Abstract
Purpose of review:
In this article, we introduce the concept of model interpretability, review its applications in deep learning models for clinical ophthalmology, and discuss its role in the integration of artificial intelligence in healthcare.
Recent findings:
The advent of deep learning in medicine has introduced models with remarkable accuracy. However, the inherent complexity of these models undermines its users’ ability to understand, debug and ultimately trust them in clinical practice. Novel methods are being increasingly explored to improve models’ “interpretability” and draw clearer associations between their outputs and features in the input dataset. In the field of ophthalmology, interpretability methods have enabled users to make informed adjustments, identify clinically relevant imaging patterns, and predict outcomes in deep learning models.
Summary:
Interpretability methods support the transparency necessary to implement, operate and modify complex deep learning models. These benefits are becoming increasingly demonstrated in models for clinical ophthalmology. As quality standards for deep learning models used in healthcare continue to evolve, interpretability methods may prove influential in their path to regulatory approval and acceptance in clinical practice.
Keywords: artificial intelligence, machine learning, deep learning, convolutional neural network, interpretability
Introduction
Deep learning has received increasing attention as a promising solution to longstanding challenges in medicine including diagnostic accuracy and optimization of clinician workflow [1–6]. Within ophthalmology, diagnostic models for age-related macular degeneration (AMD), diabetic retinopathy (DR) and retinopathy of prematurity (ROP) have demonstrated expert-level accuracy [7–16]. With some notable exceptions, these models have largely been implemented in research contexts. Numerous technical and socio-environmental factors impede real-world implementation of deep learning, including model complexity [7, 17]. As models increase in complexity, their reasoning becomes more difficult for users to understand and ultimately trust [18–20**]. To address this, varied approaches have been studied to enhance model transparency [21, 22*]. In this article, we review applications of interpretable deep learning in ophthalmology and discuss its role in the successful integration of deep learning models into healthcare.
The cost of complexity in deep learning
Machine learning is a subfield of artificial intelligence (AI) wherein algorithms learn patterns from input data and adjust their internal parameters accordingly to generate accurate predictions [1]. Deep learning is a subfield of machine learning which utilizes neural networks that process data in “hidden layers” between the inputs and outputs. Convolutional neural networks (CNNs), a key strategy in deep learning, manage hidden “convolutional” layers, each containing “filters” that can extract specific patterns from input images, creating an activation map. A convolutional layer thus outputs a set of activation maps that are passed on to the next layer. As the extracted information is passed consecutively through deeper layers, the features and their relationship with other features become hierarchically more complex. This functionality has introduced a generation of models that learn more efficiently, discover subtler patterns and analyse rich data formats like image and video. However, this multi-layered architecture substantially increases the model’s complexity and restricts the user’s control over learned features. This gives rise to the perception of deep learning models as “black boxes” that conceal how input features contribute to the final output [7, 23,24**,25].
Several problems arise from non-transparent models. Such models are not only inherently difficult to audit for quality, but also limit the user’s ability to “debug” or make informed adjustments to the underlying algorithm [26*]. “Black-box” models may also rely on misleading features in the data to generate predictions with falsely high accuracy, unbeknownst to users. Finally, models lacking ethical, human-centred monitoring may produce outputs that augment social biases or inadvertently discriminate against demographics within the dataset [27].
Interpretability as a solution
The concept of interpretability seeks to address these potential downsides. While a consensus has not been reached on a definition for the term, “interpretability” in the context of AI generally refers to the degree to which the reasoning behind a model’s outputs is evident to the user [21, 28, 29]. Interpretations refer to automated translations of a concept or methodology into a comprehensible medium like images or words ranging from heatmaps illustrating a fundus image features’ relative contribution to a diagnostic prediction, to textual explanations of a self-driving car’s actions [21,30,31*]. Focused efforts to clarify the concept within medicine have shown that clinicians generally view interpretability as transparency in model reasoning, adjustable features, and limitations [19, 32].
Applications of interpretable AI in ophthalmology
The field of ophthalmology particularly stands to benefit from deep learning, largely due to its reliance on diagnostic imaging. Toward this end, varied interpretability methods have been designed to refine deep learning models and identify relevant clinical traits imaging patterns. To review these methods, we devise an intuitive classification of the varied interpretability techniques in ophthalmology, organizing them according to two fundamental characteristics: model-centric, in which solutions operate at the level of the model itself, and data centric, which target the relationship between inputs and outputs (Figure 1).
Figure 1:
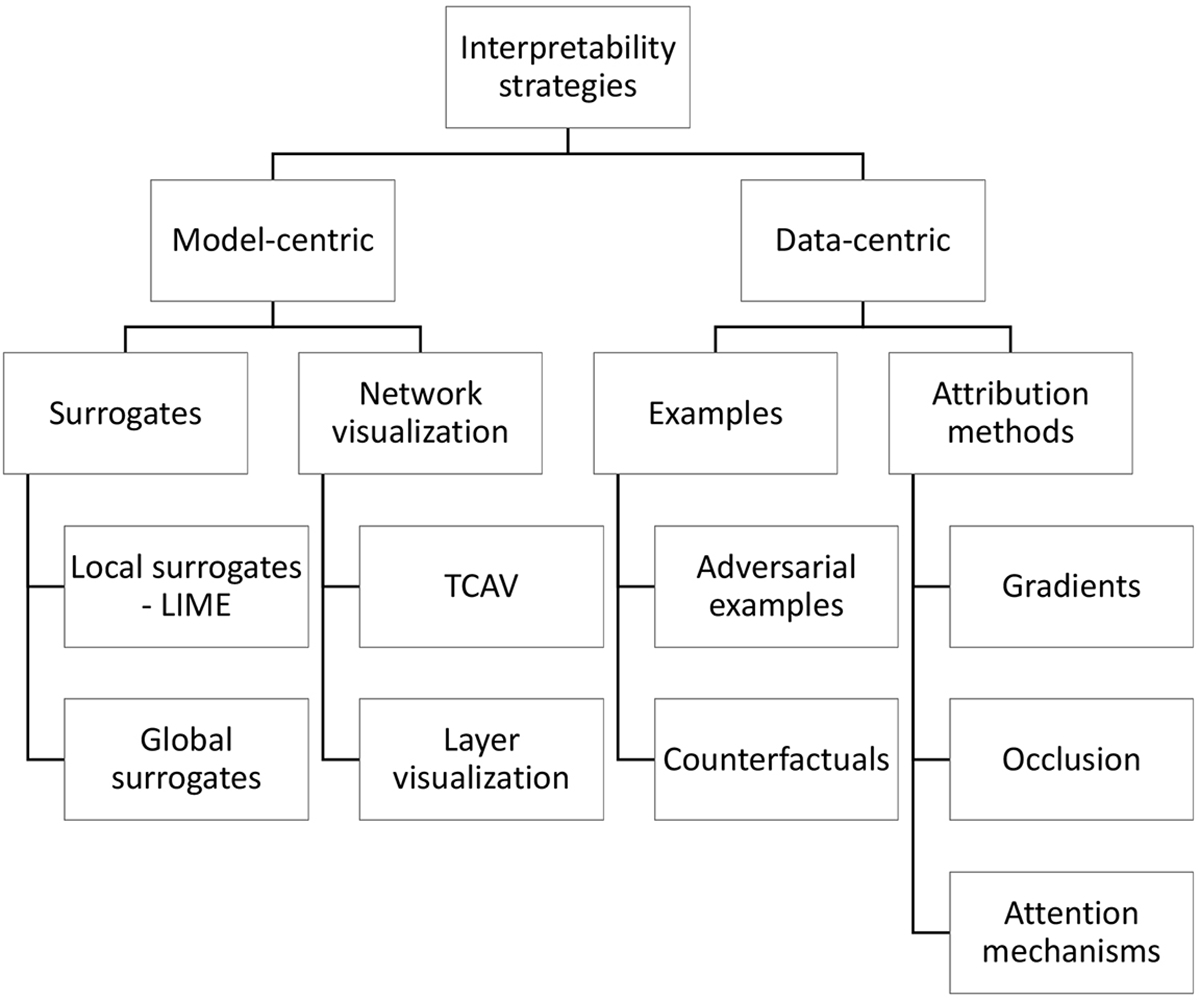
Classification of interpretability methods
Structured representation of the varied categories of interpretability methods discussed in this article. LIME = Local Interpretable Model-Agnostic Explanations; TCAV = Testing with Concept Activation Vectors; DeConvNet = Deconvolutional networks; CAM = Class activation mapping.
Model-centric: Surrogates
“Surrogates” or “proxy models” are intrinsically interpretable models designed to approximate the behaviour of a more complex, pre-existing model [33, 34]. Decision trees are classic examples of intrinsically interpretable methods. They are composed of sequential “if-then” decisions that separate data into nodes, progressively branching outward until final classifications are reached, illustrating the model’s categorical decision making. Naïve Bayes classifiers are built upon conditional probabilities. The term “naïve” refers to the assumption that features are mutually exclusive, allowing the estimation of their individual contribution to the output [35]. These models can also be used as standalone, interpretable classifiers, as has been demonstrated in DR classification through decision trees by Mane & Jadhav, and Naïve Bayes by Harangi et al [36, 37].
Surrogate methods may be sorted into two categories. Global surrogates explain a complex model’s operations on the entire dataset. Contrastingly, local surrogates such as Local Interpretable Model-Agnostic Explanations (LIME) explain decisions for individual input-output pairs [38]. Gheisari et al applied LIME to a glaucoma image classifier to illustrate the significance of vascularized regions of the superior and inferior retina, supporting previous studies on this finding [39].
Model-centric: Network visualization methods
Testing with Concept Activation Vectors (TCAV) “translates” activation maps into semantic concepts. A model is given a dataset of images, with some containing a specific feature (e.g. stripes). A simple linear classifier then isolates the activation maps associated with the images with the chosen feature and calculates its significance to the final prediction. TCAV was applied in a DR classification task to evaluate the weight of diagnostic fundus features like microaneurysms and pan-retinal laser scars. This revealed model’s tendency to overestimate DR severity by placing a high TCAV score on aneurysms, consequently allowing the experts to correct this behaviour [40].
Deconvolutional networks (DeConvNet) function in “reverse” by projecting activation maps from a convolutional layer back to the input space. Zhou et al used a model pre-trained with non-retinal images, and fine-tuned it with retinal images for a DR detection task. DeConvNet was implemented for individual layers pre- and post-training with retinal images, visualizing DR-specific features learned by the model. A focus on red and green channels in the first layer and microaneurysm-like and vessel-like structures in deeper layers was revealed [41].
Class Activation Mapping (CAM) utilizes the global pooling layer of a CNN, which “pools” each activation map into a mean value. The following layers assign a weight to that value, providing the final class probability. These weights are similarly applied to the activation maps themselves, which are then added to produce a class-specific heatmap [42]. Worrall et al incorporate CAM in a ROP classifier, revealing emphasis on vasculature, consistent with clinical practice [43]. In DR classification, Gondal et al and Gargeya et al similarly confirm appropriate focus on hard and soft exudates as well as haemorrhages [44, 45]. In glaucoma detection, Maetschke et al and Ran et al use CAM to verify model focus on clinically relevant features such as the neuroretinal rim and lamina cribrosa [46, 47].
Data-centric: Examples
Adversarial examples are created by altering input images to produce counterexamples that lead to a maximal prediction of a desired output. Comparison of the counterexample to the original illustrates what feature transformations lead to either model outcome [48]. Chang et al utilized this technique for CNN detection of glaucomatous fundus features [49]. In referrable glaucoma images, a negative model outcome resulted when lowering the cup-to-disc ratio, illustrating this feature’s role in detection.
Google’s What-If Tool generates counterfactuals mainly for tabular datasets [50]. Abbas et al implemented this for visual acuity (VA) prediction from AMD patient data. They discovered differing final VA outcome predictions for an 84-year-old British male, and a female of the same age, nationality and initial VA. This indicated the decision boundary for an individual patient level [51].
Data-centric: Attribution methods
Attribution methods interpret CNN decisions by revealing the contribution of different regions or features of an image to the output. This may manifest as heatmaps, also termed saliency maps, which highlight pixels of the image according to their significance.
Gradient methods derive heatmaps from information gathered during the second of two steps of model training: forward propagation and backpropagation. Forward propagation first calculates prediction accuracy by comparing the output to the original input labels, producing a “loss” value proportional to the difference. Backpropagation then updates its parameters to minimize this loss, guided by the gradient, or rate of change, of the loss associated with each parameter [52]. Gradient methods analyse backpropagation to calculate the gradient of the loss with respect to the input pixels, with higher values indicating higher significance of that pixel to the prediction [53].
Kuo et al utilized a specific gradient method, Gradient-weighted Class Activation Mapping (Grad-CAM) to interpret their keratoconus classifier, confirming that the model was following similar corneal topography features as ophthalmologists by focusing on the largest gradient differences in the scan [54, 55]. Medeiros et al applied Grad-CAM to a glaucoma classifier, showing significant activation in the region of the optic nerve and adjacent RNFL on fundus photography [56]. When applying Grad-CAM to DR detection, Chetoui et al confirmed a model’s focus on exudates, microaneurysms and haemorrhage, validating its conformance with clinical reasoning [57**]. They additionally employed Grad-CAM for failure cases, showing that false predictions arose from artifacts, pale areas and image noise.
Occlusion methods modify image regions and compare the resulting change in model predictions to estimate the relative importance of these selectively altered areas. Kermany et al performed occlusion testing for a classifier trained to detect drusen, choroidal neovascularization and diabetic macular oedema [58]. Localization of drusen was found to be most accurate, compared to other findings. In an AMD classification model, Grassman et al noted that masking areas of the macula and fovea led to a confidence reduction, indicating their weight in prediction-making [59].
Attention mechanisms, as employed by Poplin et al, create a heatmap by relying first on a CNN encoder to reduce the image size and extract important features, followed by a decoder which projects only these features back into the original image size. This was used to visualize the decisions of a cardiovascular risk factor classifier trained on fundus images. This revealed that prediction of risk factors such as smoking, systolic blood pressure and age relied on blood vessels, whereas gender predictions appeared to draw from areas including the optic disc, macula and vessels, suggesting patterns imperceptible by the human eye [60].
Interpretability’s role in implementing deep learning solutions in medicine
Providing insights to a model’s reasoning and shortcomings may enhance the trust necessary to ultimately accept these models in clinical practice [61,62**,63]. For example, interpretability methods enable clinicians to monitor models’ reasoning for conformance to clinical standards and principles [26]. Some methods permit quality monitoring for systemic weaknesses or biases, as demonstrated by Winkler et al, who used heat maps to discover a melanoma detection model’s errant reliance on surgical skin marker artifacts in the training images to form predictions, rather than the features of the lesion alone [64]. Clinicians may also benefit from enhanced diagnostic accuracy and specificity, as demonstrated by Rajpurkar et al in a report of human participants in a tuberculosis detection experiment using chest radiographs aided by heatmaps [65]. Finally, a common barrier to training accurate models in medicine is infrequent access to large datasets that represent a population’s demographic and clinical traits. When designing models with imperfect datasets, interpretability is essential for ensuring fairness and equitable representation of marginalized demographics in predictions [66].
The complexity of trust
Despite these benefits, model interpretability may not be a “silver bullet” for user distrust. Gaube et al describe “algorithmic aversion” in individuals who consistently rated expert advice as lower quality if labelled as AI-derived [67*]. Criticism of “black boxes” often overlooks other factors for acceptance of AI. An analysis of trust by Asan et al place robustness and fairness as pillars alongside transparency [20]. Kitamura et al argue that trust is built on prolonged experience with models that have demonstrated the ability to make consistently accurate predictions [68*]. Finally, the enhanced model performance granted by “black box” complexity may justify the distrust they engender. Deep learning models bear the potential to detect otherwise imperceptible patterns, drive new scientific discovery, and enhance clinician accuracy [69]. Thus, enhancing interpretability at the cost of model complexity may sacrifice this added benefit.
It often goes unmentioned that clinicians themselves act as “black boxes” whose experiential reasoning cannot be distilled into a list of arguments or heatmaps. This raises the question of why human black-boxes are trusted and not AI. Hatherley discusses that trust encompasses fundamentally human concepts not found in AI like goodwill and moral responsibility which could explain the phenomenon of “algorithmic aversion’ [70]. Trust as defined by human terms may therefore be an unattainable ideal. We might instead strive for model reliability by establishing standards for robustness, fairness, and high performance.
Evaluating interpretability methods
It is important to consider whether interpretability methods truly reflect image features relevant to model output, or just feature rich portions of an image. In a review of 8 common saliency map techniques, Arun et al demonstrate that none satisfied four basic criteria for localizing of image features, sensitive to model weight randomization, repeatability, and reproducibility [71*]. Adebayo et al demonstrate that gradient heatmaps can sometimes provide a seemingly correct appearance without reflecting the true internal model weights, and propose two “sanity checks” to evaluate the outputs’ correspondence to the learned weights of the model [72]. They first propose randomizing model weights so that the interpretability method output reflects no learned information relevant to the task. If the outputs reflecting randomized vs. non-randomized model weights look the same, the interpretability method may be highlighting features non-relevant features. They alternatively propose training a network on randomized input class labels. If the output of the interpretability method is invariant to the class label, then it is not valid for interpretation of a trained classification model. Further, feature rich portions of an image may be part of what a model learns, but not fully explain model reasoning, as illustrated by Chang et al using adversarial examples [49].
The most appropriate interpretability method may depend on the classification task at hand. For instance, some classification efforts rely on recognition of localizable patterns (e.g. cup-to-disc ratios), to which techniques like Grad-CAM that illustrate learned representations graphically would be more suited. Others require assessment of more global features (e.g. cardiovascular risk in fundus photos), for which a technique that maps to higher order features such as regression concept vectors would be preferred. Finally, Doshi-Velez et al propose that a formal definition of interpretability must be established so as to provide standardized context for quality when evaluating explanations [73].
Thus, with growing interest in interpretability and new global regulations mandating the “right to explanation” in AI, we advocate for qualitative and quantitative evaluation of these methodologies in the context of a given application, integrating the user’s perception of explanation quality with an objective measurement of the accuracy to holistically represent the model’s intrinsic features [32, 69].
Conclusion
Interpretability methods are powerful tools in meeting the evolving complexity and scrutiny of deep learning models in medicine. In the field of ophthalmology, model transparency has highlighted diagnostically important imaging features and contributed new clinical insights. As these methods proliferate, a clear definition of interpretability and accepted frameworks for evaluating their quality must be established. Ultimately, interpretable models may play a key role in integrating deep learning models into clinical practice.
Key Points.
Deep learning models’ inherent complexity undermines user understanding and increases risk for misinterpretation of outputs.
Interpretability methods enhance model transparency through mechanisms that provide explanations of the connection between features in input data and model output.
As interpretability methods become increasingly varied and utilized, approaches to evaluate their quality and efficacy must be established.
Interpretability methods may enhance user trust and contribute to the ultimate acceptance of deep learning models in modern healthcare.
Financial support and sponsorship:
JPC is supported by grants R01EY19474, R01 EY031331, R21 EY031883, and P30 EY10572 from the National Institutes of Health (Bethesda, MD), by unrestricted departmental funding and a Career Development Award from Research to Prevent Blindness (New York, NY). PAK is supported by a Career Development Award (R190028A) from Moorfields Eye Charity (London, UK) and a Future Leaders Fellowship (MR/T019050/1) from UK Research & Innovation (London, UK).
This work was supported by the following grants from the National Institutes of Health (Bethesda, MD): R01EY19474, R01 EY031331, R21 EY031883, and P30 EY10572
3. Conflicts of interest: JPC is a consultant for Boston AI Labs and receives research funding from Genentech. PAK has acted as a consultant for DeepMind, Roche, Novartis, Apellis, and BitFount and is an equity owner in Big Picture Medical. He has received speaker fees from Heidelberg Engineering, Topcon, Allergan, and Bayer.
References
- 1.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 2.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones LD, Golan D, Hanna SA, Ramachandran M. Artificial intelligence, machine learning and the evolution of healthcare: A bright future or cause for concern? Bone Joint Res 2018;7(3):223–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajpurkar P, Irvin J, Zhu K, et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:171105225. 2017. [Google Scholar]
- 5.Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J 2019;6(2):94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin W-C, Chen JS, Chiang MF, Hribar MR. Applications of Artificial Intelligence to Electronic Health Record Data in Ophthalmology. Transl Vis Sci Technol 2020;9(2):13–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol 2019;103(2):167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature Medicine. 2018;24(9):1342–50. [DOI] [PubMed] [Google Scholar]
- 9.Lee CS, Baughman DM, Lee AY. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol Retina. 2017;1(4):322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motozawa N, An G, Takagi S, et al. Optical Coherence Tomography-Based Deep-Learning Models for Classifying Normal and Age-Related Macular Degeneration and Exudative and Non-Exudative Age-Related Macular Degeneration Changes. Ophthalmol Ther 2019;8(4):527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402–10. [DOI] [PubMed] [Google Scholar]
- 12.Abràmoff MD, Lavin PT, Birch M, et al. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digital Medicine. 2018;1(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TY, Bressler NM. Artificial Intelligence With Deep Learning Technology Looks Into Diabetic Retinopathy Screening. Jama. 2016;316(22):2366–7. [DOI] [PubMed] [Google Scholar]
- 14.Brown JM, Campbell JP, Beers A, et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol 2018;136(7):803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyner AS, Swan R, Campbell JP, et al. Automated Fundus Image Quality Assessment in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. Ophthalmology Retina. 2019;3(5):444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell JP, Ataer-Cansizoglu E, Bolon-Canedo V, et al. Expert Diagnosis of Plus Disease in Retinopathy of Prematurity From Computer-Based Image Analysis. JAMA ophthalmology. 2016;134(6):651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beede E, Baylor E, Hersch F, et al. A Human-Centered Evaluation of a Deep Learning System Deployed in Clinics for the Detection of Diabetic Retinopathy. Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems; Honolulu, HI, USA: Association for Computing Machinery; 2020. p. 1–12. [Google Scholar]
- 18.Keel S, Lee PY, Scheetz J, et al. Feasibility and patient acceptability of a novel artificial intelligence-based screening model for diabetic retinopathy at endocrinology outpatient services: a pilot study. Sci Rep 2018;8(1):4330–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzinger A, Biemann C, Pattichis CS, Kell DB. What do we need to build explainable AI systems for the medical domain? arXiv preprint arXiv:171209923. 2017. [Google Scholar]
- 20. Asan O, Bayrak AE, Choudhury A. Artificial Intelligence and Human Trust in Healthcare: Focus on Clinicians. J Med Internet Res 2020;22(6):e15154–e. ** An analysis of the complex nature of trust in AI, with a focus on healthcare. The authors discuss the lack of trust as a crucial barrier to acceptance of the technology in practice and propose possible strategies to enhance this trust, specifically targeting transparency, robustness and fairness of models.
- 21.Montavon G, Samek W, Müller K-R. Methods for interpreting and understanding deep neural networks. Digital Signal Processing. 2018;73:1–15. [Google Scholar]
- 22. Singh A, Balaji JJ, Jayakumar V, et al. Quantitative and Qualitative Evaluation of Explainable Deep Learning Methods for Ophthalmic Diagnosis. arXiv preprint arXiv:200912648. 2020. * An evaluation of 14 different attribution interpretability approaches. The authors design a reader study, where participants are asked to provide both a quantitative rating of the explanation as well as qualitative feedback. The results suggested that the best performing method may be specific to the task considered by this analysis and may not apply as well to other AI tasks.
- 23.Coyner AS, Campbell JP, Chiang MF. Demystifying the Jargon: The Bridge between Ophthalmology and Artificial Intelligence. Ophthalmology Retina. 2019;3(4):291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi RY, Coyner AS, Kalpathy-Cramer J, et al. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol 2020;9(2):14. ** A broad introduction to key concepts in artificial intelligence aimed at professionals without a background in computer science.
- 25.Udofia U. Basic Overview of Convolutional Neural Network (CNN) medium.com2018 [Available from: https://medium.com/dataseries/basic-overview-of-convolutional-neural-network-cnn-4fcc7dbb4f17.
- 26. Reyes M, Meier R, Pereira S, et al. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol Artif Intell. 2020;2(3):e190043. * A review of interpretability as it pertains to radiology, providing an introduction into varying techniques as well as suggesting clinical areas where these could be applied to the workflow.
- 27.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern Med 2018;178(11):1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipton ZC. The Mythos of Model Interpretability: In machine learning, the concept of interpretability is both important and slippery. Queue. 2018;16(3):31–57. [Google Scholar]
- 29.Molnar C. Interpretable machine learning. A Guide for Making Black Box Models Explainable 2019. [Google Scholar]
- 30.Kim J, Rohrbach A, Darrell T, et al. , editors. Textual explanations for self-driving vehicles. Proceedings of the European conference on computer vision (ECCV); 2018. [Google Scholar]
- 31. Singh A, Sengupta S, Lakshminarayanan V. Explainable deep learning models in medical image analysis. Journal of Imaging. 2020;6(6):52. * A review of interpretability techniques bringing together examples from different specialties and imaging modalities.
- 32.Vellido A. The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Computing and Applications. 2020;32(24):18069–83. [Google Scholar]
- 33.Explainable AI: the basics. Policy briefing. The Royal Society; 2019. [Google Scholar]
- 34.Hayashi Y. The Right Direction Needed to Develop White-Box Deep Learning in Radiology, Pathology, and Ophthalmology: A Short Review. Front Robot AI. 2019;6:24–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loor M, De Tré G, editors. Contextualizing Naive Bayes Predictions. Information Processing and Management of Uncertainty in Knowledge-Based Systems; 20202020//; Cham: Springer International Publishing. [Google Scholar]
- 36.Mane VM, Jadhav DV. Holoentropy enabled-decision tree for automatic classification of diabetic retinopathy using retinal fundus images. Biomed Tech (Berl). 2017;62(3):321–32. [DOI] [PubMed] [Google Scholar]
- 37.Harangi B, Antal B, Hajdu A, editors. Automatic exudate detection with improved Naïve-bayes classifier. 2012 25th IEEE International Symposium on Computer-Based Medical Systems (CBMS); 2012 20–22 June 2012. [Google Scholar]
- 38.Ribeiro MT, Singh S, Guestrin C. Model-agnostic interpretability of machine learning. arXiv preprint arXiv:160605386. 2016. [Google Scholar]
- 39.Gheisari S, Shariflou S, Phu J, et al. A combined convolutional and recurrent neural network for enhanced glaucoma detection. Sci Rep 2021;11(1):1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim B, Wattenberg M, Gilmer J, et al. , editors. Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (tcav). International conference on machine learning; 2018: PMLR. [Google Scholar]
- 41.Zhou L, Zhao Y, Yang J, et al. Deep multiple instance learning for automatic detection of diabetic retinopathy in retinal images. IET Image Processing. 2018;12(4):563–71. [Google Scholar]
- 42.Zhou B, Khosla A, Lapedriza A, et al. , editors. Learning deep features for discriminative localization. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. [Google Scholar]
- 43.Worrall DE, Wilson CM, Brostow GJ, editors. Automated Retinopathy of Prematurity Case Detection with Convolutional Neural Networks2016; Cham: Springer International Publishing. [Google Scholar]
- 44.Gondal WM, Köhler JM, Grzeszick R, et al. , editors. Weakly-supervised localization of diabetic retinopathy lesions in retinal fundus images. 2017 IEEE International Conference on Image Processing (ICIP); 2017 17–20 Sept.2017. [Google Scholar]
- 45.Gargeya R, Leng T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology. 2017;124(7):962–9. [DOI] [PubMed] [Google Scholar]
- 46.Maetschke S, Antony B, Ishikawa H, et al. A feature agnostic approach for glaucoma detection in OCT volumes. PLOS ONE. 2019;14(7):e0219126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran AR, Cheung CY, Wang X, et al. Detection of glaucomatous optic neuropathy with spectral-domain optical coherence tomography: a retrospective training and validation deep-learning analysis. Lancet Digit Health. 2019;1(4):e172–e82. [DOI] [PubMed] [Google Scholar]
- 48.Woods W, Chen J, Teuscher C. Adversarial explanations for understanding image classification decisions and improved neural network robustness. Nature Machine Intelligence. 2019;1(11):508–16. [Google Scholar]
- 49.Chang J, Lee J, Ha A, et al. Explaining the Rationale of Deep Learning Glaucoma Decisions with Adversarial Examples. Ophthalmology. 2021;128(1):78–88. [DOI] [PubMed] [Google Scholar]
- 50.Wexler J, Pushkarna M, Bolukbasi T, et al. The What-If Tool: Interactive Probing of Machine Learning Models. IEEE Transactions on Visualization and Computer Graphics. 2020;26(1):56–65. [DOI] [PubMed] [Google Scholar]
- 51.Abbas ABS, Wagner S, Korot E, Singh R, Struyven R, Keane P. Using the What-if Tool to perform nearest counterfactual analysis on an AutoML model that predicts visual acuity outcomes in patients receiving treatment for wet age-related macular degeneration. The Association for Research in Vision and Ophthalmology; Virtual Conference2021. [Google Scholar]
- 52.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights into Imaging. 2018;9(4):611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:13126034. 2013. [Google Scholar]
- 54.Kuo B-I, Chang W-Y, Liao T-S, et al. Keratoconus Screening Based on Deep Learning Approach of Corneal Topography. Transl Vis Sci Technol 2020;9(2):53–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selvaraju RR, Cogswell M, Das A, et al. , editors. Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision; 2017. [Google Scholar]
- 56.Medeiros FA, Jammal AA, Thompson AC. From Machine to Machine: An OCT-Trained Deep Learning Algorithm for Objective Quantification of Glaucomatous Damage in Fundus Photographs. Ophthalmology. 2019;126(4):513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chetoui M, Akhloufi MA. Explainable end-to-end deep learning for diabetic retinopathy detection across multiple datasets. J Med Imaging (Bellingham). 2020;7(4):044503. ** An example of implementing interpretability for DR detection from retinal fundus images. The authors utilize a heatmap visualization approach to check that the model focuses on clinically relevant features for diagnosis, and to analyze misclassification cases.
- 58.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172(5):1122–31.e9. [DOI] [PubMed] [Google Scholar]
- 59.Grassmann F, Mengelkamp J, Brandl C, et al. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology. 2018;125(9):1410–20. [DOI] [PubMed] [Google Scholar]
- 60.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nature Biomedical Engineering. 2018;2(3):158–64. [DOI] [PubMed] [Google Scholar]
- 61.Cai CJ, Winter S, Steiner D, et al. “Hello AI”: Uncovering the Onboarding Needs of Medical Practitioners for Human-AI Collaborative Decision-Making. Proc ACM Hum-Comput Interact. 2019;3(CSCW):Article 104. [Google Scholar]
- 62. Tschandl P, Rinner C, Apalla Z, et al. Human–computer collaboration for skin cancer recognition. Nature Medicine. 2020;26(8):1229–34. ** An investigation of the impact of different AI-based support strategies on clinician performance. This includes a knowledge transfer study incorporating patterns from saliency maps into medical teaching, enhancing the diagnostic accuracy of medical students.
- 63.Miller T. Explanation in artificial intelligence: Insights from the social sciences. Artificial intelligence. 2019;267:1–38. [Google Scholar]
- 64.Winkler JK, Fink C, Toberer F, et al. Association Between Surgical Skin Markings in Dermoscopic Images and Diagnostic Performance of a Deep Learning Convolutional Neural Network for Melanoma Recognition. JAMA Dermatol 2019;155(10):1135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajpurkar P, O’Connell C, Schechter A, et al. CheXaid: deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. npj Digital Medicine. 2020;3(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lloyd K. Bias amplification in artificial intelligence systems. arXiv preprint arXiv:180907842. 2018. [Google Scholar]
- 67. Gaube S, Suresh H, Raue M, et al. Do as AI say: susceptibility in deployment of clinical decision-aids. npj Digital Medicine. 2021;4(1):31. * A study of the human-AI interaction, revealing the complex effects of human biases in the acceptance of AI support.
- 68. Kitamura FC, Marques O. Trustworthiness of Artificial Intelligence Models in Radiology and the Role of Explainability. J Am Coll Radiol 2021. * An analysis on the role of explainability in human trust and integration of AI in medical practice, suggesting that trust is multifactorial, and that model transparency may be insufficient or even unnecessary in achieving this desirable end-goal.
- 69.Holm EA. In defense of the black box. Science. 2019;364(6435):26–7. [DOI] [PubMed] [Google Scholar]
- 70.Hatherley JJ. Limits of trust in medical AI. Journal of Medical Ethics. 2020;46(7):478. [DOI] [PubMed] [Google Scholar]
- 71. Arun N, Gaw N, Singh P, et al. Assessing the (un) trustworthiness of saliency maps for localizing abnormalities in medical imaging. arXiv preprint arXiv:200802766. 2020. ** A quantitative evaluation of saliency map accuracy. The authors design a series of tests to determine and compare the performance of 8 popular saliency map algorithms in localisation, sensitivity to model weights, repeatability and reproducibility, ultimately showing that none of the techniques satisfy all the criteria. This is an impactful piece, reminding readers to be cautious of implementing interpretability techniques assessed purely by subjective visual interpretation.
- 72.Adebayo J, Gilmer J, Muelly M, et al. Sanity checks for saliency maps. arXiv preprint arXiv:181003292. 2018. [Google Scholar]
- 73.Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv preprint arXiv:170208608. 2017. [Google Scholar]


