Abstract
The expression of c-fos mRNA is an indirect marker of neuronal activity. RNAscope ACD Bio RNAscope (now Biotechne) is a proprietary in-situ mRNA detection technology using branched DNA amplification and z paired probes to deliver a robust and specific assay designed primarily for use on formalin fixed paraffin sections [1]. In the present study we adapted this technology to be used in frozen sections to allow quantitative analysis of c-fos gene expression in different mouse brain regions during neuropharmacology studies. The method was applied by Cosi et al. 2021 [2] and the image analysis is described here in details.
-
•
The patented RNAscope (ACD Bio) flourescent in-situ hybridisation technology designed primarily for use on formalin fixed paraffin sections was adapted to be used on frozen section from mouse brain.
-
•
We carefully controlled sample preparation and handling to maximise mRNA preservation and used the fluorescent properties of the fast Red substrate combined with fluorescent whole slide scanning and image analysis.
-
•
A customized algorithm was set up for image analysis
-
•
The method developed permitted the quantitative analysis of c-fos expression in specific brain regions from whole sections.
Keywords: FISH, RNAscope, Mouse, Brain
Graphic abstract
Specifications table
| Subject Area: | Neuroscience |
|---|---|
| More specific subject area: | Immediate early genes |
| Method name: | Brain c-fos RNAscope in-situ hybridisation |
| Name and reference of original method: | RNAscope Assay (in-situ hybridization), Advanced Cell Diagnostics, Inc (ACD Bio) Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, Wu X, Vo H-T, Ma X-J and Luo Y. RNAscope®: A Novel In Situ RNA Analysis Platform for Formalin-Fixed Paraffin-Embedded Tissues. J. Mol. Diagnostics, 2012, 14:22–29 |
| Resource availability: | Zeiss Axioscan Z1 https://www.zeiss.com/microscopy/int/products/imaging-systems/axio-scan-z1.html RNAscope (ACD Bio) https://acdbio.com/manual-assays-rnascope?_bk=rnascope&_bt=384508666307&_bm=e&_bn=g&gclid=Cj0KCQjw3ZX4BRDmARIsAFYh7ZIc5OBPjhNxpfm9-RquQRXj61EHMPyU_cRPjF1hyUiOjcvtXH7_shMaAgqPEALw_wcB Leica Biosystems Bond RX staining robot https://www.leicabiosystems.com/ihc-ish-fish/fully-automated-ihc-ish-instruments/bond-rx/ OracleBio Image Analysis https://oraclebio.com/image-analysis-services/ |
Method details
Introduction
The expression of c-fos mRNA is an indirect marker of neuronal activity because c-fos is often expressed when neurons fire action potentials. In the mammalian central nervous system neurons undergo profound changes in excitability and synaptic organization in response to brief periods of excitation thanks to the rapid and transient induction of a number of immediate-early genes like fos and Jun that regulate gene transcription. The study of c-fos mRNA expression in particular has been extensively applied in psychopharmacology over the past 4 decades because it is associated to the main effect of endogenous excitatory aminoacids like glutamate. The effects of glutamate are cell type dependent thanks to a variety of intracellular regulatory elements. The accessibility and the assembly of transcription factor complexes at the c-fos gene DNA regulatory regions is in fact controlled by enhancing or suppressing DNA elements modulating a variety of stimuli like cAMP or BDNF [3]. The changes in c-fos mRNA are typically followed over the period of few hours by RT-PCR or autoradiography techniques with clear limitations to the degree of definition of the neuronal effects in different neurocircuits. Here we present a fluorescent method of quantitative analysis of single c-fos mRNA molecules in situ in rodent brain frozen tissue that should allow a more precise analysis of the mechanisms controlling immediate early genes in different neurocircuitries. Following previous observations [4,5] the method has been applied to the study of the neuronal effects of the NMDA channel blocker MK-801 and the modulation by a preferential D3 receptor antagonist in mice in Cosi et al. 2021 [2].
Materials and methods
Animals
Forty male OF1 mice (Charles River Laboratories France), 20-22g at arrival were used, eight per dose group. Animals were housed and all experiments were conducted, in an AAALAC accredited facility. They were handled and cared for in accordance with the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources on Life Sciences, National Research Council, 2011) and the European Directive 2010/63/EU, and the protocol was carried out in compliance with French regulations and the local ethical committee guidelines for animal research. Animals were group housed 10 per cage (Polycarbonate Type III cages) with sawdust bedding in air-conditioned rooms (T = 21 ± 1°C, Humidity 55 ± 5%) with light on from 07:00 a.m. to 07:00 p.m. They had free access to food (A04; SAFE; Augy, France) and filtered water.
Mice were individually housed on the day of the experiment. A total five groups of mice were tested, with a 8 animals per group:
-
•
Group 1: Vehicle + vehicle
-
•
Group 2: Vehicle + MK-801 0.14 mg/kg ip
-
•
Group 3: F17464 0.04 mg/kg ip + MK-801 0.14 mg/kg ip
-
•
Group 4: F17464 0.32 mg/kg ip + MK-801 0.14 mg/kg ip
-
•
Group 5: F17464 2.5 mg/kg ip + MK-801 0.14 mg/kg ip p
Group 4, animal 3 was accidentally defrosted in testing laboratory and was therefore excluded from the study.
Brain extraction
The injection and brain extraction was spread over two days, 4 mice from each drug group being used on the first day, the other 4 on the second day. On day 1, mice were weighed by trained internal technicians, injected with F17464 or saline, and isolated in individual cages. 30 min later, mice were injected with MK-801 or saline, and move back in the same cage for another 60 min. Then, every 4 min a mouse was taken from the cage and sacrificed by dislocation of the cervical vertebrae followed by decapitation. In RNAse free conditions, the brain was extracted from skulls, quickly cleaned with a NaCl solution the excess of liquid removed by absorption on sterile gauze and frozen according to the procedure below.
Brain freezing
8 Pyrex beakers containing 250 ml isopentane each were arranged in a raw on dry ice pellets in stereo foam containers under a chemical wood. The temperature of the isopentane was monitored with an ultra low temperature thermometer. When -20°C was reached, the isopentane was quickly stirred with a spatula creating a vortex. The mouse brain was deposited on the surface of the vortex and touched the bottom of the beaker already frozen. The beaker containing the isopentane with the sample was left on the dry ice for 30 min to allow stabilization at the dry ice temperature.
This procedure allows an even and relatively quick (within 4 min after devascularization) freezing of relatively large amounts of mouse brain samples and preserve morphology and 100% of the targeted mRNA, that is important if your target mRNA is from low to moderately expressed. In fact in paraffin-embedded tissues about 25% of the mRNA signal is lost [7]. In this procedure, the mouse brains, whose weights were around 400 mg, were snap frozen by immersion into -20°C dry ice chilled isopentane [8]. They were then left to equilibrate to the dry ice temperature for 30 min. Snap freezing at lower temperatures, for example by immersion into liquid nitrogen, depending on the tissue, can produce cracks in the tissue making the sample impossible to cut at the cryostat. The procedure was performed under a chemical wood and using RNAse free materials. Brains were than stored at -80°C in 15 ml RNAse free Eppendorf tubes. The same procedures was performed on day 2 with the other 20 mice. Frozen brains were shipped to testing laboratory on dry ice, for measuring c-fos expression by RNAScope in situ hybridization staining technology.
Sectioning and optimization for RNAscope assay
The brains were stored at -80°C, then brought up to temperature of -20°C overnight for cutting.
The cryostat temperature was -20°C.
Cryostat cut coronal sections of 10 µm were collected at two levels per brain with three sections at each level. Level 1 –Thalamus and Retrosplenial Cortex [6] (Franklin and Paxinos. 2001. Mouse Brain Atlas. Academic Press. Interaural 1.86–1.98 mm) and level 2 – Nucleus Accumbens, Piriform Cortex and Cingulate Cortex (Franklin and Paxinos. 2001. Mouse Brain Atlas. Academic Press. Interaural 5.5–5.58 mm). Sections were air dried, then stored in sealed boxes at ‐80°C with desiccant. Prior to staining, slides were thawed to ‐20°C for 24 h before opening sealed boxes to avoid condensation. Prior to commencing the RNAscope protocol, slides were fixed under the following conditions:
-
•
4% PFA at 4°C
-
•
50% EtOH at room temperature (x2)
-
•
100% EtOH at room temperature (x2)
RNAscope ISH was performed on a Leica Bond RX automated platform for greater consistency and higher throughput. Optimization of hybridization conditions were performed on one brain (group 1 – animal 1) using sections out with the test region. This step was required for assessment of sample quality and mRNA integrity preservation, as well as optimal pre-hybridization conditions. This phase consisted of 9 slides (Fig. 1), which were ran with 3 different ACD Bio RNAscope probes positive housekeeping mouse (mus musculus) genes with different levels of expression as follows
-
•
UBC (ubiquitin C) – high expression level. UBC has an important role in diverse biological processes, such as innate immunity, DNA repair and kinase activity (Cat. N 310778).
-
•
PPIB (peptidylprolyl isomerase B [cyclophilin B]) mRNA – medium expression level (Cat. N 313918).
-
•
Polr2A (DNA-directed RNA polymerase II subunit RPB1) – low expression level (Cat. N 312478).
Fig. 1.
RNAscope optimisation results. Mouse brain sections were stained with different control probes to assess the best suitable conditions for consequent staining. The red staining represents the control probe mRNA, the blue represents DAPI nuclear counterstain. Conditions chosen were based on images A, B and C at mild pretreatment (5 min) conditions.
A range of 3 different ‘pre-treatment conditions’ were tested to identify the best pre-treatment condition based on the most signal and the least amount of unspecific binding, as well as preservation of tissue integrity.
-
•
Mild Pre-treatment (5 min ACD proteinase K digestion)
-
•
Standard Pre-treatment (15 min ACD proteinase K digestion)
-
•
Extended Pre-treatment (25 min ACD proteinase K digestion)
Gene expression was visualized using a Fast Red substrate. Stained slides were imaged and reviewed with the client to identify suitable pre-treatment conditions and sample suitability for RNAscope assay. All stained slides were rinsed in dH2O, dried at 60°C, cleared in xylene and mounted with Ecomount. A Zeiss LSM 7 series confocal microscope was used for image acquisition. The “mild” pre-treatment (5 min ACD proteinase K digestion) was selected on the basis of signal to noise ratio and preservation of tissue section morphology integrity.
c-fos RNAscope staining
PFA % 4 post-fixed mouse brain sections were stained using an RNAscope assay protocol with the target probe (Mm-fos), as well as a positive (Mm-PPIB) and a negative (DapB) in-house control per run (Fig. 2). A total of 4 runs were done, with brain slides from 2 animals from each group stained with the Mm-fos target probe, along with 1 animal from each group stained with positive control probe Mm-PPIB and negative control probe DapB. The runs were split using the following run layout: group 1: yellow; group 2: blue: group 3: green; group 4: purple; group 5: red. L 1 = level 1; +ve control = positive control; - ve control = negative control (Table 1).
Fig. 2.
Example images of Group 1(vehicle treated) . Images in pannels A,B,C are level 1 sections from the same animal, animal 1; image in pannel D is a level 2 section from animal 3. The red staining represents the probe mRNA, the blue represents DAPI nuclear counterstain. (A) section stained with mouse positive control probe (Mm-PPIB) showing a medium level of expression. (B) section stained with negative control probe (DapB) showing no staining as predicted. (C) (thalamus) section stained with mouse c-fos target probe (Mm-fos) showing expression. (D) (accumbens) section stained with mouse c-fos target probe (Mm-fos) showing expression.
Table 1.
c-fos RNAscope staining run layout. A total of 4 runs were done. The runs were split using the following run layout: group 1: yellow; group 2: blue: group 3: green; group 4: purple; group 5: red. L 1 = level 1; +ve control = positive control; - ve control = negative control. Animal treatment “groups” are described in the “Animals” section and brain slides levels (L 1 and L 2) in the “Sectioning and optimization for RNAscope assay” section.
 |
The following ACD Bio RNAscope probes were used for each run:
-
•
Mm-PPIB (mouse peptidylprolyl isomerase B [cyclophilin B]) mRNA – medium expression level positive control probe (Cat. N 313918).
-
•
DapB – a gene from soil bacterium Bacillus subtilis strain SMY – negative control probe (Cat N 312038)
-
•
Mm-fos (mouse c-fos) – a proto-oncogene involved in important cellular events, including cell proliferation, differentiation and survival (Cat N 316928).
The following sequential staining protocol was carried out on Leica Bond RX staining robot:
-
•
ACD Bio Proteinase K (5 min)
-
•
ACD Bio Probe (120 min at 42°C)
-
•
ACD Amp 1 Red (30 min at 42°C)
-
•
LS Rinse (2 × 5 min at room temperature)
-
•
ACD Amp 2 Red (15 min at 42°C)
-
•
ACD Amp 3 Red (30 min at 42°C)
-
•
LS Rinse (2 × 5 min at room temperature)
-
•
ACD Amp 4 Red (15 min at 42°C)
-
•
ACD Amp 5 Red (15 min at room temperature)
-
•
ACD Amp 6 Red (15 min at room temperature)
-
•
LS Rinse
-
•
Mixed Red Refine
-
•
DAPI counterstain
All stained slides were rinsed in dH2O, dried at 60°C, cleared in xylene and mounted with Ecomount. Whole slide scans, a Zeiss Axioscan Z1 was used. Images were acquired as .czi files and consequently converted to TIFF format with 40% compression. These were then provided to Oracle Bio for image analysis/quantification.
Quantitative image analysis
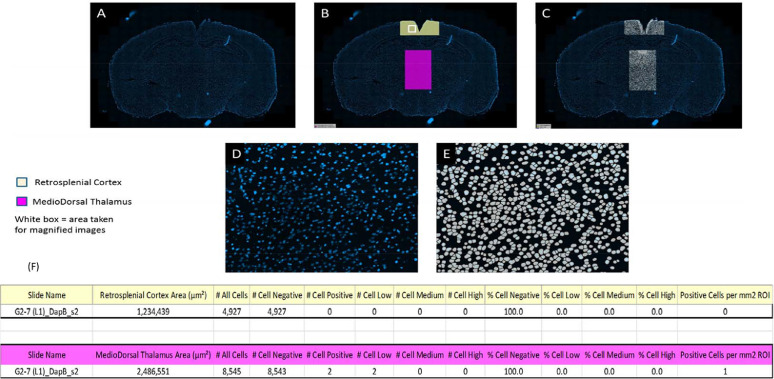
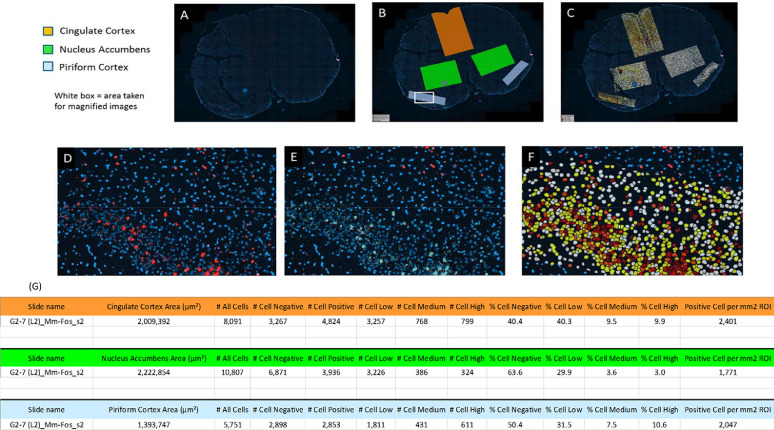
All quantitative image analysis was performed by OracleBio using Definiens Tissue Studio® software Image analysis was performed to quantify the amount and locations of mRNA Fluorescent In-Situ Hybridisation (FISH) staining within specific regions of mouse brain samples. Digital whole slide images (n = 274, x20 magnification, .tiff format) were uploaded to the OracleBio cloud server, whereupon quantitative image analysis was performed on the brain tissue sectioned at x2 different levels (L1 and L2). Within these levels, defined regions of interest (ROI) were identified by the Client and were transcribed on to each section by OracleBio (Fig. 3). At this stage, any prominent artefacts present were manually excluded per image. Two of the images provided were excluded from any further analysis as ROI could not be identified due to tissue damage. Initially, a customised algorithm was configured within Tissue Studio using a subset of the study images to detect each cell by the presence of a DAPI stained nucleus. A pseudo cytoplasm area was grown out and around the detected nucleus to form a cell object. Cellular Analysis settings within the algorithm were then established for the detection of positive mRNA signal in each cell object within the defined ROI.
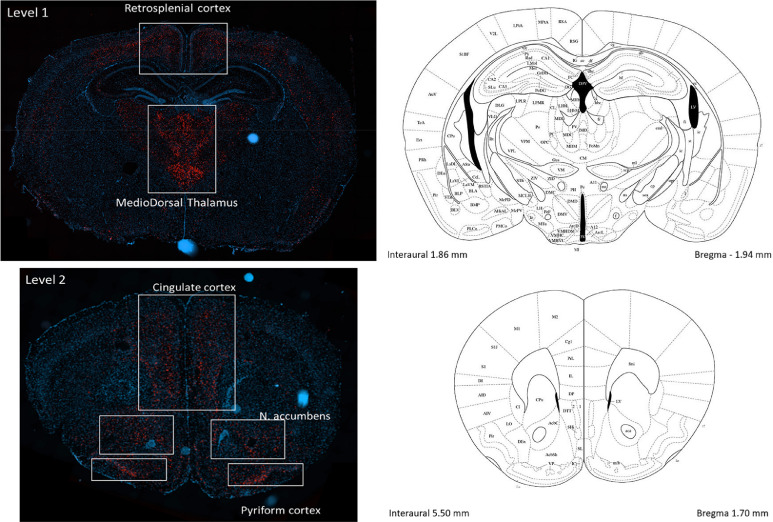
Fig. 3.
Regions of interest (ROI) per Level. Coronal section diagrams are from Franklin and Paxinos.2001. Mouse Brain Atlas in stereotaxic coordinates, with minor modifications, they are shown to indicate the approximative levels of the tissue sections.
Once developed, the algorithm was applied objectively to each image to quantify the area of detected ROI (Retrosplenial Cortex, MedioDorsal Thalamus, Cingulate Cortex, Nucleus Accumbens and Pyriform Cortex) per whole section in mm2. Total number of cells, based on DAPI stain, and total mRNA positive cells (FISH signal) were calculated within each ROI per section. Finally, of FISH positive cells were further classified based on the estimated number of spots/mRNA signal per cell, per defined ROI. Within the analysis algorithm, the area of 1 mRNA spot signal was set at 1 µm2 and each cell was then classified based on the amount of mRNA signal present within the cell using the following in-house criteria: <1 µm2 area = negative (white analysis mask overlay); ≥1 µm2 <6 µm2 area = positive low (yellow analysis mask overlay); ≥6 µm2 <11 µm2 area = positive medium (orange analysis mask overlay); ≥11 µm2 area = positive high (red analysis mask overlay).
Quantitative Mm-fos expression data per cell, per ROI, was generated for each of the sections per level (see example analysis overlays in panels G-F in Fig. 4, Fig. 5, Fig. 6, Fig. 7). Data for the positive control probe MM-PPIB and negative control probe DapB, generated from Level 1 sections slides, one section per mouse, 4 mice per treatment group were also included.
Fig. 4.
Example Analysis Images for Positive Control: G1-5 (L1)_PPIB_Run C_s2. A-C show example of whole section analysis; D-F show higher magnification of analysis performed. F shows cell detection as negative (white), low (yellow), medium (orange) and high (red). G shows analysis data generated from both ROI analysed.
Fig. 5.
Example Analysis Images for Negative Control: G2-7 (L1)_DapB_s2. A-C show example of whole section analysis; D shows a higher magnification of analysis performed. E shows cell detection as negative (white), low (yellow), medium (orange) and high (red). F shows analysis data generated from both ROI analysed.
Fig. 6.
Example Analysis Images for Level 1: G3-2 (L1)_Mm-fos _s1. A-C show example of whole section analysis; D-F show higher magnification of analysis performed. F shows cell detection as negative (white), low (yellow), medium (orange) and high (red). G shows analysis data generated from both ROI analysed.
Fig. 7.
Example Analysis Images for Level 2: G2-7 (L2)_Mm-_s3. A-C show example of whole section analysis; D-F show higher magnification of analysis performed. F shows cell detection as negative (white), low (yellow), medium (orange) and high (red). G shows analysis data generated from both ROI analysed.
Statistical analysis
Data were analyzed as “number of c-fos positive cells/mm2 ROI”, “% c-fos mRNA low, medium and high expressing cells” compared to total number of cells (c-fos mRNA positive and negative) within the ROI.
The treatment effects were analysed by the mean of a one-way ANOVA to compare the treatment effect with control within each brain structure analysed. When the effect of group was reached, the post-hoc Dunnett‘s test was performed and presented in this report. For the retro splenial cortex significant differences were detected with the t-test of student. A complementary 2 ways ANOVA Treatment x Brain structure was performed on the “Number of c-fos mRNA positive cells/mm2” and on “% c-fos mRNA medium expressing cells” to highlight a possible different response of brain structure at the different doses tested.
Results
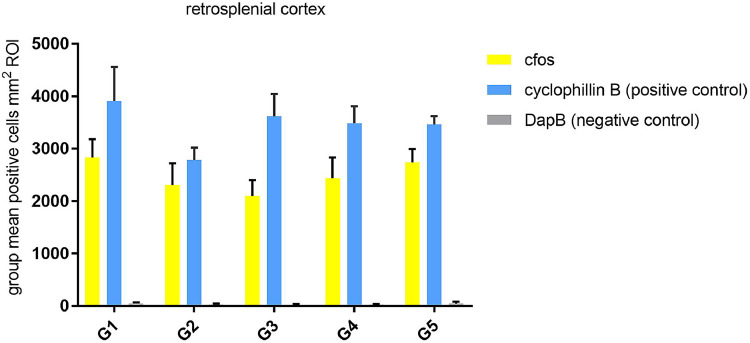
C-fos mRNA results are presented and discussed in the correlated pharmacology article [2]. Part of the data analysis is shown here in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7 to describe the validation of the method. Figs. 8 and 9 highlight a graph of the group mean Mm-fos positive cells per mm2 in the Level 1 Retrosplenial Cortex (Fig. 8) MedioDorsal Thalamus (Fig. 9) ROI, respectively. On both Level 1 graphs is the associated positive control cyclophilin B and negative control DapB data for the same ROI. The number of slides for the cyclophyllin B was n = 4 per group of treatment. The density of MmPPIB expressing cells did not change except for group G2: the density of MmPPIB expressing cells had the tendency to be lower than control (G1).
Fig. 8.
shows a graph of data of the Group (mean ± SEM) Mm-fos positive cells per mm2 in the Level 1 Retrosplenial Cortex ROI, yellow bars, n = 7–8, of Mm-PPIB (cyclophilin B), blue bars, n = 4 and DapB (gene from Bacillus subtilis) gray bars n = 4.
Fig. 9.
shows a graph of data of the Group (mean ± SEM) Mm-fos positive cells per mm2 in the Level 1 Mediodorsal Thalamus ROI, purple bars, n = 7–8, of Mm-PPIB (cyclophilin B), blue bars, n = 4 and DapB (gene from Bacillus subtilis), grey bars n = 4.
The group treated with MK-801 + vehicle (Group 2) showed a significant lower c-fos density than the veh. + veh. group (Group 1) when combining results for the cortical and sub-cortical areas. This effect was antagonized by the D3 antagonist (Fig. 10). The presence of a reduction of c-fos mRNA expression in some neurons has been previously reported in the medial prefrontal cortex after chronic MK-801 treatment, in that case the effect was also blocked by a D3 antagonist [4].
Fig. 10.
Mean number of c-fos positive cells/mm2 in cortical and subcortical regions. * p < 0.05 vs veh.+ veh., one way ANOVA (p= 0.031), Dunnet"s post test.
The two way ANOVA performed on both ”number of c-fos mRNA positive cells/mm2” and “% c-fos mRNA medium expressing cells” showed a significant main effect of Brain structures and a significant main effect of Treatment (see Fig. 11 as example). No effects on MK-801 were detected in % c-fos mRNA low and high expressing cells.
Fig. 11.
% of c-fos medium expressing cells (mean ± SEM) comparing brain structures vs treatment groups. The two way ANOVA showed very significant main effect of Brain structures (p < 0.0001), as well as a very significant main effect of Treatment (p < 0.0001).
In conclusion, the provided method allows the relative quantitative analysis of c-fos mRNA expression in specific mouse brain regions using frozen brain tissue and it was used to study dose dependent effects and brain region specific responses.
Declaration of Competing Interests
All the authors were employees of Pierre Fabre Labcooratories or Aquila Biomedical or OracleBio at the time the study was carried out.
Acknowledgements
The authors thanks Dr Frederic Longo for the two-way statistical analysis of the data. The authors also thank the technicians involved in the executions of the animal treatments and brain preparation: Veronique N’Guyen, Jerome Floutard, Pascal Petiot and Anne Marie Ormiere.
References
- 1.Wang F., Flanagan J., Su N., Wang L.-C., Bui S., Nielson A., Wu X., Vo H.-T., Ma X.-J., Luo Y. RNAscope®: A novel in situ RNA analysis platform for formalin-fixed paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosi C., Martel J.C., Auclair A.L., Collo G., Cavalleri L., Heusler P., Leriche L., Gaudoux F., Sokoloff P., Moser P.C., Gatti-McArthur S. Pharmacology Profile of F17464, a Dopamine D3 Receptor preferential Antagonist. Eur. J. Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173635. [DOI] [PubMed] [Google Scholar]
- 3.Joo J.Y., Schaukowitch K., Farbiak L., Kilaru G., Kim T.K. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat. Neurosci. 2016;19:75–83. doi: 10.1038/nn.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokoloff P., Leriche L., Diaz J., Louvel J., Pumain R. Direct and indirect interactions of the dopamine D3 receptor with glutamate pathways: implications for the treatment of schizophrenia. Naunyn Schmiedebergs Arch Pharmacol. 2013;386:107–124. doi: 10.1007/s00210-012-0797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokoloff P., Foll B.Le. The dopamine D3 receptor, a quarter century later. Eur. J.Neurosci. 2017;45:2–19. doi: 10.1111/ejn.13390. [DOI] [PubMed] [Google Scholar]
- 6.Franklin K.B.J., Paxinos G. Access Online via Elsevier; 2001. Mouse Brain Atlas in stereotaxic coordinates: hard cover edition. [Google Scholar]
- 7.Wilcox N.J. Fundamental principals of in situ hybridization. J Histochem. Cytochem. 1993;41:1725–1733. doi: 10.1177/41.12.8245419. [DOI] [PubMed] [Google Scholar]
- 8.Altar A.C., Ryan S., Abood M., Eberwine J.H. Situ mRNA hybridization: standard procedures and novel approaches. methods in neurosciences, Editor(s): P. Michael Conn. Academic Press. 1989;1:238–281. doi: 10.1016/B978-0-12-185251-1.50019-2. [DOI] [Google Scholar]