Abstract
Background
The underlying pathology of inguinal hernia is still not fully known; thus, further investigations of genetic backgrounds is needed. Here, we aimed to identify genetic factors attributing to inguinal hernias and explore the polygenic architecture of which some components are population-specific, while others are more common among populations.
Methods
We performed a genome-wide association study (GWAS) on subjects with inguinal hernias using BioBank Japan (BBJ) data with 1,983 cases and 172,507 controls, followed by a trans-ethnic meta-analysis with UK Biobank (UKBB) data. We performed downstream analyses in order to identify the mechanisms underlying inguinal hernias supported by genetic findings.
Findings
We identified a locus closest to ELN, which encodes elastin, at the GWAS significant level. The trans-ethnic meta-analysis revealed 23 additional significant loci, including five loci newly identified not significant in BBJ or UKBB GWAS: TGFB2, RNA5SP214/VGLL2, LOC646588, HMCN2, and ATP5F1CP1/CDKN3. Downstream analyses revealed the overlap of GWAS significant signals in extracellular components, including elastin fiber formation. We also found a highly shared polygenic architecture across different populations (trans-ethnic genetic-effect correlation = 0•77, standard error = 0•26) and population-specific lead variants in ELN, indicating the critical role of elastin in inguinal hernias.
Interpretation
We identified a significant locus of the ELN gene in the Japanese population and five additional loci across different populations. Downstream analyses revealed highly shared genetic architectures across populations and highlighted the important roles of extracellular components in the development of inguinal hernias. These findings deepen our understanding of the mechanisms underlying inguinal hernia.
Funding
The Japan Agency for Medical Research and Development (AMED) (Grant Number: JP19km0605001)
Keywords: inguinal hernia, genome-wide association studies, trans-ethnic meta-analysis, polygenic architecture, BioBank Japan
Research in context.
Evidence before this study
Genetic features related to inguinal hernia are still not well investigated across complete sets of DNA (genome-wide) especially in non-European populations.
Added value of this study
We conducted a genome-wide association study (GWAS) for inguinal hernia in the Japanese population, which identified a previously unreported susceptibility locus at the GWAS significant level. A trans-ethnic meta-analysis (using biobank data from the UK) revealed 23 additional significant loci. Additionally, downstream analyses revealed the overlap of GWAS significant signals in enhancers of extracellular components. Lastly, we found a highly shared polygenic architecture of inguinal hernias across different populations and the important role of elastin in both populations.
Implications of all the available evidence
This is the first study showing both population-specific and common genetic features across different populations with inguinal hernias. Our findings pave a path to future research and the potential improvement of treatment for patients with inguinal hernias.
Alt-text: Unlabelled box
1. Introduction
An inguinal hernia occurs when there is a protrusion of the abdominal contents out of the body's surface through a weak spot in the low abdominal wall [1]. A global epidemiology study estimated prevalence is 4.88% in Southeast Asia and 4.06% in Europe [2]. Severe complications of inguinal hernia are incarceration and strangulation, and currently, surgery is the only treatment option [3]. However, the reoperation rate is not negligible, a study reported to be 8•9%, [4] and chronic groin pain developed in more than 10% of the patients after surgeries [5,6]. Multiple risk factors have been reported for inguinal hernias, such as older age, [7] male sex, [7,8] smoking, [9,10] chronic obstructive pulmonary disease (COPD) [11,12] and systemic connective tissue disorders with genetic abnormalities. [[13], [14], [15]] Family history is also a risk factor, [11] and studies investigating genetic risk have been conducted mainly by candidate gene analyses [[16], [17], [18], [19], [20], [21], [22]]. The first genome-wide association study (GWAS) was conducted by Jorgenson et al. and included European populations, with 5,295 cases as a discovery cohort and 9,701 cases as replication cohort. They identified four significant loci and their functional roles were investigated [23]. However, no additional GWAS have been published, although the single nucleotide polymorphism (SNP)-heritability has not yet been fully explained by the associated SNPs in previous studies, and the genetic architecture in non-European populations remains unknown.
The BioBank Japan Project (BBJ) is a nationwide hospital-based genome cohort, which started in 2003. The BBJ has collected the data of approximately 200,000 patients with 47 target diseases including clinical data such as past medical history [24]. In this study, we aimed to identify genetics factors contributing to inguinal hernias using the data from the BBJ and explored both population-specific and common genetic architecture across populations.
2. Methods
2.1. Study Participants
The BBJ consists of DNA samples and clinical data of patients with 47 target diseases [24]. We selected cases from patients with a past medical history of inguinal hernia, documented by doctors-in-charge as previously described, [24] and controls from those with the 47 target diseases excluding COPD, which is a known risk factor of inguinal hernia [11,12].
Ethical committees at the Institute of Medical Sciences, The University of Tokyo (Tokyo, Japan) and the RIKEN Center for Integrative Medical Sciences (Yokohama, Japan) approved this study's protocol (Approval No. 17-17-16[8]). Written informed consent was provided for all patients recruited to the BBJ project. We complied with all relevant ethical regulations.
2.2. Whole-genome genotyping
We genotyped the BBJ samples by one of the following: [1] a combination of Illumina Infinium Omni Express and Human Exome, [2] Infinium Omni Express Exome v.1•0, [3] Infinium Omni Express Exome v.1•2.
2.3. Quality control (QC) of genotyping data
For QC of samples, we excluded individuals as follows: [1] sample call rates <0•98, [2] genetically identical to others, [3] genotypic and phenotypic sex mismatch and [4] outliers from the East Asian cluster identified by applying principal component analysis, using genotyped samples and the three major reference populations (Africans, Europeans and East Asians) from the International HapMap Project. This left 174,490 samples to be used for further analyses [25,26]. For QC of SNPs, we excluded SNPs as follows: [1] call rate <0•99 and [2] p-values for Hardy–Weinberg equilibrium (HWE) <1•0 × 10−6. We used Plink v.1•9 software for this QC process [27].
2.4. Whole-genome imputation
We generated a reference panel by using whole genome sequencing (WGS) data of 3,256 Japanese patients in the BBJ and 2,504 individuals in the 1000 Genomes Project (1KG; phase3v5) to achieve better imputation accuracy for the Japanese population (refer to Flanagan. et al. for more details) [28]. Briefly, samples were sequenced at high depth (15x, 30x) on various platforms (ex, 2 × 160-bp paired end reads on a HiSeq2500 platform Illumina with rapid run mode). The WGS data was processed, following the standardized best practice method, Genome Analysis Toolkit (GATK), [29] with the additional filters of approximate read depth and genotype quality before variant quality score recalibration (VQSR). The variants at multi-allelic sites were removed from the combined reference panel by vcftools (version 0•1•14). We estimated the haplotypes by SHAPEIT (version 2•778) and combined the data of the 1KG phase3v5 and the BBJ by using IMPUTE2 [30,31]. Quality control was then performed with bcftools (version 1•3•1) and vcftools (version 0•1•14). Variants at multi-allelic sites, monomorphic sites and singletons were excluded. We phased the genotyping data of autosomal chromosomes by SHAPEIT2 (version 2•837), and then imputed with minimac4 (version 2•0•1)(32) using the reference panel generated as described above. For X chromosome, first, we conducted phasing with SHAPEIT2 (version 2•837) and separated the variant call format files for males and females; second, we imputed with BEAGLE (version 4•1), and third, we merged males and females after excluding the variants at multi-allelic sites, monomorphic sites and singletons. The reference panel was finally composed of WGS data from 5,760 individuals, 72,406,123 autosomal variants and 3,252,444 X chromosome variants in total. Subsequently, individuals from BBJ without overlap with those in the reference panel were phased with EAGLE (version 2•3) using default parameters. In our study, we included only the variants imputed with R2 >0•3 after imputation by Minimac 4 [32].
2.5. GWAS
We performed GWAS by applying a generalized linear mixed model using Scalable and Accurate Implementation of GEneralized mixed model (SAIGE) (version 0•29•4•2) [33]. SAIGE is composed of two steps; in step 1, a null logistic mixed model is fit by using genotype data and added covariates, incorporating both sex and the top ten principal components (PC). In step 2, single-variant association tests were performed by using imputed variant dosages. We applied the leave-one chromosome-out approach in which the chromosome with the tested candidate SNPs is excluded from calculation of the genomic relationship. In each GWAS, we excluded the variants with minor allele frequencies of <0•01 and those imputed with R2 <0•3. We drew Manhattan plots by R (version 4•0•2). We regarded significant associations if loci showed association P values of <5•0 × 10−8. Significantly associated loci were defined as a genomic region within ±1 megabase (Mb) from lead variants. Novel locus was defined as those sites that did not include any known significant causal variants in inguinal hernias (p < 5•0 × 10−8). We generated regional association plots by LocusZoom (version 1•2) [34]. The estimated inflation factor λGC <1•05 after adjusting for sex and the top ten PCs meant little evidence of substantial inflation. For GWAS, we performed for the cases and control as described above the Study Participants sections. Additionally, we also performed GWAS, using the controls by excluding all cancer patients (with risk of muscle wasting status), fibroid patients (with risk of different sex steroid hormone levels), [35] and patients with genetic defects predisposing them to hernia (12 patients with Marfan syndrome, and one with Ehlers-Danlos syndrome, as there were no patients with cutis laxa in BBJ) so as to make sure that we excluded all possible patients with risks from controls.
2.6. Conditional analyses
Conditional analyses were performed using GCTA-COJO [36]. Additionally, stepwise conditional analyses were performed within ±1 Mb from the lead variants, and we repeated association tests by adding the dosages of the lead variants as covariates in SAIGE until no significant associations were identified.
2.7. Functional annotation of the lead variants in GWAS
Exonic variants in strong linkage disequilibrium (LD) with lead SNPs (r2 > 0•8) were annotated by ANNOVAR [37]. Similarly, we explored potential expression quantitative trait loci (eQTL) variants based on data reported by Ishigaki et al., including subgroups of immune cells, or the one from the Genotype-Tissue Expression project (GTEx) (version 8) [38,39].
2.8. Heritability enrichment analyses and genetic correlations
We estimated heritability in our GWAS results with linkage disequilibrium score regression (LDSC, version 1•0•0). We excluded variants in the human leukocyte antigen (HLA) region (chromosome 6: 26–34Mb). We also calculated heritability z scores and standard errors (SE) so as to assess the reliability of heritability estimation [40]. Inguinal hernia prevalence used for heritability estimation was 7•5%, 20% and 3% for all cases, only males and only females, respectively [7,[41], [42], [43]]. We additionally estimated genetic correlations with the BBJ's 42 target diseases [44]. We also evaluated enrichment of heritability of histone marks in 220 different cell types and 10 different tissue types and reported enrichment p-values to see enrichment correlation as described by Finucane et al. [40].
2.9. Pathway analysis
We conducted pathway analysis by Pathway scoring algorithm (Pascal), applying the corresponding LD structure [45]. Bonferroni corrections were applied. We set the statistically significant threshold as p < 0•05/1077, Bonferroni -corrected for the number of pathways tested by Pascal (REACTOME, KEGG, and BIOCARTA from Molecular Signature Database (MSigDB) version 4•0) [46,47].
2.10. Transcriptome-wide association study (TWAS)
We performed TWAS with the Multi-Tissue model in FUSION software consisting of 48 tissue types from the GTEx project (version 7) [48,49]. TWAS analysis used pre-computed gene expression weights and computed expression prediction models. Genes with nominally significant cis–SNP-heritability were used to train TWAS prediction models. FUSION fits predictive linear models for every gene. We used the summary statistics of our GWAS to estimate the associations of gene expression levels. Bonferroni corrections were applied as a stastistically significant threshold, based upon all tested genes (N=25,224).
2.11. Gene-based study
We implemented gene-based GWAS by MAGMA v1•07 software [50]. MAGMA transforms the p-values of genes in gene-sets to z-values by using an inverse normal transformation. Multiple linear principal components regression model was employed in order to account for LD between variants and to detect multi-marker associations. We used a significance threshold of p <5•0 × 10−8 in order to detect significance specifically at the single-variant level and to conservatively detect the gene-based specific loci across the genome.
2.12. Meta-analysis
We subsequently conducted a trans-ethnic meta-analysis with GWAS summary statistics from the UK Biobank (UKBB, ftp://share.sph.umich.edu/UKBB_SAIGE_HRC/ downloaded on July 25th, 2019) [33]. The UKBB GWAS was composed of 15,995 cases and 361,617 controls with 21 original significant loci. The QC for samples included removing individuals without white British genetic ancestry, closely related individuals and those with sex chromosome aneuploidies. Haplotype Reference Consortium (HRC) and UK10K haplotype resources were used for imputation. The SNPs were restricted for those with an INFO score of >0.8 and the minor allele frequency of 0.1%. GWAS was performed by UKBB using SAIGE. The summary statistics for males and females separately were not publicly available in the UKBB summary data (by SAIGE). Meta-ANalysis of Transethnic Association studies (MANTRA) software (version 2•0) was used to apply a random effect meta-analysis, taking into account the heterogeneity in allelic effect across populations [51]. We regarded the log10 Bayes factor (BF) > 6 as a significant threshold [52]. While the UKBB results are unpublished, we refrain from arguing novelty of a gene if the UKBB results contain variants in the gene exceeding a GWAS significant level.
2.13. Bayesian statistical fine-mapping analysis
Statistical fine-mapping analyses were performed by FINEMAP (version 1•4) so as to prioritise causal variants in inguinal hernia susceptibile loci [53]. The FINEMAP computes a posterior probability (PP) of causality for each SNP. Candidate putative causal variants were ranked for each association signal in a descending order of their PPs, which was followed by building 95% credible set of causal variants, including the variants ranked until their cumulative PP reached 95%. We used the default priors and parameters in FINEMAP.
2.14. Machine learning-based prediction of GWAS variants regulating ncRNA transcription
We used mutation effect prediction on ncRNA transcription (MENTR) which was developed by Koido et al., a machine-learning program trained with cell-type-specific long-ncRNA and enhancer transcription data obtained by cap analysis of gene expression (CAGE) in order to predict the effect of variants on promoter/enhancer expression [54]. All significant variants in our GWAS and meta-analysis were analyzed by MENTR.
2.15. Definition and annotation of the significant SNPs in our meta-analysis
Histone modifications (H3K4me1 mark often found near regulatory elements, H3K4me3 mark often found near promoters, and H3K27ac mark often found near active regulatory elements on 7 types of cell lines from ENCODE: GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, and NHLF cells) and DNase hypersensitive sites (DNaseI hypersensitivity clusters in 125 ENCODE cell types: the names of 125 cell lines are listed as follows, “DNaseI Hypersensitivity Uniform Peaks from ENCODE/Analysis”, http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=wgEncodeAwgDnaseUniform) defined by ENCODE (version 3), [55] and, if SNPs are intergenic, enhancers (hg19) mapped by FANTOM5 (phase2•5) [56] were extracted from the University of California Santa Cruz (UCSC) database [57].
2.16. Functional mapping and annotation (FUMA) of the genes with inguinal hernia-associated SNPs in our meta-analysis
FUMA (version 1•3•6ª: https://fuma.ctglab.nl/) performs hypergeometric tests of enrichment of the list of mapped genes in MSigDB gene sets. We used the GENE2FUNC procedure in FUMA to perform tissue specificity and pathway enrichment analyses of genes with the lead SNPs in our meta-analysis [58]. FUMA addditionally carries out gene mapping, tissue-expression analysis, and gene set enrichment analysis (GSEA).
2.17. Evidence of related gene expression of inguinal hernia risk genes newly identified in this study
We used RNA microarray data deposited by Zhao et al [35]. in the National Center for Biotechnology Information Gene Expression Omnibus database accessible under the accession no. GSE92748 (https://www.ncbi.nlm.nih.gov/geo/). With this data, we assessed the gene expression of each newly identified risk gene in this study. Total RNA was obtained from the lower abdominal muscle tissue of humanized aromatase transgenic mice, and was compared with those of wild type mice. The number of cases and controls was 6 for each. The online tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/gep2r/) was used for analysis. GEO2R performs comparisons by using the GEOquery and limma (Linear Models for Microarray Analysis) R package from the Bioconductor project [59]. Additionally, we used the Human Protein Atlas (http://www.proteinatlas.org)(60) in order to see which tissues are expressed for each gene. We considered positive RNA expression based on summary data of consensus Normalized eXpression (NX) levels, created by combining data from three transcriptomics datasets (HPA, GTEx and FANTOM5). We considered positive protein expression based on a best estimate from a knowledge-based annotation as per the Human Protein Atlas website.
2.18. Trans-ethnic genetic correlation
The python package Popcorn (ver.0•9•9) was used to estimate the genetic correlation of causal variants effect sizes across populations [61]. We used the data of 1KG for East Asians and Europeans so as to compute cross-population scores, taking into account each structure of LD [62].
2.19. Comparison of effects sizes between BBJ and UKBB data
We compared the beta coefficients and SEs of the inguinal hernia-associated SNPs in the data from UKBB with those SNPs in the data from BBJ. If the beta coefficients and SEs were plotted either in the first or third quadrant, effect size for both traits were in the same directions as both x and y take positive or negative values, which meant that inguinal hernia-associated SNPs were shared between European and Japanese populations.
We also showed the distribution of risk allele frequencies (RAF) of the inguinal hernia-associated SNPs specifically in data from UKBB and inguinal hernia-associated SNPs specifically in the data from our meta-analysis.
2.20. Role of funding source
Funders’ roles: Study design and data collection.
3. Results
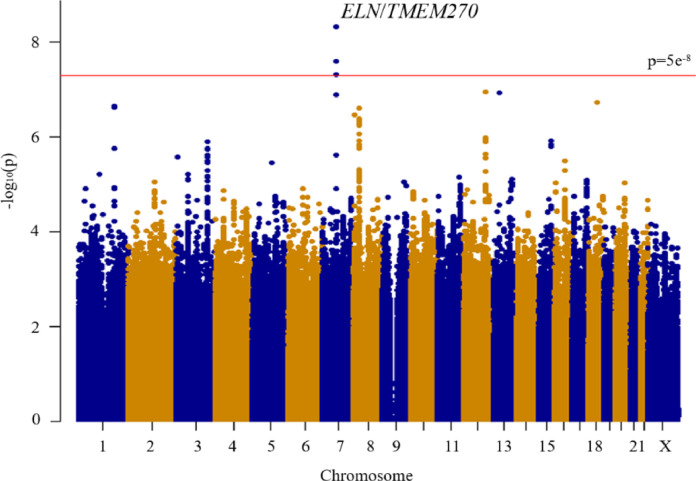
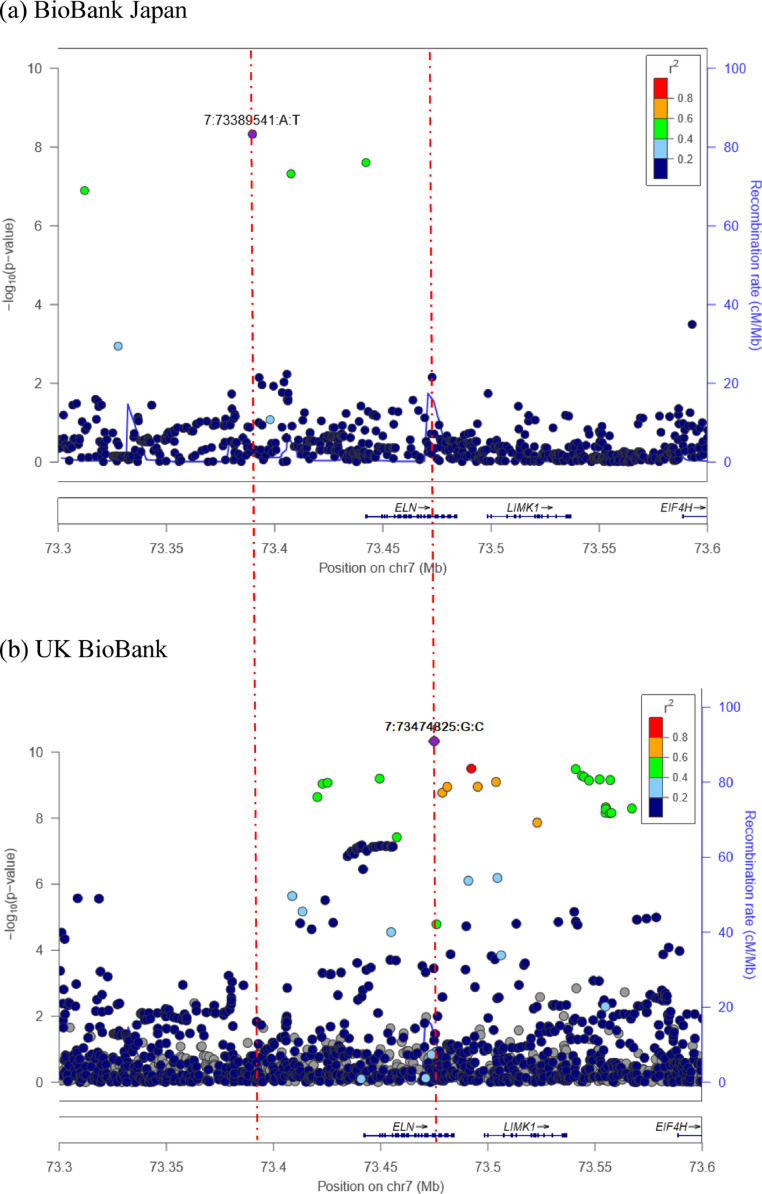
In this study, we included 1,983 cases and 172,507 controls. Baseline demographics of the subjects are shown in Table 1, and male gender was higher in the cases as shown in previous studies [7,8]. After filtering an imputed dataset, we tested 8,443,696 autosomal variants and 181,087 chromosome X variants for GWAS by SAIGE [33] (see methods). The Manhattan and Q-Q plots are shown in Fig. 1 and Supplemental Fig.1 where no evidence for inflation was observed. A significant locus, rs118109209 in the intergenic region of ELN/TMEM270 genes in chromosome 7 (the closest gene was ELN), was significantly associated with inguinal hernias (p=4•7 × 10−9, Table 2, Fig. 1 and Fig. 2a). This locus has not been reported to reach GWAS significance level in previous literature. GWAS excluding all cancer, fibroid and genetic defects predisposed to inguinal hernia from controls groups showed the same ELN locus reached GWAS significance level (p=1•6 × 10−8), confirming that the difference between controls caused no significant biases. The ELN gene encodes elastin, a protein that is one of the two components of elastic fibers, [63] and mutations of the ELN gene are observed in patients with cutis laxa, a rare connective tissue disorder, who are at a higher risk of developing inguinal hernias [64]. No exonic SNPs were in strong LD with the lead variant and no eQTL studies report the lead variant as an eQTL [39].
Table 1.
Baseline characteristics of patients included for this study (n = 174,490)
| Cases | Controls | |
|---|---|---|
| Characteristic | n = 1,983 | n = 172,507 |
| Age, mean (SD) | 64.8 (14.4) | 62.7 (14.6) |
| Men (%) | n = 1,622 (81.8) | n = 90,335 (52.4) |
| Genotyping array | (1) A combination of Illumina Infinium Omni Express and Human Exome (2) Infinium Omni Express Exome v.1.0 (3) Infinium Omni Express Exome v.1.2 |
|
n, number; SD, standard deviation.
Fig. 1.
Manhattan plot of genome-wide markers for inguinal hernia (1,983 cases and 172,507 controls). We performed genetic association tests, adjusted for sex and genetic ancestry (PCs 1 through 10). Results are plotted as –log10 p values on the y-axis by position in chromosome (x-axis) (NCBI build 37). The red line represents the genome-wide significance level. Gene name is shown next to the top locus.
Table 2.
Significant loci associated with inguinal hernia among patients at BioBank Japan Project identified by genome-wide association study
| Chr | Position | SNP id (rs) | Gene | Ref/var | Location | AF.Cases | AF.Controls | OR | 95% CI | p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 73389541 | rs118109209 | ELN/TMEM270 | A/T | Intergenic | 0.071 | 0.052 | 1.64 | 1.25, 2.15 | 4.7 × 10−9 |
Allele frequencies in the 1000 Genomes: East Asian A=0.9742, T=0.0258; Europe A=1.0000, T=0.0000.
Chr, chromosome; Ref, reference allele; Var, variant allele; AF.Cases, variant allele frequency in cases; AF.Controls, variant allele frequency in controls; OR, odds ratio; CI, confidence interval.
Fig. 2.
Locus zoom plot of results of genetic association tests at chromosome 7 regions. Locus zoom plots at significant loci in chromosome 7 are shown in the BioBank Japan (a) and UK Biobank data (b). Coloring is based on LD (genome build hg19/1KG for both populations) with the top hits in the regions for inguinal hernia. The left red dashed line represents the position of the top locus in BBJ, and the right one is for the top locus in UKBB for this region.
Conditional analyses for the significant locus by the lead SNP detected no additional independent signals. We conducted a gene-based test in order to identify the association signals of novel susceptibility genes, but no additional genes were identified (Supplemental Fig. 2). SNP heritability estimate for inguinal hernia was 25•3% (SE of 5•5%) by LD score regression, proving its firmly inherited feature. We conducted a genetic correlation analysis by bivariate LD score regression to evaluate shared polygenic architecture between inguinal hernia and BBJ target diseases, [44] showing that no traits reached Bonferroni-corrected significance (Supplemental Table 1). Additionally, we separately conducted GWAS for males and females, and estimated heritability (Table 3, Supplemental Fig. 3 and 4). We found higher heritability for males, which may be consistent with previous studies showing a higher risk of developing inguinal hernias in males [7,8].
Table 3.
Heritability
| Population | Heritability (SE) |
|---|---|
| All | 0.25 (0.055) |
| Male | 0.35 (0.098) |
| Female | 0.23 (0.23) |
SE, standard error
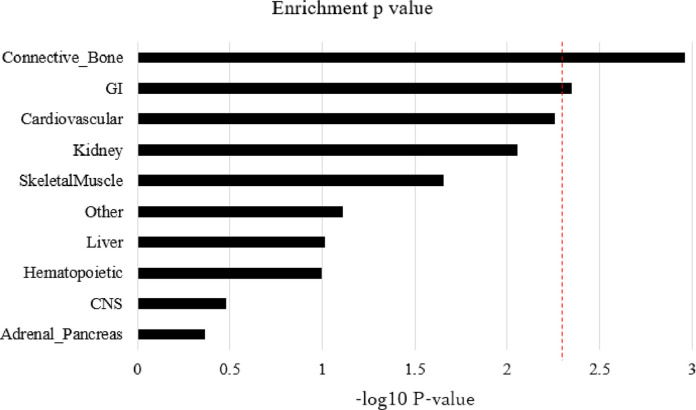
We tested pathway analysis by Pascal in order to evaluate disease-associated pathways driven by polygenic components, [45] resulting in a possibly significant signal of the focal adhesion pathway (p-value of 1•7 × 10−4, Supplemental Table 2). Given the mechanism of inguinal hernias and a previous report showing significant enrichment in the focal adhesion pathway, this finding is intriguing. [65] The partitioned heritability analysis in cell groups using LD score regression revealed enrichment in the connective/bone and gastrointestinal (GI) tissues (p-value of 0•0011 for connective/bone tissues and 0•0045 for GI tissues, see Fig. 3). Analyses of detailed cell types demonstrated significant heritability enrichment in the H3K9ac in colon smooth muscle (p-value of 1•8 × 10−4, Supplemental Table 3). Interestingly, we observed near significant enrichments in the smooth muscles of multiple GI tracts, including H3K27ac in the duodenum and H3K9ac in the stomach (p-value of 6•5 × 10−4 and 8•5 × 10−4, respectively, Supplemental Table 3). TWAS by FUSION was performed in order to investigate the transcriptional landscape regulated by genetic components of inguinal hernia. [48] We observed a significant association of the CALD1 gene in chromosome 7 with suprapubic skin (not sun exposed, p-value of 7•9 × 10−5). This is supported by CALD1’s involvement in the regulation of smooth muscle contraction (including the GI tract), [66] which is reasonable to be seen, as a network analysis investigating causative proteins related to inguinal hernia showed enrichment in the regulation of actin cytoskeleton in a previous study. [65] These findings indicate the importance of gene regulation in focal adhesion, the GI tract (the contents of protrusion), and skin covering the protrusions in the pathology of inguinal hernia.
Fig. 3.
P-value for the results of partitioned heritability analysis for the 10 different tissue types. Results are plotted as –log10 p values on the x-axis by 10 tissue types on the y-axis. The red dashed line represents the –log10 p value reaching the Bonferroni-corrected threshold.
Subsequently, we conducted a trans-ethnic meta-analysis, using the summary statistics of GWAS for inguinal hernia from UKBB. We tested 4,846,078 autosomal variants available for both the data from BBJ and UKBB. In meta-analysis, 23 significant loci were identified (Table 4). Among the 21 significant loci in the UKBB GWAS, 18 loci remained statistically significance in the meta-analysis. Most importantly, 5 out of 23 significant loci, namely, TGFB2, RNA5SP214/VGLL2, LOC646588, HMCN2, and ATP5F1CP1/CDKN3, were unreported; these were unidentified in GWAS by BBJ and UKBB, indicating a shared genetic component between these two populations. Two genes in these loci seemed relevant to inguinal hernias. First, the TGFB2 (transforming growth factor beta 2) gene in chromosome 1 encodes a secreted ligand of the transforming growth factor-beta (TGF-beta) superfamily of proteins. [67] Overexpression of one of the isoforms of TGF-beta, TGF-beta1, is reported in patients with inguinal hernias, [68] and previous GWAS also showed that another isoform, TGFB3, is regulated by WT1, the significant locus in their study. [23] Second, the VGLL2 (vestigial like family member 2) gene in chromosome 6 encodes a protein that may act as a co-factor of transcriptional enhancer factor 1 (TEF-1) regulating gene expression during skeletal muscle development. [69] When exploring potential eQTL variants based on the data reported by GTEx (version 8), [39] we found evidence suggesting that some signals in these novel loci are associated with gene regulation. Specifically, eQTL signals were noted in the lead SNP in the RNA5SP214/VGLL2 gene in the esophagus (mucosa) and in the HMCN2 gene, especially prominent in skin (not sun exposed, suprapubic, Supplemental Table 4). As per the UCSC browsers we used to define and annotate significant SNPs, the lead SNP in the TGFB2 gene has overlap in the H3K27ac and H3K4me3-marked region, while the lead SNP in the RNA5SP214/VGLL2 gene has overlap in the H3K27ac-marked region, H3K4me1-marked region, and DNaseI hypersensitivity clusters. Finally, the lead SNP in the HMCN2 gene has overlap in the H3K4me1-marked region (Supplemental Table 4). [57] We also performed statistical finemapping analyses for the five loci using both BBJ and UKBB sets of data. The lead variant in the meta-analysis in ATP5F1CP1/CDKN3 and in the TGFB2 were prioritized in both BBJ and UKBB with highly ranked PP (Supplemental Table 5), suggesting shared causal variants across populations in these loci. Lead variants in the other three loci showed top or near top PP in the UKBB, but not BBJ data. However, lead SNPs are not necessarily always the causal SNPs and variants in high LD with lead variants showing comparable PP with lead variants might be causal and functional. We discovered an additional candidate of causal variant, rs10951082 in LOC646588 in high LD with the lead SNP (R-square of 0.95), showing the 4th highest PP in UKBB and the 2nd highest PP (higher than the lead variant) in BBJ. Additionally, the variant overlapped with the H3K27ac marks.
Table 4.
Meta-analysis with UK Biobank data for inguinal hernia
| Risk allele frequency in Ctrl | BBJ | UKBB | MANTRA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | position | SNP id (rs) | Gene | Ref/var | BBJ | UKBB | Location | OR | 95% CI | p-value | OR | 95% CI | p-value | Log10 Bayes | posthg |
| 1 | 9447189 | rs2095906 | SPSB1 / LINC02606 | A/G | 0.64 | 0.62 | intergenic | 1.05 | 0.98, 1.13 | 0.14 | 1.07 | 1.04, 1.10 | 1.00 × 10−7 | 6.18 | 0.80 |
| 1 | 218524632 | rs2799097 | TGFB2 | A/G | 0.73 | 0.85 | intron | 1.07 | 0.99, 1.15 | 0.07 | 1.10 | 1.06, 1.14 | 2.01 × 10−8 | 7.01 | 0.33 |
| 1 | 219675209 | rs2820465 |
LYPLAL1-AS1 / LOC107985272 |
G/T | 0.31 | 0.59 | intron | 0.99 | 0.92, 1.06 | 0.74 | 0.93 | 0.91, 0.95 | 6.70 × 10−9 | 6.24 | -0.09 |
| 2 | 25110962 | rs6749170 | ADCY3 | A/G | 0.44 | 0.46 | intron | 0.93 | 0.88, 0.99 | 0.03 | 0.94 | 0.92, 0.96 | 1.27 × 10−7 | 6.25 | -0.42 |
| 2 | 55976844 | rs62167673 | PNPT1 / EFEMP1 | A/G | 0.45 | 0.15 | intergenic | 1.07 | 1.00, 1.14 | 0.05 | 0.91 | 0.88, 0.93 | 4.01 × 10−9 | 6.02 | 2.26 |
| 3 | 56149492 | rs13091322 | ERC2 | A/G | 0.15 | 0.31 | intron | 0.92 | 0.85, 0.9996 | 0.07 | 0.93 | 0.91, 0.96 | 2.39 × 10−7 | 6.05 | 0.31 |
| 4 | 174616822 | rs7686296 | RANP6 / LINC02269 | T/A | 0.23 | 0.30 | intergenic | 1.07 | 0.98, 1.16 | 0.10 | 1.08 | 1.05, 1.11 | 1.67 × 10−8 | 7.06 | 0.27 |
| 5 | 64451583 | rs7702887 | ADAMTS6 | C/T | 0.45 | 0.72 | intron | 1.05 | 0.98, 1.12 | 0.14 | 1.07 | 1.04, 1.11 | 7.15 × 10−8 | 6.15 | -0.08 |
| 5 | 121422560 | rs995687 | LOX / ZNF474 | A/G | 0.026 | 0.29 | intergenic | 1.05 | 0.85, 1.31 | 0.63 | 1.07 | 1.05, 1.11 | 4.68 × 10−8 | 6.13 | -0.03 |
| 6 | 6749069 | rs9504915 | LOC101928004 | A/G | 0.21 | 0.56 | intron | 1.01 | 0.93, 1.09 | 0.78 | 0.93 | 0.91, 0.95 | 7.55 × 10−9 | 6.17 | 0.20 |
| 6 | 27699581 | rs9393851 | TRV-CAC7-1 / GPR89P | A/T | 0.18 | 0.27 | intergenic | 1.03 | 0.94, 1.14 | 0.51 | 0.92 | 0.90, 0.95 | 3.48 × 10−9 | 6.00 | -0.03 |
| 6 | 117490664 | rs1405212 | RNA5SP214 / VGLL2 | T/C | 0.72 | 0.63 | regulatory region (intergenic) | 0.91 | 0.86, 0.97 | 0.01 | 0.94 | 0.92, 0.96 | 1.09 × 10−7 | 6.83 | 0.23 |
| 6 | 143556810 | rs6570551 | AIG1 | A/G | 0.93 | 0.36 | intron | 1.08 | 0.94, 1.24 | 0.23 | 0.93 | 0.91, 0.95 | 8.31 × 10−9 | 6.16 | 0.24 |
| 7 | 25692889 | rs6943068 | LOC646588 | G/A | 0.40 | 0.66 | Non coding transcript variant | 0.93 | 0.88, 0.99 | 0.03 | 0.94 | 0.92, 0.96 | 2.49 × 10−7 | 6.19 | -0.22 |
| 7 | 73503533 | rs75566398 | LIMK1 | A/G | 0.065 | 0.12 | intron | 0.99 | 0.87, 1.13 | 0.90 | 0.89 | 0.86, 0.92 | 8.00 × 10−10 | 7.02 | -0.21 |
| 8 | 25383344 | rs4872325 | CDCA2 / EBF2 | G/T | 0.77 | 0.72 | intergenic | 0.92 | 0.86, 0.99 | 0.03 | 0.93 | 0.91, 0.96 | 4.18 × 10−7 | 6.02 | 0.01 |
| 9 | 16749020 | rs4961753 | BNC2 | G/A | 0.89 | 0.92 | intron | 1.02 | 0.91, 1.14 | 0.78 | 0.88 | 0.85, 0.92 | 5.54 × 10−9 | 6.00 | 0.25 |
| 9 | 133037273 | rs4837486 | HMCN2 | T/C | 0.84 | 0.50 | intron | 0.95 | 0.88, 1.04 | 0.28 | 0.94 | 0.92, 0.96 | 7.31 × 10−8 | 6.07 | 0.16 |
| 11 | 32296455 | rs7940705 | THEM7P / WT1 | A/G | 0.82 | 0.34 | intergenic, non coding transcript variant |
0.94 | 0.87, 1.02 | 0.17 | 0.93 | 0.91, 0.96 | 1.12 × 10−7 | 6.07 | 0.04 |
| 13 | 51088052 | rs183949 | DLEU1 | A/G | 0.20 | 0.40 | intron | 1.08 | 0.99, 1.17 | 0.06 | 1.07 | 1.04, 1.10 | 9.08 × 10−8 | 6.40 | -0.01 |
| 14 | 54724054 | rs2358483 | ATP5F1CP1 / CDKN3 | C/T | 0.053 | 0.30 | regulatory region (intergenic) | 1.24 | 1.04, 1.47 | 0.00 | 1.07 | 1.04, 1.10 | 3.90 × 10−7 | 6.08 | -0.04 |
| 16 | 84856889 | rs4783079 | CRISPLD2 | C/A | 0.20 | 0.38 | intron | 1.05 | 0.96, 1.14 | 0.27 | 1.08 | 1.06, 1.11 | 5.91 × 10−11 | 8.59 | -0.15 |
| 17 | 12187295 | rs8081231 | MAP2K4 / LINC00670 | T/C | 0.13 | 0.31 | intergenic | 1.13 | 1.02, 1.25 | 0.01 | 1.08 | 1.05, 1.11 | 1.23 × 10−9 | 8.70 | 0.89 |
Unreported loci are highlighted in bold.
Ctrl, control; BBJ, BioBank Japan; UKBB, UK Biobank; MANTRA, Meta-ANalysis of Transethnic Association studies; OR, odds ratio; CI, confidence interval; Chr, chromosome; Ref, reference allele; Var, variant allele; OR, odds ratio; CI, confidence interval; log10BF, log-arithm of Bayes Factor; posthg, posterior probability of heterogeneity.
We further investigated variants among all significant loci in the meta-analysis with potential to modulate gene expression levels that are difficult to identify with conventional eQTL studies. We applied MENTR, a newly developed machine-learning model to predict an alternative allele's mutation effect, or ability to change the expression of transcribed promoter/enhancer which was defined by CAGE sequencing. [54] As a result, five SNPs in the TGFB2, LOX, and WT1-AS/WT1 genes are shown to be potential functional variants with high confidence (Supplemental Table 6). In particular, rs2234580 (chr1: 32,457,138 at hg19) in the WT1-AS/WT1 genes was predicted to increase expression of the promotors in relevant tissues, namely, epithelial folds, anatomical walls, epithelial cells of alimentary canal, and endo-epithelial cells. These findings again support the theory of involvement of extracellular components, GI tract, and dermatological pathologies in the development of inguinal hernias.
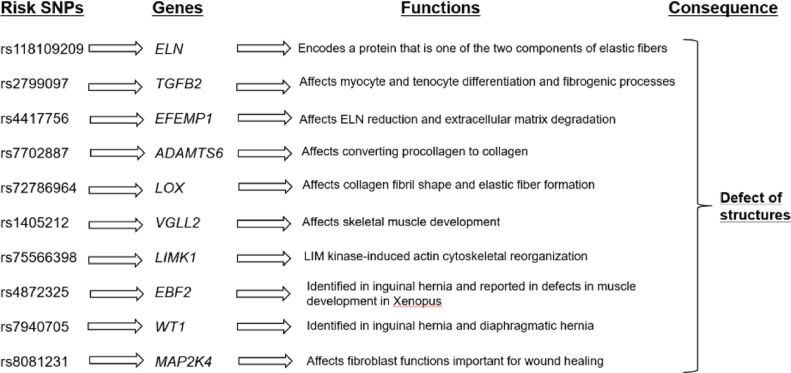
Subsequently, we explored the functional annotation of genes with lead SNPs in our meta-analysis by using FUMA. [58] Prioritized genes were overrepresented in extracellular matrix pathways (adjusted P-value of 0•036, GO cellular component (MsigDB c5), Supplemental Fig. 5a). GSEA conducted by FUMA using the curated gene sets showed enrichment in elastic fiber formation (Reactome) and NABA_MATRISOME (ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins) [46,70] (adjusted P-values of 0•018 and 0•046, respectively, Supplemental Fig. 5b), both of which are consistent with the mechanisms of inguinal hernia. These two pathways contained functionally relevant genes including TGFB2, EFEMP1, LOX, ADAMTS6, HMCN2 and CDCA2 supported by eQTL and/or histone marks described above, together with the CRISPLD2 gene. While we did not find functional evidence of the lead variant for CRISPLD2, the pathway analysis suggested this gene is causal for inguinal hernia in this region. These findings suggest that the majority of lead signals in the meta-analysis are putative causal variants, pointing to the need for future investigations. The significant loci containing genes whose biological functions relevant to inguinal hernia were shown in previous studies are illustrated in Fig. 4. Statistical finemapping analyses for the known significant loci showed rs6749170, an intronic variant of the ADCY3 gene, had the highest PP for both the BBJ and UKBB data in addition to ATP5F1CP1/CDKN3 and TGFB2 (Supplemental Table 5).
Fig. 4.
Significant loci identified in this study which can be biologically explained in development of inguinal hernia. The flows of each risk SNP, its gene name, the functions of the gene, and the consequence of the gene function which is expected to cause inguinal hernia are shown.
Subsequently, we assessed related gene expression for the five inguinal hernia risk loci newly identified in this study, in addition to the ELN. Gene expression for four of the six genes (ELN, TGFB2, VGLL and CDKN3) was available in the RNA microarray data deposited by Zhao et al. [35], which was obtained from the lower abdominal muscle tissues of humanized aromatase transgenic mice. For the ELN gene, expression was upregulated 2.3 times more compared to wild type mice (p-value of 0.0027) and for TGFB2, VGLL and CDKN3, gene expression was 1.19, 1.02, and 1.08 times more upregulated, respectively (p-values of 0.016, 0.81, and 0.023, respectively). Next, data from expressed tissues for five of the six genes (ELN, TGFB2, VGLL, HMCN2 and CDKN3) were available from the Human Protein Atlas [60]. RNA and protein expression were seen in multiple tissues related to inguinal hernia (Supplemental Table 7). These findings highlight that most of the newly identified risk genes were also supported by gene expression and/or protein expression in relevant tissues.
Of note, we also confirmed that the results of meta-analysis did not change dramatically when excluding all forms of cancer, fibroid and genetic defects predisposed to inguinal hernia from controls groups. However, additionally, an unreported locus, rs1925281, was found to be significant, an intergenic variant in EMX2/RAB11FIP2 in chromosome 10 with a 6.10 log-arithm of the Bayes Factor and posterior probability of heterogeneity of 0.10 (those were 5.93 and -0.066, respectively, in analysis with the original control samples). This locus did not reach GWAS significance level in either of the BBJ or UKBB GWAS. In GTEx data, eQTL signals were noted in this SNP in the cultured fibroblasts. This locus has overlap in the H3K27ac-marked region, according to UCSC browsers. In the finemapping analyses, this SNP ranked second with a PP of 0.30 in UKBB, and ranked third with a PP of 0.13 in BBJ. Additionally, RNA and protein expressions were seen in multiple tissues related to inguinal hernia for both EMX2/RAB11FIP2 genes as per the Human Protein Atlas. This locus could also be a causal variant, but further in-vitro studies will be necessary to understand its detailed functional role.
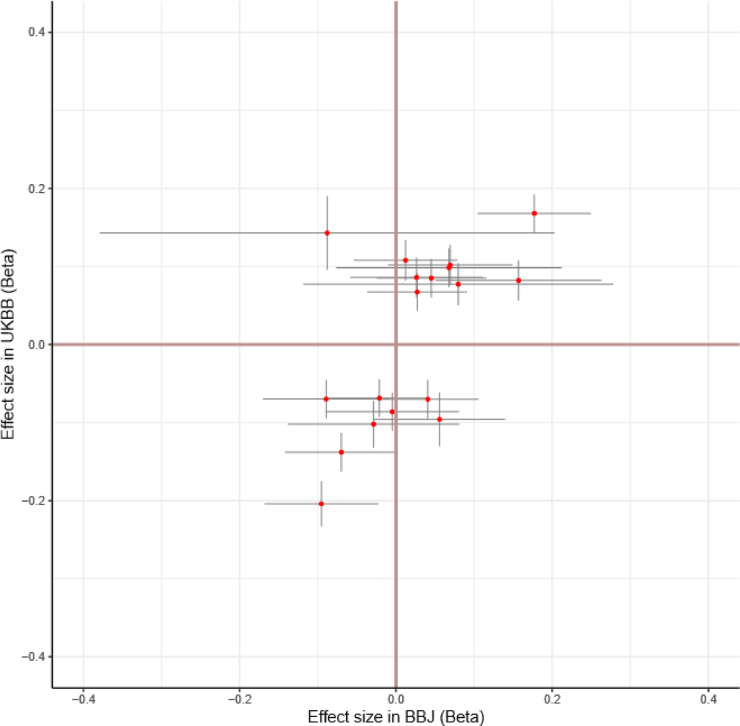
Lastly, we explored the relationship of the effect sizes of inguinal hernia-associated SNPs between BBJ and UKBB in order to investigate the differences of susceptibility across populations. We confirmed that risk alleles of inguinal hernia-associated variants in the UKBB data were shared in Japanese populations: 15 out of the 18 SNPs showed the same association directions and all 18 variants showed a strong correlation of effect size (binomial p=0•0075, Spearman rho = 0•49 with p=0•041, Fig. 5). The three variants with different directions of association between the two populations were rs2480924 in chromosome 9, rs12319548 in chromosome 12, and rs2076441 in chromosome 16. Risk alleles are rare in either population for rs2480924 and rs12319548, and the minor allele is opposite for the European and East Asian populations in the rs2076441 as per the 1000 Genomes Project. [62] We observed a strong trans-ethnic genetic-effect correlation (ρge = 0•77, SE = 0•26). These results indicate strongly shared genetic components of inguinal hernia across these populations. Meta-analysis identified the SNPs with low RAF in either population, successfully increasing the power of detecting associated SNPs (Supplemental Fig. 6a). Meta-analysis identified the SNPs with lower MAF in the data from UKBB as significant, which again increased the power of detecting associated SNPs (Supplemental Fig. 6b). Additionally, we found a population-specific variant in ELN locus in each population (Fig 2]. [62] The lead SNP in the BBJ is not seen in Europeans; A=0•9742 and T=0•0258 in East Asians but A=1•0000 and T=0•0000 in Europeans; conversely the lead SNP in the UKBB is not seen in East Asians; A=1•0000 and T=0•0000 in East Asians but A=0•9085, T=0•0915 in Europeans as per 1000 Genomes. [62] When conducting finemapping analyses for each of those loci, the lead SNP in the BBJ data was the only SNP contained in the 95% credible sets for causal signal in the region with a PP of 0.999993. For the lead locus in UKBB, two SNPs were contained in the 95% credible sets, of which the top ranked SNP with a PP of 0.94 was the lead SNP in the region. These indicate that population-specific variants in these two populations share the same associations with inguinal hernias, highlighting the critical role of elastin in inguinal hernias.
Fig. 5.
Relationship of inguinal hernia-associated SNPs reaching GWAS significance between the populations. X-axis shows the beta coefficients in the BBJ data. Y-axis shows the beta coefficients in the UKBB data. The bars crossing the red dots represent the SEs of the beta coefficients.
4. Discussion
We conducted the first large-scale GWAS for inguinal hernia in the Japanese population and successfully identified a locus, the closest gene of which was ELN at the GWAS significant level. Downstream analyses support the importance of elastin and show strong evidence of extracellular components, especially elastic fibers, involved in the development of inguinal hernias. We also found five unreported susceptibility loci for inguinal hernia in the meta-analysis and showed a strong shared genetic architecture across different populations, which implicates the generalizability of our findings.
Elastin is a component of elastic fibers in the extracellular matrix. [63] A network analysis conducted by Jorgenson et al., showed potential interactions between the ELN gene and the EFEMP1 gene(23), one of the GWAS significant loci in this current study. Additionally, a recent study also showed the decreased expression of ELN in direct inguinal hernia in the transversalis fascia [71]. In our study, the lead SNP has no overlap in H3K27ac, H3k4me1, H3k4me3 and DNaseI hypersensitivity clusters per UCSC browsers. [57] Thus, the functions of the lead variant or tightly linked variant(s) should be evaluated in hernia-specific tissues in future studies.
The encoded protein of TGFB2 is a secreted ligand of the TGF-beta, reported to be associated with inguinal hernia formation. [68] rs2799097, the lead SNP in the TGFB2 gene is the most likely causal variant, given the enrichment in histone marks as well as the results of finemapping. Additionally, previous studies show the association of the isoforms of TGF-beta with inguinal hernia and TGF-beta 2 itself with involvements in fibroblasts in various tissues and myocytes. [23,68,[72], [73], [74], [75]] The other significant loci in the meta-analysis includes the four loci identified in the previous non-UKBB GWAS, [23] namely, EFEMP1, WT1, EBF2 and ADAMTS6. The associations of those loci showed the same directions of signals as in the BBJ data though they did not reach GWAS significant levels (p-values ranged from 0•093 to 1•5 × 10−6). These indicate the shared genetic components of inguinal hernias not only between BBJ and UKBB, but across other populations. In addition, multiple genes have functions consistent with the development of inguinal hernia (Fig.4] and the enrichment in the components and curated gene sets related to extracellular matrix and elastic fiber formation. Additionally, we found enrichment in focal adhesion pathways consistent with the causal mechanism of inguinal hernia, a pathway which functions through bundles of actin filaments anchored to the integrin family's transmembrane receptors via a multi-molecular complex of junctional plaque proteins. [76] For all significant loci in our analysis, we further investigated by finemapping in order to prioritize potentially functional SNPs. By doing this, we were able to identify some loci which are not only the lead loci but also the putative causal loci. Elucidating the causative loci and mechanical pathways sow good seeds for future research.
We showed the shared genetic architecture (Fig.5], which was further supported by the trans-ethnic genetic correlation as stated above. The genetic feature of the top locus in our BBJ GWAS was similar to the UKBB; both have significant loci in or near the ELN gene. Understanding the existence of similar polygenic architecture of inguinal hernia across different populations will enhance the design of future larger studies, and facilitate further investigations of possible unknown loci in this phenotype.
Our study shows the enrichment not only in connective tissues but also GI tissues, and additionally shows strong mutation effects in the epithelial cells in the GI tract (Fig.3, Supplemental Table 3 and 6). An inguinal hernia occurs when the abdominal contents, including gastrointestinal tract, pass down through the inguinal canal; thus, there could be a tendency to have more inguinal hernias with the existence of abnormalities of the structures supporting the position of the gastrointestinal tract. Furthermore, TWAS showed overlap in the skin of suprapubic regions. These could be supported by the GWAS significant locus in our study. Mutations of the ELN gene might lead to collapse of the transcriptional network responsible to dermal elastin, loose skin, and lead to protrusion of the inguinal hernia. Additionally, our enrichment analyses showed the enrichment in the connective and GI tissues which reached the Bonnferroni-corrected threshold, but not for the skeletal musles. These could be due to the fact that the data for muscle tissues used in this analysis were from the Roadmap Epigenomics Project [77] and did not represent low abdominal skeletal muscles. [62] Further investigations are needed to confirm these results.
Our study bears multiple limitations. First, biochemical analyses using actual patient samples would make our findings much more robust; however, unfortunately the BioBank Japan Project did not possess these specimens. Thus, we utilized the results of the gene expression data which are publicly available. Future studies should definitely consider doing those valuable analyses. Second, given that the cases were selected from past medical histories of the 47 target diseases in BBJ, the details of the inguinal hernias, including direct/indirect, and medial/lateral were not available. The differences of indirect hernia and direct hernia is not only in the way the hernias occur but also in the underlying pathology as stated by Somuncu et al. [78] However, the results of previous GWAS indicated the underlying significant loci across different subtypes of inguinal hernia. [23] Thus, we believe that our conclusions should not be affected by the unavailability of such data. Also, there is the possibility of included possible future cases, those who may develop inguinal hernia in the future, as controls. However, this is the nature of case-control studies, and it does not affect our conclusions. Additionally, the closest gene of the rs118109209 was ELN, and ELN is only a gene with the SNP in the moderate LD with the lead variant (Supplemental Fig. 7). ELN could be affected by this variant, given the roles of ELN as mentioned above; however, future in-vitro studies are required in order to confirm this. Lastly, we did not investigate sex-stratified analysis due to the small sample size in females. Future studies will ideally investigate this aspect further, given the differences in heritability and prevalence of inguinal hernia between sexes.
As a summary, we have presented further insights into the genetic mechanisms of inguinal hernias and our findings pave a path to future research and a potential to improve treatment for patients with inguinal hernias.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
Contributors
K.H., M.K., and C.T. designed the study. K.H., and K.T. analyzed the data. K.H., and C.T. interpreted the data. K.H. wrote the manuscript. M.K., X.L., Yu.Mo., Ta.Mo., Yo.Mu., B.J.P., Ta.Mu., and C.T. performed critical revision. The underlying data have been verified by K.H., K.T., and C.T. All authors read and approved the final version of the manuscript.
Data Sharing Statement
Full GWAS results will be available via the website of the Japanese ENcyclopedia of GEnetic associations by Riken (JENGER, http://jenger.riken.jp/en/).
Acknowledgements
We appreciate all the patients who participated in this study. The sample and data used for this study were provided by the BBJ supported by the Japan Agency for Medical Research and Development (AMED) (Grant Number: JP19km0605001). We express our gratitude to BBJ staff for their assistance. We would like to thank Dr. Rebecca Carlson in the department of Information Arts and Sciences at Toyo University, Tokyo, Japan, for English language editing.
Footnotes
Grant support: The Japan Agency for Medical Research and Development (AMED) (Grant Number: JP19km0605001).
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103532.
Appendix. Supplementary materials
References
- 1.Fitzgibbons RJ, Jr, Forse RA. Clinical practice. Groin hernias in adults. N Engl J Med. 2015;372(8):756–763. doi: 10.1056/NEJMcp1404068. [DOI] [PubMed] [Google Scholar]
- 2.Beard JH O-YM, devries CR, et al. Hernia and Hydrocele. In: Debas HT, Donkor P, Gawande A, et al., editors. Essential Surgery: Disease Control Priorities [Third Edition (Volume 1):[Available from: https://www.ncbi.nlm.nih.gov/books/NBK333501/table/ch09.sec2.table1/. [PubMed]
- 3.Fitzgibbons RJ, Jr., Ramanan B, Arya S, Turner SA, Li X, Gibbs JO. Long-term results of a randomized controlled trial of a nonoperative strategy (watchful waiting) for men with minimally symptomatic inguinal hernias. Ann Surg. 2013;258(3):508–515. doi: 10.1097/SLA.0b013e3182a19725. [DOI] [PubMed] [Google Scholar]
- 4.Brandt-Kerkhof A, van Mierlo M, Schep N, Renken N, Stassen L. Follow-up period of 13 years after endoscopic total extraperitoneal repair of inguinal hernias: a cohort study. Surg Endosc. 2011;25(5):1624–1629. doi: 10.1007/s00464-010-1462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poobalan AS, Bruce J, Smith WC, King PM, Krukowski ZH, Chambers WA. A review of chronic pain after inguinal herniorrhaphy. Clin J Pain. 2003;19(1):48–54. doi: 10.1097/00002508-200301000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Aasvang E, Kehlet H. Chronic postoperative pain: the case of inguinal herniorrhaphy. Br J Anaesth. 2005;95(1):69–76. doi: 10.1093/bja/aei019. [DOI] [PubMed] [Google Scholar]
- 7.Ruhl CE, Everhart JE. Risk factors for inguinal hernia among adults in the US population. Am J Epidemiol. 2007;165(10):1154–1161. doi: 10.1093/aje/kwm011. [DOI] [PubMed] [Google Scholar]
- 8.Burcharth J, Pedersen M, Bisgaard T, Pedersen C, Rosenberg J. Nationwide prevalence of groin hernia repair. PLoS One. 2013;8(1):e54367. doi: 10.1371/journal.pone.0054367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen LT, Friis E, Jorgensen T, Vennits B, Andersen BR, Rasmussen GI. Smoking is a risk factor for recurrence of groin hernia. World J Surg. 2002;26(4):397–400. doi: 10.1007/s00268-001-0238-6. [DOI] [PubMed] [Google Scholar]
- 10.Jansen PL, Klinge U, Jansen M, Junge K. Risk factors for early recurrence after inguinal hernia repair. BMC Surg. 2009;9:18. doi: 10.1186/1471-2482-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau H, Fang C, Yuen WK, Patil NG. Risk factors for inguinal hernia in adult males: a case-control study. Surgery. 2007;141(2):262–266. doi: 10.1016/j.surg.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Simons MP, Aufenacker T, Bay-Nielsen M, Bouillot JL, Campanelli G, Conze J. European Hernia Society guidelines on the treatment of inguinal hernia in adult patients. Hernia. 2009;13(4):343–403. doi: 10.1007/s10029-009-0529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Judge DP, Dietz HC. Marfan's syndrome. Lancet. 2005;366(9501):1965–1976. doi: 10.1016/S0140-6736(05)67789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liem MS, van der Graaf Y, Beemer FA, van Vroonhoven TJ. Increased risk for inguinal hernia in patients with Ehlers-Danlos syndrome. Surgery. 1997;122(1):114–115. doi: 10.1016/s0039-6060(97)90273-7. [DOI] [PubMed] [Google Scholar]
- 15.Ringpfeil F. Selected disorders of connective tissue: pseudoxanthoma elasticum, cutis laxa, and lipoid proteinosis. Clin Dermatol. 2005;23(1):41–46. doi: 10.1016/j.clindermatol.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Han Q, Li C, Li W, Fan H, Xing Q. Genetic analysis of the TBX1 gene promoter in indirect inguinal hernia. Gene. 2014;535(2):290–293. doi: 10.1016/j.gene.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Han Q, Fan H, Li W, Xing Q, Yan B. Genetic analysis of the TBX2 gene promoter in indirect inguinal hernia. Hernia. 2014;18(4):513–517. doi: 10.1007/s10029-013-1199-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Tian W, Wang L, Wang H, Qin X, Xing Q. Genetic and functional analysis of the TBX3 gene promoter in indirect inguinal hernia. Gene. 2015;554(1):101–104. doi: 10.1016/j.gene.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Qiao Y, Zhang Z, Huang W, Pang S, Xing Q, Yan B. Two functional sequence variants of the GATA6 gene promoter in patients with indirect inguinal hernia. Gene. 2014;547(1):86–90. doi: 10.1016/j.gene.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Han Q, Zhang Y, Li W, Fan H, Xing Q, Pang S. Functional sequence variants within the SIRT1 gene promoter in indirect inguinal hernia. Gene. 2014;546(1):1–5. doi: 10.1016/j.gene.2014.05.058. [DOI] [PubMed] [Google Scholar]
- 21.Sezer S, Simsek N, Celik HT, Erden G, Ozturk G, Duzgun AP. Association of collagen type I alpha 1 gene polymorphism with inguinal hernia. Hernia. 2014;18(4):507–512. doi: 10.1007/s10029-013-1147-y. [DOI] [PubMed] [Google Scholar]
- 22.Antoniou GA, Lazarides MK, Patera S, Antoniou SA, Giannoukas AD, Georgiadis GS. Assessment of insertion/deletion polymorphism of the angiotensin-converting enzyme gene in abdominal aortic aneurysm and inguinal hernia. Vascular. 2013;21(1):1–5. doi: 10.1258/vasc.2011.oa0322. [DOI] [PubMed] [Google Scholar]
- 23.Jorgenson E, Makki N, Shen L, Chen DC, Tian C, Eckalbar WL. A genome-wide association study identifies four novel susceptibility loci underlying inguinal hernia. Nat Commun. 2015;6:10130. doi: 10.1038/ncomms10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol. 2017;27(3S):S2–S8. doi: 10.1016/j.je.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International HapMap C. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack Flanagan MH, Chikashi Terao. Reference panel using whole genome sequencing data in Japanese Population (Manuscript in Preparation). 2020.
- 29.Institute B. 2021 [Available from: https://gatk.broadinstitute.org/hc/en-us.
- 30.Huang J, Howie B, McCarthy S, Memari Y, Walter K, Min JL. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6) doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Zhou L, Li L, Coon VJ, Chatterton RT, Brooks DC. Shift from androgen to estrogen action causes abdominal muscle fibrosis, atrophy, and inguinal hernia in a transgenic male mouse model. Proc Natl Acad Sci U S A. 2018;115(44):E10427–E10E36. doi: 10.1073/pnas.1807765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Ferreira T, Morris AP, Medland SE. Genetic Investigation of ATC, Replication DIG, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–375. doi: 10.1038/ng.2213. S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishigaki K, Kochi Y, Suzuki A, Tsuchida Y, Tsuchiya H, Sumitomo S. Polygenic burdens on cell-specific pathways underlie the risk of rheumatoid arthritis. Nat Genet. 2017;49(7):1120–1125. doi: 10.1038/ng.3885. [DOI] [PubMed] [Google Scholar]
- 39.Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J. Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingsnorth A, LeBlanc K. Hernias: inguinal and incisional. Lancet. 2003;362(9395):1561–1571. doi: 10.1016/S0140-6736(03)14746-0. [DOI] [PubMed] [Google Scholar]
- 42.Burcharth J, Pommergaard HC, Bisgaard T, Rosenberg J. Patient-related risk factors for recurrence after inguinal hernia repair: a systematic review and meta-analysis of observational studies. Surg Innov. 2015;22(3):303–317. doi: 10.1177/1553350614552731. [DOI] [PubMed] [Google Scholar]
- 43.The Burden of Digestive Diseases in the United States 2008 [Available from: https://www.niddk.nih.gov/about-niddk/strategic-plans-reports/burden-of-digestive-diseases-in-united-states.
- 44.Ishigaki K, Akiyama M, Kanai M, Takahashi A, Kawakami E, Sugishita H. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52(7):669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and Rigorous Computation of Gene and Pathway Scores from SNP-Based Summary Statistics. PLoS Comput Biol. 2016;12(1) doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consortium GT The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4) doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Chua HX, Chen P, Ong RT, Sim X, Zhang W. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum Mol Genet. 2013;22(11):2303–2311. doi: 10.1093/hmg/ddt064. [DOI] [PubMed] [Google Scholar]
- 53.Benner C, Spencer CC, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32(10):1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.C-CH Masaru Koido, Koyama Satoshi, Kawaji Hideya, Murakawa Yasuhiro, Ishigaki Kazuyoshi, Ito Kaoru, Sese Jun, Kamatani Yoichiro, Carninci Piero. View ORCID ProfileChikashi Terao. Predicting celltype-specific non-coding RNA transcription from genome sequence: bioRxiv. 2020 [Google Scholar]
- 55.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 61.Brown BC, Asian Genetic Epidemiology Network Type 2 Diabetes C. Ye CJ, Price AL, Zaitlen N. Transethnic Genetic-Correlation Estimates from Summary Statistics. Am J Hum Genet. 2016;99(1):76–88. doi: 10.1016/j.ajhg.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genomes Project C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Library of Medicine (US), National Center for Biotechnology Information; BethesdaMD: 2004. Gene [Internet]https://www.ncbi.nlm.nih.gov/gene/2006 [Available from. [Google Scholar]
- 64.Zhang MC, He L, Giro M, Yong SL, Tiller GE, Davidson JM. Cutis laxa arising from frameshift mutations in exon 30 of the elastin gene (ELN) J Biol Chem. 1999;274(2):981–986. doi: 10.1074/jbc.274.2.981. [DOI] [PubMed] [Google Scholar]
- 65.Mao Y, Chen L, Li J, Shangguan AJ, Kujawa S, Zhao H. A network analysis revealed the essential and common downstream proteins related to inguinal hernia. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0226885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.National Library of Medicine (US), National Center for Biotechnology Information; BethesdaMD: 2004. Gene [Internet]https://www.ncbi.nlm.nih.gov/gene/800 [Available from. [Google Scholar]
- 67.National Library of Medicine (US), National Center for Biotechnology Information; BethesdaMD: 2004. Gene [Internet]https://www.ncbi.nlm.nih.gov/gene/7042 [Available from. [Google Scholar]
- 68.Pascual G, Corrales C, Gomez-Gil V, Bujan J, Bellon JM. TGF-beta1 overexpression in the transversalis fascia of patients with direct inguinal hernia. Eur J Clin Invest. 2007;37(6):516–521. doi: 10.1111/j.1365-2362.2007.01816.x. [DOI] [PubMed] [Google Scholar]
- 69.National Library of Medicine (US), National Center for Biotechnology Information; BethesdaMD: 2004. Gene [Internet]https://www.ncbi.nlm.nih.gov/gene/245806 [Available from. [Google Scholar]
- 70.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 71.Peng X, Guo Z, Zhang Y, Sun B, Zhang Q. EFEMP1 in Direct Inguinal Hernia: correlation with TIMP3 and Regulation Toward Elastin Homoeostasis as Well as Fibroblast Mobility. J Invest Surg. 2020:1–9. doi: 10.1080/08941939.2020.1811812. [DOI] [PubMed] [Google Scholar]
- 72.He W, Zhuang J, Zhao ZG, Luo H, Zhang J. miR-328 prevents renal fibrogenesis by directly targeting TGF-beta2. Bratisl Lek Listy. 2018;119(7):434–440. doi: 10.4149/BLL_2018_079. [DOI] [PubMed] [Google Scholar]
- 73.Raghavan CT, Smuda M, Smith AJ, Howell S, Smith DG, Singh A. AGEs in human lens capsule promote the TGFbeta2-mediated EMT of lens epithelial cells: implications for age-associated fibrosis. Aging Cell. 2016;15(3):465–476. doi: 10.1111/acel.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernasconi P, Carboni N, Ricci G, Siciliano G, Politano L, Maggi L. Elevated TGF beta2 serum levels in Emery-Dreifuss Muscular Dystrophy: Implications for myocyte and tenocyte differentiation and fibrogenic processes. Nucleus. 2018;9(1):292–304. doi: 10.1080/19491034.2018.1467722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parker MM, Hao Y, Guo F, Pham B, Chase R, Platig J. Identification of an emphysema-associated genetic variant near TGFB2 with regulatory effects in lung fibroblasts. Elife. 2019;8 doi: 10.7554/eLife.42720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–DD51. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Somuncu S, Somuncu OS, Ballica B, Tabandeh B. Deficiency of epithelial-mesenchymal transition causes child indirect inguinal hernia. J Pediatr Surg. 2020;55(4):665–671. doi: 10.1016/j.jpedsurg.2019.06.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.