Abstract
Cancer-associated fibroblasts (CAFs) participate in critical processes in the tumor microenvironment, such as extracellular matrix remodeling, reciprocal signaling interactions with cancer cells and crosstalk with infiltrating inflammatory cells. However, the relationships between CAFs and survival are not well known in lung cancer. The aim of this study was to reveal the correlations of CAFs with survival rates, genetic alterations and immune activities. This study reviewed the histological features of 517 patients with lung adenocarcinoma from The Cancer Genome Atlas (TCGA) database. We performed gene set enrichment analysis (GSEA), network-based analysis and survival analysis based on CAFs in four histological types of lung adenocarcinoma: acinar, papillary, micropapillary and solid. We found four hallmark gene sets, the epithelial-mesenchymal transition, angiogenesis, hypoxia, and inflammatory response gene sets, that were associated with the presence of CAFs. CAFs were associated with tumor proliferation, elevated memory CD4+T cells and high CD274 (encoding PD-L1) expression. In the pathway analyses, CAFs were related to blood vessel remodeling, matrix organization, negative regulation of apoptosis and transforming growth factor-β signaling. In the survival analysis of each histological type, CAFs were associated with poor prognosis in the solid type. These results may contribute to the development of therapeutic strategies against lung adenocarcinoma cases in which CAFs are present.
Subject terms: Cancer, Computational biology and bioinformatics, Drug discovery, Systems biology, Biomarkers, Medical research, Oncology, Pathogenesis
Introduction
Lung cancer is a major cancer and the most common cause of cancer death in the world1,2. According to the National Comprehensive Cancer Network (NCCN) guidelines in oncology, early lung cancer requires surgical procedures, but advanced cancer is treated with systemic treatment3. However, about half of the patients recur, usually within the first year after starting treatment4,5.
Genetic mutation of cells can induce cancer development, but disease progression and treatment sensitivity are affected by nonmutant cells within the tumor microenvironment6. One type of nonmutant cell within the dense collagenous stroma is fibroblast-like cells, so-called cancer-associated fibroblasts (CAFs)7. CAFs can drive cancer metastasis through remodeling of the extracellular matrix (ECM) and the production of growth factors and can affect angiogenesis, and these effects influence therapy response8. Recently, there has been a growing appreciation of the ability of CAFs to modulate the immune system6.
Several studies have reported that survival rates are associated with CAF histological features in different types of malignancy. Previous studies have demonstrated that CAFs and desmoplastic reaction are predictive of poor prognosis in colorectal cancer9. Another study suggested that adipocyte-derived fibroblasts are correlated with poor survival and desmoplastic reaction in breast cancer7. However, another study reported that histological type, specifically the desmoplastic type, is an independent predictor of favorable prognosis in colorectal cancer10.
Recently, molecular studies have utilized bioinformatic tools to find the mechanisms of CAFs. CAFs are a different cell population in terms of origin and pathobiological roles and are derived mainly from mesenchymal stromal cells that are resident in or recruited by the cancer11. CAFs are located close to tumor cells and stromal components such as lymphocytes, neutrophils, plasma cells, endothelial cells and ECM12. Fibroblasts include CAFs as well as myofibroblastic cells, quiescent fibroblastic cells and pericytic cells. The identification of fibroblasts within the cancer remains challenging due to the lack of specific biomarkers for known and still unclear subtypes12.
In recent years, big data analytics and next-generation sequencing (NGS) have allowed the analyses of genetic biomarkers, the quantification of the several types of tumor-infiltrating lymphoid cells and the molecular pathway network-based integration of multiomics data13–15. Considering the multiple gene-environment relationships of lung cancer, the clinicopathological application of gene expression data is difficult. For these reasons, we believe that analyses based on gene expression data should focus on identifying a simple, robust, and druggable biomarkers based on high-throughput experimental tools and bioinformatics to achieve accessible therapeutic strategies. The Cancer Genome Atlas (TCGA) has a big database, including digital pathologic slides, clinicopathological information, RNA sequencing, mutation, copy number variable and methylation data13. Moreover, the histological features reported in the TCGA database provide data on the presence of CAFs and the tumor microenvironment in lung cancer.
This study aimed to determine whether the presence of CAFs contributes to the clinical outcomes of lung cancer and to evaluate the prognostic value of CAFs16. We further aimed to find the gene sets related to CAFs based on gene set enrichment analysis (GSEA)14 and molecular pathway network analyses17,18. The relationships between lymphoid cells and CAFs were analyzed15.
Materials and methods
Patient selection
We obtained a total of 1,053 non-small-cell lung carcinoma (NSCLC) cases comprising 566 lung adenocarcinomas and 487 squamous cell carcinomas with known mRNA expression and mutation data from the TCGA database13. The analysis was performed on 517 cases containing both virtual histopathological slides and clinical data (from a total of 566 lung adenocarcinoma samples).
Cancer-associated fibroblasts
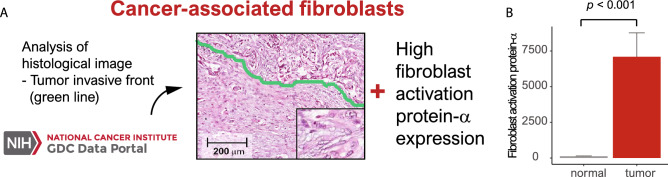
In this review, cells with both immature fibroblastic proliferation (proportion: > 10%) at the tumor invasive front and high FAP gene expression were defined as CAFs (Fig. 1A)19,20. To determine the optimal cutoff value for FAP expression, we generated receiver operating characteristic (ROC) curves comparing sensitivity versus 1–specificity. The cutoff value calculated by the ROC curve was used to evaluate the relationship between survival and FAP expression. On the basis of the ROC curve, FAP expression was classified as low (mRNA level ≤ 562.9965) or high (mRNA level > 562.9965). Of the 517 cases, CAFs were present in 101 cases (19.5%).
Figure 1.
(A) Histological images showing cancer-associated fibroblasts at the tumor invasive front (green line) (inset: immature cancer-associated fibroblasts) (original magnification × 400; inset × 1000) and high fibroblast activation protein-α (FAP-α) expression. (B) Bar plots showing the difference in fibroblast activation protein-α expression between normal and tumor tissues (p < 0.001).
Gene set enrichment analysis and pathway-based network analysis based on TCGA data
To detect significant gene sets, GSEA (version 4.1.0) was performed with 31,117 gene sets in the Molecular Signatures Database (MSigDB version 7.2) from the Broad Institute at MIT14. Specific gene sets (50 hallmark gene sets) were tested to determine which were associated with CAFs. For this analysis, 1000 permutations were utilized to calculate the p values, and the permutation type was set to phenotype. Significant gene sets were those with the following characteristics: false discovery rate < 0.001; family wise-error rate ≤ 0.001; and p < 0.001.
We analyzed tumor-infiltrating lymphocytes using deep learning-based lymphocyte classification with convolutional neural networks in whole-slide image analysis and identified immune subtypes using in silico cytometry analyses [CIBERSORT (https://cibersort.stanford.edu/) with Kallisto algorithms]15,21–23.
Pathway-based network analysis was based on the identified genes linked to CAFs using Cytoscape (version 3.8.1) network visualization software. To visualize the biological relevance of the histological subtypes and their relevant elements on the basis of GSEA, we performed functional enrichment analyses using ClueGO, an application within Cytoscape software17,18.
Machine learning algorithm for validation
We integrated CAFs with clinical risk factors (T stage, N stage, age, sex, smoking history) to composite prognostic models for survival prediction by applying machine learning (ML) algorithms in 517 cases (randomization: train set, 70%; validation set, 30%). A learning algorithm was independently applied to select and combine multiple covariates from gradient boosting machines (GBM) based on multivariate Bernoulli models. In this step, ‘‘forward” search method, which initiates on a prototype set and selects a feature if and only if the addition of the feature could increase the performance of the prognostic model, is adopted to select optimal features sequentially. Hyperparameters of the ML algorithms, such as learning rate in GBM were optimized for each combination of selected covariates and learning algorithm by grid search cross-validation through a predefined range. We searched across 81 models with varying learning rates and tree depth. The final optimal models were trained based on the selected covariates and the optimized hyperparameters24. To explore the performances of the GBM method, the receiver operator characteristic (ROC) curve was used.
Statistical analysis
Student’s t-test was used to evaluate the differences or relationships among continuous paramters. Disease-free survival (DFS) and disease-specific survival (DSS) were compared using the log rank test. Multivariate analysis was performed to identify independent prognostic markers for DFS and DSS using a Cox multistep regression model. All data were analyzed using R packages. A two-tailed p value < 0.05 was considered statistically significant.
Results
CAFs were associated with EMT, angiogenesis, hypoxia and inflammatory response
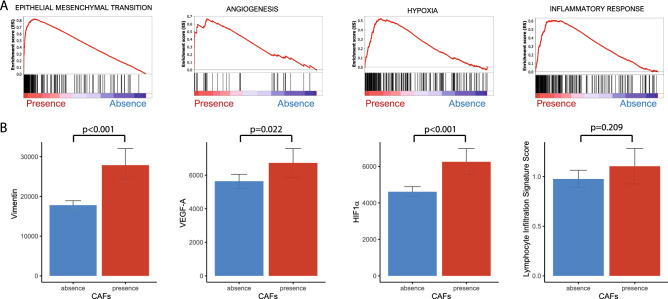
FAP was highly expressed in tumors compared with normal tissues (p < 0.001) (Fig. 1B). We performed GSEA to identify various gene sets associated with CAFs. In the analyses of hallmark gene sets, we found four gene sets (the epithelial-mesenchymal transition, angiogenesis, hypoxia and inflammatory response gene sets) that were associated with lung adenocarcinoma (Fig. 2A).
Figure 2.
(A) Four gene sets (the epithelial-mesenchymal transition, angiogenesis, hypoxia, and inflammatory response gene sets) associated with cancer-associated fibroblasts. (B) Bar plots of the relationships between cancer-associated fibroblasts and markers of the identified gene sets: vimentin, vascular endothelial growth factor-A (VEGF-A), hypoxia inducible factor 1 subunit alpha (HIF1α) and lymphocyte infiltration signature score (p < 0.001, = 0.022, < 0.001 and = 0.209, respectively).
On the basis of the GSEA results, we analyzed the association between CAFs and each gene set-related marker. Vimentin, a biomarker related to epithelial-mesenchymal transition, was highly expressed in the presence of CAFs (p < 0.001). Vascular endothelial growth factor-A (VEGF-A), as a marker related to angiogenesis, was increased in the presence of CAFs (p = 0.022). Hypoxia-inducible factor 1 subunit alpha (HIF1α), which is linked to hypoxia, was elevated in the presence of CAFs (p < 0.001). The lymphocyte infiltration signature score, which is associated with the inflammatory response, showed a tendency to increase in the presence of CAFs, but it was not statistically significant (p = 0.209) (Fig. 2B).
CAFs were related to low B cells, high CD4+T cells, high PD-L1 expression and proliferation
In the analyses of CAFs, we referred to the immune cell profiles, tumor cell proliferation parameters and biomarkers used in a study by Thorsson et al. and in silico cytometry.22
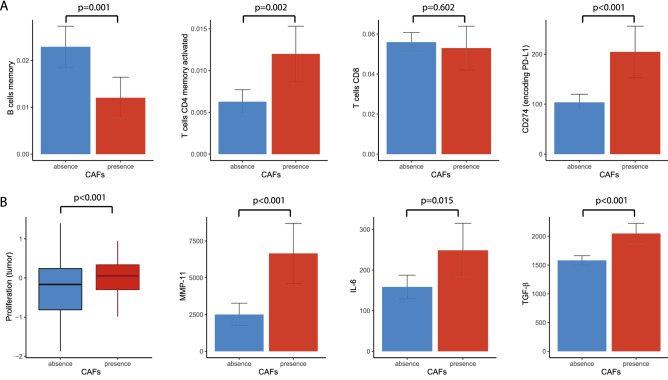
In comparing the immune cell fractions between samples with and without CAFs, memory B cells were decreased in samples with CAFs (p < 0.001), while activated memory CD4+T cells were increased in samples with CAFs (p = 0.002). CD274 (encoding PD-L1, programmed death-ligand 1) expression was more elevated in samples with CAFs than in those without CAFs (p < 0.001). CD8+T cells showed a tendency to be decreased in samples with CAFs, but this trend was not statistically significant (p = 0.602) (Fig. 3A).
Figure 3.
Bar plots of the relationships between cancer-associated fibroblasts and immune factors. (A) Memory B cells, activated CD4 + memory T cells, CD8 T cells and CD274 (encoding PD-L1) + cells (p = 0.001, 0.002, 0.602 and < 0.001, respectively). (B) Relationships between tumor proliferation, matrix metalloproteinase-11 (MMP-11), interleukin-6 (IL-6) and transforming growth factor-β (TGF-β) (p < 0.001, 0.001, = 0.015 and < 0.001, respectively).
The presence of CAFs was associated with higher proliferation and matrix metallopeptidase-11 (MMP-11), interleukin-6 (IL-6) and transforming growth factor-β (TGF-β) levels than the absence of CAFs (p < 0.001, < 0.001, = 0.015 and < 0.001, respectively) (Fig. 3B).
CAFs were linked to blood vessel remodeling, fibrosis and tissue remodeling pathway
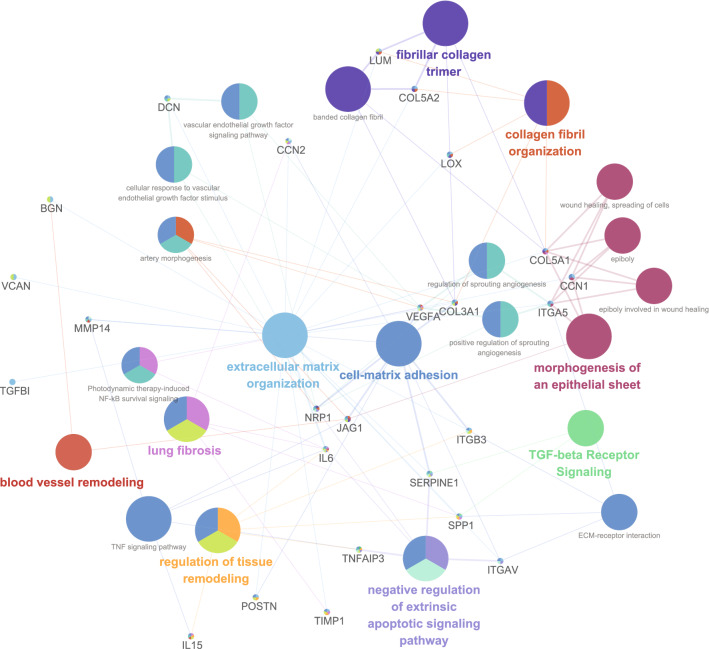
We performed pathway-based network analysis using the genes and gene sets associated with CAFs. The CAFs were linked to 10 functionally enriched Gene Ontology (GO) terms and pathways: (1) blood vessel remodeling; (2) lung fibrosis; (3) regulation of tissue remodeling; (4) extracellular matrix organization; (5) cell–matrix adhesion; (6) fibrillar collagen trimer; (7) negative regulation of extrinsic apoptosis signaling pathway; (8) collagen fibril organization; (9) morphogenesis of an epithelial sheet; and (10) TGF-β receptor signaling (Fig. 4).
Figure 4.
Grouping of networks based on functionally enriched Gene Ontology (GO) terms and pathways associated with the presence of cancer-associated fibroblasts.
CAFS improved survival prediction using machine learning.
In the TCGA data, the distributions of the five predominant histological types were as follows: 7 lepidic type (1.4%), 115 acinar type (22.2%), 107 papillary type (20.7%), 65 micropapillary type (12.6%) and 223 solid type (43.1%). There was an absence of CAFs in the lepidic cases; thus, they were not included in the survival analyses.
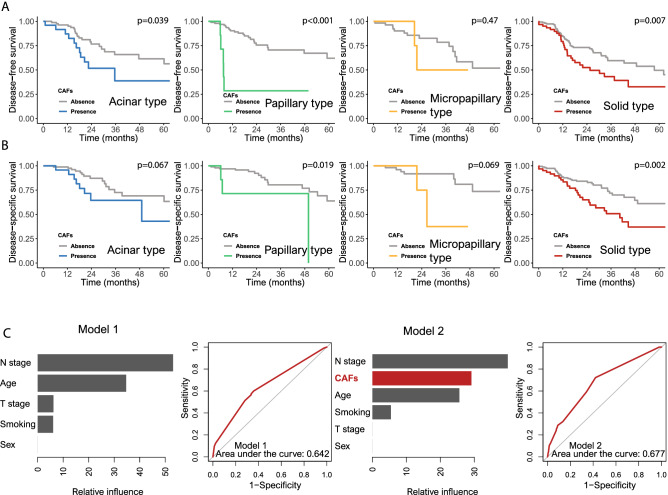
The presence of CAFs was associated with unfavorable DFS and DSS in the acinar type (p = 0.039 and 0.067, respectively), but the relationship between CAFs and DSS was not statistically significant. The presence of CAFs was significantly related to shorter DFS and DSS than the absence of CAFs in the papillary type (p < 0.001 and 0.019, respectively). The presence of CAFs correlated with worse DFS and DSS in the micropapillary type (p = 0.47 and 0.069, respectively), but the correlations were not statistically significant. In the solid type, the presence of CAFs was significantly associated with shorter DFS and DSS than the absence of CAFs (p = 0.007 and 0.002, respectively) (Fig. 5A,B). After adjustment for confounders including T stage, N stage, age, sex and smoking history, the presence of CAFs was associated with worse DFS in the papillary type and solid type than the absence of CAFs (p < 0.001 and = 0.008, respectively). There was a relationship between shorter DSS and the presence of CAFs in only solid type samples (p = 0.003) (Table 1).
Figure 5.
Survival analyses of the four types based on the absence or presence of cancer-associated fibroblasts. (A) Disease-free survival: acinar, papillary, micropapillary and solid types (p = 0.039, < 0.001, = 0.47 and 0.007, respectively). (B) Disease-specific survival: acinar, papillary, micropapillary and solid types (p = 0.067, 0.019, 0.069 and 0.002, respectively). (C) We supervised machine-learning models for prognosis prediction using gradient boosting machine (GBM). Covariates were included in the confounding factors [Model 1 (left); T stage, N stage, age, sex, smoking (pack years) versus Model 2 (right); Cancer associated fibroblasts (CAFs), T stage, N stage, age, sex, smoking (pack years)] and their relative importance using overall survival. Receiver operator characteristic curve for GBM was used based on a multivariate Gaussian model.
Table 1.
Univariate and multivariate analyses of disease-free survival and disease-specific survival based on cancer-associated fibroblasts.
| Covariate | Disease-free survival | Disease-specific survival | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |||
| Acinar type | ||||||||
| Univariate | 2.125 | 1.021 | 4.427 | 0.039 | 2.202 | 0.926 | 5.235 | 0.067 |
| Multivariate* | 1.712 | 0.775 | 3.785 | 0.184 | 1.842 | 0.698 | 4.864 | 0.217 |
| Papillary type | ||||||||
| Univariate | 5.460 | 2.031 | 14.676 | < 0.001 | 4.061 | 1.142 | 14.441 | 0.019 |
| Multivariate* | 13.630 | 4.112 | 45.174 | < 0.001 | 3.585 | 0.706 | 18.214 | 0.124 |
| Micropapillary type | ||||||||
| Univariate | 1.724 | 0.387 | 7.677 | 0.47 | 3.975 | 0.796 | 19.849 | 0.069 |
| Multivariate* | 0.535 | 0.051 | 5.642 | 0.603 | 3.844 | 0.367 | 40.251 | 0.261 |
| Solid type | ||||||||
| Univariate | 1.852 | 1.173 | 2.925 | 0.007 | 2.161 | 1.296 | 3.603 | 0.002 |
| Multivariate* | 1.980 | 1.199 | 3.269 | 0.008 | 2.339 | 1.328 | 4.119 | 0.003 |
*Adjusted for T stage, N stage, age, sex and smoking history.
We compared the performance of the two GBM models in predicting survival rates (Model 1; T stage, N stage, age, sex, smoking history versus Model 2; CAFs, T stage, N stage, age, sex, smoking history) (Supplementary information and Fig. 1). ROC curves were performed (area under the curve: Model 1, 0.642; Model 2, 0.677) (Fig. 5C). We found that the GBM algorithm performed the best while the addition of CAFs to the prediction model improved the prognostic performance. With cross-validation estimates, 7 decision trees were utilized sequentially while the maximum depth of each decision tree was optimized at 1, corresponding to one-way interactions, and the learning rate was optimized at 0.018.
Discussion
This study showed survival differences between patients with and without CAFs and analyzed genetic/molecular alterations in patients with lung adenocarcinoma. In previous studies, genetic/molecular signatures related to CAFs have been shown to correlate with prognosis in colorectal, ovarian and breast cancer25–27. Our results revealed that the presence of CAFs was associated with a shorter survival rate than the absence of CAFs in lung adenocarcinoma, especially the solid type. In this study, the machine learning model analysis which includes CAFs increased the accuracy of predicting the survival rate. A study by Marcela et al. reported that CAFs were related to increased survival in patients with diffuse large B cell lymphoma28. Moreover, another study of colorectal carcinoma demonstrated that the presence of desmoplasia and CAFs was associated with better survival than the absence of desmoplasia10. Thus, there is controversy regarding the association between CAFs and the survival of patients with cancer. It is thought that CAFs in the tumor microenvironment are phenotypically heterogeneous and may exhibit both a protumorigenic and antitumorigenic phenotypes29. We analyzed hallmark gene sets related to CAFs in lung adenocarcinoma. A total of four gene sets associated with the presence of CAFs were identified: the epithelial-mesenchymal transition (EMT), angiogenesis, hypoxia, and inflammatory response gene sets. Subsequently, we determined the correlations of representative biomarkers associated with these gene sets with to the presence or absence of CAFs. First, vimentin is an EMT biomarker and is involved in cell migration, motility and adhesion and associated with metastasis30. Second, VEGF-A is an angiogenesis biomarker and induces high microvascular density and permeability and promotes tumor expansion31. Third, HIF1α is a hypoxia biomarker and is associated with the upregulation of glycolytic genes related to oxygen deprivation with increased cancer metabolism32. Fourth, the lymphocyte infiltration signature score, which is an inflammatory response marker, is related to prognosis and host-tumor immune interactions in different types of malignancies33. Some representative markers, such as vimentin, VEGF-A and HIF1α, were elevated in the presence of CAFs compared to the absence of CAFs. There was no significant difference in the lymphocyte infiltration signature score between samples with and without CAFs. These results suggest that the presence or absence of CAFs has minimal effect on the host-tumor immune response in lung adenocarcinoma.
In addition, we analyzed the changes in immune cell subtypes according to the presence or absence of CAFs by considering the characteristics of different immune cells. Memory B cells were decreased in the presence of CAFs, but activated memory CD4+T cells were increased in the presence of CAFs. CD274 was elevated in the presence of CAFs. There was no significant difference in CD8+T cells between samples with or without CAFs. A study by Costa et al. demonstrated that FAP-high fibroblasts, such as CAFs, are correlated with Treg cell-mediated immunosuppression and poor outcome in breast cancer34. Our results showed that, in lung adenocarcinoma, CAFs have little effect on the immunomodulation associated with CD8+T cells in the tumor microenvironment.
CAFs can induce increased levels of growth factors, matrix remodeling and increased levels of numerous cytokines related to immunomodulation6. In our results, the proliferation index was increased in the presence of CAFs. TGF-β and IL-6 are related to tumor growth and/or immunosuppression and were increased in the presence of CAFs. A representative marker of ECM and cancer invasion, MMP-11, was elevated in the presence of CAFs. The pathway-based network analysis showed biological functions related to CAFs, such as blood vessel remodeling, extracellular matrix organization, negative regulation of the extrinsic apoptotic signaling pathway and TGF-β receptor signaling.
This study has several limitations that should be acknowledged. First, because this is a cross-sectional study and the in silico analyses with TCGA did not show sustained relationships over time, it is difficult to reach a definitive conclusion. Second, experimental data allowing for novel biological insights into CAFs were not obtained in our study. Further in vitro and/or in vivo studies may be necessary to clarify the molecular mechanisms of CAFs in solid lung adenocarcinoma. Third, CAF function may be highly heterogeneous in solid lung adenocarcinoma patients, as many components of signaling pathways are affected by disease status, microenvironment, and immunity. Fourth, the difficulty in identifying CAFs results largely from the lack of unique markers6. In our study, CAFs were defined by a combination of the histological features of fibroblasts and high FAP gene expression. Fifth, we used machine learning and general statistical methods to predict survival differences between patient with or without CAFs. Because CAFs were identified as important factors in predicting survival in both methods, our study based on limited data could not explain the difference between machine learning, which focuses on prediction, and statistical analysis, which focuses on inference. Discussion of the issues will require future research.
This study demonstrated that CAFs are associated with increased tumor cell growth, angiogenesis, and ECM remodeling, effects that produce an unfavorable prognosis in patients with solid lung adenocarcinoma. CAFs were found to be associated with enhanced recruitment of activated memory CD4+T cells with high CD274 expression. The presence of CAFs was related to decreased numbers of CD8+T cells, but the relationship was not statistically significant. Patients with CAFs with high CD274 expression without elevated CD8+T cells might develop resistance to anti-PD-L1 therapies. Our workflow results regarding CAFs will contribute to designing future clinical and experimental studies for patients with solid lung adenocarcinoma.
Supplementary Information
Acknowledgements
The results <published or shown> here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. Y.-K. Noh was partly supported by NRF/MSIT (No. 2018R1A5A7059549, 2021M3E5D2A01019545), IITP/MSIT Artificial Intelligence Graduate School Program for Hanyang University (2020-0-01373).
Abbreviations
- CAFs
Cancer-associated fibroblasts
- ECM
Extracellular matrix
- TCGA
The Cancer Genome Atlas
- GSEA
Gene set enrichment analysis
- FAP
Fibroblast activation protein-α
- GBM
Gradient Boosting Machine
- NSCLC
Non-small-cell lung carcinoma
- ROC
Receiver operating characteristic
- ML
Machine Learning
- MSigDB
Molecular signatures database
- DFS
Disease-free survival
- DSS
Disease-specific survival
- VEGF-A
Vascular endothelial growth factor-A
- HIF1α
Hypoxia-inducible factor 1 subunit alpha
- MMP-11
Matrix metallopeptidase-11
- IL-6
Interleukin-6
- TGF-β
Transforming growth factor-β
- GO
Gene ontology
- BRAF
V-raf murine sarcoma viral oncogene homolog B
Author contributions
Conceptualization: K.-W.M. Data curation: D-H.K. and B.K.S. Formal analysis: K.-W.M. Investigation: K.-W.M. and M.J.K. Funding acquisition: K.-W.M. Methodology: K.-W.M, Y.-K.N and J.-Y.M. Supervision: K.-W.M. Validation: K.-W.M. Writing—original draft: K.-W.M. Writing—review & editing: K.-W.M.
Data availability
The authors declare that all data supporting the findings of this study are available within the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kyueng-Whan Min, Email: kyueng@gmail.com.
Dong-Hoon Kim, Email: idavid.kim@samsung.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96344-1.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv. Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Wood DE, et al. Lung cancer screening, version 3.2018, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2018;16:412–441. doi: 10.6004/jnccn.2018.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossi A, et al. New targeted therapies and small-cell lung cancer. Clin. Lung Cancer. 2008;9:271–279. doi: 10.3816/CLC.2008.n.042. [DOI] [PubMed] [Google Scholar]
- 5.Merk J, Rolff J, Dorn C, Leschber G, Fichtner I. Chemoresistance in non-small-cell lung cancer: Can multidrug resistance markers predict the response of xenograft lung cancer models to chemotherapy? Eur. J. Cardiothorac. Surg. 2011;40:e29–33. doi: 10.1016/j.ejcts.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Sahai E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochet L, et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013;73:5657–5668. doi: 10.1158/0008-5472.CAN-13-0530. [DOI] [PubMed] [Google Scholar]
- 8.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020;11:5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin N, et al. Cancer-associated fibroblasts and desmoplastic reactions related to cancer invasiveness in patients with colorectal cancer. Ann. Coloproctol. 2019;35:36–46. doi: 10.3393/ac.2018.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporale A, et al. Is desmoplasia a protective factor for survival in patients with colorectal carcinoma? Clin. Gastroenterol. Hepatol. 2005;3:370–375. doi: 10.1016/S1542-3565(04)00674-3. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida GJ. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020;39:112. doi: 10.1186/s13046-020-01611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg TP, et al. Increased expression of cancer-associated fibroblast markers at the invasive front and its association with tumor-stroma ratio in colorectal cancer. BMC Cancer. 2019;19:284. doi: 10.1186/s12885-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng X, et al. Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep. 2018;23:255–269.e4. doi: 10.1016/j.celrep.2018.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Vega F, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bindea G, et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts EW, et al. Depletion of stromal cells expressing fibroblast activation protein-α from skeletal muscle and bone marrow results in cachexia and anemia. J. Exp. Med. 2013;210:1137–1151. doi: 10.1084/jem.20122344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 21.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saltz J, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji G-W, et al. Integrating machine learning and tumor immune signature to predict oncologic outcomes in resected biliary tract cancer. Ann. Surg. Oncol. 2020 doi: 10.1245/s10434-020-09374-w. [DOI] [PubMed] [Google Scholar]
- 25.Calon A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 26.Cheon D-J, et al. A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin. Cancer Res. 2014;20:711–723. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farmer P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 28.Haro M, Orsulic S. A paradoxical correlation of cancer-associated fibroblasts with survival outcomes in B-CELL LYMPHOMAS AND CARCINOMAS. Front. Cell Dev. Biol. 2018;6:98. doi: 10.3389/fcell.2018.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 2014;4:62. doi: 10.3389/fonc.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S, Du Y, Beckford J, Alachkar H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J. Transl. Med. 2018;16:170. doi: 10.1186/s12967-018-1539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J. Intern. Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi Y, Yokota A, Harada H, Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci. 2019;110:1510–1517. doi: 10.1111/cas.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendry S, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: Part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv. Anat. Pathol. 2017;24:311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa A, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article.