPFC to trajectories of Parkinsonian signs, global motor decline or, or to APOE genotype.
Table 2.
Relation of Epigenetic Clocks in Dorsolateral Prefrontal Cortex to Clinical Phenotypes
| EPIGENETIC CLOCKS | |||||
|---|---|---|---|---|---|
| Outcomes 1 | Hannum | Horvath | PhenoAge | GrimAge | Cortical |
| CLINICAL DIAGNOSES | Odds Ratio 2 per SD increase in clock age OR (95% CI) | ||||
| Alzheimer’s Dementia | 1.14 (0.93,1.41) | 1.03 (0.83,1.29) | 0.89 (0.74,1.07) | 0.62 (0.39,0.98) | 1.54 (1.14,2.08) |
| Any Dementia | 1.13 (0.92,1.38) | 1.00 (0.81,1.24) | 0.93 (0.78,1.12) | 0.61 (0.39, 0.96) | 1.50 (1.12,2.02) |
| COGNITIVE DECLINE |
Mean difference in annual rate of decline per SD increase in clock age2 β (p-value) |
||||
| Global cognition | −0.001 (0.9) | 0.007 (0.3) | 0.004 (0.5) | 0.036 (0.01) | −0.03 (0.003) |
| Episodic memory | −0.004 (0.6) | 0.010 (0.2) | 0.005 (0.4) | 0.031 (0.06) | −0.03 (0.02) |
| Semantic memory | −0.004 (0.6) | 0.008 (0.4) | −0.002 (0.8) | 0.041 (0.04) | −0.03 (0.04) |
| Perceptual speed | −0.002 (0.7) | 0.007 (0.3) | 0.002 (0.7) | 0.027 (0.1) | −0.03 (0.04) |
| Working memory | 0.002 (0.7) | 0.004 (0.5) | 0.002 (0.7) | 0.041 (0.01) | −0.03 (0.004) |
| Visuospatial ability | −0.004 (0.5) | −0.002 (0.7) | −0.002 (0.8) | 0.026 (0.07) | −0.03 (0.003) |
| OTHER NEUROLOGIC PHENOTYPES | |||||
| Parkinsonian signs | 0.003 (0.7) | −0.004 (0.7) | −0.003 (0.7) | −0.017 (0.4) | 0.03 (0.05) |
| Motor function | 0.001 (0.4) | 0.002 (0.4) | 0.001 (0.6) | 0.003 (0.5) | −0.001 (0.7) |
| GENETIC FACTORS |
Odds Ratio
2
per SD increase in clock age
OR (95% CI) |
||||
| ApoE e4 genotype (e3e4/e4e4) | 1.13 (0.90, 1.42) | 1.12 (0.88, 1.43) | 1.11 (0.90, 1.36) | 1.02 (0.62,1.70) | 1.49 (1.06,2.09) |
For Horvath, Hannum, PhenoAge, GrimAge clocks n=680 for global cognition, episodic memory, semantic memory, working memory; n=661 for perceptual speed and visuospatial ability; n=677 for Parkinsonian signs; n=635 for motor function; n=702 for Alzheimer’s dementia and 721 for dementia; n=697 for apoE. For Cortical clock we further excluded 88 participants who had been part of the original training set for the Cortical clock.
Odds ratios (OR) and mean differences adjusted for age at death, sex, education, and baseline levels of depressive symptoms
However, we found clear associations of Cortical age with virtually every clinical phenotype we considered, after controlling for chronologic age as well as sex, educational attainment, and depressive symptoms (Table 2). For example, each standard deviation increment in Cortical age was related to over 50% greater odds of Alzheimer dementia (OR=1.54, 95% CI 1.14, 2.08), and Cortical age was also significantly associated with worse trajectories of decline on our global composite score, as well as on all five specific cognitive systems. Moreover, beyond cognition, we observed a borderline significant association of older Cortical age with worse increases in Parkinsonian signs (p=0.05). Finally, there was a 50% greater likelihood of APOE e4 genotype with older Cortical age (OR=1.49, 95% CI 1.06, 2.09).
Sensitivity Analyses
We examined associations of epigenetic age acceleration to each phenotype; age acceleration represents the extent to which each individual’s epigenetic age is higher (or lower) than expected based on chronologic age. All findings were virtually identical to those we found here for epigenetic age (data not shown in Tables). Additionally, in analyses controlling for neuron proportion, using the two different approaches to estimating neuron proportion, all results remained generally similar; that is, control for neuron proportion did not meaningfully change any findings (data not shown in Tables).
DISCUSSION
In this study of postmortem DLPFC from over 700 older women and men, we found that only an epigenetic clock trained in brain tissue14 had clear and compelling relations with neurodegenerative pathologies as well as clinical neurologic phenotypes, from dementia to cognitive decline to Parkinsonian symptoms. Further, older Cortical age was associated with APOE e4 genotype, consistent with a large body of research identifying APOE as a robust genetic marker not only of dementia30 but also of longevity.31 In contrast, several blood-based and multi-tissue epigenetic clocks that we examined in brain specimens demonstrated limited ability to identify neuropathologic profiles or clinical neurologic diseases/conditions – although we specifically selected four different clock markers to span both traditional clocks designed to predict chronologic age and second generation clocks designed to predict biologic/health profiles of aging. Our findings provide evidence of the critical necessity for bespoke clocks32 of brain aging for advancing research to understand, and eventually prevent, neurodegenerative diseases of aging.
Despite the importance of finding molecular markers which can represent biologic aging in the brain, there is limited existing literature on DNAm clocks in brain tissue. In our own previous research examining basic characteristics of several epigenetic clocks in the ROSMAP DLPFC, we reported reasonable correlations of clock age with chronologic age at death;12 Pearson correlations were 0.64 for the Hannum clock, 0.69 for the Horvath clock, and 0.51 for the PhenoAge clock, with a higher correlation of 0.83 for the Cortical clock. We observed similar correlations in a smaller set of posterior cingulate cortex specimens, and comparable findings across several brain regions have been reported by others.11 While this would appear supportive of the value of epigenetic clocks in brain tissue, including those clocks developed in blood or other tissues, nonetheless, the Hannum, Horvath, and PhenoAge clocks primarily predicted amyloid burden, and few other phenotypes here, while older GrimAge was largely unrelated to the variety of neurologic phenotypes we examined (GrimAge is the only clock which includes CpG sites for cigarette smoking, which has modest relation with dementia33 and is inversely related to Parkinson Disease,34 perhaps explaining its poor performance here). These findings are in contrast to previous research in systemic diseases, in which these four clocks were calculated in blood samples, and uniformly predicted risks of cancer, cardiovascular diseases, and mortality.35 Our results together with this previous research underscores the importance of now tailoring clocks to specific disease processes by developing clocks in specific tissues (ie, brain tissue for research in neurodegeneration). Interestingly, we had previously found generally low correlations between the epigenetic clock ages (or clock age acceleration) calculated in 41 paired DLPFC and blood specimens,12 which could indicate that the blood-based and multi-tissue clocks may not represent congruous concepts in their tissue of origin as in brain tissue specifically. That may be one possible explanation for their larger inability to broadly identify neuropathology or neurologic outcomes. Indeed, this may also emphasize the potential value of future clocks which target CpG sites that are conserved across brain and other tissues, potentially yielding clocks which better represent aging processes in the brain and are still applicable across more accessible tissues (eg, blood).
There are substantial strengths of this study. We had an extremely well-characterized population, in terms of both neuropathologic and clinical indices, providing the ability to examine epigenetic clock ages in relation to a range of age-related neuropathologic and neurologic phenotypes. Further, we had very high follow-up and autopsy rates, reducing bias due to missing data.
There are also limitations to consider. First, cerebrovascular pathologies were less prevalent than neurodegenerative pathologies, thus we had better ability to identify relations of the epigenetic clocks to neurodegenerative (especially AD pathology, which utilized continuous measurement scales) than to cerebrovascular pathology. This could explain our mostly null findings for the associations of epigenetic clock age to several measures of cerebrovascular neuropathology. While it remains possible that the epigenetic clocks are simply not related to cerebrovascular pathology, this is inconsistent with evidence that DNAm (in peripheral blood) appears to predict development of vascular diseases.36 Second, cellular composition of cortical tissue changes with aging and disease,16 and DNAm states may also differ across cell types; thus the cell type proportion may confound relations of epigenetic clock age and the phenotypes we examined. We used several different approaches to estimating neuron proportion in each specimen, and found that none of these covariates meaningfully changed our results when added to statistical models. However, we cannot rule out the possibility that more accurate measures of neuron proportion may have produced different findings. In addition, our clocks were generated from one cortical region and many of the pathologies are widespread or elsewhere in the brain. It is possible that other brain regions would yield different results. However, in a smaller study (n=180), we previously reported reasonable correlations of epigenetic clock ages measured in DLPFC and in posterior cingulate cortex (r from 0.4 to 0.9 across the four clocks). Similarly, Lu et al.11 also found good correlations of the Horvath clock across three different cortical brain regions. Thus, there is no clear basis for believing that findings would be substantially different in other cortical regions, although this certainly deserves additional research. Further, our research considered associations of epigenetic clock ages to neurologic phenotypes; findings indicate specific associations but cannot establish epigenetic modifications as causal in the process of brain aging. Finally, our study population included exclusively older persons. Most of these epigenetic clocks were designed in participants with a wide range of ages, and limited representation of the oldest ages.2–5 Interestingly, the Cortical clock is not only the first clock trained in brain specimens, but also included substantially larger samples of older age groups in the training set than previous clocks. There is evidence that the Horvath, Hannum, and PhenoAge clocks may function worse in the oldest age groups;14,37 thus it is possible that the Cortical clock performed better in our population due at least in part to the older age of our participants, and that the other clocks could be somewhat better markers of brain aging in populations with younger age ranges. Still, the most common neurodegenerative diseases occur in older persons, thus effective biomarkers of brain aging are crucial within older demographic groups (even the Cortical clock was developed to predict chronologic ages from 1 through 108 years, and a clock trained exclusively in older persons might produce even better performance for the oldest ages). Overall, our findings in these ROSMAP DLPFC data strongly support the value of epigenetic clocks designed specifically in brain tissue in older women and men. Such work may provide important new biomarkers which can broadly subserve research to prevent or delay age-related neurodegeneration and neurodegenerative diseases.
Highlights.
We calculated five epigenetic clocks in 721 postmortem cortical specimens
Epigenetic clocks trained in blood samples did not predict most neurologic phenotypes
One epigenetic clock, trained in cortex, identified neurodegenerative phenotypes
Findings provide convincing evidence for clocks developed in tissues of interest
Acknowledgements
We thank the participants in ROSMAP and the staff of the Rush Alzheimer’s Disease Center. ROSMAP data can be requested at https://www.radc.rush.edu.
Funding:
This work was supported by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG36042, and U01AG61356. The funding sources had no role in the conduct of the research or preparation of the article.
Abbreviations:
- AD
Alzheimer’s disease
- Aβ
amyloid beta
- APOE
apolipoprotein E
- CES-D
Center for Epidemiologic Studies Depression
- CAA
cerebral amyloid angiopathy
- CI
confidence interval
- DNAm
DNA methylation
- DLPFC
dorsolateral pre-frontal cortex
- LATE-NC
limbic-predominant age-related TDP-43 encephalopathy neuropathological change
- OR
odds ratio
- ROS
Religious Orders Study
- MAP
Rush Memory and Aging Project
- TDP-43
transactive response DNA binding protein 43
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests:
The authors have no competing interests to declare.
REFERENCES
- 1.Jylhava J, Pedersen NL, Hagg S. Biological Age Predictors. EBioMedicine 2017;21, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of aging rates. Molecular Cell 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine ME, Lu AT, Quach A. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10: 573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019January21;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kane AE, Sinclair DA. Epigenetic changes during aging and their reprogramming potential. Crit Rev Biochem Mol Biol 2019;54: 61–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belsky DW, Caspi A, Arseneault L, et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife 2020May5;9:e54870. doi: 10.7554/eLife.54870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics 2019;11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marras C, Beck JC, Bower JH, et al. Prevalence of Parkinson’s disease across North America. NPJ Parkinsons Dis 2018;4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alzheimer’s Association. 2019 Alzheimer’s Disease Facts and Figures. Alzheimer’s Dement 2019; 15: 321–387 [DOI] [PubMed] [Google Scholar]
- 11.Lu AT, Hannon E, Levine ME, et al. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun 2017;8: 15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grodstein F, Lemos B, Yu L, Iatrou A, De Jager PL, Bennett DA. Characteristics of Epigenetic Clocks Across Blood and Brain Tissue in Older Women and Men. Front Neurosci. 2021January7;14:555307. doi: 10.3389/fnins.2020.555307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY) 2015;7: 1198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shireby GL, Davies JP, Francis PT, et al. Recalibrating the epigenetic clock: implications for assessing biological age in the human cortex, Brain 2020; awaa334, 10.1093/brain/awaa334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer’s Disease 2018; 64(s1):S161–S189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jager PL, Srivastava G, Lunnon K, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17(9):1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013May1;14:293. doi: 10.1186/1471-2164-14-293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson RS, Arnold SE, Schneider JA, Tang Y, Bennett DA. The relationship between cerebral Alzheimer’s disease pathology and odour identification in old age. J Neurol Neurosurg Psychiatry. 2007;78(1):30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Institute on Aging and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. Neurobiol Aging 1997;18:S1–2. [PubMed] [Google Scholar]
- 20.McKeith IG, Galasko D, Kosaka K, et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology 1996;47: 1113–1124. [DOI] [PubMed] [Google Scholar]
- 21.Wilson RS, Yu L, Schneider JA, et al. Lewy bodies and olfactory dysfunction in old age. Chem Senses 2011;36:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson RS, Beckett LA, Barnes LL, et al. Individual differences in rates of change in cognitive abilities of older persons. Psychol Aging. 2002;17(2):179–193. [PubMed] [Google Scholar]
- 23.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984;34: 939–944. [DOI] [PubMed] [Google Scholar]
- 24.Jack CR Jr, Bennett DA, Blennow K, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018April;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett DA, Shannon KM, Beckett LA, Goetz CG, Wilson RS. Metric properties of nurses’ ratings of parkinsonian signs with a modified Unified Parkinson’s Disease Rating Scale. Neurology 1997;49, 1580–1587. [DOI] [PubMed] [Google Scholar]
- 26.Buchman AS, Wilson RS, Boyle PA, Bienias JL, Bennett DA. Change in motor function and risk of mortality in older persons. J Am Geriatr Soc. 2007;55(1):11–19. [DOI] [PubMed] [Google Scholar]
- 27.Bennett DA, Schneider JA, Wilson RS, Bienias JL, Berry-Kravis E, Arnold SE. Amyloid mediates the association of apolipoprotein E e4 allele to cognitive function in older people. J Neurol Neurosurg Psychiatry. 2005;76(9):1194–1199. doi: 10.1136/jnnp.2004.054445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8(3):290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein HU, McCabe C, Gjoneska E, et al. Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat Neurosci. 2019January;22(1):37–46. doi: 10.1038/s41593-018-0291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013;9: 106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastiani P, Gurinovich A, Nygaard M, et al. APOE Alleles and Extreme Human Longevity, The Journals of Gerontology: Series A, 2019;74: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell CG, Lowe R, Adams PD, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol 2019;20, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livingston G, Huntley J, Sommerlad A, et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet. 2020August8;396(10248):413–446. doi: 10.1016/S0140-6736(20)30367-6.Epub 2020 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breckenridge CB, Berry C, Chang ET, Sielken RL Jr, Mandel JS. Association between Parkinson’s Disease and Cigarette Smoking, Rural Living, Well-Water Consumption, Farming and Pesticide Use: Systematic Review and Meta-Analysis. PLoS One. 2016April7;11(4):e0151841. doi: 10.1371/journal.pone.0151841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oblak L, van der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021April28;69:101348. doi: 10.1016/j.arr.2021.101348.Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Westerman K, Sebastiani P, Jacques P, et al. DNA methylation modules associate with incident cardiovascular disease and cumulative risk factor exposure. Clin Epigenet 2019;11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Khoury LY, Gorrie-Stone T, Smart M, et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol 2019; 20:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ROSMAP data can be requested at https://www.radc.rush.edu.


