Abstract
Androgen receptor (AR) is the primary oncogenic driver of prostate cancer (PC), including aggressive castration-resistant prostate cancer (CRPC). The molecular mechanisms controlling AR activation in general and AR reactivation in CRPC remain elusive. Here we report that monoamine oxidase A (MAOA), a mitochondrial enzyme that degrades monoamine neurotransmitters and dietary amines, reciprocally interacts with AR in PC. MAOA was induced by androgens through direct AR binding to a novel intronic androgen response element of the MAOA gene, which in turn promoted AR transcriptional activity via upregulation of Shh/Gli-YAP1 signaling to enhance nuclear YAP1-AR interactions. Silencing MAOA suppressed AR-mediated PC development and growth, including CRPC, in mice. MAOA expression was elevated and positively associated with AR and YAP1 in human CRPC. Finally, genetic or pharmacologic targeting of MAOA enhanced the growth-inhibition efficacy of enzalutamide, darolutamide, and apalutamide in both androgen-dependent and castration-resistant PC cells. Collectively, these findings identify and characterize a MAOA-AR reciprocal regulatory circuit with coamplified effects in PC. Moreover, they suggest that co-targeting this complex may be a viable therapeutic strategy to treat PC and CRPC.
Significance
MAOA and AR comprise a positive feedback loop in androgen-dependent and castration-resistant prostate cancer, providing a mechanistic rationale for combining MAOA inhibition with AR-targeted therapies for prostate cancer treatment.
Keywords: monoamine oxidase A, androgen receptor, castration-resistant prostate cancer, antiandrogen drugs, YAP1, Shh/Gli
Introduction
Prostate cancer (PC) is the second commonest cancer and the fifth leading cause of cancer death in men globally (1). Androgen receptor (AR) is considered the primary oncoprotein governing PC, making AR-targeted therapy currently the principal treatment regimen in PC. Although initial response rates to androgen deprivation therapy (ADT) exceed 90%, PC eventually transitions from a hormone-dependent to a castration-resistant disease (CRPC). Most patients develop recurrent tumors 2-3 years after ADT when AR is reactivated despite the low-androgen environment, followed by fatal CRPC (2). There is an unmet clinical need for new molecularly targeted therapies to complement current AR-targeted therapy to improve survival.
PC depends exquisitely on AR activity for survival, growth and progression. In agreement with restored AR activity in CRPC, preclinical studies suggest that AR upregulation alone is sufficient to drive progression to CRPC. A significant portion of CRPCs demonstrate AR upregulation without gene amplification (3). In disease progression, AR is regulated at many levels and cooperates with other genes and signal transduction mechanisms (4). Understanding the mechanisms of AR activation in general and AR reactivation in CRPC specifically will help uncover new druggable molecular targets for rational combination strategies synergizing with AR-targeted therapy. Despite great efforts to identify and characterize genes either regulating AR (AR regulator genes) or affected by AR (AR target genes), molecular targets linking both AR regulators and AR targets for bidirectional cooperative promotion of AR effects are little studied.
Monoamine oxidase A (MAOA), a mitochondrial membrane-bound enzyme, degrades a number of biogenic and dietary monoamines and generates hydrogen peroxide (H2O2), a major source of reactive oxygen species (ROS), as a byproduct (5). We and others demonstrated that MAOA is clinically associated with PC disease progression (6-8). We showed that MAOA induces epithelial-mesenchymal transition and tumor-stromal cell interaction through a ROS-Twist1-Shh/Gli signaling axis to promote PC metastasis (7,9). However, the functional and mechanistic link between MAOA and AR in PC cells, in the AR-driven PC disease trajectory and treatment response, and in the CRPC setting remains unclear. Filling this knowledge gap will allow precise application of MAOA inhibitors already clinically used as antidepressants as a potential PC and CRPC therapy synergizing with AR-targeted therapy. This would be especially valuable given PC’s heterogeneous and variable AR status and androgen responsiveness. This study explored previously undiscovered reciprocal crosstalk between MAOA and AR that amplifies the effects of both to promote PC and CRPC.
Materials and Methods
Clinical specimens
Hormone-sensitive and castration-resistant PC tissue microarrays, including 16 prostate adenocarcinoma specimens from each disease subtype, were provided by the Biobank of Taipei General Veterans Hospital. The study was reviewed and approved by the IRB of Taipei General Veterans Hospital, and written informed consent was provided for human samples.
Cells lines
Human PC LNCaP, VCaP, 22Rv1 and human embryonic kidney 293T cell lines were obtained from American Type Culture Collection. The enzalutamide (Enz)-resistant human PC C4-2B (C4-2BENZR) cell line was generated as described previously (10). The human PC C4-2 cell line was provided by Leland W.K. Chung (Cedars-Sinai Medical Center). The human PC LAPC4 cell line was provided by Michael Freeman (Cedars-Sinai Medical Center). All cell lines were authenticated by short tandem repeat profiling, regularly tested for Mycoplasma by the MycoProbe Mycoplasma Detection Kit (R&D Systems) and used with the number of cell passages below 10.
Plasmids and reagents
A human MAOA lentiviral expression construct was generated by inserting the human MAOA coding region at EcoRI/XbaI sites in pLVX-AcGFP1-N1 vector (Clontech) containing a puromycin-resistant gene. A Dox-inducible MAOA shRNA expression construct was generated by inserting a human MAOA shRNA sequence at NheI/EcoRI sites in EZ-Tet-pLKO-Puro vector (Addgene) containing a puromycin-resistant gene as described previously (11). Primer sequences for constructing MAOA shRNA oligomers are forward 5’-CTAGCCGGATATTCTCTGTCACCAATTACTAGTATTGGTGACAGAGAATATCCGTTTTTG-3’ and reverse 5’-AATTCAAAAACGGATATTCTCTGTCACCAATACTAGTAATTGGTGACAGAGAATATCCGG-3’). A MAOA intron androgen response element (ARE) luciferase reporter construct (MAOA ARE-luc) was generated by inserting the MAOA ARE-centric intronic sequence upstream of a minimal promoter and the Firefly luciferase gene of pGL4.26 vector (Promega). Primer sequences for cloning the MAOA intronic sequence from LNCaP genomic DNA are forward 5’-AAAGGTACCTCTCCAACGTGCCAATCAGG-3’ and reverse 5’-GGGCTCGAGGCAGTTTCTCAATACTAAGCCACT-3’. Human MAOA and non-target control shRNA lentiviral particles were purchased from Sigma-Aldrich. R1881 was purchased from PerkinElmer or Sigma-Aldrich. Clorgyline, phenelzine and doxycycline were purchased from Sigma-Aldrich. Verteporfin was purchased from Santa Cruz. Cyclopamine and enzalutamide were purchased from Selleckchem. Darolutamide was purchased from MedKoo Biosciences. Apalutamide was purchased from Toronto Research Chemicals. The Supplementary Materials and Methods provides details on additional plasmids and reagents used in this study.
Biochemical analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse-transcribed to cDNA by M-MLV reverse transcriptase (Promega) following the manufacturer’s instructions. For immunoblots, cells were extracted with RIPA buffer in the presence of a protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific). Blots were performed as described previously (12). The Supplementary Materials and Methods provides details on primary antibodies used for immunoblots. Nuclear and cytoplasmic extracts used for immunoblots were prepared with a NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific). PSA levels in cell culture media or mouse sera were quantified by ELISA (GenWay Biotech or Enzo Life Sciences).
Site-directional mutational analysis of MAOA intron ARE and YAP1 promoter
Site-directed mutagenesis was used to mutate or delete the MAOA ARE cloned in the pGL4.26 vector and mutate the Gli-binding site (GliBS) identified in the 1.6-kb YAP1 promoter, with wild-type (WT) luciferase reporter constructs used as templates. Mutagenesis was carried out by QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) following the manufacturer’s instructions. Primer sequences used for mutating or deleting the MAOA ARE were 5’-GCACGGTTCCAGGGAAATTGCGTTCTGCTTG-3’ (Mut 1), 5’-CACGGTTCCAGGGACATTGCATTTTGCTTGACATAAACAATTTC-3’ (Mut 2), and 5’-GCAGAAATTGTTTATGTCAAGGAACCGTGCCCCAAAACA-3’ (Del), with mutated nucleotides underlined. Primer sequence used for mutagenesis of YAP1 promoter was 5’-AGGGATAGCAGGGGTAGGGTGGGAGCTCCTTGAGGATGAAAG-3’ (mutated nucleotides underlined). Mutated or deleted nucleotides were verified by DNA sequencing.
Chromatin immunoprecipitation (ChIP)-qPCR assays
ChIP-qPCR assays were used to determine the association of endogenous AR protein with a MAOA ARE in LNCaP cells grown in phenol red-free medium containing 5% CSS for 72 hours and then treated with R1881 or ethanol for another 24 hours, endogenous AR protein with two known AREs of the PSA and FKBP5 genes in LNCaP cells (shCon and shMAOA), and endogenous Gli1 and Gli2 proteins with a GliBS in YAP1 promoter in LNCaP cells (shCon and shMAOA) by a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) following the manufacturer’s instructions. Briefly, the chromatin was crosslinked with nuclear proteins, enzymatically digested with micrococcal nuclease followed by sonication, and immunoprecipitated with anti-AR (PG-21, Millipore, RRID: AB_310214; or D6F11, Cell Signaling, RRID: AB_10691711), anti-Gli1 (H-300, Santa Cruz, RRID: AB_2111764), or anti-Gli2 (Cat# ab26056, Abcam, RRID: AB_2111901) antibody. Normal IgG included in the kit was used as a negative control for IP. The immunoprecipitates were pelleted with agarose beads, purified, and subjected to qPCR with primers specifically targeting the ARE-centric MAOA, PSA and FKBP5 genomic sequences or the GliBS-centric YAP1 promoter region. Details on primers used for qPCR are provided in Supplementary Materials and Methods.
Proximity ligation assay (PLA)
Cells were seeded on chamber slides and fixed with 4% formaldehyde for 10 min at room temperature (RT), washed twice with PBS containing 0.02% Tween 20, and permeabilized with 0.5% Triton X-100/PBS solution (blocking solution) for 30 min at RT. Primary antibodies against AR (N-20, rabbit IgG, Santa Cruz, RRID: AB_1563391) or YAP1 (63.7, mouse IgG, Santa Cruz, RRID: AB_1131430) were incubated in blocking solution at 4°C overnight. Assay was then performed with the Duolink In Situ Red Starter Kit Mouse/Rabbit (Duolink, Sigma-Aldrich) according to the manufacturer’s instructions using anti-mouse MINUS and anti-rabbit PLUS PLA probes (Duolink). Images were acquired by a Nikon Ti-E inverted microscope or a Zeiss Axio Imager M2 upright microscope using a x40 objective and analyzed for fluorescence per nucleus with inForm (PerkinElmer) or HALO (Indica Labs) software.
Animal studies
All animal studies received prior approval from the Washington State University IACUC and complied with IACUC recommendations. Male 4- to 6-week-old SCID, SCID/beige and NSG mice were purchased from Envigo or Jackson Laboratory and housed in the animal research facility at Washington State University. To determine MAOA’s effect on AR-dictated PC development and growth, 4 x 106 LNCaP cells expressing Dox-inducible MAOA shRNA were mixed 1:1 with Matrigel (BD Biosciences) for bilateral subcutaneous injection into SCID/beige mice. One week after tumor inoculation, mice were randomized into 2 groups (20 mice/group) and fed a diet with 625 mg/kg Dox (Dox+, Envigo) or a normal diet (Dox-) ad libitum. Two weeks later, tumor-bearing mice of both groups were randomly separated into 2 sub-groups to undergo surgical castration or not, followed by tumor size measurement by caliper 3 times a week. To determine MAOA’s effect on CRPC development and growth, 4 x 106 C4-2BENZR or 22Rv1 cells, both expressing Dox-inducible MAOA shRNA, were mixed 1:1 with Matrigel and bilaterally injected subcutaneously into NSG or SCID mice respectively. Mice implanted with C4-2BENZR cells were orally administered with Enz (10 mg/kg) every other day after tumor inoculation, while mice implanted with 22Rv1 cells received castration 2 weeks prior to tumor inoculation. One week after tumor inoculation, mice were randomly divided into 2 groups (12 mice/group or 7 mice/group for C4-2BENZR or 22Rv1 cells respectively) and fed a Dox+ or a Dox− diet. Tumor size was measured every other day by caliper after the formation of palpable tumors. Tumor volume was calculated by as length x width2 x 0.52. At the endpoints, tumors were dissected and weighed. Tumor samples and mouse sera were collected for subsequent biochemical and immunohistochemical analyses.
Immunohistochemical (IHC) and quantum dot (QD) labeling analysis
IHC analysis of xenograft and clinical tumor samples was performed using antibodies against MAOA (H-70, Santa Cruz, RRID: AB_2137260), YAP1 (63.7, Santa Cruz), AR (N-20, Santa Cruz), Ki-67 (D2H10, Cell Signaling, RRID: AB_2636984) or cleaved caspase 3 (Asp175, Cell Signaling, RRID: AB_2341188) following a published protocol (7). Cell-based IHC staining intensity and percentage of positive expression for individual proteins were analyzed by HALO software. MAOA IHC staining in human tissue microarrays was scored by a semi-quantitative method taking into account both staining intensity (I) and quantity based on the proportion of tumor cells stained (q) to obtain a final score defined as the product of I x q in the range of 0-12 as described previously (7). All scoring was performed by a pathologist. The IHC staining protocol was modified for double QD labeling as described previously (13). The human CRPC tissue microarray was stained with antibodies against MAOA (H-70, Santa Cruz) and YAP1 (63.7, Santa Cruz) sequentially by single QD labeling. Cell-based averages of QD intensity counts for MAOA and nuclear YAP1 expression in each sample were analyzed by inForm software after areas of interest were defined using manual tissue segmentation by a pathologist.
Bioinformatics analysis
The human PC datasets used for co-expression correlation studies were downloaded from the Oncomine database by licensed access or the cBioPortal for Cancer Genomics database. For analysis of ChIP-seq datasets GSE43720, GSE55062 and GSE65478 available in Gene Expression Omnibus database, Bowtie was used to map the human hg19 genome and unique mapped reads were used for peak calling, using MACS2 to perform the peak calling and ChIPseeker for peak annotation.
Statistics
Data are presented as the mean ± SEM as indicated in figure legends. Comparisons were analyzed by unpaired 2-tailed Student’s t test. Correlations were determined by Pearson correlation. A p value less than 0.05 was considered statistically significant.
Results
MAOA reciprocally interacts with AR in PC cells
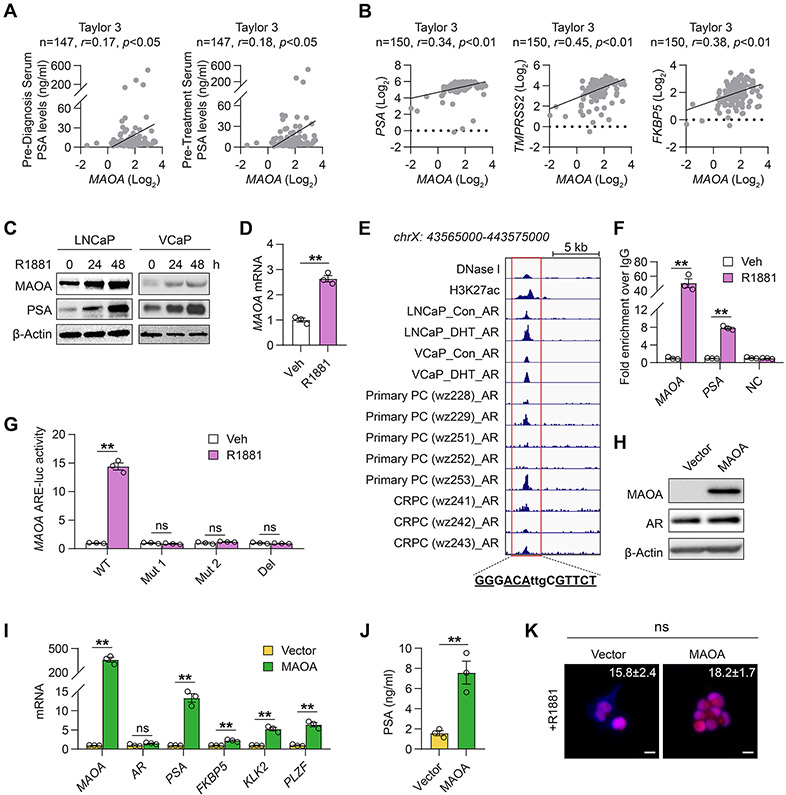
To seek initial evidence of MAOA-AR crosstalk in PC, we analyzed multiple clinical datasets for a MAOA association with AR. We found that MAOA mRNA expression was positively correlated with pre-diagnosis and pre-treatment serum PSA levels in the Taylor 3 dataset (14) (Fig. 1A). MAOA was also positively co-expressed with three bona fide AR target genes, including PSA, TMPRSS2 and FKBP5, at the transcript level in the same dataset (Fig. 1B), as corroborated by a similar mRNA co-expression pattern from additional publicly available datasets (Supplementary Table S1).
Figure 1. MAOA reciprocally interacts with AR in PC cells.
(A, B) Pearson correlation analysis of MAOA with pre-diagnosis/-treatment serum PSA levels (A) or PSA, TMPRSS2 and FKBP5 (B) in the Taylor 3 dataset. (C) Western blot of MAOA and PSA upon R1881 stimulation (10 nM) at indicated times in LNCaP and VCaP cells. (D) qPCR of MAOA by R1881 (10 nM, 24 hrs) in LNCaP cells (n=3). (E) Genomic browser representation of DNase I, H3K27ac, and AR binding in ARE-centric MAOA intron 3, with the nucleotides identical to the canonical ARE underlined, in GSE43720, GSE55062 and GSE65478. (F) ChIP-qPCR of AR occupancy at the MAOA and PSA AREs and an irrelevant sequence (NC) by R1881 (10 nM, 24 hrs) in LNCaP cells. Fold enrichment of AR was normalized to IgG in each group (n=3). (G) Determination of MAOA ARE-luc activity in WT, mutated (Mut1 and Mut2) or deleted (Del) forms by R1881 (10 nM, 24 hrs) in LNCaP cells. (H) Western blot of MAOA and AR in control and MAOA-OE LAPC4 cells. (I) qPCR of MAOA, AR and AR target genes in control and MAOA-OE LAPC4 cells (n=3). (J) ELISA of PSA secretion in culture media from control and MAOA-OE LAPC4 cells (n=3). (K) Representative nuclear AR staining and quantification of per-nucleus intensity in control (n=33) and MAOA-OE (n=91) LAPC4 cells upon R1881 stimulation (10 nM, 6 hrs). Scale bars: 20 μm. Data represent the mean ± SEM. *p<0.05, **p<0.01; ns, not significant.
To understand the regulatory relationship between MAOA and AR, we first investigated whether AR controls MAOA in PC cells. The androgen-dependent LNCaP and castration-resistant VCaP human PC cell lines, which both express AR and respond to androgens, showed time-dependently increased MAOA and PSA protein expression under R1881 synthetic androgen treatment (Fig. 1C), and a 1.5-fold increase of MAOA mRNA expression by R1881 in LNCaP cells (Fig. 1D), suggesting androgenic regulation of MAOA at the transcriptional level. To determine whether AR directly binds to the MAOA gene locus for transcriptional activation, we analyzed three ChIP-seq datasets with subjects assembled from cultured human PC cells and clinical prostate tumors. We found enriched AR occupancy at a site downstream from the transcription start site (TSS) of MAOA across most samples. Examining the AR-bound sequence, we further identified a consensus androgen response element (ARE), GGGACAttgCGTTCT (+53,107~+53,121 with the MAOA TSS set as +1) in MAOA intron 3 with high homology (10 out of 12 bp) with the canonical ARE GGT/AACAnnnTGTTCT for AR binding (15) (Fig. 1E). ChIP-qPCR assays validated direct AR interaction with this sequence, which showed significant AR association with the intronic ARE of MAOA gene as well as a known ARE in the distal enhancer of PSA gene in LNCaP cells upon R1881 stimulation, paralleled by minimal AR binding at both AREs in the absence of R1881. An irrelevant genomic sequence used as a negative control demonstrated no AR occupancy regardless of R1881 induction (Fig. 1F). To determine whether the MAOA ARE is functional, we inserted the corresponding MAOA ARE-centric intron sequence upstream of the minimal promoter-driven luciferase gene to construct a MAOA ARE-luc reporter. Compared to the WT MAOA ARE-luc notably inductive to R1881, mutation of select nucleotides in the ARE (Mut 1 and Mut 2) or deletion of the ARE (Del) made the reporter no longer responsive to R1881 (Fig. 1G).
Next, we examined whether MAOA influences AR in PC cells. Stably enforced expression of MAOA in AR-positive androgen-responsive human PC LAPC4 cells with low levels of MAOA resulted in elevated expression of MAOA at both the protein and mRNA levels (Figs. 1H and 1I). Forced MAOA expression led to mRNA activation of several AR target genes (Fig. 1I) and a more than 4-fold increase in PSA protein secretion (Fig. 1J). Interestingly, MAOA overexpression (OE) caused no changes in AR expression at the protein and mRNA levels (Figs. 1H and 1I), or the extent of AR nuclear translocation upon R1881 stimulation, a regulatory mechanism necessary for AR activation, as examined by quantitating nuclear AR staining levels (Fig. 1K).
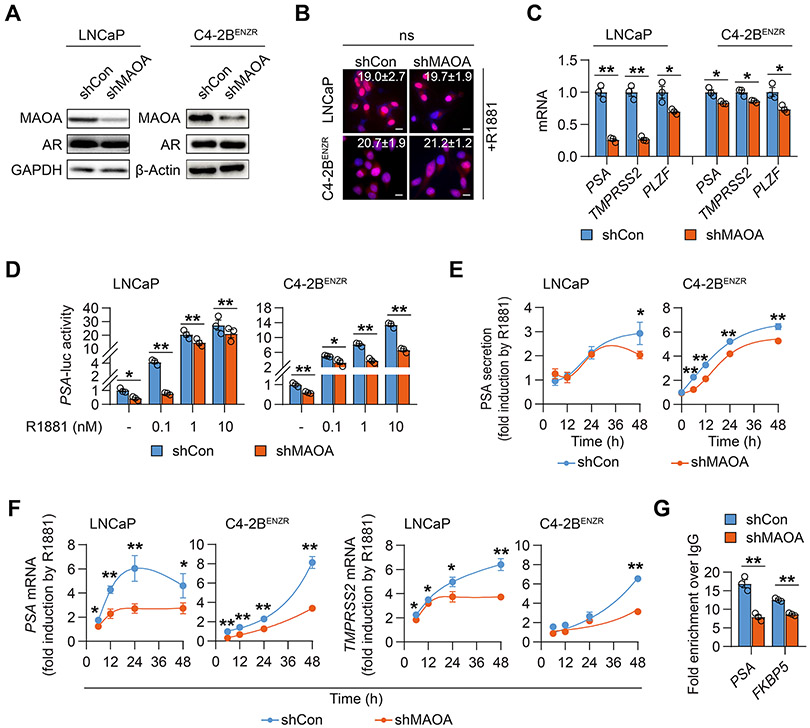
Conversely, we stably silenced MAOA expression with a shRNA in both LNCaP and castration-resistant, enzalutamide (Enz)-resistant C4-2B (C4-2BENZR) human PC cells. The C4-2BENZR cell line was established by chronic exposure of LNCaP-derived human CRPC C4-2B cells to Enz, a second-generation antiandrogen drug for CRPC, at gradually increasing doses to develop resistance (10). Consistent with observations in MAOA-OE LAPC4 cells, MAOA knockdown (KD) did not affect AR protein expression or the extent of AR nuclear translocation by R1881 in either LNCaP and C4-2BENZR cells (Figs. 2A and 2B) or 2 additional AR-positive human CRPC cell lines, C4-2 and 22Rv1 (Supplementary Fig. S1A). In contrast, MAOA KD prevalently decreased the mRNA levels of a panel of AR target genes, including PSA, TMPRSS2 and PLZF, in LNCaP and C4-2BENZR cells (Fig. 2C). In the absence or presence of various doses of R1881, MAOA KD reduced the activity of an AR-dependent luciferase reporter (PSA-luc), an androgen-responsive PSA enhancer-promoter fused sequence placed upstream of the luciferase gene, in LNCaP and C4-2BENZR cells (Fig. 2D). Moreover, MAOA KD attenuated the R1881 responsiveness of PSA and TMPRSS2 at the protein secretion and/or mRNA levels within 48 hours in LNCaP and C4-2BENZR cells (Figs. 2E and 2F). Similarly, MAOA silencing repressed induction of PSA protein secretion and PSA/TMPRSS2 mRNA by R1881 in C4-2 cells as well as the response of KLK2 mRNA to R1881 in 22Rv1 cells where KLK2 was reportedly more androgen responsive than PSA in 22Rv1 cells (16) (Supplementary Figs. S1B-S1D). ChIP-qPCR assays then demonstrated lower AR occupancy at individual AREs of PSA and FKBP5 gene loci in MAOA-KD LNCaP cells compared to controls (Fig. 2G). Collectively, these findings imply reciprocal crosstalk between MAOA and AR in both androgen-dependent and castration-resistant PC cells.
Figure 2. MAOA silencing suppressed AR transcriptional activity in PC cells.
(A) Western blot of MAOA and AR in control (shCon) and MAOA-KD (shMAOA) LNCaP and C4-2BENZR cells. (B) Representative nuclear AR staining and quantification of per-nucleus intensity in the indicated control (LNCaP, n=160; C4-2BENZR, n=663) and MAOA-KD (LNCaP, n=154; C4-2BENZR, n=760) cells upon R1881 stimulation (10 nM, 6 hrs). Scale bars: 20 μm. (C) qPCR of AR target genes in the indicated control and MAOA-KD cells (n=3). (D) Determination of PSA-luc activity by R1881 at indicated concentrations for 24 hrs in the indicated control and MAOA-KD cells (n=3). (E) ELISA of time-dependent fold induction of PSA by R1881 (10 nM) in the indicated control and MAOA-KD cells (n=3). (F) qPCR of time-dependent fold induction of PSA and TMPRSS2 by R1881 (10 nM) in the indicated control and MAOA-KD cells (n=3). (G) ChIP-qPCR of AR occupancy at PSA and FKBP5 AREs in control and MAOA-KD LNCaP cells. Fold enrichment of AR was normalized to IgG in each group (n=3). Data represent the mean ± SEM. *p<0.05, **p<0.01; ns, not significant.
MAOA activates AR-interacting YAP1 in a Gli1/2-dependent manner
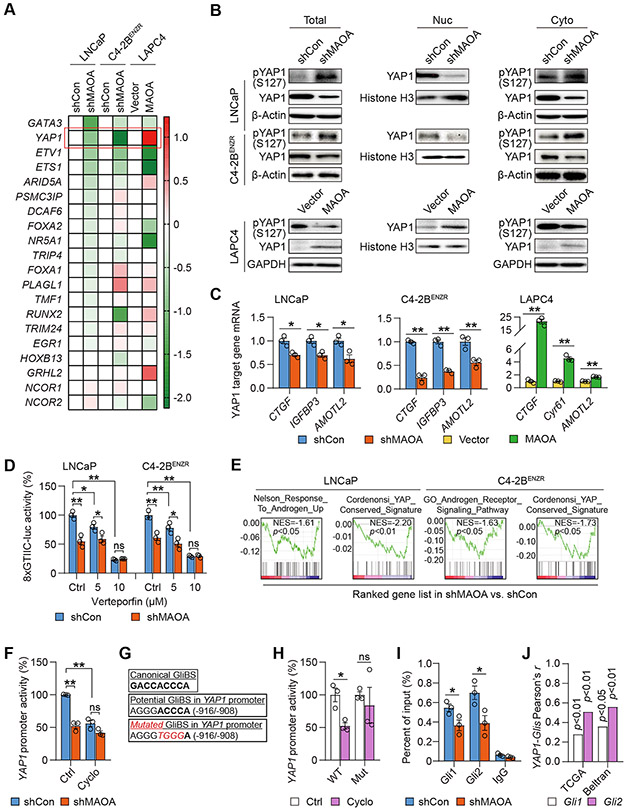
Since MAOA activates AR transcriptional activity without affecting AR mRNA/protein expression and nuclear translocation, we speculated that MAOA may regulate AR through alternative mechanisms. We first investigated whether MAOA induces the expression levels of AR-interacting transcription factors and cofactors to modulate AR effects. We conducted qPCR-based screening of a group of transcription factors and cofactors known as nuclear AR interactors controlling AR transcriptional activity in MAOA-manipulated cells (17). We identified YAP1 as the top candidate because of its relatively larger differences in expression levels between control and MAOA-manipulated cells in each cell pair as well as its consistent pattern of expression-level changes in response to MAOA across all cell pairs compared to the other genes (Fig. 3A). YAP1, a transcriptional coactivator and Hippo pathway effector, was recently reported to physically interact with AR to promote PC and CRPC growth and invasion (18-20). Using both MAOA-KD and -OE cell lines, we showed that MAOA increased YAP1 protein expression while decreasing phospho-YAP1 (Ser127) protein expression in whole cell lysates, congruent with the observations that MAOA caused higher YAP1 and lower phospho-YAP1 protein levels in nuclear and cytoplasmic fractions respectively (Fig. 3B; Supplementary Fig. S2A). Immunofluorescence assays also revealed less YAP1 protein accumulation in the nucleus in response to MAOA inactivation in C4-2BENZR cells (Supplementary Fig. S2B). We found reduced mRNA expression of multiple YAP1 target genes (CTGF, IGFBP3 and AMOTL2) in MAOA-KD LNCaP and C4-2BENZR cells compared to controls, which corresponded to elevated mRNA expression of several YAP1 target genes (CTGF, Cyr61 and AMOTL2) in MAOA-OE LAPC4 cells relative to controls (Fig. 3C). Using the 8xGTIIC luciferase reporter in which luciferase expression is driven by a YAP1-responsive synthetic promoter, we demonstrated a decrease of reporter activity when MAOA was silenced in both LNCaP and C4-2BENZR cells. This decrease weakened until abolished dose-dependently when MAOA-KD cells were treated with verteporfin, a small-molecule inhibitor of YAP1 (21), suggesting the specificity of this reporter to YAP1 (Fig. 3D). Bioinformatics analysis further indicated downregulation of androgen-responsive/AR-dependent and YAP1-directed gene signatures enriched in MAOA-KD LNCaP and C4-2BENZR cells compared to controls using RNA-seq coupled with gene set enrichment analysis (GSEA) (Fig. 3E). Importantly, MAOA and YAP1 demonstrated a significant positive co-expression correlation in multiple PC clinical datasets (Supplementary Fig. S2C).
Figure 3. MAOA activates AR-interacting YAP1 in a Gli1/2-dependent manner.
(A) qPCR-based heatmap depicting differential expressions of AR-interacting transcription factors and cofactors in the indicated cell pairs. Log2 scale was used to indicate relative gene expression from an average of 3 replicates, with gene expression in controls set as 1. (B) Western blot of YAP1 and phospho-YAP1 (S127) in total cell lysates along with nuclear and cytoplasmic fractions of the indicated cells. (C) qPCR of YAP1 target genes in the indicated control and MAOA-KD cells (n=3). (D) Determination of YAP1-responsive 8xGTIIC-luc activity by verteporfin (24 hrs) in the indicated control and MAOA-KD cells (n=3). (E) GSEA of the indicated gene sets for the comparisons of MAOA-KD versus control LNCaP and C4-2BENZR cells. (F) Determination of YAP1 promoter activity by cyclopamine (20 μM, 48 hrs) in control and MAOA-KD LNCaP cells (n=3). (G) Sequences of the canonical GliBS (top), a putative GliBS in YAP1 promoter (middle), and introduced point mutations (bottom, italic and red) to inactivate the GliBS, with the YAP1 TSS set as +1. (H) Determination of WT and mutant (Mut) YAP1 promoter activity by cyclopamine (20 μM, 48 hrs) in 293T cells (n=3). (I) ChIP-qPCR of Gli1 and Gli2 occupancy at the GliBS-centric YAP1 promoter in control and MAOA-KD LNCaP cells (n=3). (J) Pearson correlation analysis of YAP1-Gli1/Gli2 in the TCGA (n=498) and Beltran (n=49) datasets. Data represent the mean ± SEM. *p<0.05, **p<0.01; ns, not significant.
In addition to the qPCR array approach, we also examined whether MAOA regulates the assembly and stability of AR complexes, especially through the AR-interacting proteins affected by MAOA, likely more for the degree of AR binding than their expression levels. We carried out proteomic analysis of AR-bound proteins with quantitation of their AR-binding affinity in control and MAOA-KD LNCaP cells. Mass spectrometry following AR pull-down revealed 491 AR-bound proteins expressed in the nucleus and enriched in LNCaP cells, which were narrowed down to 33 high-confidence hits with differential degrees of AR binding between control and MAOA-KD cells by statistical analysis. Literature mining further filtered out a PC growth-activating protein (LMNA) with lower binding affinity to AR and 5 PC growth-repressed proteins (PDCD4, ENDOD1, DHRS7, FUS and HDAC1) with higher binding affinity to AR in MAOA-KD cells compared to controls (Supplementary Fig. S3A; Supplementary Dataset S1) (22-27). To determine whether these proteins mediated MAOA’s effect on AR, we first showed that their corresponding gene expression levels were barely changed in MAOA-KD LNCaP cells compared to controls (Supplementary Fig. S3B). Surprisingly, we found that after individual KD none of these genes affected several canonical AR target gene expressions or PSA protein secretion in LNCaP cells (Supplementary Figs. S3C-S3E), implying that MAOA is not likely to regulate AR activity by modulating the interactions of these proteins with AR. In addition, we demonstrated the negligible effect of MAOA on AR protein stability by similar AR protein levels in response to cycloheximide to inhibit protein synthesis or MG132 to inhibit protein degradation in control and MAOA-KD LNCaP cells (Supplementary Figs. S4A-S4C). Based on these findings, we decided to focus on YAP1 as a possible mediator of MAOA’s action on AR.
Next, we sought to find out how MAOA activates YAP1 in PC cells. Given the induction of YAP1 mRNA by MAOA, we surmised that MAOA might regulate YAP1 at the transcriptional level and investigated the underlying mechanism. We previously demonstrated, using AR-negative PC-3 cells as the principal model, that MAOA elicits a ROS-Twist1-Shh/Gli signaling cascade to promote PC metastasis (7,9). Mechanistically, MAOA generates ROS via oxidative deamination to stabilize HIF1α protein and subsequently induce the VEGF-A-mediated AKT/FOXO1 pathway, resulting in the nuclear export of transcription repressor FOXO1 to activate Twist1 transcription and gene expression (7). In turn, Twist1 upregulates the transcription of Shh through direct interaction with an E-box on the Shh promoter (9). Subsequent to binding to transmembrane protein Ptch1 and relieving the downstream depression of SMO, Shh activates Gli1 and Gli2 transcription factors, facilitating Gli translocation to the nucleus and occupancy at target gene promoters for transactivation (28). These findings led us to speculate that Shh/Gli signaling might be a candidate mechanism for MAOA’s transcriptional activation of YAP1, which is coincidentally supported by a recent study revealing that hedgehog pathway enhances YAP1 at both the mRNA and protein levels during liver regeneration (29).
To prove this idea, we first examined whether MAOA could induce the ROS-Twist1-Shh/Gli signaling axis in AR-positive androgen-dependent or castration-resistant PC cells as MAOA did in AR-negative PC-3 cells. We demonstrated MAOA’s ability to promote intracellular ROS production, Twist1 and Twist1’s upstream regulators (HIF1α, AKT and FOXO1) in AR-expressing cells (Supplementary Figs. S5A-S5C). Moreover, we observed that the antioxidant N-acetylcysteine (NAC) decreased MAOA-induced Twist1 expression in LAPC4 cells while addition of H2O2, the byproduct released from MAOA-mediated enzymatic reactions, diminished Twist1 repression caused by MAOA silencing in LNCaP and C4-2BENZR cells, suggesting ROS-dependent MAOA activation of Twist1 in AR-positive cells (Supplementary Fig. S5D). Then we assessed whether MAOA could further induce Shh/Gli signaling via ROS and Twist1 in AR-positive cells. To this end, we modulated the intracellular ROS and Twist1 expression levels in MAOA-manipulated AR-expressing cells, and examined Shh and Gli1 (a direct transcriptional target of Gli1 itself) expression as well as Gli-luc reporter activity, in which 8 copies of the Gli-binding site (GliBS) upstream of the luciferase gene drive luciferase expression to indicate Gli transcriptional activity. We showed that NAC treatment or siRNA-mediated Twist1 KD reduced MAOA-induced Shh/Gli1 expression and Gli-luc activity in LAPC4 cells, while H2O2 treatment or forced expression of Twist1 restored Shh/Gli1 levels and Gli-luc activity in MAOA-KD LNCaP and C4-2BENZR cells (Supplementary Figs. S6A-S6C). We also found that Twist1 is positively co-expressed with Shh, Gli1 and Gli2 at the transcript level in clinical samples from the Taylor 3 dataset (Supplementary Fig. S6D). Collectively, these data as in line with our prior findings reveal that MAOA upregulates Shh/Gli signaling through ROS and Twist1 in AR-positive androgen-dependent or castration-resistant PC cells.
Since MAOA induced Shh mRNA expression and Gli-luc activity in multiple AR-positive PC cell lines (Supplementary Figs. S7A and S7B), we then determined whether Shh/Gli signaling mediates MAOA’s control of YAP1 transcription by directly regulating YAP1 promoter. We used a human 1.6-kb YAP1 promoter Gaussia luciferase reporter with concurrent expression of secreted alkaline phosphatase (SEAP) as internal control. We observed a 48% decrease of normalized YAP1 promoter activity in MAOA-KD LNCaP cells compared to controls, which was diminished upon cyclopamine treatment, an inhibitor of SMO for blockade of Shh/Gli signaling (Fig. 3F). After testing a series of truncated YAP1 promoter reporter constructs, we found the middle ~500 bp of the 1.6-kb promoter most responsive to MAOA silencing. Examining the ~500-bp promoter sequence, we identified a consensus GliBS with the nucleotides of the half-site identical to the canonical ones (30) (Fig. 3G). We generated a YAP1 promoter reporter construct with a mutant GliBS and found that the mutated (Mut) reporter was no longer suppressed by cyclopamine, unlike its WT counterpart which showed a 47% decrease by cyclopamine (Fig. 3H). ChIP-qPCR assays then demonstrated 33% and 44% lower association of endogenous Gli1 and Gli2 proteins with the GliBS respectively after MAOA KD in LNCaP cells compared with controls (Fig. 3I). There was a significant positive correlation between YAP1 and Gli1/Gli2 mRNA expression in both the TCGA primary PC and the Beltran CRPC datasets (31), as clinical evidence supporting Gli-dependent MAOA activation of YAP1 expression (Fig. 3J; Supplementary Fig. S7C). These data in aggregate indicate MAOA’s ability to induce YAP1 through downstream ROS/Twist1-dependent activation of Shh/Gli signaling for direct Gli1/2 interaction with a GliBS in YAP1 promoter.
MAOA promotes AR transcriptional activity by enhancing nuclear YAP1-AR interaction
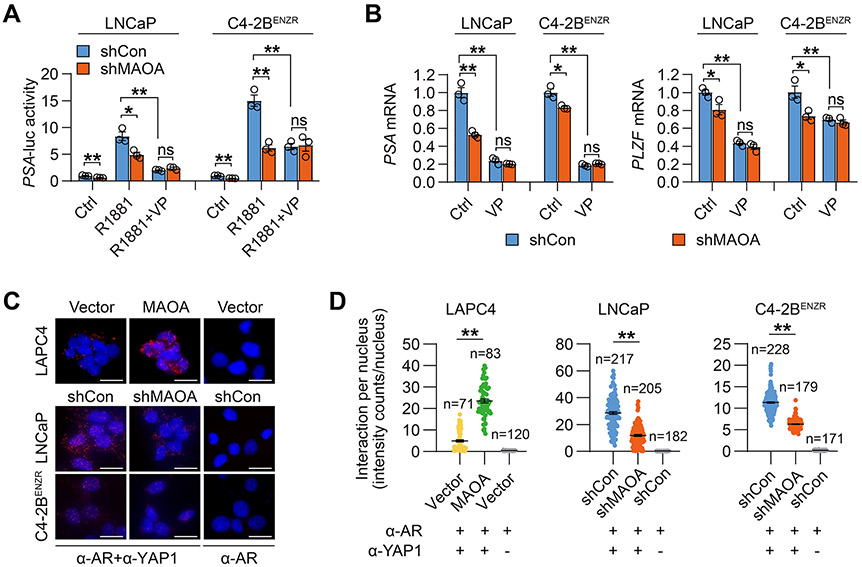
Investigating whether YAP1 mediates MAOA’s effect on AR transactivation, we showed 41% and 59% decreases of R1881-induced PSA-luc activity in LNCaP and C4-2BENZR cells respectively, where MAOA expression was silenced compared with controls. These decreases were abolished by YAP1 inhibition through verteporfin (Fig. 4A). A similar pattern of PSA and PLZF mRNA expression in the absence or presence of verteporfin was also observed in MAOA-KD LNCaP and C4-2BENZR cells compared with controls (Fig. 4B). Next, we examined the direct YAP1-AR interaction, reportedly the mechanism by which YAP1 modulates AR activity (19). In situ proximity ligation assay visualized endogenous YAP1-AR protein complexes in both the nucleus and cytoplasm of cells, in agreement with the fact that YAP1 and AR can reside in the nucleus as well as the cytoplasm, making their interaction possible in both compartments. Quantitative analysis of the fluorescence restricted to the nucleus revealed a 3.8-fold increase in nuclear YAP1-AR interaction in MAOA-OE LAPC4 cells and a 59% and 44% reduction in MAOA-KD LNCaP and C4-2BENZR cells respectively, compared to controls. Parallel incubation of an AR antibody only as a negative control in the assay demonstrated undetectable fluorescence in all cell lines (Figs. 4C and 4D). A co-immunoprecipitation assay also revealed less YAP1 bound with AR protein in MAOA-KD LNCaP whole cell lysates compared to controls (Supplementary Fig. S8).
Figure 4. MAOA promotes nuclear YAP1-AR interaction for enhanced AR transcriptional activity.
(A) Determination of PSA-luc activity by R1881 (10 nM, 24 hrs) ± verteporfin (5 μM, 24 hrs) in the indicated control and MAOA-KD cells (n=3). (B) qPCR of PSA and PLZF by verteporfin (5 μM, 24 hrs) in the indicated control and MAOA-KD cells (n=3). (C) Representative PLA staining of YAP1-AR interaction in the indicated cell pairs. AR antibody incubation alone served as negative control. Scale bars: 20 μm. (D) Quantitation of nuclear YAP1-AR interaction by per-nucleus fluorescence intensity in the indicated cell pairs. Data represent the mean ± SEM. *p<0.05, **p<0.01; ns, not significant.
Silencing MAOA suppressed AR-dictated prostate tumor development and growth
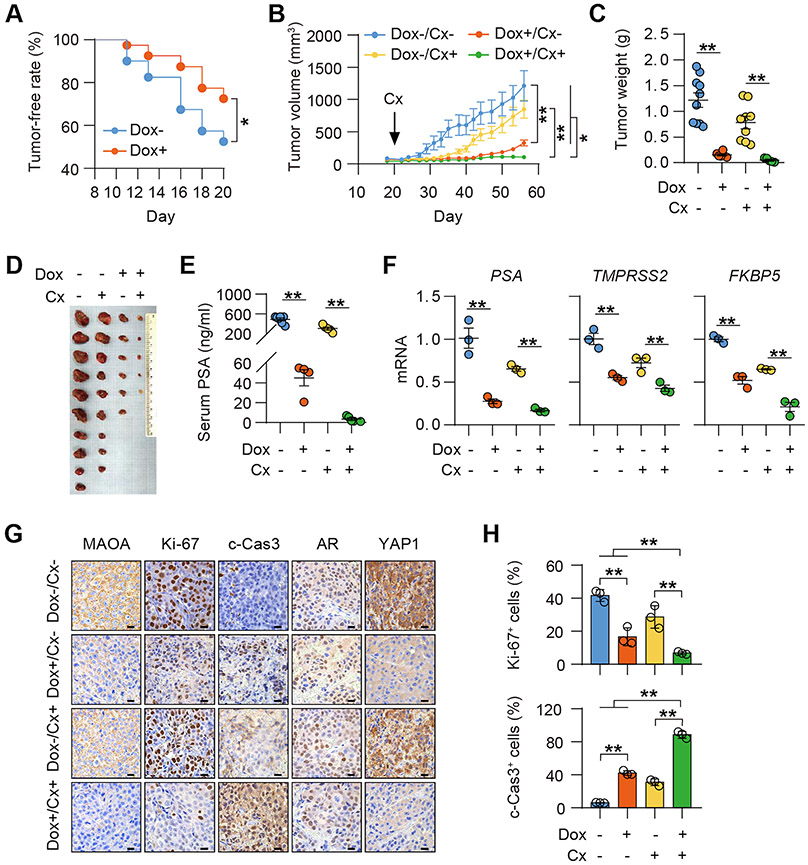
We demonstrated the necessity of MAOA for maintaining AR activity in PC cultures, which sheds light on MAOA’s supportive role in AR-dictated prostate tumor behavior in vivo, including castration-resistant tumors where functional reactivated AR provides survival and growth signals (32). To test this idea, we established multiple PC xenograft mouse models to assess MAOA’s function in controlling AR-governed PC and CRPC development and growth. Our findings of MAOA’s effect on AR signaling suggested that suppressing MAOA might reduce the viability of AR-dependent xenografts pre-injection, rendering baseline growth inequivalent at the starting point. This is especially likely based on previous studies, including ours, indicating MAOA’s ability to promote AR-dependent PC cell growth (7,8,33). To address this concern, we generated a Dox-dependent inducible MAOA shRNA expression construct and stably expressed it in LNCaP, C4-2BENZR and 22Rv1 cells. We observed a notable reduction of MAOA protein expression as well as cell growth upon Dox stimulation consistently across all cell lines, validating the efficacy of induced MAOA KD (Supplementary Figs. S9A and S9B).
To examine the effect of MAOA on AR-directed prostate tumor development and growth, we subcutaneously implanted LNCaP cells stably expressing Dox-inducible MAOA shRNA into mice. One week after inoculation, mice were randomly separated into 2 groups and given either a Dox-containing (Dox+) diet or a normal (Dox−) diet. Mice fed a Dox+ diet leading to induced tumor MAOA KD formed fewer tumors compared to control mice over a 2-week observation period after Dox administration (Fig. 5A). Two weeks after treatment, tumor-bearing mice from both groups were further randomized into 2 sub-groups to receive surgical castration (Cx+) or not (Cx−). Castration stopped tumor growth for 2-3 weeks followed by regrowth of castration-resistant tumors (Fig. 5B). Dox-induced tumor MAOA KD markedly slowed tumor growth, evidenced by smaller tumor volumes and lower tumor weights and serum PSA levels at the endpoint, in both castrated and intact mice compared to controls (Figs. 5B-5E). Strikingly, silencing tumor MAOA expression suppressed tumor growth more dramatically than castration, implying the superiority of MAOA ablation over castration for limiting AR signaling. Combining castration and tumor MAOA inactivation retarded tumor growth the most among all groups and nearly halted the growth of relapsed LNCaP tumors after castration in mice (Fig. 5B). Even more appealing, further quantitative analysis revealed that MAOA silencing resulted in a significantly greater fold reduction in average endpoint tumor volume, tumor weight and serum PSA level in castrated mice compared to intact mice (Supplementary Figs. S10A-S10C), suggesting MAOA’s particular importance in regulating CRPC tumor growth. Similar to in vitro findings, silencing MAOA inhibited AR activity by reduced expression of several canonical AR target genes in both hormone-naïve and castration-resistant LNCaP xenograft samples (Fig. 5F). By characterizing tumor samples, we demonstrated decreased MAOA protein expression in Dox-treated tumors in both castrated and intact mice, indicating effective and sustainable Dox-induced MAOA KD under in vivo conditions. Ki-67 staining of tumor samples revealed an average 76% and 60% drop in Ki-67+ cells in MAOA-KD tumors compared to controls in castrated and intact mice, respectively. Higher cleaved caspase-3 staining correlating to increased apoptosis was also shown in MAOA-KD tumors compared to controls regardless of castration status. Consistent with in vitro findings, we found minimal changes in the percentage of nuclear AR+ cells and nuclear AR expression, parallel with reduced nuclear YAP1 levels, in MAOA-KD tumors in both castrated and intact mice compared to controls (Figs. 5G and 5H; Supplementary Fig. S11A).
Figure 5. MAOA is essential for driving AR-dictated PC development and growth in mice.
(A) Kaplan-Meier tumor-free curves of mice inoculated with LNCaP cells expressing a Dox-inducible MAOA shRNA and fed a Dox− or a Dox+ diet (n=40). (B) Tumor growth curves of mice fed a Dox− or a Dox+ diet in combination with the state of castration (Cx+) or not (Cx−) (n=5-10, which applies to C and D). (C, D) Tumor weights (C) and anatomic tumor images (D) at the endpoint. (E) ELISA of endpoint serum PSA levels (n=4-7). (F) qPCR of indicated genes in tumor samples (n=3). (G) Representative MAOA, Ki-67, cleaved caspase 3 (c-Cas3), AR and YAP1 IHC staining in tumor samples. Scale bars: 20 μm. (H) Quantification of % of Ki-67+ and c-Cas3+ cells in tumor samples (n=3). Data represent the mean ± SEM. *p<0.05, **p<0.01.
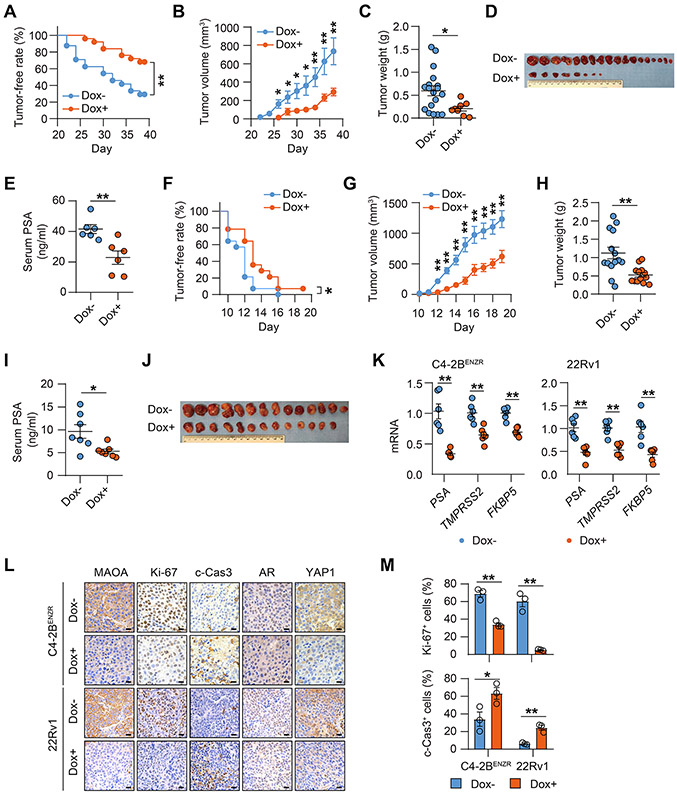
Next, we used two CRPC cell lines, C4-2BENZR and 22Rv1, both stably expressing Dox-inducible MAOA shRNA, to establish subcutaneous xenografts in mice to assess MAOA’s effect on CRPC tumor development and growth. To mimic the real CRPC tumor environment, mice implanted with C4-2BENZR cells were administered Enz continuously after inoculation to create an AR-repressed host environment analogous to a castrate environment as well as to maintain Enz resistance as the tumors developed, while mice implanted with 22Rv1 cells received prior surgical castration. Mice were randomized into 2 groups to receive either a Dox+ or a Dox− diet to induce tumor MAOA KD or not, one week after inoculation in both the C4-2BENZR and 22Rv1 xenograft models. We demonstrated a substantial reduction in C4-2BENZR tumor formation frequency and tumor growth, including smaller tumor sizes and lower tumor weights and serum PSA levels at the endpoint, in Dox-fed mice compared to controls (Figs. 6A-6E). In 22Rv1 xenografts, Dox treatment remarkably delayed the onset of tumor formation and repressed tumor growth compared to controls (Figs. 6F-6J). Tumor samples showed downregulated expression of several canonical AR target genes, indicating decreased AR transactivation, in both MAOA-silenced C4-2BENZR and 22Rv1 tumors (Fig. 6K). We also confirmed continued Dox-induced MAOA KD in both C4-2BENZR and 22Rv1 tumors by IHC analysis. Ki-67 staining of tumor samples revealed an average 51% and 91% drop of Ki-67+ cells in MAOA-KD C4-2BENZR and 22Rv1 tumors respectively compared to controls. A roughly 2- and 4-fold increase of cleaved caspase-3 staining indicative of enhanced tumor cell apoptosis was shown in MAOA-silenced C4-2BENZR and 22Rv1 tumors compared to controls. In line with the in vitro observations, we detected no changes in the percentage of nuclear AR positivity and nuclear AR staining levels, but reduced nuclear YAP1 expression, in both MAOA-KD C4-2BENZR and 22Rv1 tumors compared to controls (Figs. 6L and 6M; Supplementary Figs. S11B and S11C). Based on these findings, we firmly concluded that MAOA is essential for AR-dictated PC and CRPC development and growth in mice.
Figure 6. MAOA silencing suppressed CRPC development and growth in mice.
(A) Kaplan-Meier tumor-free curves of mice inoculated with C4-2BENZR cells expressing a Dox-inducible MAOA shRNA and fed a Dox− or a Dox+ diet (n=24). (B) C4-2BENZR tumor growth curves of mice fed a Dox− or a Dox+ diet (Dox-, n=18; Dox+, n=8 tumors/group; which applies to C and D). (C, D) C4-2BENZR tumor weights (C) and anatomic tumor images (D) at the endpoint. (E) ELISA of endpoint serum PSA levels from C4-2BENZR tumor-bearing mice (n=6). (F) Kaplan-Meier tumor-free curves of mice inoculated with 22Rv1 cells expressing a Dox-inducible MAOA shRNA and fed a Dox− or a Dox+ diet (n=14). (G) 22Rv1 tumor growth curves of mice fed a Dox− or a Dox+ diet (Dox-, n=14; Dox+, n=13; which applies to H and I). (H, I) 22Rv1 tumor weights (H) and anatomic tumor images (I) at the endpoint. (J) ELISA of endpoint serum PSA levels from 22Rv1 tumor-bearing mice (n=7). (K) qPCR of indicated genes in tumor samples (n=6). (L) Representative MAOA, Ki-67, c-Cas3, AR and YAP1 IHC staining in tumor samples. Scale bars: 20 μm. (M) Quantification of % of Ki-67+ and c-Cas3+ cells in tumor samples (n=3). Data represent the mean ± SEM. *p<0.05, **p<0.01.
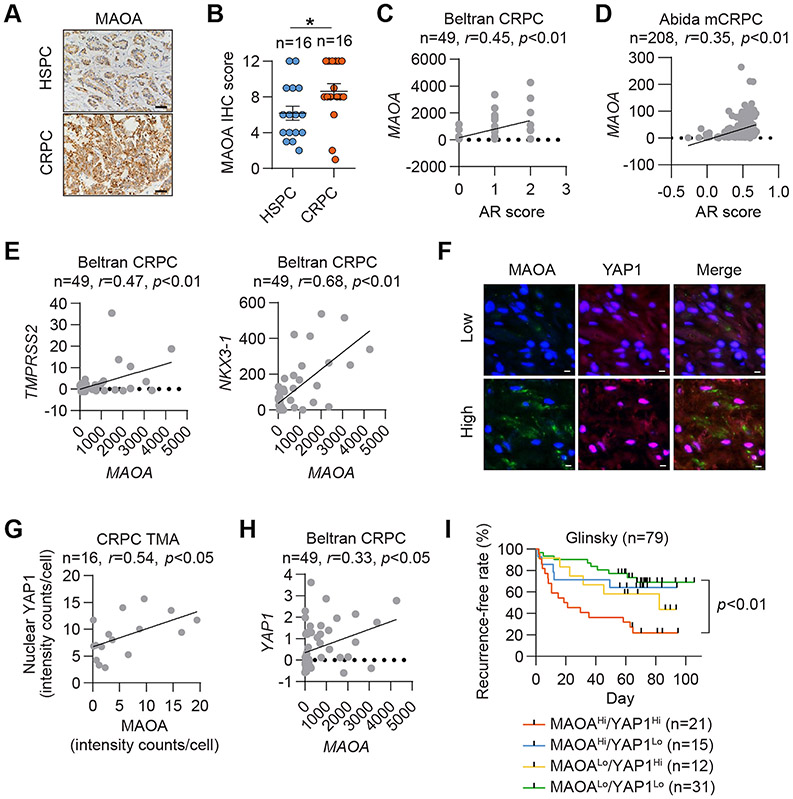
MAOA expression is elevated and associated with AR and YAP1 in human CRPC
CRPC is a fatal PC disease stage with the remarkable feature of near-universal AR reactivation (34,35). This prompted us to examine MAOA expression and its clinical association with AR alongside the CRPC characteristics developed during disease progression. We first performed histological analysis of a tissue panel of hormone-sensitive PC (HSPC) and CRPC and found elevated MAOA protein expression in CRPC relative to HSPC (Figs. 7A and 7B). Two CRPC datasets, Beltran and Abida (36), showed a positive association of MAOA mRNA expression with AR score defined by overall assessment of a group of AR target genes (Figs. 7C and 7D). We also demonstrated a positive mRNA co-expression correlation between MAOA and two AR target genes, TMPRSS2 and NKX3-1, in the Beltran dataset (Fig. 7E). Then we analyzed our CRPC cohort to address whether YAP1 is associated with MAOA in the clinical CRPC setting. Using double quantum dot labeling analysis, we found that MAOA and active YAP1, which reside in the cytoplasm and nucleus respectively, were positively co-expressed on a single-cell basis in CRPC (Figs. 7F and 7G). This was corroborated by a positive correlation between MAOA and YAP1 mRNA expression in the Beltran dataset (Fig. 7H). Further, higher mRNA expression of both MAOA and YAP1 was associated with disease recurrence in the Glinsky dataset (37) (Fig. 7I). These results strongly support the clinical significance of MAOA-AR crosstalk in CRPC.
Figure 7. MAOA expression associated with AR and YAP1 is elevated in human CRPC.
(A, B) Representative MAOA IHC staining (A) and quantification (B) in a PC cohort containing HSPC (n=16) and CRPC (n=16). Scale bars: 20 μm. Data represent the mean ± SEM. *p<0.05. (C, D) Pearson correlation analysis of MAOA with AR score in the Beltran (C) and Abida (D) datasets. (E) Pearson correlation analysis of MAOA with TMPRSS2 and NKX3-1 in the Beltran dataset. (F, G) Representative MAOA and YAP1 double QD staining (F) and corresponding Pearson correlation analysis (G) in a CRPC cohort. Scale bars: 20 μm. (H) Pearson correlation analysis of MAOA and YAP1 in the Beltran dataset. (I) Kaplan-Meier recurrence-free curves of PC patients categorized by MAOA/YAP1 mRNA levels from the Glinsky dataset.
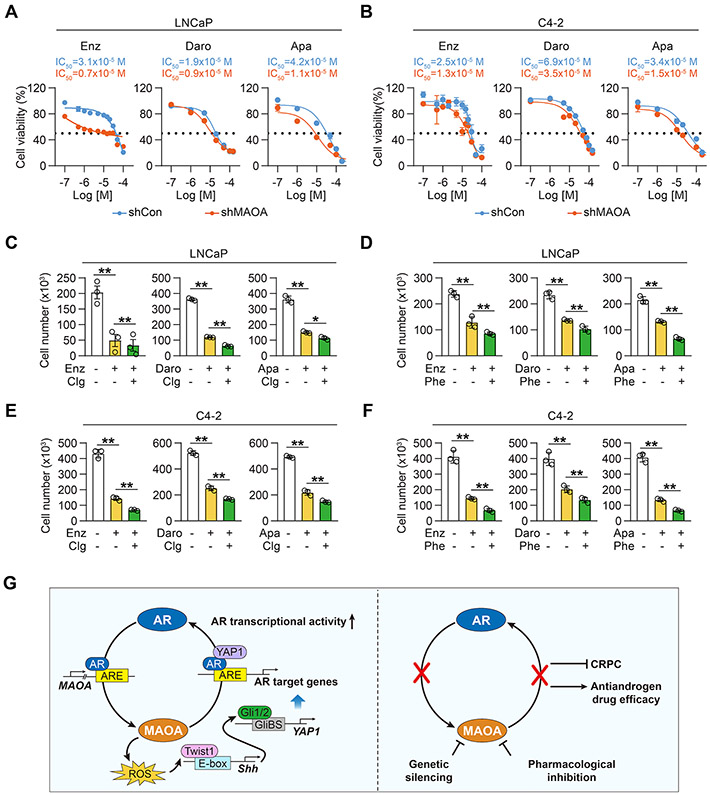
MAOA inhibition enhances antiandrogen drug efficacy
Given the current clinical use of MAOA inhibitors (38), we evaluated the therapeutic potential of synergizing MAOA inhibition with antiandrogen drugs to suppress PC cell growth. We tested the combinational effects of MAOA inactivation by either shRNA or small-molecule inhibitors with three antiandrogen drugs, Enz, darolutamide (Daro) and Apalutamide (Apa), all second-generation antiandrogen drugs currently used clinically to treat CRPC (39), in both LNCaP and its lineage-related castration-resistant C4-2 PC cell lines. We first showed that shRNA-mediated MAOA KD enhanced Enz, Daro and Apa growth inhibition in both LNCaP and C4-2 cells with up to 4- and 2-fold decreases of IC50 values respectively (Figs. 8A and 8B). Next, we evaluated the effectiveness of two conventional MAOA inhibitors, clorgyline and phenelzine, in promoting antiandrogen drug efficacy. Phenelzine is clinically prescribed as an antidepressant in the United States (40). We found that Enz, Daro and Apa suppressed LNCaP and C4-2 cell survival further by up to 50% and 53% respectively in the presence of clorgyline or phenelzine compared to antiandrogen drug treatment alone (Figs. 8C-8F). These findings suggest that MAOA targeting has significant potential for combinational use with AR-targeted therapy to treat AR-driven PC and CRPC.
Figure 8. Genetically and pharmacologically inhibiting MAOA enhanced antiandrogen drug efficacy.
(A, B) MTS cell proliferation assays of control and MAOA-KD LNCaP (A) and C4-2 (B) cells by Enz, Daro or Apa at various doses for 5 days (n=3). (C-F) Cell counting assays of LNCaP (C, D) and C4-2 (E, F) cells by Enz, Daro or Apa (5 μM for each) together with clorgyline (1 μM) or phenelzine (2 μM) for 5 days (n=3). Average cell numbers in control group with no treatment were set as 100%. (G) Schematic summarizing MAOA-AR reciprocal interaction in PC. Data represent the mean ± SEM. *p<0.05, **p<0.01.
In summary, our data show that AR promotes MAOA through direct binding to an intronic ARE of MAOA, and in a reciprocal manner MAOA induces Shh/Gli signaling via ROS-dependent Twist1, which activates YAP1 to enhance nuclear YAP1-AR interaction, thereby upregulating AR transcriptional activity in AR-dominant PC cells (Fig. 8G).
Discussion
This study showed that MAOA synergizes with AR through reciprocal crosstalk to amplify AR-directed PC disease progression, including the aggressive castration-resistant variant. Our previous study revealed that MAOA upregulation could be under the concerted control of aberrant oncogenic signaling, including activation of c-Myc and loss of PTEN and p53, at different disease stages (7). This study provides an additional regulatory mechanism for elevated MAOA expression in PC through androgenic signaling. MAOA activation by androgens in neuroblastoma cells through direct AR interaction with an ARE in the proximal promoter of MAOA has been reported (41). However, we identified a previously undescribed functional intronic ARE in the MAOA gene locus, demonstrated in both PC cultured cells and clinical datasets, suggesting that AR may regulate MAOA in a cell context-dependent manner.
CRPC is a significant clinical challenge mainly due to AR reactivation after escape from ADT. We found increased MAOA expression and MAOA association with AR activity in human CRPC. In line with clinical re-expression of most if not all of the genes known to be under AR regulation in castration-resistant tumors (34,35), our observations support the role of MAOA as an AR target gene in CRPC. Compelling evidence indicates that reactivated AR is functional and fuels castration-resistant tumor regrowth after a period of regression, rendering AR a viable therapeutic target even after castration resistance develops (3). Our discovery of a new mechanism where MAOA is regulated by AR and in turn controls AR transcriptional activity in PC cells may reveal a positive feed-forward loop augmenting AR signaling in CRPC. This provides a rationale for targeting MAOA to untangle the crosstalk between MAOA and AR as a potential CRPC therapy. Indeed, we demonstrated in preclinical xenograft mouse models that MAOA inactivation significantly impeded CRPC development and growth. Pharmacological inhibition of MAOA also enhanced the efficacy of three second-generation antiandrogen drugs in CRPC cells. These findings call for further evaluation of MAOA inhibitors for clinical application in CRPC to complement current therapies targeting the AR axis.
Our data indicate that MAOA promotes AR transactivation through upregulation of YAP1 and enhanced nuclear YAP1-AR interaction. YAP1, a transcriptional coactivator that regulates diverse cellular processes, was recently reported to act as a physiological binding partner and positive regulator of AR in PC through both androgen-dependent and -independent mechanisms in different disease states. YAP1-AR interaction also contributes to the switch from androgen-dependent to castration-resistant growth in PC (19). These mechanistic details support YAP1-dependent MAOA upregulation of AR in both androgen-dependent and castration-resistant PC cells. YAP1 utilizes the WW/SH3 domain to interact specifically with the N-terminal transactivation domain (NTD) of AR (19). This protein-protein interaction mode makes YAP1 interaction with AR variants possible, especially those lacking a C-terminal ligand-binding domain (LBD) while maintaining NTD (e.g., AR-V7) as observed in PC following ADT (42,43). Thus, it seems likely that MAOA might have an activating effect on AR variants through the same YAP1-dependent mechanism. This provides a rationale for antagonizing MAOA in CRPC, which is prone to develop resistance to the more highly potent antiandrogen drugs like Enz owing to the emergence of LBD-deficient AR variants for constitutive activation of AR signaling.
One of the salient mechanistic findings of our study is that MAOA activates YAP1 through downstream ROS/Twist1-mediated Shh/Gli signaling, wherein Gli1/2 directly binds to a GliBS on the YAP1 promoter to activate YAP1 transcription. YAP1 is amplified and upregulated in hedgehog-associated medulloblastomas and was also recently found to have functional interplay with hedgehog signaling in different development and disease states (44,45). Despite these mechanistic advances, this study provides an alternative mechanism to YAP1 regulation by hedgehog signaling. In addition, numerous studies have demonstrated crosstalk between hedgehog signaling and androgen signaling under certain conditions depending on the tumor microenvironment (46-48). In this study, we demonstrated that MAOA-dependent autocrine Shh/Gli signaling activates YAP1 to enhance AR transactivation, distinct from the paracrine signaling we previously found supporting MAOA-elicited tumor-stromal cell interaction to promote metastasis (9), suggesting that MAOA/Shh signaling might be context-dependent in regulating different aspects of prostate tumor behavior. Hedgehog signaling has also been shown to support androgen signaling and the growth of androgen-deprived and -independent PC cells (46), which might sustain YAP1-mediated MAOA upregulation of AR and the resulting AR-driven phenotype in CRPC.
In conclusion, this study uncovered MAOA’s reciprocal crosstalk with AR, amplifying the effects of both to promote PC development and growth dictated by AR signaling. This provides new insights into the mechanistic basis of AR regulation and functions in PC. We also provided strong pre-clinical evidence for targeting MAOA, alone or in combination with AR-targeted therapy, to disengage the MAOA/AR complex as a potential therapy for PC and CRPC.
Supplementary Material
Acknowledgements
We thank Xiangyan Li (Cedars-Sinai Medical Center) for technical help, Jen-Ming Huang (Cedars-Sinai Medical Center) for insightful discussion, Leland W.K. Chung (Cedars-Sinai Medical Center) for comprehensive support of this study, and Gary Mawyer for editorial assistance. This work was supported by NIH/NCI grant R37CA233658, DOD Prostate Cancer Research Program grant W81XWH-19-1-0279, and WSU start-up funding (to B. J. Wu).
Footnotes
Conflict of interest statement: The authors declare no potential conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 2018;68:394–424 [DOI] [PubMed] [Google Scholar]
- 2.Saad F, Fizazi K. Androgen Deprivation Therapy and Secondary Hormone Therapy in the Management of Hormone-sensitive and Castration-resistant Prostate Cancer. Urology 2015;86:852–61 [DOI] [PubMed] [Google Scholar]
- 3.Graham L, Schweizer MT. Targeting persistent androgen receptor signaling in castration-resistant prostate cancer. Medical oncology 2016;33:44. [DOI] [PubMed] [Google Scholar]
- 4.Culig Z, Santer FR. Androgen receptor signaling in prostate cancer. Cancer metastasis reviews 2014;33:413–27 [DOI] [PubMed] [Google Scholar]
- 5.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annual review of neuroscience 1999;22:197–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America 2006;103:10991–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JB, Shao C, Li X, Li Q, Hu P, Shi C, et al. Monoamine oxidase A mediates prostate tumorigenesis and cancer metastasis. J Clin Invest 2014;124:2891–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flamand V, Zhao H, Peehl DM. Targeting monoamine oxidase A in advanced prostate cancer. Journal of cancer research and clinical oncology 2010;136:1761–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu JB, Yin L, Shi C, Li Q, Duan P, Huang JM, et al. MAOA-Dependent Activation of Shh-IL6-RANKL Signaling Network Promotes Prostate Cancer Metastasis by Engaging Tumor-Stromal Cell Interactions. Cancer cell 2017;31:368–82 [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Lou W, Zhu Y, Yang JC, Nadiminty N, Gaikwad NW, et al. Intracrine Androgens and AKR1C3 Activation Confer Resistance to Enzalutamide in Prostate Cancer. Cancer research 2015;75:1413–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank SB, Schulz VV, Miranti CK. A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector. BMC Biotechnol 2017;17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu JB, Chen K, Ou XM, Shih JC. Retinoic acid activates monoamine oxidase B promoter in human neuronal cells. The Journal of biological chemistry 2009;284:16723–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu P, Chu GC, Zhu G, Yang H, Luthringer D, Prins G, et al. Multiplexed quantum dot labeling of activated c-Met signaling in castration-resistant human prostate cancer. PloS one 2011;6:e28670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell 2010;18:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche PJ, Hoare SA, Parker MG. A consensus DNA-binding site for the androgen receptor. Molecular endocrinology 1992;6:2229–35 [DOI] [PubMed] [Google Scholar]
- 16.Denmeade SR, Sokoll LJ, Dalrymple S, Rosen DM, Gady AM, Bruzek D, et al. Dissociation between androgen responsiveness for malignant growth vs. expression of prostate specific differentiation markers PSA, hK2, and PSMA in human prostate cancer models. The Prostate 2003;54:249–57 [DOI] [PubMed] [Google Scholar]
- 17.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev 2007;28:778–808 [DOI] [PubMed] [Google Scholar]
- 18.Jiang N, Hjorth-Jensen K, Hekmat O, Iglesias-Gato D, Kruse T, Wang C, et al. In vivo quantitative phosphoproteomic profiling identifies novel regulators of castration-resistant prostate cancer growth. Oncogene 2015;34:2764–76 [DOI] [PubMed] [Google Scholar]
- 19.Kuser-Abali G, Alptekin A, Lewis M, Garraway IP, Cinar B. YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat Commun 2015;6:8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Yang S, Chen X, Stauffer S, Yu F, Lele SM, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Molecular and cellular biology 2015;35:1350–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Zhu X, Feng W, Yu Y, Jeong K, Guo W, et al. Verteporfin inhibits YAP function through up-regulating 14-3-3sigma sequestering YAP in the cytoplasm. Am J Cancer Res 2016;6:27–37 [PMC free article] [PubMed] [Google Scholar]
- 22.Araya S, Kratschmar DV, Tsachaki M, Stucheli S, Beck KR, Odermatt A. DHRS7 (SDR34C1) - A new player in the regulation of androgen receptor function by inactivation of 5alpha-dihydrotestosterone? The Journal of steroid biochemistry and molecular biology 2017;171:288–95 [DOI] [PubMed] [Google Scholar]
- 23.Brooke GN, Culley RL, Dart DA, Mann DJ, Gaughan L, McCracken SR, et al. FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer research 2011;71:914–24 [DOI] [PubMed] [Google Scholar]
- 24.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. The Journal of biological chemistry 2002;277:25904–13 [DOI] [PubMed] [Google Scholar]
- 25.Zennami K, Choi SM, Liao R, Li Y, Dinalankara W, Marchionni L, et al. PDCD4 Is an Androgen-Repressed Tumor Suppressor that Regulates Prostate Cancer Growth and Castration Resistance. Molecular cancer research : MCR 2019;17:618–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu J, Peng S, Si-Tu J, Hu C, Huang W, Mao Y, et al. Identification of endonuclease domain-containing 1 as a novel tumor suppressor in prostate cancer. BMC Cancer 2017;17:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L, Schafer G, Bu H, Zhang Y, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 2012;33:751–9 [DOI] [PubMed] [Google Scholar]
- 28.McMillan R, Matsui W. Molecular pathways: the hedgehog signaling pathway in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2012;18:4883–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swiderska-Syn M, Xie G, Michelotti GA, Jewell ML, Premont RT, Syn WK, et al. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver. Hepatology 2016;64:232–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Molecular and cellular biology 1990;10:634–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine 2016;22:298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweizer MT, Yu EY. Persistent androgen receptor addiction in castration-resistant prostate cancer. J Hematol Oncol 2015;8:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon RR, Wu M, Huang CY, Harris WP, Sim HG, Lucas JM, et al. Chemotherapy-induced monoamine oxidase expression in prostate carcinoma functions as a cytoprotective resistance enzyme and associates with clinical outcomes. PloS one 2014;9:e104271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl Androl Urol 2015;4:365–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katzenwadel A, Wolf P. Androgen deprivation of prostate cancer: Leading to a therapeutic dead end. Cancer letters 2015;367:12–7 [DOI] [PubMed] [Google Scholar]
- 36.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 2019;116:11428–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bortolato M, Chen K, Shih JC. Monoamine oxidase inactivation: from pathophysiology to therapeutics. Advanced drug delivery reviews 2008;60:1527–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice MA, Malhotra SV, Stoyanova T. Second-Generation Antiandrogens: From Discovery to Standard of Care in Castration Resistant Prostate Cancer. Frontiers in oncology 2019;9:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shulman KI, Herrmann N, Walker SE. Current place of monoamine oxidase inhibitors in the treatment of depression. CNS Drugs 2013;27:789–97 [DOI] [PubMed] [Google Scholar]
- 41.Ou XM, Chen K, Shih JC. Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. The Journal of biological chemistry 2006;281:21512–25 [DOI] [PubMed] [Google Scholar]
- 42.Haile S, Sadar MD. Androgen receptor and its splice variants in prostate cancer. Cell Mol Life Sci 2011;68:3971–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du K, Hyun J, Premont RT, Choi SS, Michelotti GA, Swiderska-Syn M, et al. Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells. Gastroenterology 2018;154:1465–79 e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes & development 2009;23:2729–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M, Feuerstein MA, Levina E, Baghel PS, Carkner RD, Tanner MJ, et al. Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Molecular cancer 2010;9:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng YC, Levine CM, Zahid S, Wilson EL, Joyner AL. Sonic hedgehog signals to multiple prostate stromal stem cells that replenish distinct stromal subtypes during regeneration. Proceedings of the National Academy of Sciences of the United States of America 2013;110:20611–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamichi F, Shigemura K, Behnsawy HM, Meligy FY, Huang WC, Li X, et al. Sonic hedgehog and androgen signaling in tumor and stromal compartments drives epithelial-mesenchymal transition in prostate cancer. Scand J Urol 2014;48:523–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.