Abstract
Purpose:
The potential to compute volume metrics of emphysema from planar scout images was investigated in this study. The successful implementation of this concept will have a wide impact in different fields, and specifically, maximize the diagnostic potential of the planar medical images.
Methods:
We investigate our premise using a well-characterized chronic obstructive pulmonary disease (COPD) cohort. In this cohort, planar scout images from computed tomography (CT) scans were used to compute lung volume and percentage of emphysema. Lung volume and percentage of emphysema were quantified on the volumetric CT images and used as the “ground truth” for developing the models to compute the variables from the corresponding scout images. We trained two classical convolutional neural networks (CNNs), including VGG19 and InceptionV3, to compute lung volume and the percentage of emphysema from the scout images. The scout images (n=1,446) were split into three subgroups: (1) training (n=1,235), (2) internal validation (n=99), and (3) independent test (n=112) at the subject level in a ratio of 8:1:1. The mean absolute difference (MAD) and R-square (R2) were the performance metrics to evaluate the prediction performance of the developed models.
Results:
The lung volumes and percentages of emphysema computed from a single planar scout image were significantly linear correlated with the measures quantified using volumetric CT images (VGG-19: R2= 0.934 for lung volume and R2 = 0.751 for emphysema percentage, and InceptionV3: R2 = 0.977 for lung volume and R2 = 0.775 for emphysema percentage). The mean absolute differences (MADs) for lung volume and percentage of emphysema were 0.302 ± 0.247L and 2.89 ± 2.58%, respectively, for VGG-19, and 0.366 ±0.287L and 3.19±2.14, respectively, for InceptionV3.
Conclusions:
Our promising results demonstrated the feasibility of inferring volume metrics from planar images using CNNs.
Keywords: quantitative analysis, planar images, emphysema, artificial intelligence
I. INTRODUCTION
Medical imaging is one of the most widely used clinical modalities to screen, diagnose, and manage patients. In essence, there are two types of images, including two-dimensional (2-D) (or planar imaging) and three-dimensional (3-D) (or volumetric images). Planar images (e.g., chest x-ray (CXR), mammogram) usually are less burdensome to the patient, less time-intensive to interpret, and more widely available when compared to volumetric imaging (e.g., a computed tomography (CT) scan, magnetic resonance imaging (MRI)). Specifically, a patient’s exposure to ionizing radiation is orders of magnitude less during CXR compared to a CT scan. The primary issue with planar images is the superposition of tissues that limits the ability to unambiguously isolate and visualize individual anatomical structures. This inherence characteristic makes it challenging to reliably detect small abnormalities and quantify the volumetric characteristics of normal or abnormal anatomy depicted on planar images.
When a radiologist or clinician orders a radiographic examination, the balance between the type of radiographic examination and the benefit or detriment to a patient is often considered. Is a specific type of planar imaging (e.g., chest x-ray (CXR) or mammogram) sufficient to reach a diagnosis, or is a volumetric examination (e.g., a computed tomography (CT) scan or magnetic resonance imaging (MRI)) required? The choice for a specific imaging modality often evolves over time through clinical experience and scientific advancement. For example, whether a CXR or a CT scan should be used to screen individuals at risk of lung cancer has been addressed by the National Lung Screening Trial (NLST) [1–3]. NLST reported that the benefit of low-dose CT lung cancer screening (i.e., the reduction of lung cancer mortality by 20%) outweighed the potential deleterious effects associated with increased exposure to ionizing radiation when compared to screening individuals using CXR. Another example is the use of mammography and MRI for breast cancer screening among women. Although MRI can enable a sensitive and accurate diagnosis of breast cancer, its cost and burden to the patient outweigh its benefit when compared to mammography. Hence, two-view planar mammography remains the initial and ubiquitous examination for breast cancer screening, while MRI is often ordered to manage women who are at higher than average risk for developing breast cancer and/or women with heterogeneous to extremely dense breasts [4]. In both examples, significant improvement in the diagnostic performance of planar images (CXR or mammograms) could potentially obviate the need for a volumetric radiographic examination.
A significant amount of research has been dedicated to developing computer tools to facilitate the detection and diagnosis of a variety of lung diseases depicted on CXR images in an effort to improve radiologists’ performances and ease the workload. Recently the use of deep learning technology, namely convolutional neural network (CNN), has demonstrated remarkable performance in medical image analysis [5–12]. In research involving CXR, most investigations leveraged publicly available CXR datasets that only have bimodal disease assessment as positive or negative [13–17]. The disease type and location were typically manually labeled by a clinician to establish “ground truth,” which is a time- and cost-consuming endeavor. The accuracy and consistency of these datasets can be significantly compromised by reader variability.
In this study, we attempted to explore whether the planar images can be used to quantitatively assess the volume metrics of the disease or other important regions of interest by leveraging the state of the art of Artificial Intelligence (AI). Chronic obstructive pulmonary disease (COPD) was used as an example. COPD is a leading cause of disability and death in the United States and worldwide [18, 19], which is typically caused by chronic bronchitis and/or emphysema. Emphysema typically involves the physical damage of lung parenchyma (alveoli), which can be visualized and accurately quantified using chest CT scans. Many investigations have been performed to quantify the percentage of emphysema and its morphological characteristics using chest CT examinations [20–24]. However, chest CT scans are not typically used in clinical practice to confirm a clinician’s suspicion for emphysema, to assess the percentage of emphysema, or to observe the progress of emphysema until late in the disease process. A clinician often relies on a CXR to assist in diagnosing emphysema despite the poor sensitivity of a CXR to detect early disease and the inability to quantify the presence of emphysema. In practice, a CXR is the most common radiographic exam and used to diagnose the presence of many other lung diseases [25], such as pneumonia and interstitial lung disease (ILD), because of its low level of radiation exposure, straightforward implementation, low cost, and portability. However, the sensitivity of a CXR to detect and quantify the presence of emphysema is suboptimal based on the limitation described above.
We believe that AI software has the potential to increase the information that can be extracted from planar images beyond a subjective assessment or past research effort that focused on only the planar images. Our novel approach is to use volumetric image datasets to train software to extract similar 3-D information from planar images. Chest CT scans and CXR were used to evaluate this novel concept. Specifically, AI software was developed to compute lung volume and percentage of emphysema from planar scout images, which are part of a routine chest CT scan. If successful, this research could not only significantly increase the utility of a CXR for the clinical management of COPD patients but would also provide the “proof of concept” for developing AI software for 2-D images by using 3-D images as the gold standard.
II. MATERIALS AND METHODS
A. Scheme overview
Chest CT scans from the participants in a COPD cohort were used to compute lung volume and the percentage of emphysema, which serve as the “ground truth” for machine learning and validation. Emphysema was quantified as the percentage of lung voxels with a Hounsfield (HU) value less than <−950 HU. The scout images acquired as part of the CT scans were used as an alternative to planar CXR images. Two classical convolutional neural networks (CNNs) were used to compute the lung volume and the percentage of emphysema present in the lungs. The performance of the CNN models was evaluated by the agreement between the values computed using the scout images and the “ground truth” from the corresponding CT scans.
B. Study population
The study cohort consisted of 753 participants in an NIH-sponsored Specialized Center for Clinically Oriented Research (SCCOR) diagnosed with COPD at the University of Pittsburgh. The inclusion criteria for enrollment were age > 40 years and current or former smokers with at least a 10 pack-year history of tobacco exposure. The SCCOR participants completed pre- and post-bronchodilator spirometry and plethysmography, measurement of lung diffusion capacity, a chest CT examination, and demographic and medical history questionnaires. All subjects had a baseline CT scan. A subset of the subjects had repeat chest CT exams. Specifically, 385 subjects had a 2-year follow-up CT scan, 313 subjects had a 6-year follow-up CT scan, and 75 subjects had a 10-year follow-up CT scan. The dataset included 495 participants diagnosed with COPD as defined by the Global Initiative for Obstructive Lung Disease (GOLD) [19, 26] and 258 participants without airflow obstruction (Table 1). All study procedures were approved by the University of Pittsburgh Institutional Review Board (#0612016). Written informed consent was obtained.
Table 1:
Subject demographics and dataset distribution
| Our Cohort | SCCOR (n=753) | |
|---|---|---|
| Age, year (SD) | 64.7 (7) | |
| Male, n (%) | 414 (54.7) | |
| Race | ||
| White, n (%) | 708 (94.0) | |
| Black, n (%) | 34 (4.5) | |
| Other, n (%) | 11 (1.5) | |
| FEV1, % predicted (SD) | 82.3 (21.3) | |
| FEV1/FVC, % (SD) | 60.6 (17.9) | |
| Five-category classification | ||
| Sub-groups of the scout images | before data balancing | after data balancing |
| 1235 | 3924 | |
Abbreviations: SD – standard deviation, FEV1 - forced expiratory volume in one second, FVC - functional vital capacity, T – training, V – internal validation, I – independent test. GOLD – Global Initiative for Obstructive Lung Disease [26].
C. Acquisition of thin-section CT examinations and scout images
The chest CT exams were acquired on a 64-detector CT scanner (LightSpeed VCT, GE Healthcare, Waukesha, WI, USA) with subjects holding their breath at end inspiration without the use of radiopaque contrast. Scans were acquired using a helical technique at the following parameters: 32×0.625 mm detector configuration, 0.969 pitch, 120 kVp tube energy, 250 mA tube current, and 0.4 sec gantry rotation (or 100 mAs). Images were reconstructed to encompass the entire lung field in a 512×512 pixel matrix using the GE “bone” kernel at 0.625-mm section thickness and 0.625-mm interval. Pixel dimensions ranged from 0.549 to 0.738 mm, depending on participant body size. Planar scout images were acquired to set the frame of reference for the helical CT scan. In our cohort, the scout images had a consistent matrix of 888×733 and a pixel size of 0.5968×0.5455 mm. Only the CT scans with an available scout image were used in the study. In total, there were 1,446 paired CT scans and scout images that included both baselines and follow-up exams.
D. Compute lung volume and percentage of emphysema depicted on CT images
To compute lung volume and percentage of emphysema in the lung, the lung was automatically segmented, and a threshold of −950 Hounsfield unit (HU) was applied to the segmented lung region to identify voxels associated with emphysema [20, 22]. Emphysema was quantified as the volume of voxels below the emphysema threshold as a percentage of the total lung volume. To reduce the effects caused by image noise or artifact, small clusters of voxels less than 3 mm2 (4 ~ 5 pixels) were removed from the emphysema computation [20, 27].
E. CNN models for computing lung volume and percentage of emphysema on scout images
The unique strength of a CNN is its ability to automatically learn specific features (or feature map) by repeatedly applying the convolutional layers to an image [11, 12, 28–32]. A CNN is typically formed by several building blocks, including convolution layers, activation functions, pooling layers, batch normalization layers, flatten layers, and fully connected (FC) layers. Two classical CNN models called “VGG19” and “InceptionV3”, which have been widely used for 2-D image classification purposes [10, 33–39], were used to perform logistic regression [35, 36]. The objective of this study is to predict the continuous values of lung volume and emphysema percentage from scout images. In architecture, a CNN used for regression is almost the same as a CNN used for classification. The differences are: (1) a CNN-based regression uses a fully connected regression layer with linear or sigmoid activations, while a CNN-based classification network uses the Softmax activation function and (2) a CNN-based regression network typically uses the mean absolute error (MAE) or mean squared error (MSE) as the loss function, while a CNN-based classification network uses binary or categorical cross-entropy as the loss function [40]. In the implementation, other classical models for classification purposes (e.g., DenseNet or ResNet [37, 41]) can also be used for the regression purpose by adjusting activation and loss functions in the last layers [42, 43].
Several procedures were implemented to alleviate the requirement of deep learning for a large dataset: (1) The normalization of the scout image matrix. The objective of this normalization is to (1) limit the machine learning to the lung regions and feed the CNN models with a consistent size of images (e.g., 512×512) that is required by CNN. In the implementation, a deep learning-based algorithm described in [32] was used to segment the lung regions depicted on the scout images. Then, based on the center of the identified lung regions, a square box with a dimension of 400×400 mm2 was used to crop the lung regions. The cropped lung regions were resized to a 512×512 pixel matrix. (2) The normalization of image intensity. Based on the maximum and the minimum values of the scout images, the intensity of the images was scaled into a range of [0, 255]. (3) The application of transfer learning. The pre-trained ImageNet weights for VGG19 and InceptionV3 were used to transfer the learning [44]. Transfer learning can improve learning efficiency and also reduce the requirement for a large and diverse dataset. To use the pre-trained ImageNet weight, we converted the grayscale images into an RGB form by simply replacing the red, green, and blue channels with the grayscale images.
To develop the deep learning-based regression models, the cases were split into three subgroups at the patient level: (1) training, (2) internal validation, and (3) independent test sets in a ratio of 8:1:1 (Table 1). The minority groups were oversampled to reduce the effect of data imbalance, which could significantly lead to bias and incorrect assessment of the prediction model. First, the values were binned based on emphysema percentage by 5% in the training and internal validation sets (Table 1). Second, the cases in the bins were oversampled with a lower number of cases to ensure their equal distribution within each bin relative to the largest bin.
The batch size was set at 8 for training the models. To improve the data diversity and the reliability of the models, the scout images were augmented using geometric and intensity transformations, such as rotation, translation, vertical/horizontal flips, intensity shift [−10, 10], smoothing (blurring) operation, and Gaussian noises. The initial learning rate was set to 0.0001 and would be reduced by a factor of 0.5 if the validation performance did not increase in two epochs. An Adam optimizer was used in the training. The training procedure would stop when the validation performance of the current epoch did not improve compared to the previous fifteen epochs. Based on these parameters, we trained the VGG19 and InceptionV3 models for quantifying the lung volume and the percentage of emphysema from the scout images separately.
F. Performance validation
Two performance metrics were used to assess the performance on the independent testing dataset (scout images not used in training). Standard and adjusted R-squared (R2) were used to assess the linear relation between lung volume and percentage of emphysema computed from the CT scans and scout images. R2, namely the coefficient of determination, is an important and useful concept in statistics indicating the goodness of fit of a model. In regression, R2 measures how well the regression predictions approximate the real data points. If R2 is 1, it suggests that the regression predictions perfectly fit the data. Mean absolute difference (MAD) was computed as the average absolute difference between the two methods. MAD describes the absolution errors between the regression predictions and the data and gives us a very straightforward concept about the performance of the regression model. A p-value of less than 0.05 was considered statistically significant. The performance was stratified based on COPD severity. IBM SPSS v.25 was used for the analyses.
III. RESULTS
By testing the developed CNN model on an independent test set (Table 1), we found that lung volume and percentage of emphysema computed by the model using the planar scout images were significantly, linearly correlated with the values computed from chest CT scans (Table 2 and Fig. 1). For the VGG-19 model, the slope, intercept, R2, and adjusted R2 of the linear regression between the scout image and CT scan for the percentage were 0.870, 2.312%, 0.751, and 0.749, respectively (Fig. 1(a)); and for lung volume of emphysema were 0.934, 0.333L, 0.924, and 0.923, respectively (Fig. 1(b)). The mean absolute differences (MADs) between the two approaches were 0.302 ± 0.247 L for computing lung volume and 2.89 ± 2.58% for computing the percentage of emphysema. For the InceptionV3 model, the slope, intercept, R2, and adjusted R2 of the linear regression between the scout image and CT scan for the percentage were 1.069, 0.310%, 0.775, and 0.773, respectively (Fig. 1(c)); and for lung volume of emphysema were 0.977, 0.406L, 0.927, and 0.926, respectively (Fig. 1(b)). The mean absolute differences (MADs) between the two approaches were 0.366 ± 0.287L for computing lung volume and 3.19 ± 2.14% for computing the percentage of emphysema.
Table 2:
Agreement for computing emphysema and lung volume using scout images versus CT scans
| CNN models | MAD | R-square | Adjusted R-square | Slope | Intercept |
|---|---|---|---|---|---|
| VGG-19 | |||||
| Emphysema (%) | 2.89 ± 2.58 | 0.751* | 0.749* | 0.870 | 2.312 |
| Lung volume (L) | 0.302 ± 0.247 | 0.924* | 0.923* | 0.934 | 0.333 |
| InceptionV3 | |||||
| Emphysema (%) | 3.19 ± 2.14 | 0.775* | 0.773* | 1.069 | 0.310 |
| Lung volume (L) | 0.366 ± 0.287 | 0.927* | 0.926* | 0.977 | 0.406 |
MAD: mean absolute difference;
p≤0.01
Fig. 1.
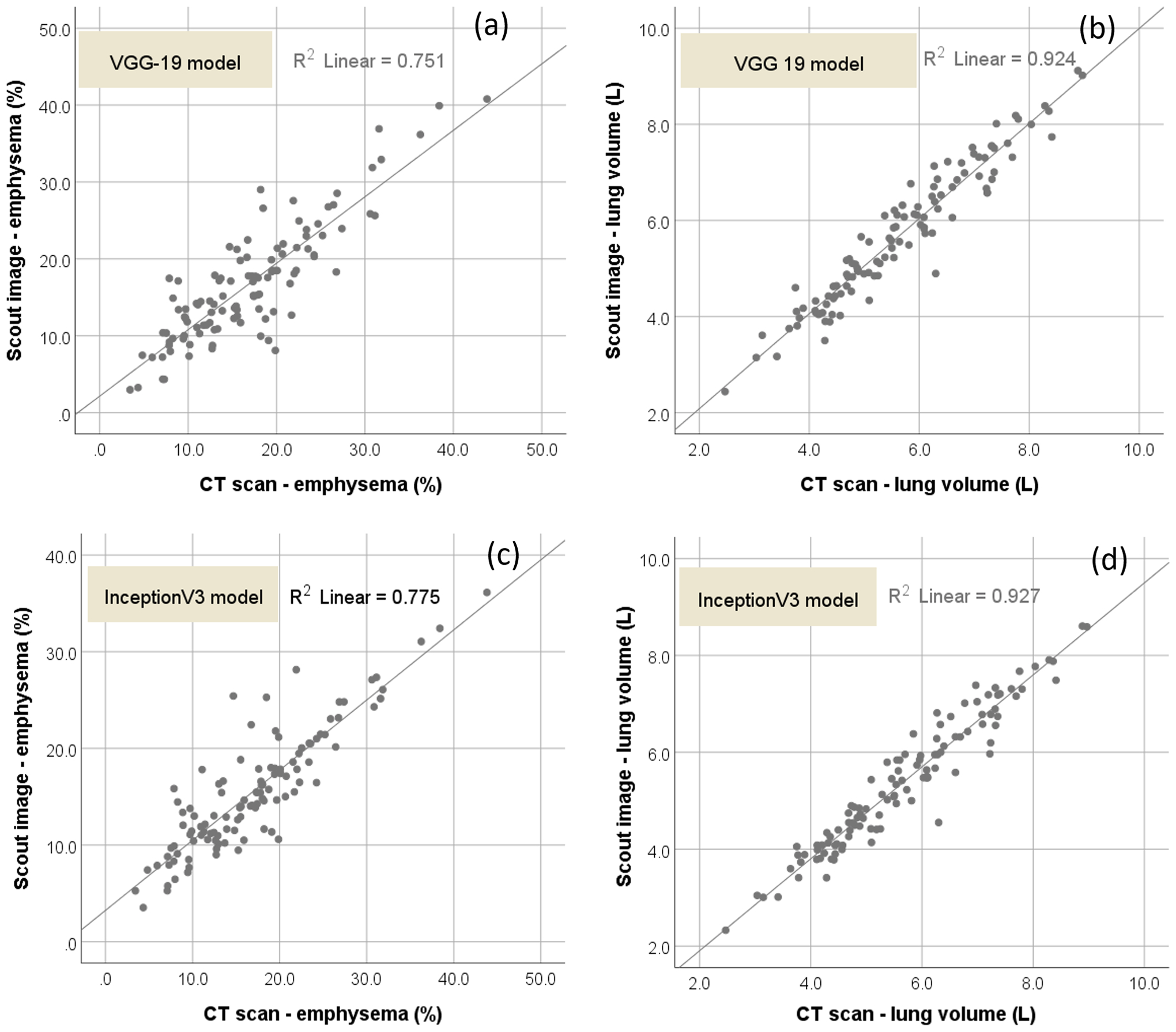
Scatter plots of the lung volume and percentage of emphysema computed from CT scans versus scout image with the linear regression lines. (a) and (b) showed the scatter plots by the VGG-19 model, and (c) and (d) showed the scatter plots by the InceptionV3 model.
On the whole, the VGG-19 model demonstrated a better performance than the IncepitonV3 in inferring emphysema extent (Tables 2–4). For the cases without COPD, the VGG-19 model showed MAD of 2.30 ± 1.80%, R2 of 0.577, and the adjusted R2 of 0.566, while the InceptionV3 model showed MAD of 2.09 ± 1.37%, R2 of 0.710, and the adjusted R2 of 0.703. For the VGG19 model, the strongest agreement was observed for the cases with severe COPD (GOLD IV) with a MAD = 1.39 ± 0.89%, R2 of 0.956, and adjusted R2 of 0.947. For the InceptionV3 model, the strongest agreement was observed for the cases with mild COPD with a MAD = 3.46 ± 2.05%, R2 of 0.489, and adjusted R2 of 0.463. The one-way analyses of variance (ANOVA) showed that the linear trend was statistically significant (p < 0.01). Several examples with different levels of COPD severity were shown in Fig. 2 to demonstrate the performance of the developed algorithm.
Table 4:
Agreement for computing the percentage of emphysema using scout images versus CT scans stratified by COPD severity when using the InceptionV3 model.
| COPD severity | MAD (%) | R-square | Adjusted R-square |
|---|---|---|---|
| Without COPD | 2.09 ± 1.37 | 0.710* | 0.703* |
| GOLD I | 3.46 ± 2.05 | 0.489* | 0.463* |
| GOLD II | 3.67 ± 2.40 | 0.548* | 0.532* |
| GOLD III | 5.06 ± 2.01 | 0.467* | 0.391* |
| GOLD IV | 4.40 ± 2.47 | 0.969* | 0.962* |
GOLD: Global Initiative for Obstructive Lung Disease; MAD: mean absolute difference; COPD chronic obstructive pulmonary disease
p≤0.01
Fig. 2.
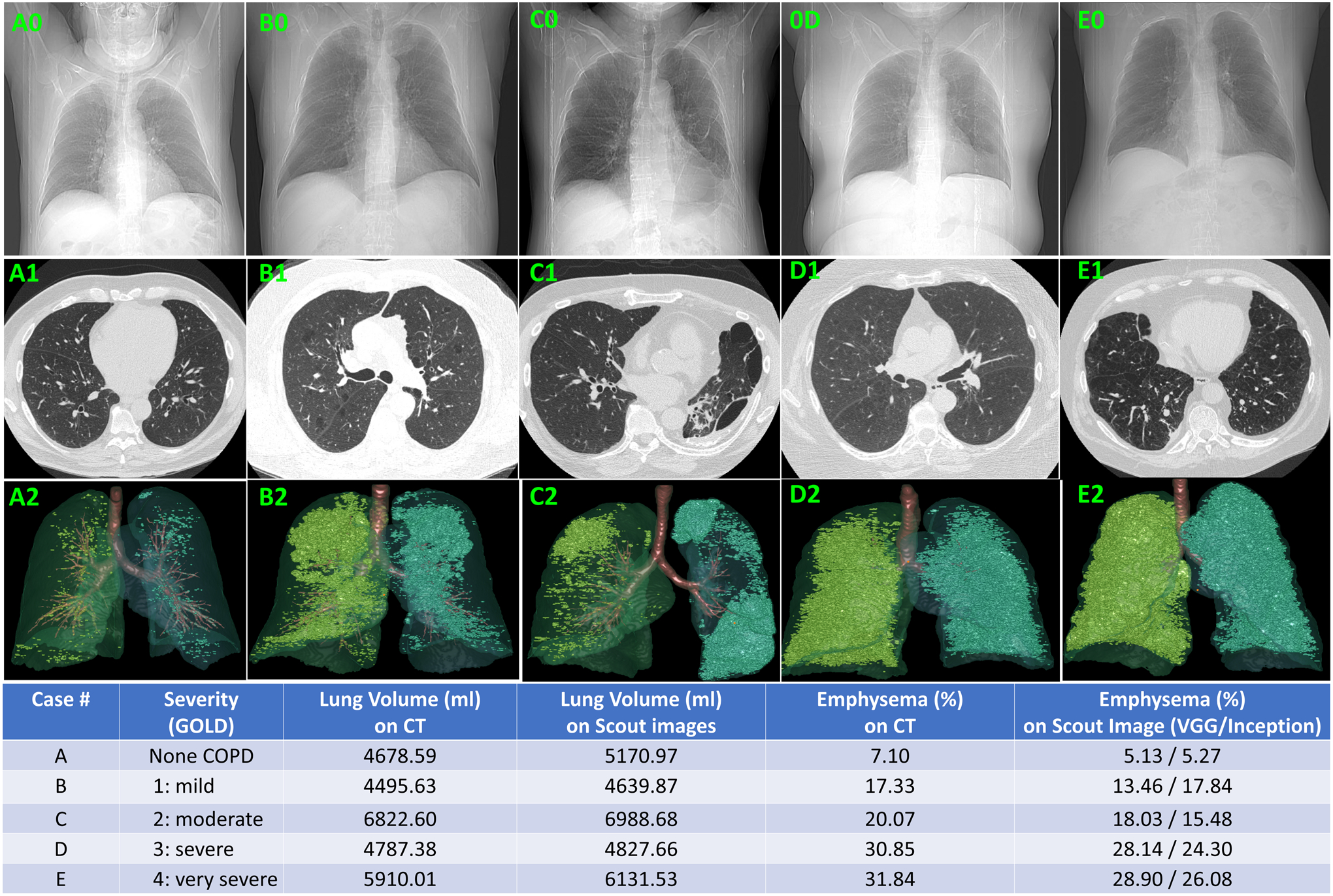
Examples of different levels of emphysema and the volume metrics computed from planar scout images (first row), the CT images (second row), the 3-D visualization of the emphysema quantified from the volumetric CT images (third row).
IV. DISCUSSION
This study demonstrated the feasibility of using artificial intelligence to compute volume metrics from planar images by using 3-D datasets to initially train the algorithm. This concept was supported by the significant linear relationship between the lung volumes and percentages of emphysema computed separately using planar scout images and 3-D CT scan images with a slope close to unity and an average difference of less than 0.4 L and 3.0%, respectively. We believe that the use of a 3-D dataset as the reference standard (or ground truth) to train 2-D CNN-based prediction models for computing volume metrics is a novel approach. Notably, our primary objective is not to develop novel computer algorithms but to verify the feasibility of the premise we proposed in this study.
Typically, when applying machine learning technology to medical imaging, the creation of “ground truth” for training a machine requires time-intensive, manual labeling of a large number of images by an expert human reader, which is often vulnerable to inaccuracy and variability. In this study, the lung volume and percentage of emphysema that served as the “ground truth” for developing the 2-D models were automatically computed from CT scans. This strategy relieved the need for a human reader to outline the lung and regions of emphysema depicted on CT scans and thus significantly reduced the inherent variability between human readers. The underlying concept is applicable to planar images used to assess other diseases, such as pneumonia, interstitial lung disease (ILD), lung cancer, and breast cancer. In addition, the application of artificial intelligence to compute volume metrics from planar images will add a valuable quantitative diagnostic ability to planar images. This quantitative capability will enable a precise assessment of disease severity and accurate monitoring of disease progression, such as Covid-19, using a 2-D imaging modality that is typically less burdensome on a patient, easier to perform, less timely to interpret, and less costly. As demonstrated by the examples in Fig. 2, although it is straightforward to visually assess and quantify emphysema depicted on CT images, it is more challenging to visually assess emphysema depicted on planar scout images and particularly challenging to quantify the presence of emphysema depicted on planar scout images.
A number of classical CNN models have been developed and demonstrated exciting performance, including VGG16, VGG19, InceptionV3, ResNet, DenseNet, InceptionResNet, Xception, and NASNetMobile [45, 46]. We implemented and tested two classical CNN models, namely VGG19 and InceptionV3, in this study, which have been widely used in many applications [10, 33–39]. The promising performance suggested the unique strength of the deep learning technology in inferring 3-D measures from planar images. We note that the emphasis of this study is not to develop novel deep learning architecture but to test the feasibility of the proposed novel idea. If there is one CNN model that can demonstrate a reasonable performance for our specific problem, it will verify our hypothesis. When training the CNN model, we used a data over-resampling strategy to deal with data imbalance. The objective was to alleviate the bias of the prediction models caused by the data imbalance. In addition, several strategies, such as FOV normalization and ImageNet-based transfer learning, were used to reduce the requirement of deep learning for a relatively large dataset.
Notably, the objective of this study is not to replace 3-D imaging modality (e.g., CT) with 2-D imaging modality (e.g., CXR) but to improve the diagnostic performance of 2-D images and thus reduce the use of the relatively expensive CT and the exposure to unnecessary radiation. The relatively small errors (<3.0%), as demonstrated by the VGG-19 model, suggest the feasibility of inferring volume measures from 2-D images. Of course, to justify the error acceptance range and usability of such an AI model in clinical practice, additional investigative effort, such as an observer study, is needed. In this study, our emphasis is the technical feasibility of inferring volume measures from 2-D images.
Our experiments showed that the VGG-19 model had a better overall performance than the InceptionV3 model. The two models demonstrated different performances for cases with different levels of severity. For example, the VGG-19 model had the best performance for cases with severe COPD (MAD = 1.39%) and the worst performance for cases with mild COPD (MAD = 3.55%). In contrast, the InceptionV3 model had a worse performance for the cases with severe COPD (MAD = 4.4%). At the same time, as shown in Fig. 1, the developed regression models have somewhat large prediction errors for some cases (i.e., outliers). Unlike traditional methods based on handcrafted features, where we can identify the samples with large errors and then analyze the features of these samples, a CNN model does not involve any explicit image features, making it extremely challenging to identify the reason behind the decision of the prediction model. This “unexplainable” characteristic is an open problem associated with CNN-based deep learning. In the past years, a concept termed “Explainable AI” has been drawing significant investigative interest in machine learning [47, 48]. Unfortunately, this concept has not demonstrated its feasibility at this time. In our opinion, a possible reason for the cases with large errors may be that the characteristics associated with the relevant cases were not well learned based on the associated CNN architecture.
Notably, the primary emphasis of this study is to address a regression problem, namely inferring numeric measures from 2-D images, not a classification problem, namely classifying images into different categories or subgroups in terms of COPD severity. In clinical practice, the COPD severity level is determined based on the lung function measures (i.e., FEV1/FVC) [26], not the extent of emphysema. The extent of emphysema depicted on CT images only contributes partially to the COPD severity. In order to develop a prediction model for assessing COPD severity from planar images, a separate classification scheme should be used.
We are fully aware that there are differences between the scout images and CXR images, albeit the underlying imaging mechanisms are similar. The primary differences lie in (1) the CXR images have much higher resolution than the scout images, and (2) the CXR images are the projections on a flat surface, while scout images are the projections on a somewhat curved surface. Our preliminary results on scout images and CT images suggest the feasibility of this idea, namely inferring 3-D measures from 2-D images. However, we cannot make a claim that a CNN model trained with scout images could be directly applied to CXR images. To implement this idea in practice, the CNN models need to be trained on CXR images separately.
There are some limitations with this study. First, the CNN models were developed and validated using a COPD cohort from a single institution using the same CT protocols. As a result, the diversity of the cases in the cohort is limited. However, we believe that our experimental results on an independent test set confirmed the feasibility of the proposed idea. Second, scout images were used instead of CXR images because populating a well-characterized database of paired CXR and chest CT scans acquired on a patient within days is challenging. In clinical practice, a CXR often includes both posteroanterior (PA) and lateral views, which have an order of magnitude finer spatial resolution compared to scout images. Hence, it is desirable to combine PA and lateral views of a CXR to maximize the performance of assessing volume metrics from planar images. At this time, the available dataset of scout images and CT scans provided the opportunity to test the feasibility of our novel concept. Additional investigative efforts are needed to further verify the idea and its performance using CXR images and justify how it will help radiologists or clinicians diagnose diseases and assess their severity and progression.
V. CONCLUSION
Our preliminary results demonstrated that artificial intelligence tools have the potential to compute volume metrics from planar images. The significant linear correlation and the relatively small absolute difference between the two approaches in the computation of lung volume and percentage of emphysema in our well-characterized COPD cohort serve as a “proof of concept” that AI can be used to compute volumetric measures from planar images. Our novel concept is applicable beyond emphysema assessment and can be applied to other diseases and planar image types. In clinical practice, the implementation of our concept should significantly increase the functionality of planar images, which are less burdensome on a patient, easier to perform, less timely to interpret, and less costly.
Table 3:
Agreement for computing the percentage of emphysema using scout images versus CT scans stratified by COPD severity when using the VGG-19 model.
| COPD severity | MAD (%) | R-square | Adjusted R-square |
|---|---|---|---|
| Without COPD | 2.30 ± 1.80 | 0.577* | 0.566* |
| GOLD I | 3.55 ± 3.05 | 0.324* | 0.290* |
| GOLD II | 3.72 ± 3.16 | 0.464* | 0.445* |
| GOLD III | 2.32 ± 2.14 | 0.832* | 0.808* |
| GOLD IV | 1.39 ± 0.89 | 0.956* | 0.947* |
GOLD: Global Initiative for Obstructive Lung Disease; MAD: mean absolute difference; COPD chronic obstructive pulmonary disease
p≤0.01
ACKNOWLEDGEMENT
This work is supported by the National Institutes of Health (NIH) (Grant No. R01CA237277) and the UPMC Hillman Developmental Pilot Program. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Nvidia Titan Xp GPU for this research.
Footnotes
Disclosure Statement:
The authors have no conflicts of interest to declare.
Data availability statement:
The data will be available upon request and the data transfer agreement as required by the University of Pittsburgh.
REFERENCES
- 1.National Lung Screening Trial Research T, Aberle DR, Berg CD, Black WC, Church TR, Fagerstrom RM, Galen B, Gareen IF, Gatsonis C, Goldin J, Gohagan JK, Hillman B, Jaffe C, Kramer BS, Lynch D, Marcus PM, Schnall M, Sullivan DC, Sullivan D, Zylak CJ. The National Lung Screening Trial: overview and study design. Radiology. 2011;258(1):243–53. Epub 2010/11/04. doi: 10.1148/radiol.10091808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aberle DR, DeMello S, Berg CD, Black WC, Brewer B, Church TR, Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gatsonis CA, Gierada DS, Jain A, Jones GC, Mahon I, Marcus PM, Rathmell JM, Sicks J, National Lung Screening Trial Research T. Results of the two incidence screenings in the National Lung Screening Trial. N Engl J Med. 2013;369(10):920–31. Epub 2013/09/06. doi: 10.1056/NEJMoa1208962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gawlitza J, Trinkmann F, Scheffel H, Fischer A, Nance JW, Henzler C, Vogler N, Saur J, Akin I, Borggrefe M, Schoenberg SO, Henzler T. Time to Exhale: Additional Value of Expiratory Chest CT in Chronic Obstructive Pulmonary Disease. Can Respir J. 2018;2018:9493504. Epub 2018/04/25. doi: 10.1155/2018/9493504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert M, Schnabel F, Chun J, Schwartz S, Lee J, Klautau Leite AP, Moy L. The relationship of breast density in mammography and magnetic resonance imaging in high-risk women and women with breast cancer. Clin Imaging. 2015;39(6):987–92. Epub 2015/09/10. doi: 10.1016/j.clinimag.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, Naidich DP, Shetty S. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25(6):954–61. Epub 2019/05/22. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Chen X, Lu H, Zhang L, Pan J, Bao Y, Su J, Qian D. Feature-shared adaptive-boost deep learning for invasiveness classification of pulmonary subsolid nodules in CT images. Med Phys. 2020;47(4):1738–49. Epub 2020/02/06. doi: 10.1002/mp.14068. [DOI] [PubMed] [Google Scholar]
- 7.Xie Y, Xia Y, Zhang J, Song Y, Feng D, Fulham M, Cai W. Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Trans Med Imaging. 2019;38(4):991–1004. Epub 2018/10/20. doi: 10.1109/TMI.2018.2876510. [DOI] [PubMed] [Google Scholar]
- 8.Wang XYJ, Zhu Q, Li S, Zhao Z, Yang B, Pu J. . The potential of deep learning in assessing pneumoconiosis depicted on digital chest radiography Occupational and Environmental Medicine. 2020:In press. [DOI] [PubMed]
- 9.Hwang EJ, Park S, Jin KN, Kim JI, Choi SY, Lee JH, Goo JM, Aum J, Yim JJ, Cohen JG, Ferretti GR, Park CM, Development D, Evaluation G. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw Open. 2019;2(3):e191095. Epub 2019/03/23. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402–10. Epub 2016/11/30. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 11.Falk T, Mai D, Bensch R, Cicek O, Abdulkadir A, Marrakchi Y, Bohm A, Deubner J, Jackel Z, Seiwald K, Dovzhenko A, Tietz O, Dal Bosco C, Walsh S, Saltukoglu D, Tay TL, Prinz M, Palme K, Simons M, Diester I, Brox T, Ronneberger O. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70. Epub 2018/12/19. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhen Y, Chen H, Zhang X, Meng X, Zhang J, Pu J. Assessment of Central Serous Chorioretinopathy Depicted on Color Fundus Photographs Using Deep Learning. Retina. 2019. Epub 2019/07/10. doi: 10.1097/IAE.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers R. ChestX-ray14: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases 2017.
- 14.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong D, Halabi S, Sandberg J, Jones R, Larson D, Langlotz C, Patel B, Lungren M, Ng A. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison 2019.
- 15.RSNA Pneumonia Detection Challenge (2018) 2018. Available from: https://www.rsna.org/en/education/ai-resources-and-training/ai-image-challenge/RSNA-Pneumonia-Detection-Challenge-2018.
- 16.Mittal A, Kumar D, Mittal M, Saba T, Abunadi I, Rehman A, Roy S. Detecting Pneumonia using Convolutions and Dynamic Capsule Routing for Chest X-ray Images. Sensors (Basel). 2020;20(4). Epub 2020/02/23. doi: 10.3390/s20041068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.JI Pranav Rajpurkar, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, Ding Daisy, Bagul Aarti, Langlotz Curtis, Shpanskaya Katie, Lungren Matthew P., Ng Andrew Y.. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv:171105225. 2017. [Google Scholar]
- 18.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. Epub 2006/11/30. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collaborators GBDCRD. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5(9):691–706. Epub 2017/08/22. doi: 10.1016/S2213-2600(17)30293-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Gu S, Leader JK, Kundu S, Tedrow JR, Sciurba FC, Gur D, Siegfried JM, Pu J. Optimal threshold in CT quantification of emphysema. Eur Radiol. 2013;23(4):975–84. Epub 2012/11/01. doi: 10.1007/s00330-012-2683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konietzke P, Wielputz MO, Wagner WL, Wuennemann F, Kauczor HU, Heussel CP, Eichinger M, Eberhardt R, Gompelmann D, Weinheimer O. Quantitative CT detects progression in COPD patients with severe emphysema in a 3-month interval. Eur Radiol. 2020;30(5):2502–12. Epub 2020/01/23. doi: 10.1007/s00330-019-06577-y. [DOI] [PubMed] [Google Scholar]
- 22.Lynch DA, Austin JH, Hogg JC, Grenier PA, Kauczor HU, Bankier AA, Barr RG, Colby TV, Galvin JR, Gevenois PA, Coxson HO, Hoffman EA, Newell JD Jr., Pistolesi M, Silverman EK, Crapo JD. CT-Definable Subtypes of Chronic Obstructive Pulmonary Disease: A Statement of the Fleischner Society. Radiology. 2015;277(1):192–205. Epub 2015/05/12. doi: 10.1148/radiol.2015141579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Hobbs BD, Crapo JD, Make BJ, Regan EA, Humphries S, Carey VJ, Lynch DA, Silverman EK, Investigators CO. Subtyping COPD by Using Visual and Quantitative CT Imaging Features. Chest. 2020;157(1):47–60. Epub 2019/07/10. doi: 10.1016/j.chest.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Occhipinti M, Paoletti M, Bartholmai BJ, Rajagopalan S, Karwoski RA, Nardi C, Inchingolo R, Larici AR, Camiciottoli G, Lavorini F, Colagrande S, Brusasco V, Pistolesi M. Spirometric assessment of emphysema presence and severity as measured by quantitative CT and CT-based radiomics in COPD. Respir Res. 2019;20(1):101. Epub 2019/05/28. doi: 10.1186/s12931-019-1049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wielputz MO, Heussel CP, Herth FJ, Kauczor HU. Radiological diagnosis in lung disease: factoring treatment options into the choice of diagnostic modality. Dtsch Arztebl Int. 2014;111(11):181–7. Epub 2014/04/05. doi: 10.3238/arztebl.2014.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel AR, Patel AR, Singh S, Singh S, Khawaja I. Global Initiative for Chronic Obstructive Lung Disease: The Changes Made. Cureus 2019;11(6):e4985. Epub 2019/08/28. doi: 10.7759/cureus.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kundu S, Gu S, Leader JK, Tedrow JR, Sciurba FC, Gur D, Kaminski N, Pu J. Assessment of lung volume collapsibility in chronic obstructive lung disease patients using CT. Eur Radiol. 2013;23(6):1564–72. Epub 2013/03/16. doi: 10.1007/s00330-012-2746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Lei Y, Wang T, Higgins K, Bradley JD, Curran WJ, Liu T, Yang X. LungRegNet: An unsupervised deformable image registration method for 4D-CT lung. Med Phys. 2020. Epub 2020/02/06. doi: 10.1002/mp.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, Peng L, Webster DR. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2(3):158–64. Epub 2019/04/25. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol. 2018;73(5):439–45. Epub 2017/12/23. doi: 10.1016/j.crad.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham). 2019;6(1):014006. Epub 2019/04/05. doi: 10.1117/1.JMI.6.1.014006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Wang L, Nan Y, Jin F, Wang Q, Pu J. SDFN: Segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput Med Imaging Graph. 2019;75:66–73. Epub 2019/06/08. doi: 10.1016/j.compmedimag.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S K, Z A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv:14091556. 2014. [Google Scholar]
- 34.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z, editors. Rethinking the Inception Architecture for Computer Vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 201627–30June2016. [Google Scholar]
- 35.Masumoto H, Tabuchi H, Yoneda T, Nakakura S, Ohsugi H, Sumi T, Fukushima A. Severity Classification of Conjunctival Hyperaemia by Deep Neural Network Ensembles. J Ophthalmol. 2019;2019:7820971. Epub 2019/07/06. doi: 10.1155/2019/7820971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Asnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020:1–12. Epub 2020/05/14. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Nóbrega RVM, Rebouças Filho PP, Rodrigues MB, da Silva SPP, Dourado Júnior CMJM, de Albuquerque VHC. Lung nodule malignancy classification in chest computed tomography images using transfer learning and convolutional neural networks. Neural Computing and Applications. 2018. doi: 10.1007/s00521-018-3895-1. [DOI] [Google Scholar]
- 38.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–67. Epub 2018/09/19. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Yu J, Zhu Q, Li S, Zhao Z, Yang B, Pu J. Potential of deep learning in assessing pneumoconiosis depicted on digital chest radiography. Occup Environ Med. 2020;77(9):597–602. Epub 2020/05/31. doi: 10.1136/oemed-2019-106386. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–29. Epub 2018/06/24. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibali A, He Z, Wollersheim D. Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg. 2017;12(10):1799–808. Epub 2017/05/16. doi: 10.1007/s11548-017-1605-6. [DOI] [PubMed] [Google Scholar]
- 42.Ren X, Li T, Yang X, Wang S, Ahmad S, Xiang L, Stone SR, Li L, Zhan Y, Shen D, Wang Q. Regression Convolutional Neural Network for Automated Pediatric Bone Age Assessment From Hand Radiograph. IEEE J Biomed Health Inform. 2019;23(5):2030–8. Epub 2018/10/23. doi: 10.1109/JBHI.2018.2876916. [DOI] [PubMed] [Google Scholar]
- 43.Reddy NE, Rayan JC, Annapragada AV, Mahmood NF, Scheslinger AE, Zhang W, Kan JH. Bone age determination using only the index finger: a novel approach using a convolutional neural network compared with human radiologists. Pediatr Radiol. 2019. Epub 2019/12/22. doi: 10.1007/s00247-019-04587-y. [DOI] [PubMed] [Google Scholar]
- 44.Mazo C, Bernal J, Trujillo M, Alegre E. Transfer learning for classification of cardiovascular tissues in histological images. Comput Methods Programs Biomed. 2018;165:69–76. Epub 2018/10/20. doi: 10.1016/j.cmpb.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. Epub 2017/08/05. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical Image Analysis using Convolutional Neural Networks: A Review. J Med Syst. 2018;42(11):226. Epub 2018/10/10. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 47.Amann J, Blasimme A, Vayena E, Frey D, Madai VI, Precise Qc. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak. 2020;20(1):310. Epub 2020/12/02. doi: 10.1186/s12911-020-01332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linardatos P, Papastefanopoulos V, Kotsiantis S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy (Basel). 2020;23(1). Epub 2020/12/31. doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]


