Abstract
The SARS-CoV-2 S protein on the membrane of infected cells can promote receptor-dependent syncytia formation, relating to extensive tissue damage and lymphocyte elimination. In this case, it is challenging to obtain neutralizing antibodies and prevent them through antibodies effectively. Considering that, in the current study, structural domain search methods are adopted to analyze the SARS-CoV-2 S protein to find the fusion mechanism. The results show that after the EF-hand domain of S protein bound to calcium ions, S2 protein had CaMKII protein activities. Besides, the CaMKII_AD domain of S2 changed S2 conformation, facilitating the formation of HR1-HR2 six-helix bundles. Apart from that, the Ca2+-ATPase of S2 pumped calcium ions from the virus cytoplasm to help membrane fusion, while motor structures of S drove the CaATP_NAI and CaMKII_AD domains to extend to the outside and combined the viral membrane and the cell membrane, thus forming a calcium bridge. Furthermore, the phospholipid-flipping-ATPase released water, triggering lipid mixing and fusion and generating fusion pores. Then, motor structures promoted fusion pore extension, followed by the cytoplasmic contents of the virus being discharged into the cell cytoplasm. After that, the membrane of the virus slid onto the cell membrane along the flowing membrane on the gap of the three CaATP_NAI. At last, the HR1-HR2 hexamer would fall into the cytoplasm or stay on the cell membrane. Therefore, the CaMKII_like system of S protein facilitated membrane fusion for further inducing syncytial multinucleated giant cells.
Keywords: Cell syncytia, Syncytial multinucleated giant cells, Ca2+-ATPase, HR1-HR2 hexamer, Calcium homeostasis
Background
COVID-19 is a unique disease characterized by extensive pulmonary thrombosis, the presence of viral RNA in lung cells and endothelial cells, and the formation of infected cell syncytia [1]. Histopathological results show that the patient’s lungs are featured with interval lymphocyte infiltration and macrophage alveolar exudate. The cells are enlarged, while the cytoplasm and lysosomes significantly grow [2]. Besides, there is focal type II pulmonary cell hyperplasia and multinucleated syncytial cell-induced cytopathic changes [3]. Atypical mesothelial cell aggregates with enlarged nuclei and multinucleated syncytial cells are also found in the pleural effusion cell by blocking agent preparations [4]. Alveolar syncytial cells have large nucleoli, granular cytoplasm, and prominent nucleoli [5]. Syncytial respiratory virus, metapneumovirus, paravirus, adenovirus, boca virus, coronavirus, and mycoplasma pneumoniae infection are tested negative for respiratory secretions on admission [6]. The SARS-CoV-2 S protein on the membrane of infected cells can initiate receptor-dependent syncytia formation, related to extensive tissue damage [7]. However, the mechanism of the SARS-CoV-2 S protein regulating the formation of these syncytia is not fully clear. In addition, from a study, it has been found that SARS-CoV-2 S protein mediated lymphocyte elimination through syncytia [8]. Apart from that, as infected cells form syncytia through membrane fusion with minimal S protein, it is difficult to obtain neutralizing antibodies and prevent them through antibodies in an effective way [9]. Thus, understanding the mechanism of SARS-CoV-2 S protein driving membrane fusion and inducing syncytia is of great value for immune prevention and treatment.
Some viral fusion proteins have apparent similarities to cell membrane fusion proteins [10]. The fusion of lipid bilayers to transmit genetic information is a standard process of viral infection and fertilization, since both have a common molecular mechanism [11]. The requirement for enveloped virus infection, including membrane fusion, may be related to the origin of sexual reproduction [12]. Research on fertilization reveals that a widely conserved protein HAP2/GCS1 is critical in the conservative membrane fusion reaction (13). As for HAP2/GCS1, it is an ancient gamete fusion protein [14], and also a structural homolog of the class II fusion protein used by many viruses [15] (including dengue fever and Zika virus) and parasitic microorganisms [16] (including Plasmodium (malaria), Toxoplasma gondii, and Trypanosoma). Actually, enveloped viruses use virus-encoded class II fusion proteins to fuse with host cells, while the decrease of the pH near the fusion protein causes the exposure of the HAP2/GCS1 structure that triggers membrane fusion and later forms a fusion hole [11], which then expands to complete the complete coalescence of the two membrane-bound compartments. Furthermore, it should be mentioned that HAP2/GCS1 in the extracellular domain has a cysteine-rich region [17], while the SARS-CoV-2 S protein possesses a cysteine-rich region across the viral membrane and cytoplasmic domain, namely CAF-motif (cysteine aggregation fusion). If CAF-motif belongs to the HAP2/GCS1 domain, the SARS-CoV-2 S2 protein has the characteristics of a class II fusion protein, whereas the endogenous retrovirus and the SNARE protein are featured with a class I virus fusion protein [10]. In terms of class I virus fusion protein, CoV S protein is the typical one, and protease cleavage is required to activate the fusion potential of S protein [18]. The SARS-CoV-2 S protein also has a protease cleavage site and a strong membrane fusion ability. Besides, the S protein possesses the characteristics of both class I and class II virus fusion proteins. Therefore, it can be believed that SARS-CoV-2 membrane fusion has events similar to fertilization.
Ion currents control SARS-CoV-2 virus particle entry, host membrane fusion, and subsequent virus replication in host cells [19], while Ca2+ and other ions play an important role in the sorting and delivery during virus infection [20]. To be specific, Ca2+ triggers membrane fusion between the virus and the infected cell and is like the early stage of fertilization [21]. The entry of sea urchin sperm into eggs is a relatively primitive fertilization process and pH changes throughout the entire process that includes depolarization of the egg cell membrane (Na + influx) [22], and an increase in membrane voltage to prevent polyfertilization [23]. Apart from that, there is Ca2+ release in egg cells, cortical response and H+ excretion without Na+ (acid discharge) [24], while respiration associated with the production of H2O2 increased, as well as oxygen consumption [25]. The increase of Ca2+ concentration will cause a series of chain reactions. For example, when NAD kinase is activated, NAD and NADH are converted into NADP and NADPH [26]; lipoxygenase is activated, and fat is oxidized [27]. In the later stage, H+ excretion is carried out through the NA+-H+ exchange system (needing Na+ to excrete acid) [28], which increases intracellular Na+ and reduces H+ concentration.
The enhanced cardiac late Na current (late INa) in host cell channels regulates SARS-CoV-2 infection. To be specific, the cardiac sodium channel Na v1.5 produces a rapidly inactivated depolarizing current I Na, responsible for the initiation and propagation of cardiac action potentials [29]. The dysfunction of Na v1.5 causes a decrease in peak I Na or an increase in residual late I Na (I Na, L). I NaL prolongs the increase in action potential (AP), leading to arrhythmias and sudden cardiac death [30]. Here, it should be mentioned that some ECG markers in patients with COVID-19 characterize that SARS-CoV-2 infection is associated with abnormal ion channel function, including Tp-e/QTc, Tp-e interval, and QTd increasing significantly [31], as well as QRS and QTc [32], while TDR decreasing, and iCEB value changing dramatically [32]. The epithelial sodium channel α subunit (ENaC-α) is often found in the same human lung and respiratory cells as ACE2. SARS-CoV-2 may employ the same protease that cleaves and activates ENaC-α to enter human respiratory cells [33]. Besides, SARS-CoV S downregulates ENaC channel activity at the whole-cell and single-channel level and affects the rate of cell exocytosis and endocytosis [34]. Moreover, late sodium current (INaL) increases in cardiomyocytes under many pathological conditions [35] and can cause Ca2+ overload [36]. An increase in I NaL leads to that in the intracellular sodium concentration, which is then increased by the reverse mode of the Na+/Ca2+ exchanger (NCX) [Ca2+]i [37]. Inhibition of INaL can reduce intracellular [Ca2+]i overload [38]. At the same time, Na+/K+-ATPase is used as a cardiac glycoside target to treat SARS-CoV-2 infection [39] and clear the symptoms of pulmonary edema. Respiratory viruses such as the influenza virus and coronavirus have downregulated the expression and activity of NKA [39]. However, the failure of the NA–K pump also caused an increase in NA-CA exchange, thus further resulting in calcium overload [40]. In addition, the IVS3-S4 extracellular loop concerning the α subunit of the extracellular Na + channel is an important component of α-scorpion and anemone toxin receptors. Then, the binding of toxins in this area activates and rapidly gets out of control of the Na+ channel [41]. Although the amino acid residues of the Na+ channel in the brain and the heart are slightly different, they will not affect the binding of the toxin to the Na + channel to a great extent. Therefore, after the SARS-CoV-2 S protein binds to the receptor, how the S protein binds to the Na+ channel extracellular α-scorpion and anemone toxin receptors will produce a cascade effect similar to the sodium influx in sea urchin fertilization.
The fusion peptide of S leads to the fusion of the viral membrane and the cell membrane at the initial stage and increases the influx of Na+ into the cell, while the insertion concerning the fusion peptide (FP) of S2 into the cell membrane starts an HR1-HR2 six-helix bundle conformation [42] and prompts membrane fusion [43]. According to calculation studies, it has been found that residues 816–855 of Spike are fusion peptides, when the combined peptide sequence in the trimer conformation contains Spike protein FP1 and FP2, activating the double bridge membrane system [44]. This computational simulation has revealed that the trimer (six-helix structure) interacts with the double cell membrane, causing lipids to migrate from the upper leaf of the lower bilayer to the lower leaf of the upper bilayer, thus forming a structural unit similar to a membrane bridge [44]. Furthermore, the long-trimeric double-stranded fusion peptide (FP-L) brings about lipid conformation and membrane bending, triggering the membrane bridge group instead of the fusion bridge group [44], and besides, it can pull the double membranes to an average distance of 1.8 nm within 200 ns [44], which will undoubtedly violently stimulate the cell membrane. In addition, noxious stimuli depolarize the cell membrane, instantaneously open voltage-gated sodium channels, and induce a large flow of sodium ions along a concentration gradient [45]. When the stimulation causes the depolarization of the cell membrane to exceed the excitation threshold, an action potential will be formed. In this case, all sodium ion channels will open, and extracellular sodium will quickly flow into the cell. The FP-L study finds that pH conditions and the presence of Ca2+ ions also affect the activity of S fusion peptides [44]. Moreover, calcium triggers protease-mediated membrane fusion activation of SARS-CoV-2 [46], while phosphorylated residues Y789 and T791 [47] near the N-terminus of FP may increase the negative charge and facilitate the accumulation of Ca2+ ions. Besides, in the absence of Ca2+ ions, membrane binding of FP will be reduced drastically [48], along with the fusion peptide binding two Ca2+ ions whose binding sites show greater synergy [49]. Therefore, it can be seen that the combination of Ca2+ ions with the FP can enhance penetration that changes the molecular organization of a specific area on the double layer to a greater extent.
The endolysosome is a huge calcium store in the cell. Many viruses have adapted to using the ion balance of the endolysosome to enter the cell [50]. Considering that, high [Na+] pretreatment increases the infectivity of Bunyavirus [51], while inhibition of K+ channels changes the distribution of K+ through the endosomal system and prevents the transport of viruses in the endosome [51]. However, the K+/Na + concentration status in the endolysosome has been reported to the contrary. Some studies have found that [K+] is high and [Na+] is low in the lysosome cavity [52]. In contrast, other studies have found that the content of [K+] is lower, while that of [Na+] is higher [53]. Cl− is an antagonist of H+ and other positively charged ions. In this case, Cl− channel modulators prevent viruses from entering cells by inhibiting virus binding and penetration and destroying Ca2+ homeostasis [54]. TPC dual-porous channels (TPC1 and TPC2) are part of the effective Ca2+ release channel family of endosomes and lysosomes—nicotinic acid-adenine dinucleotide phosphate (NAADP) receptors [55]. Besides, they have also been lysosomal Na + selective cation channels [53], whose opening induces strong sodium-driven depolarization in the endolysosome membrane to enhance membrane fusion [56]. The dual-porous channel coordinated the trafficking of endosomes from early to late periods, and late endosomes fuse with lysosomes to promote the release of viral RNA into the cytoplasm [57]. Thus, dual-porous channel is necessary for viruses such as EBOV (Ebola virus), MERS-CoV, and SARS-CoV, coordinating the interaction between the virus and the endolysosome environment [48]. As is known that the Middle East Respiratory Syndrome Coronavirus (MERS) relies on TPC to escape the endosome [58], Knockdown of TPC1 (for endosome) or TPC2 (for lysosome) will reduce furin activity and inhibit the fusion of MERS-CoV with the cell membrane [59]. The double-hole channel is also related to the invasion of SARS-CoV-2 [60], and the blockade of TPC also inhibits SARS-CoV-2 infection [50], when the flavonoid naringenin blocks the intracellular pathway of virus infection by targeting TPC [56]. Besides, JPT2 is a component of the NAADP receptor complex and is essential for TPC-dependent Ca2+ signaling and coronavirus entry control [61]. When the intracellular messenger NAADP (niacin adenine dinucleotide phosphate) activates TPC, Ca2+ is released through the TPC channel. During the process of SARS-CoV-2 fusions, NAADP mediates Ca2+ signaling, while acidic endosomes and TPC promote the release of Ca2+ from the endolysosome system [62]. Interestingly, NAADP is a derivative of NADP that adjusts to calcium storage, which is insensitive to inositol three phosphoric acid and cyclic ADP-ribose [63]. Besides, it also mobilizes Ca2+ from the reserve particles and lysosome-related organelles in sea urchin eggs [64]. In other words, the injection of sodium in the cell causes calcium overload and induces the production of NAADP that mobilizes the lysosome to unlock the double-pore channel and release calcium.
Ca2+ is released from the lysosome cavity and discharged to the outside of the cell through a calcium pump, which mediates membrane fusion. The interaction of Ca2+ ions with the fusion peptide induces spatial changes in the S protein, while the structure of the changed fusion peptide interacts with the cell membrane to promote the entry of MERS-CoV, SARS-CoV, and SARS-CoV-2 [65]. Besides, it should be mentioned that the multi-base cleavage site allows SARS-CoV-2 to mediate virus entry in a pH-independent manner [66]. When the protease of S is not bound to the surface of the plasma membrane, the coronavirus enters endocytic vesicles through endocytosis [67]. Unless A lysosomal protease cleaves the virus’s S protein, the virus will not escape into the cytoplasm by fusing with the inner lysosomal membrane [68], since the lysosome is an acidic lumen [69], and the proton pump V-ATPase complex maintains its low pH [70], which activates endolysosomal proteases to cleave the virus’s S glycoprotein that induces conformational changes in the virus fusion loop inserted into the endosomal membrane, pulling the virus and endolysosome membrane together. In the later stage of endocytic vesicles maturing into lysosomal endosomes, the coronaviruses get Ca2+ in mature endocytic vesicles or lysosomes [71]. Apart from that, the fusion of the viral envelope with the endolysosome membrane creates a fusion hole in the endosomal membrane, allowing its genome to be released into the cytoplasm for downstream viral replication [68]. Then, it can be seen from the above that membrane fusion between intracellular compartments and between lipid-binding structures (such as virus particles and cell membranes) follows the similar principle [72]; besides, the plasma membrane fusion (or lysosomal membrane fusion) mediated by SARS-CoV-2 S protein is like the membrane fusion of sea urchin fertilization as the fusion is regulated by the Na+, H+, and Ca2+ plasma and ion channels, and the Ca2+ sensor protein is actively involved.
Ca2+ sensing proteins include calmodulin and Ca2+/calmodulin-dependent protein kinase II (CaMKII), while EBOV virus protein possesses PI3 Kinase and CaMKII activities, which mediate the process of EBOV virus infection and membrane fusion [73], and are regulated by lysosomal Ca2+ flux [74]. The eukaryotic serine/threonine-protein kinase PfPK2 is expressed in the inner membrane of Plasmodium falciparum [75], and its catalytic domain partly similar to rat calmodulin-dependent protein kinase, human protein kinase C and bovine cAMP-dependent protein kinase, is essential for the invasion of P. falciparum merozoites into red blood cells. In terms of the CaMK-related kinase (TgCaMKrk) of Toxoplasma gondii, it is a CaMK homolog [76], and also a crucial regulator of the parasite’s invasion. In addition to that, the regulatory domain of PfPK2 comprises a self-inhibitory domain and a calmodulin-binding domain. However, TgCaMKrk is independent of calmodulin’s autophosphorylation and substrate phosphorylation activity. The calmodulin antagonists do not affect its kinase activity, while TgCaMKrk is at the top of the extracellular and intracellular tachyzoites of Toxoplasma gondii and is related to the glideosome. Then, Toxoplasma gondii tachyzoites directly invade nucleated cells and enter cells through phagocytic cells and phagocytosis. As mentioned earlier, the infection of cells by the EBOV virus and SARS-CoV-2 are both regulated by the TPC two-hole channel. Therefore, it is not surprising if the SARS-CoV-2 S protein possesses CaMKII activity that mediates the fusion of the virus with the cell membrane.
CaMKII is a vital calmodulin effector for endosomal fusion [77], and it is also a kind of multifunctional serine/threonine kinase expressed abundantly in the heart. Besides, CAMKII has a self-associative domain (CaMKII_AD) found at the C-terminus, which helps to assemble a single protein into two stacked hexameric holoenzymes composed of two monomers [78], each of which comprises an N-terminal catalytic domain and a C-terminal association domain. Between the two, it also involves the auto-regulatory domain (auto-inhibitory peptide, AIP) of the Ca2+/CaM binding site, which regulates the activation state through Ca2+/CaM binding and autophosphorylation [79]. Autophosphorylation in AIP prevents CaM binding and subsequent kinase reactivation [79],while phosphorylation at this site will permanently activate the CaMKII enzyme. Even without calcium and calmodulin, CamKII can remain active. After that, during the catalytic domain, there are several binding sites for ATP and other substrate-anchored proteins that are responsible for transferring phosphate from ATP to Ser or Thr residues in the substrate. A variety of upstream signals activates CaMKII, including Ca2 + and ROS [43]. Long-term Ca2+ pulses and ROS convert CaMKII into autonomous enzymes that do not rely on Ca2+ and calmodulin. Then, it should be noticed that the kinase families involved in Ca2+ signaling (CDPK, CRK, CCaMK, and SnRK3) all use Ca2+ sensors, combining Ca2+ with multiple EF-hands, with CDPK as a calmodulin-dependent protein kinase, a multifunctional protein kinase existing in plants and some protozoa. Apart from that, they are directly activated by binding to Ca2+ by the EF-hand of its C-terminal regulatory domain [80]. The EF-hand proteins of the Ca2+ sensor all have a high-speed turn-on rate, and the Ca2+ binding is only limited by the diffusion of Ca2+ [81]. Therefore, the CaMKII structure of SARS-CoV-2 S protein may have an EF-hand, which senses and responds to Ca2+.
When the two membranes are extremely close, the repulsive force of the thin water layer is stronger [82], preventing the two double layers from merging into a continuous sheet. In vesicle fusion, SNAREs interact with vesicle tethers in a calcium-dependent manner [83], while in the presence of Ca2+, t-SNARE, and v-SNARE in the opposite bilayer interact. In addition, they self-assemble into a ring conformation to form the conductive and leak-proof channel—Ca2+ bridges. Double-layer calcium bridging tightly binds bilayers, releases water from hydrated Ca2+ ions and displaces loosely coordinated water on the phosphate groups in the lipid membrane [84]. Therefore, it discharges water at the contact site between the bilayers, causing membrane instability: lipid mixing and membrane fusion. Beyond that, t-/v-SNARE is a tight complex, and its disassembly requires an NSF-ATPase, which acts as a right-hand molecule for movement [85]. In this case, it is speculated that SARS-CoV-2 membrane fusion was also calcium-induced and a similar calcium bridge was formed. Thus, the transport of Ca2+ across the membrane should precede the fusion and fission of various lipid bilayers, when Ca2+-ATPase is a prerequisite for membrane fusion in the process of exocytosis [86] and vacuolar fusion [87], and it is also involved in the Chlorella's virus infection [88]. Besides, the Ca2+-ATPase pump (IIB group of P-type ATPase) takes part in membrane fusion [89] in the fertilization process of sea urchin [90], guinea pig [91], hamster [92], bull [93], and Arabidopsis [94], regulates the dynein ATPase and controls the concentration of Ca2+[95], while animal Ca2+-ATPase (PMCA) [96] is located almost entirely on the plasma membrane, triggering Ca2+-mediated membrane fusion [97].
The animal Ca2+-ATPase (PMCA) is characterized with a C-terminal self-inhibitory extension, which can be released after binding Ca/calmodulin [98], while homologous proteins from fungi [96], plants [99], and protozoa lack this C-terminal extension. They are often in vacuoles or other intracellular organelles. CaATP_NAI is a unique N-terminal domain of the ACA2 pump (Arabidopsis IIB Ca2+-ATPase), being short of a long C-terminal calmodulin binding regulatory domain [100]. Whereas, CaM only acts on the full-length calcium pump in a concentration-dependent manner [101]. Moreover, the N-terminal truncation mutant restores the endogenous Ca pump, and the truncated pump is fully active and insensitive to calmodulin [100]. The pump with truncated N-terminal residues (ACA2-2) is 4 to 10 times more active than the full-length pump [102]. Moving the N-terminal region of ACA8 (Arabidopsis Ca2+-ATPase) to the C-terminus of the protein resulted in an utterly auto-suppressive mutant [103], while the virus AR158 of Chlorella NC64A encodes the Ca2+-ATPase of M535L, lacking the calmodulin-binding domain [88]. In the absence of CaM, the regulatory domain of type IIB Ca2+-ATPase interacts with one or more sites in the cytoplasmic head, hindering the enzyme’s catalytic activity [104] since its activation is aroused by Ca2+ and its binding of transporter sites [105]. Furthermore, environmental stress causes an increase in cytoplasmic Ca2+, and submicromolar Ca2+ concentrations trigger the activation of Ca2+-ATPases [106]. In addition, the S45/A (at N-terminal inhibitory or regulatory domain) substitution at the phosphorylation site of Ca2+-ATPase completely prevented the inhibition of pump activity by phosphorylation, and the S45/D (at N-terminal inhibitory or regulatory domain) substitution mimics the inhibition of pump activity phosphorylation [80].
Two types of Ca2+-activated kinases, namely PKCs [107] and CaMKs [108], regulate the activity of the Ca2+ pump. Specifically, IIB Ca2+-ATPase contains a single membrane-like Ca2+ binding site and directly undergoes ATP hydrolysis after binding to Ca2+ [101], while the sarcoplasmic reticulum of skeletal muscle keeps the membrane-bound Ca2+-ATPase, which mutually converts different energy. The initiation step before the final fusion step triggered by Ca2+ requires MgATP [109], while ATP takes part in biochemical reactions, mainly as the ATP-Mg complex. ATP is very stable, and motor protein catalyzes its decomposition (or hydrolysis). For example, myosin speeds up the rate of hydrolysis by 107 times [110]. The Ca2+-ATPases region of SARS-CoV-2 S may overlap with the structure of CaMKII function and motor protein function. Apart from that, Ca2+ activates the CaMKII kinase domain, promotes conformational changes, and arouses the activation of the Ca2+-ATPases domain. Then, the motor structure catalyzes the hydrolysis of ATP and pulls the viral membrane and cell membrane closer.
The SARS-CoV-2 E protein is an ion channel [111] that can mediate the permeable transmission of Ca2+, Na+, K+, and Cl− ions into the viral membrane [112]. SARS-CoV-2 may be a calcium reservoir, and the transmembrane trimer of S protein has Ca2+-ATPase activity. The time for the virus to infect cells is short. Calcium ions from cell efflux and the environment may accumulate in the membrane integration area for a long time or low [Ca2+]. Therefore, the virus quickly squeezes out the calcium in the virus cytoplasm through the Ca2+-ATPase enzyme of the S protein to participate in the rapid membrane fusion reaction. In addition to that, suppose in this short time, the calcium content pumped by the virus cannot meet the requirements of membrane fusion, the virus can only enter the endosomal vesicles of the cell through endocytosis.
In the current study, bioinformatics techniques such as conserved domain search were adopted to study the CaMKII regulation-related functions of the SARS-CoV-2 virus protein. The results showed that the S protein had conserved domains of CaMKII and Ca2+-ATPase. Besides, the EF-hand domain (“YEQYIKWPWYIWLGF”) of S protein bound to calcium ions; S2 protein had CaMKII protein and Ca2+-ATPase activities. After the S protein fusion peptide was inserted into the infected cell membrane and fixed the S2 protein on the cell membrane, the CaMKII_AD domains prompted the S2 protein to form HR1-HR2 six-helix bundles. To be specific, there are three Ca2+ATPase pumps with CaATP_NAI domain on the membrane contact surface of the six-helix. They are powered by ATP to promote the irreversible folding of the six helixes. Thus, the precise fusion of the viral membrane and the cell membrane is promoted. Therefore, the CaMKII-like system of S protein stimulated membrane fusion to induce syncytial multinucleated giant cells.
Method
Dataset
The sequences of SARS-CoV-2 proteins
The SARS-CoV-2 S protein sequences came from the NCBI database. In addition, the S protein sequence of SARS-CoV and MERS virus was downloaded from NCBI for comparison with SARS-CoV-2 protein.
The total length of SARS-CoV-2 S is 1273 amino acids, consisting of signal peptide (1–13 residues), S1 subunit (14–685 residues), and S2 subunit (686–1273 residues) [113], when the last two subunits are responsible for receptor binding and membrane fusion, respectively. In the S1 subunit, there is an N-terminal domain (14–305 residues) and a receptor-binding domain (RBD, 319–541 residues); besides, fusion peptide (FP) (788–806 residues), heptapeptide repeat 1 (HR1) (912–984 residues), HR2 (1163–1213 residues), TM domain (1213–1237 residues), and cytoplasmic domain (1237–1273 residues) contain S2 subunit.
Related sequences
The related sequence was downloaded from UniProt data set (Table 1).
Table 1.
Related sequences are used to search for conserved domains
| No | Related protein | Keywords | Count |
|---|---|---|---|
| 1 | Calmodulin | Bacteria + calmodulin | 6,386 |
| 2 | Serine/threonine kinase | Bacteria + serine + threonine + protein + kinase | 281,290 |
| 3 | Tyrosine kinase | Tyrosine + protein + kinase | 256,057 |
| 4 | Fertilization | Fertilization | 25,009 |
| 5 | ATPase | plant + atpase | 262,494 |
| 6 | Na ion channel | Na + ion + channel | 99,756 |
| 7 | Calcium ion channel | calcium + ion + channel | 130,875 |
| 8 | Hydrogen ion channel | Hydrogen + ion + channel | 443,511 |
| 9 | Potassium ion channel | Potassium + ion + channel | 172,110 |
| 10 | Ryanodine | ryanodine | 8,931 |
| 11 | IP3 | IP3 | 22,602 |
| 12 | Lysosomal ion channel | Lysosomal + ion + channel | 3,304 |
Crystal structures of HR1-HR2
The HR1-HR2 (PDB ID:6m3w) protein was downloaded from the PDB database.
The localized MEME tool of scanning for conserved domains
The analysis steps are listed as follows:
Downloaded MEME from the official Web site and subsequently installed it in the virtual machine ubuntu operating system. The virtual machine was VM 15.2.
Downloaded the SARS-CoV-2 S protein sequence from NCBI official Web site.
Downloaded the fasta format sequence such as calmodulin-related ones from Uniprot official website, respectively. The search keyword was “bacteria + calmodulin.”
For each sequence in all calmodulin-related protein, paired with each SARS-CoV-2 S protein sequence to generate fasta format files for MEME analysis.
For the files generated in step 4, a batch of 50,000 was used to create several batches, and it was considered as the limited space of the virtual ubuntu system.
In ubuntu, searched the conserved domains (E-value ≤ 0.05) of SARS-CoV-2 S protein and calmodulin-related with MEME tools in batches.
Collected the result files of conserved domains. Then, found the domain name corresponding to the motif from the uniprot database.
The domains’ activity of each SARS-CoV-2 S protein was analyzed according to the characteristics of the calmodulin-related protein domains.
Multiple sequence alignment
The local version of Clustal X 2.1 [114] was used to perform multiple sequence alignments on the S proteins of SARS-CoV-2, SARS-CoV, and MERS, when the parameters were the default values.
Results
SARS-CoV-2 S protein featured with ion channel regulation function
In order to determine the ion channel regulation function of S protein, Ca2+ ion channel (keyword “calcium + ion + channel”), K+ ion channel (keyword “Potassium + ion + channel”), H+ ion channel (keyword “Hydrogen + ion + channel”), Na+ ion channel (keyword “Na + ion + channel”), lysosome ion channel (keyword “Lysosomal + ion + channel”), IP3 channel (keyword “IP3”) and ryanodine channel (keyword “ryanodine”) related sequences of ion channels were downloaded from the UniProt database. Besides, the local MEME tool was adopted to compare these sequences with the S protein to search for ion channel regulatory domains. Then, the motif sequences were merged according to the domain of the search results. Furthermore, it should be mentioned that the current study focuses on the relevant domains of Na+ and Ca2+ ion channel regulation. The results are that S protein can regulate Na+ ion channels, and some regions of S protein have functions similar to Ca2+ ion channels, as shown in Table 2.
Table 2.
Na+ and Ca2+ ion channel regulation functions of SARS-CoV-2 S protein
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| LCN-type CS-alpha/beta | A | YYHKNNKSWMESEFR | 144 | 158 |
| B | ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSC | 1202 | 1243 | |
| RyR | α1 | VYYHKNNKSWMESEFRVY | 143 | 160 |
| α2 | VYAWNRKRI | 350 | 358 | |
| β1 | FPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPRE | 1052 | 1092 | |
| β2 | QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHY | 1201 | 1272 | |
| B30.2/SPRY | β1 | CHDGKAHFPRE | 1082 | 1092 |
| β2 | YEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCC | 1206 | 1248 | |
| Ion_trans | α1 | WFHAIHVSGTNGTKRFDNPVLPF | 64 | 86 |
| α2 | VCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREF | 130 | 192 | |
| α3 | RGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAW | 403 | 436 | |
| α4 | GAEHVNNSYECDIPIGAGIC | 652 | 671 | |
| β1 | DCTMYICG | 737 | 744 | |
| β2 | WTFGAGAALQIPFAMQMAYRF | 886 | 906 | |
| β3 | SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLH | 1021 | 1064 | |
| β4 | PAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTD | 1069 | 1118 | |
| β5 | ESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHY | 1195 | 1272 |
The LCN-type CS-alpha/beta domain (IPR044062, PS51863) is a stable α/β (CS-α/β) domain of LCN-type cysteine. In addition, scorpion venom is a kind of neurotoxin and shares the CS-α/β structural motif, which represents a cysteine-stabilized α-helix/β-sheet. Long-chain or voltage-gated sodium channel scorpion neurotoxins (LCN, NaScTxs) can be classified as: α toxins, which can slow down the inactivation of Na+ channels; and β toxins, which can affect the channel activation process. Moreover, the LCN-type CS-alpha/beta A domain of S protein is located in the S1 protein. After S1 is bound to the ACE2 receptor, the LCN-type CS-alpha/beta A bound to the Na+ channel extracellular α-scorpion and anemone toxin receptors [41], triggering the inactivation of the Na+ channel and the infusion of a large number of sodium ions into the cell. Beyond that, the LCN-type CS-alpha/beta B domain exists in the S2 protein, close to the virus’s outer membrane, transmembrane, and cytoplasmic regions. In the late stage of membrane fusion, LCN-type CS-alpha/beta B also bound to the Na + channel extracellular α-scorpion and anemone toxin receptors [41], causing Na+ channel inactivation.
The RyR domain (PF02026) is the Ryanodine receptor. Ryanodine receptors are calcium-release channels. The B30.2/SPRY domain (IPR001870) can appear after the RYanodine receptor. The Ion_trans domain (PF00520) is the sodium, potassium, and calcium ion channel. Table 2 shows that S1 and S2 can respectively construct calcium release ion channels. Ion_trans α1 to Ion_trans α4 constitutes the calcium channel of S1, while Ion_trans β1 to Ion_trans β5 is composed of the calcium channel of S2. However, these two calcium-releasing ion channels are far apart and may be connected to different calcium sources. Besides, the calcium-releasing ion channel of S1 mainly comes from the environment or the efflux of cells. Due to the limited literature published, its role is not yet clear. In addition to that, the calcium-releasing ion channels of S2 are close to the virus’s outer membrane, transmembrane, and cytoplasmic regions, while the calcium-releasing ion channel of S2 provides Ca2+ for the membrane fusion stage. As for the calcium source, it should come from the virus cytoplasm, while the calcium-release ion channel of S2 may squeeze out the calcium in the virus cytoplasm through the nearby Ca2+-ATPase.
S protein with conserved domains of CaATP_NAI and CaMKII_AD
CaMKII-Like domains of S protein
The bacterial calmodulin-related protein sequence was downloaded from the UniProt Web site (search keyword “bacteria + calmodulin”). Then, the MEME local tool was used to compare the S protein sequence with the calmodulin-related protein sequence, and search for conserved domains. Furthermore, the S protein motif sequences were integrated by domains and continuity (Table 3), and then the motifs were coded according to S1 corresponding to alpha, S2 corresponding to beta, and the sequence number of the site. Table 3 shows that S protein has CaMKII_AD, CaATP_NAI, DUF4440, EF-hand, and SnoaL-like domains, while the catalytic domain of CaMKII protein binds ATP. From the KEGG official Web site, it was found that the Mus musculus CaMKII protein is featured with entry = 12,323, and its motifs are Protein Kinase, CaMKII_AD, PK_Tyr_Ser-Thr, DUF4440, SnoaL_3, and SnoaL_2 domains. Table 3 shows that S has other domains except for the PK_Tyr_Ser-Thr domain. Besides, the PK_Tyr_Ser-Thr domain would be searched through the kinase sequences later.
Table 3.
CaMKII-like related motifs possessed by S protein of SARS-CoV-2
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| CaATP_NAI | β | APAICHDGKAHFPRE | 1078 | 1092 |
| CaMKII_AD | α1 | CNDPFLGVYYHKNNKSWMESEFR | 136 | 158 |
| α2 | RFASVYAWNRKRI | 346 | 358 | |
| β1 | MQMAYRF | 900 | 906 | |
| β2 | RVDFCGKGYHLMSFPQSAPH | 1039 | 1058 | |
| β3 | AQEKNFTTAPAICHDGKAHFPREGVFVSNGTHW | 1070 | 1102 | |
| β4 | ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF | 1202 | 1256 | |
| DUF1338 | β | KWPWYIW | 1211 | 1217 |
| DUF4440 | β | QYIKWPWYIW | 1208 | 1217 |
| EF-hand | β | YEQYIKWPWYIWLGF | 1206 | 1220 |
| SnoaL-like | α | YHKNNKSWMES | 145 | 155 |
| β | WPWYIW | 1212 | 1217 |
Table 3 shows that CaMKII-like related domains are mainly in and near the S2 protein region. However, there was no finding that these two cleavage sites are located in the CaMKII-like related domains of the S protein, which indicates that CaMKII-like related domains may have relatively conservative functions.
X. Fan et al. determined the S2 structure before and after the fusion [115]. The transition from the pre-fusion state to the post-fusion state triggers the drastic structural rearrangement and conformational changes of SARS-CoV S glycoprotein, and the largest structural rearrangement is located in the HR1-CH helix region. By comparing the sequence’s range, it was found that the CaMKII_AD and CaATP_NAI domains play an essential role in the fusion. In addition, the CaMKII_AD motif “MQMAYRF” (900–906) is in the HR1 motif (894–966) region, and the CaMKII_AD motif “RVDFCGKGYHLMSFPQSAPH” (1039–1058) exists in the BH motif (1017–1053) region, while the CaMKII_AD motif “AQEKNFTTAPAICHDGKAHFPREGVFVSNGTHW” (1070–1102) appears in the SD3 motif (1054–1104) region, and the CaATP_NAI domain “APAICHDGKAHFPRE” (1078–1092) is also in the SD3 motif. HR1 is a crucial component of HR1-HR2 six-helix, while the pan-coronavirus fusion inhibitor EK1 peptide variant EK1C4 can inhibit SARS-CoV-2 fusion in a dose-dependent manner by binding to HR1 [116], along with the BH motif generating β-hairpin. X. Fan et al. believed that the linker region and the SD3 domain are involved in the folding and extension actions during fusion. The disulfide bond (C1064–C1108) between the linker region upstream of the HR2 motif and the SD3 domain stabilizes the linker region during the fusion transition. Furthermore, the disulfide bond is in the overlapping region of the CaMKII_AD and CaATP_NAI domains. Except the FP motif (789–809, fusion peptide), CR motif (823–893, connecting region), CH motif (967–1016, central helix region), L motif (688–711, 1105–1143), UH motif (712–753, upper spiral region), and HR2 motif (1145–1178), the CaMKII_AD domain makes up three motifs of HR1, BH and SD3, which means that HR1, BH, and SD3 mediate the membrane fusion process through activity of CaMKII_AD and CaATP_NAI.
CaMKII_AD domain
HR2 is at residues 1163–1213, and part of the fragment is in the C-terminal CaMKII_AD domain of the S protein (“ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF,” 1202–1256) region. Besides, the C-terminal CaMKII_AD domain is in and near the virus cytoplasmic domain [117], while the cytoplasmic domain region is close to the viral membrane-proximal part of the viral membrane CD domain region (1234–1273). There are also three adjacent CaMKII_AD domains in the connecting region upstream of the HR2 motif: (“MQMAYRF,” 900–906), (“AQEKNFTTAPAICHDGKAHFPREGVFVSNGTHW,” 1070–1102) and (“RVDFCGKGYHLMSFPQSAPH,” 1039–1058). In this case, it is shown that the CaMKII_AD domain of the S protein is involved in the formation of the HR1-HR2 six-helix bundle, holding the function of the self-association domain in the CAMKII protein. Besides, the formation of multimers through the CaMKII_AD domain may be an essential way for the SARS-CoV-2 S protein.
CaATP_NAI domain
The CaATP_NAI motif of S is “APAICHDGKAHFPRE,” which overlaps with the CaMKII_AD motif “AQEKNFTTAPAICHDGKAHFPREGVFVSNGTHW.” Obviously, there is no threonine (L-threonine) in the CaATP_NAI motif and the overlapped CaMKII_AD motif. Therefore, Ca2+-ATPase of S protein may not depend on threonine phosphorylation for permanent activation.
Related domains of CaMKII-like system activation
Table 3 shows that the motifs of DUF4440, EF-hand, protein kinase, and SnoaL-like domains of S protein are all shorter and have a common motif “WPWYIW” (1212–1217). In those domains, the consensus motif is the C-terminal CaMKII_AD domain (1202–1256), outside the viral transmembrane domain. In addition to that, the highly conserved 8-residue sequence (KWPWY/WVWL) is necessary for membrane fusion but not crucial for S protein incorporation into particles [118]. Moreover, the specific motif also mediates the particle assembly of coronavirus spikes [118], which mainly related the functions of these domains to the activation of the CaMKII_AD domain function, when these domains may be similar to the Ca2+/CaM binding region.
SnoaL-like domain
It is a polyketide cyclase biosynthesized by Nogalamycin, which uses a twisted alpha–beta barrel fold [119]. In addition to that, SnoaL-like catalyzes the loop-closing step in the biosynthesis of polyketide antibiotics produced by Streptomyces, and it also plays an important role in the binding of aglycones. The SnoaL-like domain motifs of S are “WPWYIW” and “YHKNNKSWMES,” while the “WPWYIW” motif is in the S2 protein, along with the “YHKNNKSWMES” motif in the S1 protein and the motif “YHKNNKSWMES” in the N-terminal CaMKII_AD domain (136–158 residues), which represents that the SnoaL-like domain is related to CaMKII_AD function.
EF-hand domain
The EF-hand motif of S protein is “YEQYIKWPWYIWLGF”. In the current study, it was believed that the region of EF-hand motif bound to Ca2+, had a calmodulin-like role and promoted the S conformation change.
DUF4440 domain
The domain superfamily (IPR032710) is similar to NTF2 [149]. The NTF2 protein has a beta-alpha(2)-beta insertion behind the primary helix, and it promotes protein transport to the nucleus. Besides, the protein of this domain also includes calcium/calmodulin-dependent protein kinase II and the association domain (IPR013543). Furthermore, the S protein DUF4440 motif “QYIKWPWYIW” may mainly play the role of CaMKII_AD, assisting with the formation of HR1–HR2 six-helix bundles.
PK_Tyr_Ser-Thr domain and autophosphorylation
Protein kinases fall into three broad classes: serine/threonine-protein kinases, tyrosine-protein kinases, and dual specificity protein kinases, while CaMKII mainly involves serine/threonine-protein kinases (i.e., PK_Tyr_Ser-Thr domain).
The protein sequences related to serine/threonine kinase were downloaded from the uniprot database (keyword “Bacteria + serine + threonine + protein + kinase”) as well as protein sequences related to tyrosine kinase from the uniprot database (keyword “Tyrosine + protein + kinase”). Then, the local MEME tool was used to search for the conserved result domains of S and two types of kinase proteins. Furthermore, the searched motif sequences were merged according to structural domain names, and besides, aliases were given to the motifs. The PK_Tyr_Ser-Thr domains and models owned by S are shown in Table 4.
Table 4.
PK_Tyr_Ser-Thr domains of SARS-CoV-2 S protein
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| PK_Tyr_Ser-Thr | ɑ1 | KVCEFQFCNDPFLGVYYHKNNKSWMESEFRVY | 129 | 160 |
| ɑ2 | YAWNRKRI | 351 | 358 | |
| β1 | MSECVL | 1029 | 1034 | |
| β2 | CHDGKAHFPREGVFVSNGTHW | 1082 | 1102 | |
| β3 | QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLH | 1180 | 1271 |
PK_Tyr_Ser-Thr represents the catalytic domain [120] found in much serine/threonine and tyrosine protein kinases, excluding the catalytic domain of dual-specificity kinases. Besides, it is responsible for the reversible process mediated by protein kinase and phosphoprotein phosphatase. Furthermore, phosphorylation usually changes the target protein’s function by changing the activity of the enzyme, the location of the cell, or the association with other proteins.
Table 4 shows that both S1 and S2 proteins have serine/threonine-protein kinases domains. Then, it can be seen that PK_Tyr_Ser-Thr ɑ1 is in the N-terminal domain, and PK_Tyr_Ser-Thr ɑ2 is in the receptor-binding domain, while PK_Tyr_Ser-Thr β1 and β2 are in the BH and SD3 domains. In addition, PK_Tyr_Ser-Thr β3 is in the HR2, TM, and cytoplasmic domains, with PK_Tyr_Ser-Thr ɑ1 being in the CaMKII_AD ɑ1 domain, and PK_Tyr_Ser-Thr ɑ1 in the CaMKII_AD ɑ2 domain. Apart from that, PK_Tyr_Ser-Thr β2 and CaMKII_AD β3 overlap, as well as PK_Tyr_Ser-Thr β3 and CaMKII_AD β4 (1202–1256). Furthermore, PK_Tyr_Ser-Thr’s ɑ1, ɑ2, β2 and β3 are all in the auto-inhibitory domain of CaMKII.
PK_Tyr_Ser-Thr ɑ1, β1, β2 and β3 all have serine and threonine. To determine whether these four domains are featured with the function of catalyzing autophosphorylation, the S protein phosphorylation sites [121] found in the global phosphorylation spectrum experiment were compared after infection: S31, S349, S459, S637, S640, S816, S1161, S1196, S1261, T29, T240, and T791. Only the two phosphorylation sites of S1196 (CK2_group) and S1261 (CK2_group) are in the PK_Tyr_Ser-Thr β3 domain. In addition to that, none of the other phosphorylation sites are in the PK_Tyr_Ser-Thr domain. However, as neither S1196 nor S1261 is in the CaMKII_AD domain, these two sites are not phosphorylation concerning the CaMKII self-inhibitory domain of S2. Besides, PK_Tyr_Ser-Thr β3 is the most extended motif PK_Tyr_Ser-Thr, and is the region with Serine/threonine-protein kinases activity. Whereas, PK_Tyr_Ser-Thr β3 spans the calcium-binding region of the EF-hand. Therefore, S1196 and S1261 are two autophosphorylation sites for the entire CaMKII-like structure. Since these two phosphate sites are measured after cell infection [122] and are not dephosphorylated, phosphorylation of S1196 and S1261 sites is very important for viral infection and membrane integration. After the EF-hand of S2 engages with Ca2+, the CaMKII-like activity of S2 is initially activated. Besides, the PK_Tyr_Ser-Thr β3 domain is also activated. After PK_Tyr_Ser-Thr β3 domain autophosphorylated the S1196 and S1261 sites, the CaMKII-like structure of S2 may be in a self-activated state. In other words, Ca2+ no longer regulates the activated state.
The Ca2+-ATPase and motor of the SARS-CoV-2 S protein
Position of Ca2+-ATPase and motor of SARS-CoV-2 S
The Ca2+-ATPase pump with the CaATP_NAI domain is the IIB P-ATPase. The Ca2+ pump can hydrolyze ATP, while the motor protein is located near the ATP binding zone and can catalyze the hydrolysis of ATP. Unlike that, the ACA2 pump found in Arabidopsis has the CaATP_NAI domain. In order to determine the position of the ATPase and motor structure of SARS-CoV-2 S, the plant ATPase sequence (keyword “plant + atpase”) was downloaded. Then, these sequences were compared with the SARS-CoV-2 S protein to search for the relevant domains and motifs of ATPase and motors. In addition to that, the motif sequences were merged by domains. The results (Table 5) show that SARS-CoV-2 S contains PhoLip_ATPase, Cation_ATPase, Kinesin motor, and Myosin motor domains.
Table 5.
ATPase and motor of the SARS-CoV-2 S protein
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| PhoLip_ATPase_N | β1 | QEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEP | 1071 | 1112 |
| β2 | WPWYIW | 1212 | 1217 | |
| PhoLip_ATPase_C | α | AWNRKRISNC | 352 | 361 |
| β1 | DFCGKGYHLMSFPQSAPH | 1041 | 1058 | |
| β2 | KYEQYIKWPWYIWLGF | 1205 | 1220 | |
| Cation_ATPase_N | α | VYYHKNNKSWME | 143 | 154 |
| β | QELGKYEQYIKWPWYIWLGFIAGLIAIVM | 1201 | 1229 | |
| Cation_ATPase_C | β1 | HWFVTQRNFYEPQII | 1101 | 1115 |
| β2 | YEQYIKWPWYIW | 1206 | 1217 | |
| Kinesin motor | α1 | WFHAIH | 64 | 69 |
| α2 | YHKNNKSWM | 145 | 153 | |
| β1 | IPFAMQMAYR | 896 | 905 | |
| β2 | HFPREGVFVSNGTHWFVTQRNFYE | 1088 | 1111 | |
| β3 | QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF | 1201 | 1256 | |
| Myosin motor | β | YIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCK | 1209 | 1255 |
PhoLip_ATPase is a phospholipid transport ATPase, while PhoLip_ATPase_N (PF16209) and PhoLip_ATPase_C (PF16212) are the N-terminal and C-terminal of PhoLip_ATPase, respectively. Furthermore, it is also called ATP phospholipid-flipping (P-ATPase) that catalyzes the inward movement of phospholipids from the extracellular/intraluminal lobules to the cytoplasmic lobules and the outward movement of other lipids [123]. In addition to that, the membrane fusion of S2 protein involves the generation of fusion pores. PhoLip_ATPase flipped the phospholipids of the membrane during the formation of the fusion channel, and the hydrophilic end of the inner membrane faced the center of the pore, when the hydrophobic end of the inner membrane faced the outside of the pore. Therefore, the inner membrane phospholipids were spliced together to form the inner membrane of the pore. Meanwhile, outer membrane phospholipids are spliced together to form the outer membrane of the pore. PhoLip_ATPase_C α is in the S1 protein, being short and having no corresponding N-terminal, which may not be functional. Here, it is noted that PhoLip_ATPase_N β1, PhoLip_ATPase_N β2, PhoLip_ATPase_C β1, and PhoLip_ATPase_C β2 are all in the S2 protein. Taking the smallest N-terminus and the largest C-terminus, the region of PhoLip_ATPase can be determined to be PhoLip_ATPase_N β1 to PhoLip_ATPase_C β2 or the region of 1071–1220 that includes EF-hand (“YEQYIKWPWYIWLGF”) calcium-binding and other activation areas, which indicates that the ATPase is also calcium-activated.
Cation_ATPase participates in the transmission of Na+/K+, H+/K+, Ca2+, and Mg2+, while Cation_ATPase_N (PF00690) and Cation_ATPase_C (PF00689) are the N-terminal and C-terminal of Cation_ATPase, respectively. Besides, Cation_ATPase_N α is in the S1 protein, featured with shortness and no corresponding N-terminal, which may not be functional. In addition, Cation_ATPase_N β, Cation_ATPase_C β1, and Cation_ATPase_C β2 are all in the S2 protein. Taking the smallest N-terminal and the largest C-terminal as an example, the region of Cation_ATPase can be determined to be Cation_ATPase_N β, that is, the region of 1201–1229, which also contains the EF-hand (“YEQYIKWPWYIWLGF”) calcium-binding area, indicating that the ATPase is also calcium-activated.
The CaATP_NAI domain (1078–1092) is in PhoLip_ATPase_N β1, which indicates that PhoLip_ATPase_N β1 is an extended region of CaATP_NAI. Combining CaATP_NAI domain, Cation_ATPase_N β and Cation_ATPase_C β2 of SARS-CoV-2 S, it shows that the Ca2+-ATPase region of SARS-CoV-2 S is 1071–1229. Besides, the sites of PhoLip_ATPase and Cation_ATPase overlap, demonstrating that the region 1071–1229 is the ATPase of S protein involved in Ca2+ ion transport and phospholipid turnover.
Kinesin motor (cd00106) is the kinesin motor domain, which has ATPase activity, while myosin motor (PS51456) is the myosin motor domain and contains the ATPase domain. As 1071–1229 is the ATPase of S protein, kinesin motor β2, and kinesin motor β3, myosin motor β has motor functions. Furthermore, since kinesin motor β3 and Myosin motor β overlap, the region 1088–1255 is the motor structure of S protein.
The ATPase of S protein overlaps with the motor structure area.
Activation and inhibition of Ca2+-ATPase of SARS-CoV-2 S
The initial analysis indicated that S1196 and S1261 are the autophosphorylation sites of CaMKII of S protein that both are not in the CaATP_NAI domain of the S protein (“APAICHDGKAHFPRE,” 1078–1092). Apart from that, the protein name corresponding to the CaATP_NAI domain of S protein was found from the domain search result file: A0A365XXB7 CaATP_NAI domain-containing protein (Chitinophaga Flava). Besides, the motif sequence corresponding to this protein is “WRHVVHKNMSHRWRM” [2–31], while the S11 of the A0A365XXB7 protein is mutated to A1087 on the S protein. A study shows that due to S/A mutation in N-terminal inhibitory (or the regulatory domain), Ca2+-ATPase activity is not inhibited by phosphorylation [80]. Therefore, the CaATP_NAI domain of S protein cannot be phosphorylated, and the Ca2+-ATPase activity of S protein cannot be inhibited.
The phosphorylation site of S1196 is in the Ca2+-ATPase of S protein (1071–1229). Therefore, the phosphorylation site of Ca2+-ATPase is S1196 that is also one of the autophosphorylation activation sites of CaMKII. Since S1196 is phosphorylated during all infection processes [122], Ca2+-ATPases of S protein actives for a long time.
No CaM protein binding region in the CaATP_NAI domain region of the S protein was found through the domain search mentioned above. In the absence of CaM, the regulatory domain of type IIB Ca2+-ATPase interacts with one or more sites in the cytoplasmic head, hindering the enzyme’s catalytic activity [104], while the activation of Ca2+-ATPases results from Ca2+ and its binding of transporter sites [105]. Moreover, environmental stress causes an increase in cytoplasmic Ca2+, and submicromolar Ca2+ concentrations trigger the activation of Ca2+-ATPases [106]. Therefore, the CaATP_NAI domain combines with TM and cytoplasmic head domains to inhibit the Ca2+-ATPase active of S protein. Besides, the Ca2+-ATPase of S protein is activated by EF-hand (“YEQYIKWPWYIWLGF”) binding to Ca2+, while because of the autophosphorylation of S1196 and S1261, CaMKII and Ca2+-ATPase is always active.
CaATP_NAI of SARS-CoV-2 S, SARS-CoV S, and MERS S. For SARS-CoV S and MERS S protein, the same domain search method of SARS-CoV-2 S protein was adopted. Since many motif fragments were not conducive to analysis, the motif sequences of the search results were merged. Furthermore, the relevant structural domains of the CaMKII_Like system of SARS-CoV are shown in Table 6, and those of the CaMKII_Like system of MERS are displayed in Table 7. Then, it can be seen from the two tables that SARS-CoV and MERS also have CaATP_NAI and CaMKII_AD domains, but their motifs are different from SARS-CoV-2. Besides, SARS-CoV’s motif “RVDFCGKGYHLMSFPQAAPH” (CaMKII_AD) is similar to the SARS-CoV-2’s motif “RVDFCGKGYHLMSFPQSAPH” (CaMKII_AD), while SARS-CoV-2’s motif “ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF” (CaMKII_AD) is originated from SARS-CoV’s motif “NLNESLIDLRELGKYEQYIKWPWYVWLGFIAGLIAIVMVT” (CaMKII_AD) and MERS’s motif “CCTGCGTNCMGKLKCNRCCDRYEEYDLEPHKVHVH” (CaMKII_AD), when this SARS-CoV motif is at the N-terminal, and this MERS motif at the C-terminal. Both CaATP_NAI and CaMKII_AD also overlap, but the CaATP_NAI motifs of the three species are different, which may be a significant cause of SARS-CoV-2 specific fusion events. Besides, DUF4440, EF-hand, protein kinase, and SnoaL-like domains all include the “WPWY[I/V]W” motif, while the position and function of this motif are similar, showing that the site is highly conservative.
Table 6.
CaMKII-like related motifs possessed by S protein of SARS-CoV
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| CaATP_NAI | α | YRYLRHGKLRPFERDI | 440 | 455 |
| CaMKII_AD | α | YNYKYRYLRHGKLRPFERDISNVP | 436 | 459 |
| β1 | TKMSECVLGQSKRVDFC | 1009 | 1025 | |
| β2 | RVDFCGKGYHLMSFPQAAPH | 1021 | 1040 | |
| β3 | HEGKAYFPR | 1065 | 1073 | |
| β4 | NLNESLIDLRELGKYEQYIKWPWYVWLGFIAGLIAIVMVT | 1174 | 1213 | |
| DUF4440 | β | QYIKWPWYVW | 1190 | 1199 |
| EF-hand | α | DDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHGK | 414 | 447 |
| Protein kinase | β | GKYEQYIKWPWYVW | 1186 | 1199 |
| β | WPWYVWLGF | 1194 | 1202 | |
| SnoaL-like | β | RELGKYEQYIKWPWYVW | 1183 | 1199 |
Table 7.
CaMKII-like related motifs possessed by S protein of MERS
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| CaATP_NAI | β | CCTGCGTNCMGKLKCNRC | 1319 | 1336 |
| CaMKII_AD | α1 | CTFMYTYNITEDEILEWF | 237 | 254 |
| α2 | PLEGGGWLVASGSTVAMTEQLQMGF | 547 | 571 | |
| β1 | PNGLYFMHVGYYPSNHIE | 1131 | 1148 | |
| β2 | KELGNYTYYNKWPWYIWLGFIAG | 1284 | 1306 | |
| β3 | CCTGCGTNCMGKLKCNRCCDRYEEYDLEPHKVHVH | 1319 | 1353 | |
| DUF4440 | β | KWPWYIW | 1294 | 1300 |
| EF-hand | β1 | WPWYIWLGF | 1295 | 1303 |
| β2 | KLKCNRCCDRYEEYDLEPHKVHVH | 1330 | 1353 | |
| Protein kinase | β1 | KWPWYIW | 1294 | 1300 |
| β2 | NCMGKLKCNRCCDRYEEYDLEPHKVHVH | 1326 | 1353 | |
| SnoaL-like | β1 | KWPWYIW | 1294 | 1300 |
| β2 | RCCDRYEEYDLEPHKVHVH | 1335 | 1353 |
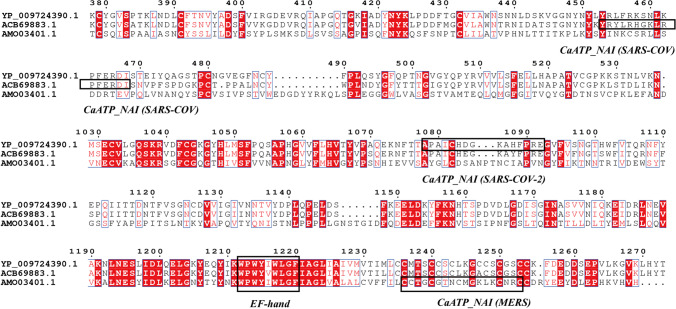
In addition to what was mentioned above, the local version of Clustal X 2.1 [114] was employed to perform multiple sequence alignments of the S proteins in the three species. Based on this comparison result, the online ENDscript server [124] was adopted to plot the comparison result and besides, the EF-hand and CaATP_NAI domains were marked on the result map (Fig. 1). Since the original image is huge, only the sequence near the EF-hand and CaATP_NAI domains was intercepted. Then, the comparison result of S proteins showed that SARS-CoV-2 and SARS protein were very similar, with more point mutation differences. However, MERS and SARS-CoV-2 (or SARS) were pretty different. In addition to some mutations, there were many insertion mutations. Therefore, it can be seen that the three species have a similar EF-hand domain sequence “WPWY[I/V]WLGF.” Apart from that, the motif “KWPWY/WVWL” has been confirmed by experiments reported in the literature to participate in membrane fusion [118].
Fig. 1.
CaATP_NAI domains of S protein among SARS-CoV-2, SARS, and MERS; YP_009724390.1 is the S protein of SARS-CoV-2. ACB69883.1 is the S protein of SARS-CoV. AMO03401.1 is the S protein of MERS. This figure is drawn by ENDscript server [124]
The positions of the CaATP_NAI domains belonging to the three species are quite different. To be specific, the CaATP_NAI of MERS is located at the end of C-terminal, the CaATP_NAI of SARS-CoV-2 at the C-terminal (upstream of MERS), and the CaATP_NAI of SARS-CoV at the N-terminal, which is caused by insertion mutations and point mutations, while the difference in SARS-CoV is caused by point mutations. Beyond that, the difference in position reflects the different roles of CaATP_NAI in the three species. As for the CaATP_NAI of SARS-CoV, located in the S1 protein, it may be mainly responsible for participating in receptor binding and may contribute less to the fusion of S2 protein. Apart from that, the CaATP_NAI of MERS is at the C-terminus of S2 protein, close to the cytoplasmic region of the virus, and the fragment near the C-terminus is also the CaMKII_AD region of SARS-CoV-2. Therefore, CaATP_NAI of MERS may be able to drive S2 conformational changes, and less contribute to the fusion too. Besides, the CaATP_NAI of SARS-CoV-2 is at the C-terminal of the S2 protein, outside the virus membrane, and at the interface between the virus and the cell. Then, an ATP motor at the position will undoubtedly give a strong driving force to achieve precise membrane positioning and fusion, which however doesn’t exist in the S2 proteins of SARS-CoV and MERS. Therefore, cell infections of SARS-CoV and MERS are often manifested in the form of endocytosis, while SARS-CoV-2 cell infections are not only in the formation of endocytosis but also in membrane fusion.
The S2 protein generating membrane fusion pores through the HAP2/GCS1 structure
To study whether the S protein has a HAP2/GCS1 activity, the protein sequences related to fertilization were downloaded from the UniProt database (keyword “Fertilization”). Then, the local MEME tool was adopted to search for the conserved result domains of S and fertilization-related proteins. After that, the searched motif sequences were merged according to structural domain names, and aliases were provided for the motifs. The fertilization-related domains and motifs owned by S are shown in Table 8.
Table 8.
HAP2/GCS1 and Izumo-Ig domains possessed by SARs-CoV-2 S protein
| Domain | Alias | Motif | Start | End |
|---|---|---|---|---|
| HAP2/GCS1 | α | YYHKNNKSWME | 144 | 154 |
| β | KYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT | 1205 | 1273 | |
| Izumo-Ig | β1 | GKYEQYIKWPWYIWL | 1204 | 1218 |
| β2 | MVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVK | 1229 | 1269 |
HAP2/GCS1 is a cysteine-rich structure, which is necessary for gamete fusion during fertilization. Besides, its specific protein binds the male and female gamete membranes together. Then, the second widely conserved protein directly or indirectly causes the two membranes to fuse. Inside this fusion channel, it is a channel with water molecules attached, while outside the channel, there is a pore structure formed by HAP2/GCS1. After that, it can be seen from Table 8 that the HAP2/GCS1 α domain belongs to the S1 protein. Furthermore, HAP2/GCS1 α participates in the binding of S protein and ACE2 receptor, and without cysteine, it is impossible to have HAP2/GCS function. In addition to that, the β domain of HAP2/GCS1 is at cytoplasm, transmembrane, and extracellular regions, containing the CAF-motif (cysteine aggregation fusion), while HAP2/GCS1 β participates in the formation of membrane fusion pores. Here, it is noted that the “WPWYIW” of HAP2/GCS1 β is in the trans-outer membrane region and is an essential associated region of EF-hand, kinase, phosphorylation, and Ca2+-ATPase.
Izumo-Ig is the Izumo protein of the immunoglobulin domain [125]. Only after the exocytosis (acrosome reaction) occurs, the Izumo-Ig structure is exposed, and besides Izumo-Ig β1 and Izumo-Ig β2 are in the cytoplasm, transmembrane and extracellular regions too. Actually, the Izumo-Ig domain of S protein highly overlaps with the HAP2/GCS1 β domain, while the acrosome reaction occurs after the sperm acrosome structure is bound to the egg cell, which signifies that only after S1 is bound to the cell receptor, the HAP2/GCS1 β structure of the S2 protein undergoes a conformational change. Then, it exposes the “WPWYIW” and nearby important structural regions.
CaMKII-Like system mediating the fusion of viral membranes and cell membranes
Schematic diagram of functional domains related to membrane fusion
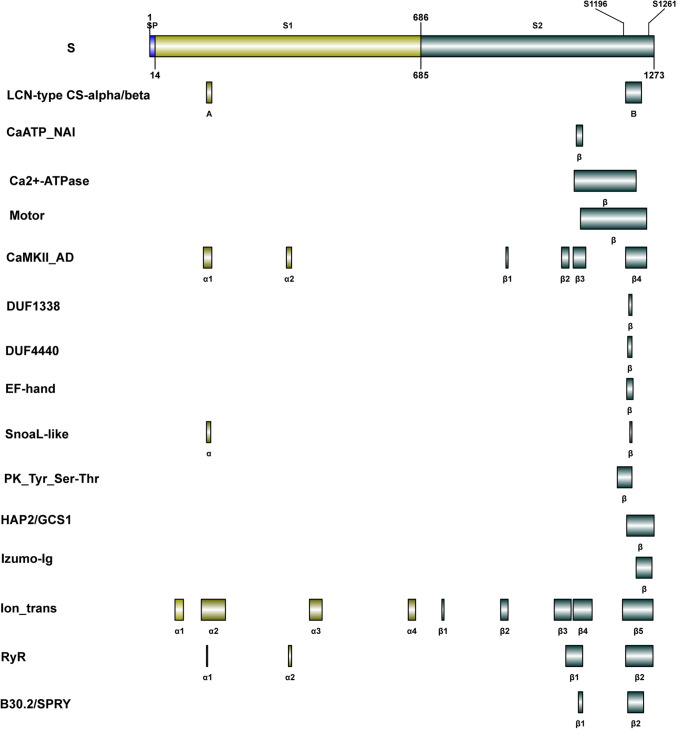
The IBS tool [126] was used to draw a schematic diagram of functional domains related to membrane fusion of S protein (Fig. 2). The autophosphorylation activation sites are S1196 and S1261. CaMKII_AD and ion channel domains are distributed on S1 and S2, while Ca2+-ATPase, CaATP_NAI, motor, EF-hand, HAP2/GCS1, PK_Tyr_Ser-Thr, and other domains related to the activation of CaMKII-like activity and membrane fusion events are mostly concentrated on the C-terminal of S2, and they overlap to a great extent. In addition, CaATP_NAI is located at the N-terminus of Ca2+-ATPase, and CaATP_NAI has no phosphorylation sites, indicating that Ca2+-ATPase is not inhibited by phosphorylation. Beyond that, CaATP_NAI also appears in CaMKII_AD, displaying that CaMKII_AD causes the S2 conformational rearrangement, which will expose CaATP_NAI and Ca2+-ATPase, thus the EF-hand structure of Ca2+-ATPase being able to bind Ca2.
Fig. 2.
S protein-membrane fusion-related functional domains and autophosphorylation activation sites. The autophosphorylation activation sites are S1196 and S1261. CaMKII_AD and ion channel domains are distributed on S1 and S2. Ca2 + -ATPase, CaATP_NAI, motor, EF-hand, HAP2/GCS1, and other structural domains related to the activation of CaMKII-like activity and membrane fusion events are mostly concentrated on the C-terminal of S2. This figure is drawn by IBS tool [126]
Ca2+-ATPase and CaMKII_AD
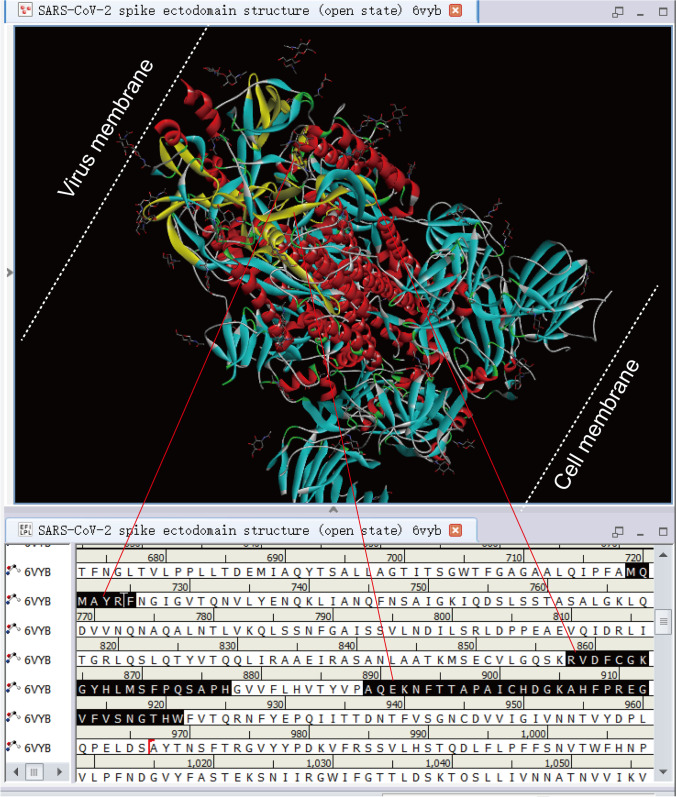
The SARS-CoV-2 S open state crystal structure 6vyb was obtained from the PDB database, and then the main CaATP_NAI and CaMKII_AD domains were marked on the structure (Fig. 3). Figure 3 shows that the CaATP_NAI and CaMKII_AD domains are both near the outer membrane of the virus. The CaMKII_AD domain motif “MQMAYRF” is a helical structure, while the CaMKII_AD domain motifs “RVDFCGKGYHLMSFPQSAPH” and “AQEKNFTTAPAICHDGKAHFPREGVFVSNGTHW” are folded structures. However, the CaMKII_AD domain motif “ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF” is a transmembrane structure and does not exist in the 6vyb structure diagram.
Fig. 3.
The position of some CaATP_NAI and CaMKII_AD domains of SARS-CoV-2 S (PDBID: 6vyb). The bottom right corner of the picture belongs to the S1 protein, close to the cell. The upper left corner of the picture belongs to the S2 protein, close to the viral membrane
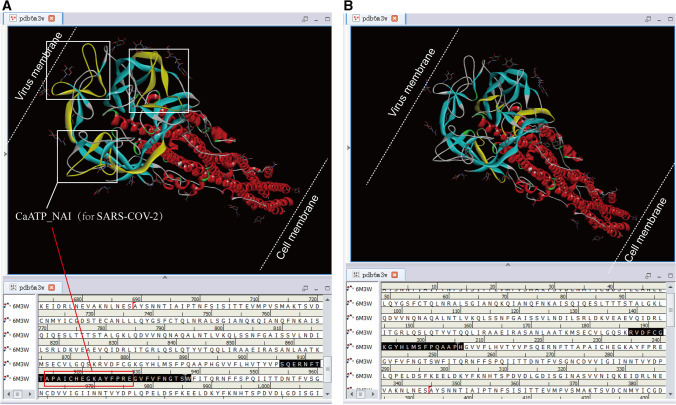
There is no SARS-CoV-2 HR1-HR2 hexamer in the PDB database. As the structure of the HR1-HR2 hexamer is relatively conservative, the position concerning the CaATP_NAI domain of SARS-CoV-2 was marked on the HR1-HR2 hexamer of SARS-CoV (Fig. 4). In this case, Fig. 4 shows that CaATP_NAI and CaMKII_AD overlap so that the folded sheet is close to the outer membrane of the virus. Furthermore, there is also a conservative CaMKII_AD domain motif “RVDFCGKGYHLMSFPQSAPH” near CaATP_NAI, while the motif of SARS-CoV here is “RVDFCGKGYHLMSFPQAAPH.” CaATP_NAI may drive the HR1-HR2 hexamer to fold irreversibly toward the cell membrane, thus promoting the proximity of the virus membrane to the cell membrane. About the CaATP_NAI position difference between SARS-CoV-2 and SARS-CoV, it is similar to the rear-wheel drive and front-wheel drive of a car.
Fig. 4.
The position of the CaATP_NAI of SARS-CoV-2 S in the HR1-HR2 hexamer (PDBID: 6m3x). R1-HR2 hexamer belongs to S2 protein. There is no S1 protein in the lower right corner of the picture, close to the cell membrane. The upper left corner of the image is near the viral membrane. 6m3x is the SARS-CoV crystal structure, and there is no SARS-CoV-2 HR1-HR2 hexamer in the PDB database. The HR1-HR2 hexamer structures of SARS-CoV-2 and SARS-CoV are similar. A The CaATP_NAI motif of SARS-CoV-2 (three white boxes in the figure) is “APAICHDGKAHFPRE,” which overlaps with the CaMKII_AD domain there. Besides, there are point mutations in the sequences between SARS-CoV-2 and SARS-CoV. Therefore, SARS-CoV fails to form a CaATP_NAI domain in the white box. B Both SARS-CoV-2 and SARS-CoV have a conserved CaMKII_AD domain “RVDFCGKGYHLMSFPQ[S/A]APH,” which is close to CaATP_NAI. It is worth noting that the three boxes are marked with the CaATP_NAI domain, which is only found on SARS-CoV-2 instead of SARS-CoV. These marked sequence residues are identical. However, in these locations, there are point mutations in the residues between SARS-CoV-2 and SARS-CoV, so that the sequences are inconsistent
Membrane fusion mechanism
After the receptor-binding domain of the S protein bound to the ACE2 receptor, the LCN-type CS-alpha/beta A domain of the S1 protein bound to the Na+ channel extracellular α-scorpion and anemone toxin receptors [41], triggering Na+ channel inactivation. Then, a large number of sodium ions were perfused into the cell through the inactivated sodium channel, while the Na+/Ca2+ exchanger (NCX) discharged sodium and took in calcium, causing calcium overload. Apart from that, excessive calcium caused a chain reaction in the cell. After that, NAD kinase was activated, NAD and NADH were converted into NADP and NADPH. A derivative of NADP NAADP activated the double-pore channel of the lysosome and promoted the release of calcium into the cytoplasm by the lysosome, followed by the cells releasing acid (H+) through the NA+-H+ exchange system and other channels. At the same time, the cells released calcium through the calcium pump. Besides, sodium channel inactivation could prevent multiple viruses from simultaneously infecting the bound cells.
Furin cleaved S1 and S2 proteins
The HAP2/GCS1 β domain of S2 protein broke away from the regulatory domain in the transmembrane region, exposing the EF-hand domain. The EF-hand domain bound to calcium ions, and the S2 protein had CaMKII-like protein activity, when the S2 protein rearranged through the CaMKII_AD domain to form a HR1-HR2 six-helix bundle. At the same time, Ca2+-ATPase in the HAP2/GCS1 β region of S2 was formed and then activated by calcium ions, while the calcium ion channel of Ion_trans β1 to Ion_trans β5 was also formed, and the S2 protein kinase domain and phosphorylation sites S1196 and S1261 were also exposed. The S2 kinase structure autophosphorylates S1196 and S1261, when the CaMKII-like activity of S2 protein was permanently activated. Furthermore, calcium ion regulation was no longer required. Then, the motor structure of the Ca2+-ATPase region catalyzed the hydrolysis of ATP, while Ca2+-ATPase pumped calcium ions from the virus cytoplasm to the calcium ion channel of S2. After that, the Ca2+ released by the calcium ion channel of S2 helped the fusion peptide FP insert into the cell membrane and anchored the S2 protein on the cell membrane. In addition to that, the violent pulling and stimulation of the cell membrane by FP depolarized the cell membrane rapidly, causing more sodium channels to open. Of course, it should also be mentioned that inactivation of many sodium channels could prevent the insertion concerning fusion peptides of other viruses into binding cells.
The CaATP_NAI domain could be anchored on and move on the cell (or virus) membrane. The motor structure drove the three CaATP_NAI domains to move in a specific direction of rotation, which forced the HR1-HR2 hexamer to fold irreversibly toward the viral membrane, thus bringing the virus membrane closer to the cell membrane. When the viral membrane was very close to the cell membrane, the CaATP_NAI and CaMKII_AD domains at the contact site of the HR1-HR2 hexamer extended outward so as to form a groove-like barrel structure. Besides, the cylinder was connected to the virus membrane and the cell membrane at the same time, while Ca2+ accumulated in the barrel-like structure and combined with water molecules to form hydrated Ca2+ ions, further forming a calcium bridge between the contact surfaces of the two membranes. At the same time, the LCN-type CS-alpha/beta B domain of the S2 protein was bound to the nearby Na+ channel extracellular α-scorpion and anemone toxin receptors, which caused the inactivation and opening of sodium channels. In this step, the inactivation of sodium channels could prevent other viruses from infecting the bound cells and membrane fusion.
The Ca2+-ATPase region was also ATP phospholipid-flipping. The phospholipid flippase released water from hydrated Ca2+ ions, and then displaced loosely coordinated water from the phosphate groups of the lipid membrane. When moisture was discharged from the contact position between the double membranes, the membrane became unstable, triggering the lipid mixing and fusion of the membrane lobules. Besides, the phospholipid flippase rapidly flipped and spliced the inner membrane to form the inner membrane of the fusion hole. Simultaneously, the outer membrane was spliced to form the outer membrane of the fusion hole, while the pore side of the inner membrane and the outer side of the outer membrane were the hydrophilic ends of phospholipids. At this time, the Ca2+ ion channel of the S2 protein evolved into a fusion hole.
The motor structure facilitated the CaATP_NAI and CaMKII_AD structures on the periphery of the fusion hole to continue to expand, which accelerated the expansion of the fusion hole. Then, the cytoplasmic contents of the virus (such as the nucleocapsid and RNA complex) were injected into the infected cell cytoplasm along with [H+], while the membrane and membrane proteins of the virus slid onto the cell membrane along the flowing membrane on the gap of the three CaATP_NAI (and CaMKII_AD). When most cytoplasmic content of the virus was injected into the cell, and most of the virus’s membrane and membrane proteins were fused to the cell membrane, the fusion hole would gradually shrink. After the cytoplasmic contents of the virus were completely discharged into the cell cytoplasm and the virus membrane was completely fused to the cell membrane, the motor structure drove the conformational change of the HR1-HR2 hexamer, decomposing the barrel structure and canceling the fusion hole. Therefore, the HR1-HR2 hexamer would fall into the cytoplasm or stay on the cell membrane.
The S protein of the infected cell may bind to the ACE2 receptor of another cell (or also an infected cell) and then achieve membrane fusion through a similar principle, forming cell syncytia, including syncytial multinucleated giant cells.
Discussion
Ca2+-ATPase’s power and membrane fusion time
The [Ca2+] on both sides of the sarcoplasmic reticulum membrane is kept under micromolar. Of the total chemical energy (~ 30 kcal/mol) released from the ATP hydrolysis process, 2/3 (~ 20 kcal/mol) is used for the membrane transferring Ca2+(127), and the remaining of about 1/3 is converted into heat (~ 10 kcal). It should be noted that the amount of heat released depends on whether the transmembrane Ca2+ gradient forms a trans-sarcoplasmic reticulum membrane [128]. Besides, the S2 trimer in the SARS-CoV fusion takes the shape of a baseball bat, with a width of 20–50 Å and a length of 185 Å. ATP hydrolysis energy can drive the high-speed movement of two motor molecules, while myosin ranges from about 0.2 to 60 μm/s, and kinesin varies from about 0.02 to 2 μm/s [129]. Suppose the distance between the virus membrane and the cell membrane is a trimer length of 185 Å, and the S2 motor structure pulls the two membranes closer at the slowest speed of 0.02 μm/s, the membrane fusion time is 18.5 nm/20 nm/s = 0.925 s ≈ 1 s. When the motor structure maintains the fastest speed of 60 microns/s, the membrane fusion time is 3.08 × 10−4 s. Considering the time for aerosol transmission and ACE2 receptor binding, this membrane fusion time is in line with epidemiological investigations. In this case, it can be found that SARS-CoV-2 will infect people who have not taken protective measures in public places within over 10 s.
The increased calcium demand during membrane fusion possibly disrupting the calcium homeostasis
Most viral infections can cause changes in intracellular Ca2+ concentration. Besides, Ca2+ plays a vital role in the process of virion structure formation, virus entry, virus gene expression, virus protein translation, virus particle maturation, and release [130]. Most known mechanisms indicate viruses interfere with Ca2+ stability and facilitate virus replication by interfering with Ca2+ and Ca2+ binding proteins [131], while viral proteins interfere with Ca2+ homeostasis by changing membrane permeability or manipulating key components of the Ca2+ signaling pathway [130], when viruses provide enough space for processing the gradient of the membrane interval, and can easily convert various signals. To live in the host cell, the virus hijacks important Ca2+ signaling pathways [132], and it also speeds up or induces cell apoptosis by interfering with the Ca2+ pathway, thus promoting the release of virus particles and spreading the virus to the greatest extent [133].
If the S protein mediates the SARS-CoV-2 virus and cell membrane fusion in a calcium-dependent manner, the demand for Ca2+ is enormous. Hence, the Ca2+ homeostasis in the human body is easily destroyed. For COVID-19 patients, calcitonin (PCT), C-reactive protein (CRP), and neutrophil to lymphocyte ratio (NLR) indicators are elevated in moderate and severe patients [134]. Calcitonin is a hormone that regulates blood calcium secreted by the thyroid and bronchi, and it responds to hypercalcemia, when quickly lowering blood calcium by inhibiting bone resorption [135, 136]. There is also a report showing that the total serum calcium and the actual ionized calcium levels of the whole blood of COVID-19 patients are lower [137]. In addition, COVID-19 patients have low serum zinc, calcium, and vitamin D levels [138]. Hypocalcemia is an essential and reliable indicator of disease severity and progression, but vitamin D levels cannot reflect the correlation with the severity of COVID-19 infection [139], which indicates that there may be a disorder in the calcium regulation system of COVID-19 patients [140].
The protein-bound calcium in plasma is called non-diffusible calcium and cannot penetrate the capillary wall. Ionized calcium [141] and calcium compounds such as calcium citrate in plasma [142] are called diffusible calcium, which can penetrate the capillary wall [143]. In plasma calcium, only ionized calcium directly plays a physiological role, and calcium ions play an essential role in the excitability and contraction of vascular smooth muscle. Non-diffusible calcium in plasma has no direct physiological effect, is in a dynamic balance with ionized calcium, and is affected by blood pH. When the blood pH decreases, it promotes the dissociation of bound calcium and increases Ca2+; conversely, when the pH increases, the bound calcium increases, and Ca2+ decreases. In terms of Vitamin D, it is combined in active form with calcitonin from thyroid C cells and parathyroid hormone (PTH) from parathyroid glands. Besides, vitamin D also keeps circulating calcium levels in the body within a concentration range [144]. As for the increase in plasma calcium, it is because of the mobilization of calcium in the bones, the increase in calcium absorption, and the stimulation of vitamin D3 (calcitriol) synthesis by the kidneys. Furthermore, calcitriol increases calcium absorption from the intestines and works synergistically with PTH to absorb calcium from the bones. Together, these hormone-mediated events normalize serum calcium levels.
In the current study, it was found that the CaMKII-like domains of the S protein mediated the fusion of virus and infected cells, and it involved calcium ions in this process. In membrane fusion, calcium ions near the interface between S protein and cell membrane quickly accumulated, which produced abnormal calcium ion currents. Besides that, abnormal flow of calcium ions caused depolarization of cells near the infected cell, prompting many calcium ions to flush into nearby cells. Finally, the calcium ion demand in serum accelerated in a short time, thus facilitating the release of calcium in the body. If the SARS-CoV-2 virus infection area was large, it encouraged the abnormality of serum calcium levels and disrupted the body’s calcium homeostasis.
Membrane fusion increasing the risk of coagulation and vascular calcification
S protein has a conserved cysteine abundance, and “CSCLKGCCSCGSC” is the consensus motif region [118]. The consensus pattern is C-[TS]-C-h–X-G-X(4,6)-C. It is named CAF-motif (cysteine aggregation fusion), where h represents a hydrophobic residue and X refers to any other residue. This specific domain mediates the particle assembly of coronavirus spikes [118], while the cysteine-rich region facilitates the formation of fusion pores [145]. In addition, the importance of its membrane fusion has also been confirmed [146]. The palmitoylation of the cysteine and its internal domain is also the rate-limiting step for changing the membrane fusion reaction (147). Besides, similar functional cysteine structures are found in the proteins of influenza virus HA, vesicular stomatitis virus, influenza virus and murine leukemia virus [118], and the coagulation of COVID-19 patients may have many factors. Still, it is unclear whether clotting or hemolysis must occur during membrane fusion between the SARS-CoV-2 virus and the infected cell. In membrane fusion, the cytoplasmic domain of the S protein and the surface CAF-motif (cysteine aggregation fusion) of the infected cell may assist the formation of blood clots [118].
It was also found that the S protein has a CaMKII_Like conserved domain. There is an EF-hand domain “YEQYIKWPWYIWLGF” upstream of CAF-motif, that is, in the N-terminal direction, a region that binds calcium ions. CAF-motif belongs to the C-terminal of the CaMKII_AD (“ELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF”) domain, and EF-hand belongs to N-terminal of the CaMKII_AD domainto. The CaMKII_AD is at C-terminal of S protein, promoting the formation of the HR1-HR2 hexamer. In addition to that, the CaMKII-like system of S protein induced the fusion of virus and infected cells, while many calcium ions were consumed during the fusion process that caused many calcium ions near the interface between the virus and the infected cell to accumulate in a short time. As calcium ion was a coagulation factor, clots may occur near the contact surface. Moreover, SARS-CoV-2 infection depends on cell heparan sulfate [148] and ACE2, as heparan sulfate and dermatan sulfate have anticoagulant effects [149]. If lower levels of heparan sulfate cannot neutralize coagulation factors in infection and fusion, it will risk clotting. Therefore, if cytokines and oxidative stress factors were not considered, abnormal calcium release and collection mainly induced the coagulation reaction of COVID-19 patients because of a viral infection and membrane fusion. According to the heme theory, it was held that the SARS-CoV-2 virus could attack the hemoglobin in red blood cells [150]. Considering that, the SARS-CoV-2 virus could infect red blood cells through the CD147 receptor, while calcium ions triggered coagulation during the membrane fusion process. Therefore, hemolysis in membrane fusion was an infrequent phenomenon.
In addition, calcium attached to the endothelium of blood vessels easily to form atherosclerotic plaques. The systemic inflammatory response caused by acute infection intensified the inflammatory activity in the coronary atherosclerotic plaque, making it prone to rupture and leading to acute coronary syndrome [151]. As is known, vascular calcification was the pathological basis of cardiovascular disease, and it also correlated vascular calcification with the severity of cardiovascular disease. Among them, intimal calcification was the most dangerous, since atherosclerotic calcification easily increased blood pressure, heart failure, myocardial ischemia, and even myocardial infarction.
Conclusion
COVID-19 is a unique disease characterized by extensive pulmonary thrombosis, and the formation of infected cell syncytia, including syncytial multinucleated giant cells. However, the mechanism by which the SARS-CoV-2 S protein regulates the construction of these syncytia is still ambiguous. Under this circumstance, in the current study, structural domain search methods were adopted to analyze the S protein of the SARS-CoV-2 virus. Then, the results showed that after the EF-hand domain of S protein bound to calcium ions, S2 protein had CaMKII protein activities. Besides, the CaMKII_AD domain changed S2 conformation, tending to form HR1-HR2 six-helix bundles. Furthermore, the Ca2+-ATPase of S pumped calcium ions from the virus cytoplasm to help fusion peptide insert into the cell membrane, and then mediate membranes' fusion. Beyond that, motor structures of S drove the CaATP_NAI and CaMKII_AD domains to extend to the outside and combined the viral membrane and the cell membrane, thus forming a calcium bridge. Then, the phospholipid-flipping ATPase released water, triggering lipid mixing and fusion of the membrane and generating fusion pore. At the same time, motor structures were conducive to fusion pore extension, with the cytoplasmic contents of the virus being discharged into the cell cytoplasm, and the membrane of the virus sliding onto the cell membrane along the flowing membrane on the gap of the three CaATP_NAI. In this case, the HR1-HR2 hexamer would fall into the cytoplasm or stay on the cell membrane.
The S protein of the infected cell could bind to the ACE2 receptor of another cell (or also an infected cell) and then achieve membrane fusion through a similar principle, further forming cell syncytia, including syncytial multinucleated giant cells, by the manner.
Author contribution
Funding was obtained by LWZ. Besides, design, analysis, and writing are finished by LWZ, while data curation and manuscript checking are undertaken by LHL. Both authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a grant from the Talent Introduction Project of Sichuan University of Science and Engineering (award number: 2018RCL20, grant recipient: LWZ).
Data availability
The datasets and results supporting the conclusions of this article are available at: https://pan.baidu.com/s/1OBS_AKjMZGWJhPfS5T_hXQ; code: 5kvg.
Declarations
Ethics approval and consent to participation
Not applicable.
Consent for publication.
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bussani R, Schneider E, Zentilin L, Collesi C, Ali H, Braga L, Volpe MC, Colliva A, Zanconati F, Berlot G, Silvestri F, Zacchigna S, Giacca M. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61:103104. doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santana MF, Pinto RAdA, Marcon RAdA, Medeiros LCASd, Morais TBdNd, Dias LC, Souza LPd, Melo GCd, Monteiro WM, Lacerda MVG. Pathological findings and morphologic correlation of the lungs of autopsied patients with SARS-CoV-2 infection in the Brazilian Amazon using transmission electron microscopy. Revista da Sociedade Brasileira de Medicina Tropical. 2021;54. [DOI] [PMC free article] [PubMed]
- 3.Keresztesi A-A, Perde F, Ghita-Nanu A, Radu C-C, Negrea M, Keresztesi G. Post-Mortem Diagnosis and Autopsy Findings in SARS-CoV-2 Infection: Forensic Case Series. Diagnostics. 2020;10:1070. doi: 10.3390/diagnostics10121070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calabrese F, Pezzuto F, Fortarezza F, Hofman P, Kern I, Panizo A, von der Thüsen J, Timofeev S, Gorkiewicz G, Lunardi F. Pulmonary pathology and COVID-19: lessons from autopsy. The experience of European Pulmonary Pathologists. Virchows Archiv. 2020;477(3):359–372. [DOI] [PMC free article] [PubMed]
- 5.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duarte-Neto AN, Caldini EG, Gomes-Gouvêa MS, Kanamura CT, de Almeida Monteiro RA, Ferranti JF, Ventura AMC, Regalio FA, Fiorenzano DM, Gibelli MABC, Carvalho WBd, Leal GN, Pinho JRR, Delgado AF, Carneiro-Sampaio M, Mauad T, Ferraz da Silva LF, Saldiva PHN, Dolhnikoff M. An autopsy study of the spectrum of severe COVID-19 in children: From SARS to different phenotypes of MIS-C. E Clinical Medicine. 2021;35:100850. doi: 10.1016/j.eclinm.2021.100850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Chen C-H, Badeti S, Cho JH, Naghizadeh A, Wang Z, Liu D. Deletion of ER-retention motif on SARS-CoV-2 spike protein reduces cell hybrid during cell–cell fusion. Cell & Bioscience. 2021;11(1):114. [DOI] [PMC free article] [PubMed]
- 8.Zhang Z, Zheng Y, Niu Z, Zhang B, Wang C, Yao X, Peng H, Franca DN, Wang Y, Zhu Y, Su Y, Tang M, Jiang X, Ren H, He M, Wang Y, Gao L, Zhao P, Shi H, Chen Z, Wang X, Piacentini M, Bian X, Melino G, Liu L, Huang H, Sun Q. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death & Differentiation. 2021:1–213 [DOI] [PMC free article] [PubMed]
- 9.Theuerkauf SA, Michels A, Riechert V, Maier TJ, Flory E, Cichutek K, Buchholz CJ. Quantitative assays reveal cell fusion at minimal levels of SARS-CoV-2 spike protein and fusion from without. iScience. 2021;24(3):102170. [DOI] [PMC free article] [PubMed]
- 10.Doms RW. What Came First—the Virus or the Egg? Cell. 2017;168:755–757. doi: 10.1016/j.cell.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Pinello JF, Snell WJ. Plant sperm need a little help. Nature plants. 2019;5:247–248. doi: 10.1038/s41477-019-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miner JJ. Gamete fusion gone viral. Sci Trans Med. 2017;9(381):eaam9866. [DOI] [PubMed]
- 13.Clark T. HAP2/GCS1: Mounting evidence of our true biological EVE? PLoS Biol. 2018;16:e3000007. doi: 10.1371/journal.pbio.3000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valansi C, Moi D, Leikina E, Matveev E, Graña M, Chernomordik LV, Romero H, Aguilar PS, Podbilewicz B. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J Cell Biol. 2017;216:571–581. doi: 10.1083/jcb.201610093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modis, Y. Class II Fusion Proteins. In Viral Entry into Host Cells. Edited by Pöhlmann S, Simmons G. New York, NY. Springer New York. 2013: 150–166.
- 16.Fédry J, Liu Y, Péhau-Arnaudet G, Pei J, Li W, Tortorici MA, Traincard F, Meola A, Bricogne G, Grishin NV. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 2017;168:904–915.e910. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinello JF, Lai AL, Millet JK, Cassidy-Hanley D, Freed JH, Clark TG. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr Biol. 2017;27:651–660. doi: 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarese EP, Musci RL, Frediani L, Gurbel PA, Kubica J. Ion channel inhibition against COVID-19: A novel target for clinical investigation. Cardiol J. 2020;27:421–424. doi: 10.5603/CJ.a2020.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luzio J, Bright N, Pryor P. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem Soc Trans. 2007;35:1088–1091. doi: 10.1042/BST0351088. [DOI] [PubMed] [Google Scholar]
- 21.Tulsiani DR, Abou-Haila A. Is sperm capacitation analogous to early phases of Ca2+-triggered membrane fusion in somatic cells and viruses? Bio Essays. 2004;26:281–290. doi: 10.1002/bies.20003. [DOI] [PubMed] [Google Scholar]
- 22.Steinhardt R, Lundin L, Mazia D. Bioelectric responses of the echinoderm egg to fertilization. Proc Natl Acad Sci. 1971;68:2426–2430. doi: 10.1073/pnas.68.10.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe LA. Fast block to polyspermy in sea urchin eggs is electrically mediated. Nature. 1976;261:68–71. doi: 10.1038/261068a0. [DOI] [PubMed] [Google Scholar]
- 24.Paul M, Epel D. Formation of fertilization acid by sea urchin eggs does not require specific cations. Exp Cell Res. 1975;94:1–6. doi: 10.1016/0014-4827(75)90524-8. [DOI] [PubMed] [Google Scholar]
- 25.Boldt J, Schuel H, Schuel R, Dandekar PV, Troll W. Reaction of sperm with egg-derived hydrogen peroxide helps prevent polyspermy during fertilization in the sea urchin. Gamete Res. 1981;4:365–377. doi: 10.1002/mrd.1120040502. [DOI] [Google Scholar]
- 26.Epel D, Patton C, Wallace RW, Cheung WY. Calmodulin activates NAD kinase of sea urchin eggs: an early event of fertilization. Cell. 1981;23:543–549. doi: 10.1016/0092-8674(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 27.Perry G, Epel D. Characterization of a Ca2+-stimulated lipid peroxidizing system in the sea urchin egg. Dev Biol. 1985;107:47–57. doi: 10.1016/0012-1606(85)90374-4. [DOI] [PubMed] [Google Scholar]
- 28.Johnson JD, Epel D, Paul M. Intracellular p H and activation of sea urchin eggs after fertilisation. Nature. 1976;262:661–664. doi: 10.1038/262661a0. [DOI] [PubMed] [Google Scholar]
- 29.Matasic DS, Yoon J-Y, McLendon JM, Mehdi H, Schmidt MS, Greiner AM, Quinones P, Morgan GM, Boudreau RL, Irani K. Modulation of the cardiac sodium channel NaV1. 5 peak and late currents by NAD+ precursors. J Mol Cell Cardiol. 2020;141:70–81. doi: 10.1016/j.yjmcc.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antzelevitch C. Electrical heterogeneity, cardiac arrhythmias, and the sodium channel. Circ Res. 2000;87:964–965. doi: 10.1161/01.RES.87.11.964. [DOI] [PubMed] [Google Scholar]
- 31.Tondas AE, Mulawarman R, Trifitriana M, Nurmaini S, Irfannuddin I. Arrhythmia risk profile and ventricular repolarization indices in COVID-19 patients: a systematic review and meta-analysis. J Infect Dev Count. 2021;15:224–229. doi: 10.3855/jidc.13922. [DOI] [PubMed] [Google Scholar]
- 32.Alareedh M, Nafakhi H, Shaghee F, Nafakhi A. Electrocardiographic markers of increased risk of sudden cardiac death in patients with COVID-19 pneumonia. Annals of Noninvasive Electrocardiology. 2021;26(3):e12824. [DOI] [PMC free article] [PubMed]
- 33.Anand P, Puranik A, Aravamudan M, Venkatakrishnan AJ, Soundararajan V. SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. eLife. 2020;9:e58603. [DOI] [PMC free article] [PubMed]
- 34.Ji H-L, Song W, Gao Z, Su X-F, Nie H-G, Jiang Y, Peng J-B, He Y-X, Liao Y, Zhou Y-J. SARS-CoV proteins decrease levels and activity of human ENaC via activation of distinct PKC isoforms. Am J Physiol-Lung Cell Mol Physiol. 2009;296:L372–L383. doi: 10.1152/ajplung.90437.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammarström AK, Gage PW. Hypoxia and persistent sodium current. Eur Biophys J. 2002;31:323–330. doi: 10.1007/s00249-002-0218-2. [DOI] [PubMed] [Google Scholar]
- 36.Undrovinas AI, Maltsev VA, Kyle JW, Silverman N, Sabbah HN. Gating of the late Na+ channel in normal and failing human myocardium. J Mol Cell Cardiol. 2002;34:1477–1489. doi: 10.1006/jmcc.2002.2100. [DOI] [PubMed] [Google Scholar]
- 37.Imahashi K, Kusuoka H, Hashimoto K, Yoshioka J, Yamaguchi H, Nishimura T. Intracellular sodium accumulation during ischemia as the substrate for reperfusion injury. Circ Res. 1999;84:1401–1406. doi: 10.1161/01.RES.84.12.1401. [DOI] [PubMed] [Google Scholar]
- 38.Luo A-T, Cao Z-Z, Xiang Y, Zhang S, Qian C-P, Fu C, Zhang P-H, Ma J-H. Ketamine attenuates the Na+-dependent Ca 2+ overload in rabbit ventricular myocytes in vitro by inhibiting late Na+ and L-type Ca 2+ currents. Acta Pharmacol Sin. 2015;36:1327–1336. doi: 10.1038/aps.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souza E Souza KFC, Moraes BPT, Paixão ICNdP, Burth P, Silva AR, Gonçalves-de-Albuquerque CF. Na(+)/K(+)-ATPase as a Target of Cardiac Glycosides for the Treatment of SARS-CoV-2 Infection. Front Pharmacol. 2021;12:624704–624704. [DOI] [PMC free article] [PubMed]
- 40.Skogestad J, Aronsen JM. Hypokalemia-induced arrhythmias and heart failure: new insights and implications for therapy. Front Physiol. 2018;9:1500. doi: 10.3389/fphys.2018.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. Molecular determinants of high affinity binding of α-Scorpion toxin and sea anemone toxin in the S3–S4 extracellular loop in domain IV of the Na+ channel α subunit. J Biol Chem. 1996;271(27):15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Pan X, Huang Y, Cheng C, Xu X, Wu Y, Xu Y, Shang W, Niu X, Wan Y, Li Z, Zhang R, Liu S, Xiao G, Xu W. Drug repurposing of itraconazole and estradiol benzoate against COVID-19 by blocking SARS-CoV-2 spike protein-mediated membrane fusion. Adv Ther. 2021;4(5):2000224. [DOI] [PMC free article] [PubMed]
- 43.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borkotoky S, Dey D, Banerjee M. Computational Insight Into the Mechanism of SARS-CoV-2 Membrane Fusion. J Chem Inf Model. 2021;61:423–431. doi: 10.1021/acs.jcim.0c01231. [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran VS. Encyclopedia of human behavior. Academic Press; 2012. [Google Scholar]
- 46.Cashman D. Dominance of SARS-CoV-2 D614G variant explained by the requirement of COVID-19 for calcium; proximate therapeutic implication (s) for COVID-19. J Clin Immunol Immunother. 2020;6:2. doi: 10.24966/CIIT-8844/1000048. [DOI] [Google Scholar]
- 47.Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ, Zambon M, Ellis J, Lewis PA, Hiscox JA. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome medicine. 2020;12:1–15. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khelashvili G, Plante A, Doktorova M, Weinstein H. Ca2+-dependent mechanism of membrane insertion and destabilization by the SARS-CoV-2 fusion peptide. Biophys J . 2021;120:1105–1119. doi: 10.1016/j.bpj.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai AL, Freed JH. SARS-CoV-2 fusion peptide has a greater membrane perturbating effect than SARS-CoV with highly specific dependence on Ca2+. J Mol Biol. 2021;433(10). [DOI] [PMC free article] [PubMed]
- 50.Charlton FW, Pearson HM, Hover S, Lippiat JD, Fontana J, Barr JN, Mankouri J. Ion channels as therapeutic targets for viral infections: further discoveries and future perspectives. Viruses. 2020;12:844. doi: 10.3390/v12080844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hover S, Foster B, Fontana J, Kohl A, Goldstein SA, Barr JN, Mankouri J. Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Path. 2018;14:e1006845. doi: 10.1371/journal.ppat.1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinberg BE, Huynh KK, Brodovitch A, Jabs S, Stauber T, Jentsch TJ, Grinstein S. A cation counterflux supports lysosomal acidification. J Cell Biol. 2010;189:1171–1186. doi: 10.1083/jcb.200911083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Zhang X, Dong X-P, Samie M, Li X, Cheng X, Goschka A, Shen D, Zhou Y, Harlow J. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng K, Chen M, Xiang Y, Ma K, Jin F, Wang X, Wang X, Wang S, Wang Y. Inhibition of herpes simplex virus type 1 entry by chloride channel inhibitors tamoxifen and NPPB. Biochem Biophys Res Commun. 2014;446:990–996. doi: 10.1016/j.bbrc.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 55.Galione A, Morgan AJ, Arredouani A, Davis LC, Rietdorf K, Ruas M, Parrington J. NAADP as an intracellular messenger regulating lysosomal calcium-release channels. Portland Press Ltd; 2010. [DOI] [PubMed] [Google Scholar]
- 56.Filippini A, D'Amore A, Palombi F, Carpaneto A. Could the inhibition of endo-lysosomal two-pore channels (TPCs) by the natural flavonoid naringenin represent an option to fight SARS-CoV-2 infection? Front Microbiol. 2020;11:970. doi: 10.3389/fmicb.2020.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grimm C, Tang R. Could an endo-lysosomal ion channel be the Achilles heel of SARS-CoV2? Cell Calcium. 2020;88:102212. doi: 10.1016/j.ceca.2020.102212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Zhang L-K, Li S-F, Zhang S-F, Wan W-W, Zhang Y-L, Xin Q-L, Dai K, Hu Y-Y, Wang Z-B. Calcium channel blockers reduce severe fever with thrombocytopenia syndrome virus (SFTSV) related fatality. Cell Res. 2019;29:739–753. doi: 10.1038/s41422-019-0214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunaratne GS, Yang Y, Li F, Walseth TF, Marchant JS. NAADP-dependent Ca2+ signaling regulates Middle East respiratory syndrome-coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium. 2018;75:30–41. doi: 10.1016/j.ceca.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gunaratne GS, Brailoiu E, He S, Unterwald EM, Patel S, Slama JT, Walseth TF, Marchant JS. Essential requirement for JPT2 in NAADP-evoked Ca2+ signaling. Sci. Signal. 2021;14(675):eabd5605. [DOI] [PMC free article] [PubMed]
- 62.Petersen OH, Gerasimenko OV, Gerasimenko JV. Endocytic uptake of SARS-CoV-2: the critical roles of pH, Ca2+, and NAADP. Function. 2020;1(1):zqaa003. [DOI] [PMC free article] [PubMed]
- 63.Lee HC, Aarhus R. A derivative of NADP mobilizes calcium stores insensitive to inositol trisphosphate and cyclic ADP-ribose (∗) J Biol Chem. 1995;270:2152–2157. doi: 10.1074/jbc.270.5.2152. [DOI] [PubMed] [Google Scholar]
- 64.Churchill GC, Okada Y, Thomas JM, Genazzani AA, Patel S, Galione A. NAADP mobilizes Ca2+ from reserve granules, lysosome-related organelles, in sea urchin eggs. Cell. 2002;111:703–708. doi: 10.1016/S0092-8674(02)01082-6. [DOI] [PubMed] [Google Scholar]
- 65.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winstone H, Lista MJ, Reid AC, Bouton C, Pickering S, Galao RP, Kerridge C, Doores KJ, Swanson CM, Neil SJD. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol. 2021;95(9):e02422–02420. [DOI] [PMC free article] [PubMed]
- 67.Inoue Y, Tanaka N, Tanaka Y, Inoue S, Morita K, Zhuang M, Hattori T, Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Z, Qin P, Huang Y-W. Lysosomal ion channels involved in cellular entry and uncoating of enveloped viruses: Implications for therapeutic strategies against SARS-CoV-2. Cell Calcium. 2021;94. [DOI] [PMC free article] [PubMed]
- 69.Johnson DE, Ostrowski P, Jaumouillé V, Grinstein S. The position of lysosomes within the cell determines their luminal pH. J Cell Biol. 2016;212:677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Z, Qin P, Huang Y-W. Lysosomal ion channels involved in cellular entry and uncoating of enveloped viruses: Implications for therapeutic strategies against SARS-CoV-2. Cell Calcium. 2021;94:102360. doi: 10.1016/j.ceca.2021.102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. The role of intraorganellar Ca2+ in late endosome–lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 73.Kolokoltsov AA, Saeed MF, Freiberg AN, Holbrook MR, Davey RA. Identification of novel cellular targets for therapeutic intervention against Ebola virus infection by siRNA screening. Drug Dev Res. 2009;70:255–265. doi: 10.1002/ddr.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medina DL, Ballabio A. Lysosomal calcium regulates autophagy. Autophagy. 2015;11:970–971. doi: 10.1080/15548627.2015.1047130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Y, Kappes B, Yang J, Franklin RM. Molecular cloning, stage-specific expression and cellular distribution of a putative protein kinase from Plasmodium falciparum. Eur J Biochem. 1992;207:305–313. doi: 10.1111/j.1432-1033.1992.tb17051.x. [DOI] [PubMed] [Google Scholar]
- 76.Kato K, Sugi T, Takemae H, Takano R, Gong H, Ishiwa A, Horimoto T, Akashi H. Characterization of a Toxoplasma gondii calcium calmodulin-dependent protein kinase homolog. Parasit Vectors. 2016;9:405. doi: 10.1186/s13071-016-1676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colombo MI, Beron W, Stahl PD. Calmodulin regulates endosome fusion. J Biol Chem. 1997;272:7707–7712. doi: 10.1074/jbc.272.12.7707. [DOI] [PubMed] [Google Scholar]
- 78.Chao LH, Stratton MM, Lee I-H, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, Kuriyan J. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hudmon A, Schulman H. Structure–function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochemical Journal. 2002;364:593–611. doi: 10.1042/bj20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hwang I, Sze H, Harper JF. A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci. 2000;97:6224–6229. doi: 10.1073/pnas.97.11.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falke JJ, Drake SK, Hazard AL, Peersen OB. Molecular tuning of ion binding to calcium signaling proteins. Q Rev Biophys. 1994;27:219–290. doi: 10.1017/S0033583500003012. [DOI] [PubMed] [Google Scholar]
- 82.Harrison SC. Viral membrane fusion. Virology. 2015;479:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Haro L, Quetglas S, Iborra C, Lévêque C, Seagar M. Calmodulin-dependent regulation of a lipid binding domain in the v-SNARE synaptobrevin and its role in vesicular fusion. Biol Cell. 2003;95:459–464. doi: 10.1016/S0248-4900(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 84.Leabu M. Membrane fusion in cells: molecular machinery and mechanisms. J Cell Mol Med. 2006;10:423–427. doi: 10.1111/j.1582-4934.2006.tb00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jena BP. Membrane fusion: role of SNAREs and calcium. Protein Pept Lett. 2009;16:712–717. doi: 10.2174/092986609788681869. [DOI] [PubMed] [Google Scholar]
- 86.Tiggemann R, Plattner H. Possible involvement of a calmodulin regulated Ca2+-ATPase in exocytosis performance in Paramecium tetraurelia cells. FEBS Lett. 1982;148:226–230. doi: 10.1016/0014-5793(82)80812-0. [DOI] [PubMed] [Google Scholar]
- 87.Miner GE, Sullivan KD, Zhang C, Rivera-Kohr D, Guo A, Hurst LR, Ellis EC, Starr ML, Jones BC, Fratti RA. Phosphatidylinositol 3,5-bisphosphate regulates Ca2+ transport during yeast vacuolar fusion through the Ca2+ ATPase Pmc1. Traffic. 2020;21(7):503–517. doi: 10.1111/tra.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonza MC, Martin H, Kang M, Lewis G, Greiner T, Giacometti S, Van Etten JL, De Michelis MI, Thiel G, Moroni A. A functional calcium-transporting ATPase encoded by chlorella viruses. J Gen Virol. 2010;91:2620–2629. doi: 10.1099/vir.0.021873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunaratne HJ, Vacquier VD. Evidence for a secretory pathway Ca2+-ATPase in sea urchin spermatozoa. FEBS Lett. 2006;580:3900–3904. doi: 10.1016/j.febslet.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 90.Gunaratne HJ, Neill AT, Vacquier VD. Plasma membrane calcium ATPase is concentrated in the head of sea urchin spermatozoa. J Cell Physiol. 2006;207:413–419. doi: 10.1002/jcp.20575. [DOI] [PubMed] [Google Scholar]
- 91.Roldan E, Fleming A. Is a Ca2+-ATPase involved in Ca2+ regulation during capacitation and the acrosome reaction of guinea-pig spermatozoa? Reproduction. 1989;85:297–308. doi: 10.1530/jrf.0.0850297. [DOI] [PubMed] [Google Scholar]
- 92.Feng H, Hershlag A, Han Y, Zheng L. Localizations of intracellular calcium and Ca2+-ATPase in hamster spermatogenic cells and spermatozoa. Microsc Res Tech. 2006;69:618–623. doi: 10.1002/jemt.20329. [DOI] [PubMed] [Google Scholar]
- 93.Vijayasarathy S, Shivaji S, Balaram P. Plasma membrane bound Ca2+-ATPase activity in bull sperm. FEBS Lett. 1980;114:45–47. doi: 10.1016/0014-5793(80)80857-X. [DOI] [PubMed] [Google Scholar]
- 94.Lucca N, León G. ArabidopsisACA7, encoding a putative auto-regulated Ca2+-ATPase, is required for normal pollen development. Plant Cell Rep. 2012;31:651–659. doi: 10.1007/s00299-011-1182-z. [DOI] [PubMed] [Google Scholar]
- 95.Gordon M, Morris EG, Young RJ. The localization of Ca2+-ATPase and Ca2+ binding proteins in the flagellum of guinea pig sperm. Gamete Res. 1983;8:49–55. doi: 10.1002/mrd.1120080106. [DOI] [Google Scholar]
- 96.Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takita Y, Engstrom L, Ungermann C, Cunningham KW. Inhibition of the Ca2+-ATPase Pmc1p by the v-SNARE protein Nyv1p. J Biol Chem. 2001;276:6200–6206. doi: 10.1074/jbc.M009191200. [DOI] [PubMed] [Google Scholar]
- 98.Penniston J, Enyedi A. Modulation of the plasma membrane Ca 2+ pump. J Membr Biol. 1998;165:101–109. doi: 10.1007/s002329900424. [DOI] [PubMed] [Google Scholar]
- 99.Geisler M, Axelsen KB, Harper JF, Palmgren MG. Molecular aspects of higher plant P-type Ca2+-ATPases. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2000;1465(1):52–78. [DOI] [PubMed]
- 100.Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H. A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem. 1998;273:1099–1106. doi: 10.1074/jbc.273.2.1099. [DOI] [PubMed] [Google Scholar]
- 101.Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89:1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- 102.Hwang I, Sze H, Harper JF. A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci. 2000;97:6224–6229. doi: 10.1073/pnas.97.11.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bonza MC, Luoni L. Plant and animal type 2B Ca2+-ATPases: evidence for a common auto-inhibitory mechanism. FEBS Lett. 2010;584:4783–4788. doi: 10.1016/j.febslet.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 104.Luoni L, Meneghelli S, Bonza MC, DeMichelis MI. Auto-inhibition of Arabidopsis thaliana plasma membrane Ca2+-ATPase involves an interaction of the N-terminus with the small cytoplasmic loop. FEBS Lett. 2004;574:20–24. doi: 10.1016/j.febslet.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 105.Bose J, Pottosin I, Shabala SSS, Palmgren MG, Shabala S. Calcium efflux systems in stress signaling and adaptation in plants. Front Plant Sci. 2011;2:85. doi: 10.3389/fpls.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Møller JV, Olesen C, Winther A-ML, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43:501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 107.Enyedi A, Verma AK, Filoteo AG, Penniston JT. Protein kinase C activates the plasma membrane Ca2+ pump isoform 4b by phosphorylation of an inhibitory region downstream of the calmodulin-binding domain. J Biol Chem. 1996;271:32461–32467. doi: 10.1074/jbc.271.50.32461. [DOI] [PubMed] [Google Scholar]
- 108.Odermatt A, Kurzydlowski K, MacLennan DH. The vmax of the Ca2+-ATPase of cardiac sarcoplasmic reticulum (SERCA2a) is not altered by Ca2+/calmodulin-dependent phosphorylation or by interaction with phospholamban. J Biol Chem. 1996;271:14206–14213. doi: 10.1074/jbc.271.24.14206. [DOI] [PubMed] [Google Scholar]
- 109.Banerjee A, Barry VA, DasGupta BR, Martin TF. N-Ethylmaleimide-sensitive factor acts at a prefusion ATP-dependent step in Ca2+-activated exocytosis. J Biol Chem. 1996;271:20223–20226. doi: 10.1074/jbc.271.34.20223. [DOI] [PubMed] [Google Scholar]
- 110.Kiani FA, Fischer S. Catalytic strategy used by the myosin motor to hydrolyze ATP. Proc Natl Acad Sci. 2014;111:E2947–E2956. doi: 10.1073/pnas.1401862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh Tomar PP, Arkin IT. SARS-CoV-2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem Biophys Res Commun. 2020;530:10–14. doi: 10.1016/j.bbrc.2020.05.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McClenaghan C, Hanson A, Lee SJ, Nichols CG. Coronavirus proteins as ion channels: current and potential research. Front Immunol. 2020;11. [DOI] [PMC free article] [PubMed]
- 113.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immun. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clustal W, Clustal X. version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 115.Fan X, Cao D, Kong L, Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat Commun. 2020;11:3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu C, Feng Y, Gao F, Zhang Q, Wang M. Characterization of HCoV-229E fusion core: implications for structure basis of coronavirus membrane fusion. Biochem Biophys Res Commun. 2006;345:1108–1115. doi: 10.1016/j.bbrc.2006.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buonvino S, Melino S. New Consensus pattern in Spike CoV-2: potential implications in coagulation process and cell–cell fusion. Cell Death Discovery. 2020;6:134. doi: 10.1038/s41420-020-00372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sultana A, Kallio P, Jansson A, Wang J-S, Niemi J, Mäntsälä P, Schneider G. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 2004;23:1911–1921. doi: 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 121.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Marrero MC, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e619. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ, Zambon M, Ellis J, Lewis PA, Hiscox JA, Matthews DA. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG. P4-ATPases: lipid flippases in cell membranes. Pflugers Arch. 2014;466:1227–1240. doi: 10.1007/s00424-013-1363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rubinstein E, Ziyyat A, Wolf JP, Le Naour F, Boucheix C. The molecular players of sperm-egg fusion in mammals. Semin Cell Dev Biol. 2006;17:254–263. doi: 10.1016/j.semcdb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 126.Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meis Ld. Energy interconversion by the sarcoplasmic reticulum Ca2+-ATPase: ATP hydrolysis, Ca2+ transport, ATP synthesis and heat production. An Acad Bras Ciênc. 2000;72:365–79. [DOI] [PubMed]
- 128.de Meis L. Role of the sarcoplasmic reticulum Ca2+-ATPase on heat production and thermogenesis. Biosci Rep. 2001;21:113–137. doi: 10.1023/A:1013640006611. [DOI] [PubMed] [Google Scholar]
- 129.Roberts K, Alberts B, Johnson A, Walter P, Hunt T. Molecular biology of the cell. New York: Garland Science; 2002. [Google Scholar]
- 130.Zhou Y, Frey TK, Yang JJ. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium. 2009;46:1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van Kuppeveld FJ, de Jong AS, Melchers WJ, Willems PH. Enterovirus protein 2B po (u) res out the calcium: a viral strategy to survive? Trends Microbiol. 2005;13:41–44. doi: 10.1016/j.tim.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 132.Carretero M, Gómez-Gonzalo M, Lara-Pezzi E, Benedicto I, Aramburu J, Martinez-Martinez S, Redondo JM, López-Cabrera M. The hepatitis B virus X protein binds to and activates the NH2-terminal trans-activation domain of nuclear factor of activated T cells-1. Virology. 2002;299:288–300. doi: 10.1006/viro.2002.1526. [DOI] [PubMed] [Google Scholar]
- 133.Kruman II, Nath A, Mattson MP. HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol. 1998;154:276–288. doi: 10.1006/exnr.1998.6958. [DOI] [PubMed] [Google Scholar]
- 134.Hu R, Han C, Pei S, Yin M, Chen X. Procalcitonin levels in COVID-19 patients. Int J Antimicrob Agents. 2020;56:106051. doi: 10.1016/j.ijantimicag.2020.106051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Austin LA, Heath H., III Calcitonin: physiology and pathophysiology. N Engl J Med. 1981;304:269–278. doi: 10.1056/NEJM198101293040505. [DOI] [PubMed] [Google Scholar]
- 136.Foster GV. Calcitonin (thyrocalcitonin) N Engl J Med. 1968;279:349–360. doi: 10.1056/NEJM196808152790704. [DOI] [PubMed] [Google Scholar]
- 137.Cappellini F, Brivio R, Casati M, Cavallero A, Contro E, Brambilla P. Low levels of total and ionized calcium in blood of COVID-19 patients. Clin Chem Lab Med (CCLM) 2020;58:e171–e173. doi: 10.1515/cclm-2020-0611. [DOI] [PubMed] [Google Scholar]
- 138.Elham AS, Azam K, Azam J, Mostafa L, Nasrin B, Marzieh N. Serum vitamin D, calcium, and zinc levels in patients with COVID-19. Clinical Nutrition ESPEN. 2021;43:276–282. doi: 10.1016/j.clnesp.2021.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Osman W, Al Fahdi F, Al Salmi I, Al Khalili H, Gokhale A, Khamis F. Serum Calcium and Vitamin D levels. Correlation with severity of COVID-19 in hospitalized patients in Royal Hospital, Oman. International Journal of Infectious Diseases. 2021;107:153–163. [DOI] [PMC free article] [PubMed]
- 140.Gittoes NJ, Criseno S, Appelman-Dijkstra NM, Bollerslev J, Canalis E, Rejnmark L, Hassan-Smith Z. ENDOCRINOLOGY IN THE TIME OF COVID-19: management of calcium metabolic disorders and osteoporosis. Eur J Endocrinol. 2020;183:G57–G65. doi: 10.1530/EJE-20-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tai C-Y, Smith QR, Rapoport SI. Calcium influxes into brain and cerebrospinal fluid are linearly related to plasma ionized calcium concentration. Brain Res. 1986;385:227–236. doi: 10.1016/0006-8993(86)91068-1. [DOI] [PubMed] [Google Scholar]
- 142.Kozik-Jaromin J, Nier V, Heemann U, Kreymann B, Böhler J. Citrate pharmacokinetics and calcium levels during high-flux dialysis with regional citrate anticoagulation. Nephrol Dial Transplant. 2009;24:2244–2251. doi: 10.1093/ndt/gfp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zweifach BW. The structural basis of permeability and other functions of blood capillaries. In Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press. 1940:216–223.
- 144.Means AR. Calcium and calmodulin-mediated regulatory mechanisms. In Principles of Molecular Regulation. Edited by Conn PM, Means AR. Totowa, NJ. Humana Press. 2000:187–204.
- 145.Chang KW, Sheng Y, Gombold JL. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology. 2000;269:212–224. doi: 10.1006/viro.2000.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bos EC, Heijnen L, Luytjes W, Spaan WJ. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology. 1995;214:453–463. doi: 10.1006/viro.1995.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shulla A, Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J Biol Chem. 2009;284:32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, Thacker BE, Glass CA, Yang Z, Torres JL, Golden GJ, Bartels PL, Porell RN, Garretson AF, Laubach L, Feldman J, Yin X, Pu Y, Hauser BM, Caradonna TM, Kellman BP, Martino C, Gordts PLSM, Chanda SK, Schmidt AG, Godula K, Leibel SL, Jose J, Corbett KD, Ward AB, Carlin AF, Esko JD. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e1015. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Teien AN, Abildgaard U, Höök M. The anticoagulant effect of heparan sulfate and dermatan sulfate. Thromb Res. 1976;8:859–867. doi: 10.1016/0049-3848(76)90014-1. [DOI] [PubMed] [Google Scholar]
- 150.Wenzhong Liu, Hualan Li. COVID-19: attacks the 1-Beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism. ChemRxiv. Cambridge: Cambridge Open Engage. 2020. 10.26434/chemrxiv.11938173.v9.
- 151.Madjid M, Vela D, Khalili-Tabrizi H, Casscells SW, Litovsky S. Systemic infections cause exaggerated local inflammation in atherosclerotic coronary arteries: clues to the triggering effect of acute infections on acute coronary syndromes. Tex Heart Inst J. 2007;34:11. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and results supporting the conclusions of this article are available at: https://pan.baidu.com/s/1OBS_AKjMZGWJhPfS5T_hXQ; code: 5kvg.