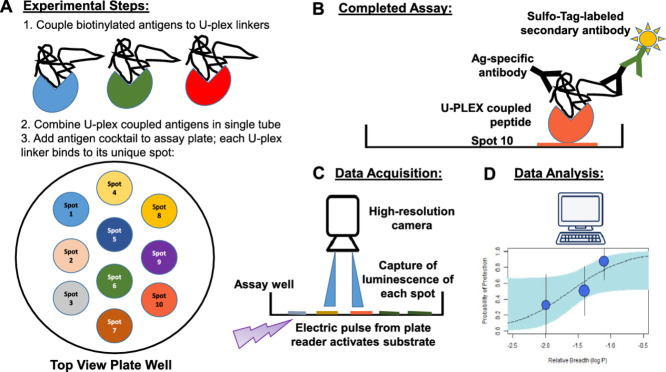
Abstract
Profiling of serological responses to establish the landscape of antibody specificities in individuals exposed to pathogens or vaccines is crucial for (a) revealing humoral immune correlates of protection; (b) uncovering markers of pathogen exposure; and (c) identifying antigens and epitopes associated with disease vs. protection. Establishing the antigenic profile of serological responses requires either expensive microarrays or labor- and time-intensive ELISA assays. Multiplex assay platforms are increasingly being evaluated for their usefulness for high-throughput testing of sera or plasma. The methodology described here utilizes a plate-based assay that allows the simultaneous detection of up to ten antigens per well in a 96 well format using an electrochemiluminescence immunoassay (ECLIA).
-
•
The newly developed protocol outlines high-throughput profiling of serological responses using a multiplex testing platform with subsequent computational analysis.
-
•
The protocol is a modification of the basic assay development manual from the manufacturer of the MESO QuickPlex SQ 120 instrument (MSD, Gaithersburg, MD) and can be used for synthetic peptides as well as full length proteins.
-
•
The protocol can be applied to map serological responses to pathogens or pathogen-derived antigens to establish serological profiles in search for biomarkers or immune correlates.
Keywords: Electro-chemiluminescence, Multiplex immune assay, Immunoprofiling, Antibody, Computational analysis, Immune correlates, Biomarker
Graphical abstract
Specifications table
| Subject area: | Immunology and Immunoassays |
| More specific subject area: | Profiling serological responses to closely related, polymorphic antigens |
| Method name: | Sero-surveillance multiplex assay |
| Name and reference of original method: | MesoScale Discovery, basic manual |
| Resource availability: | All reagents and equipment are listed with the name of the suppliers |
Method details
Reagents
-
1.
Serum, plasma or culture supernatant of stimulated B cells/hybridoma
-
2.
Diluent for antigens 500 mL 1x DPBS (Life Technologies, Waltham, MA) 0.5% Bovine Serum Albumin (BSA) (Thermo Fisher Scientific, Waltham, MA)
-
3.
Peptides (CS Bio Co, Menlo Park, CA)
-
4.
Water, Cell Culture (Quality Biological, Gaithersburg, MD)
-
5.U-PLEX assay platform, 1/5/25 plate packs, (MesoScale Discovery (MSD) Inc, Gaithersburg, MD)
-
-U-PLEX Plates-10 spot, 96 wells (MSD)
-
-Linker Set (1-10) (MSD)
-
-Stop Solution (MSD)
-
-Read Buffer T (4x) (MSD)
-
-
-
6.
MSD Wash Buffer (20x) (MSD)
-
7.MSD Diluents (MSD)
-
-Diluent 2
-
-Diluent 3
-
-
-
8.
MSD SULFO-TAG goat-anti-human IgG (H+L) (MSD)
Equipment
-
1.
NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) to validate peptide concentrations
-
2.
Titramax plate shaker (Heidolph, Schwabach, Germany) for vigorous plate shaking, per manufacturer's instructions
-
3.
MESO QuickPlex SQ 120 (MSD) for data acquisition
-
4.
Fisherbrand™ Analog Vortex Mixer (Thermo Fisher Scientific)
Procedure
Peptide dilution
-
1.
Reconstitute lyophilized peptides (3D7, H12, H13, H18, H1, H234, H3, H50 – see Table 1 for sequences) based on solubility notes provided by manufacturer . The tested peptides (93AA in length, >95% purity) were synthesized with an N-terminal biotin-tag to facilitate binding to Streptavidin-coated plates or the MSD linkers.
-
2.
Monitor concentrations of reconstituted peptides using the NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific).
-
3.Calculate the required concentrations using molarity; concentrations can be converted from µg/ml to nM by using this formula:
-
4.
Dilute peptides using the antigen diluent (1x DPBS, 0.5% BSA).
Table 1.
Sequences of CSP C-terminal peptides (biotinylated 93-mer peptides, CSP residues 283 – 375).
| 3D7 | HNMPNDPNRNVDENANANSAVKNNNNEEPSDKHIKEYLNKIQNSLSTEWSPCSVTCGNGIQVRIKPGSANKPKDELDYANDIEKKICKMEKCS |
| H3 | HNMPNDPNRNVDENANANNAVKNNNNEEPSDQHIEKYLKTIKNSLSTEWSPCSVTCGNGIQVRIKPGSANKPKDQLDYANDIEKKICKMEKCS |
| H12 | HNMPNDPNRNVDENANANNAVKNNNNEEPSDQHIEKYLQKIQNSLSTEWSPCSVTCGNGIQVRIKPGSANKPKDQLNYENDIEKKICKMEKCS |
| H13 | HNMPNDPNRNVDENANANNAVKNNNNEEPSDKHITEYLKRIQNSLSTEWSPCSVTCGNGIQVRIKPGSAGKSKNELDYENDIEKKICKMEKCS |
| H18 | HNMPNDPNRNVDENANANNAVKNNNNEEPSDKHIKEYLNKIQNSISTEWSPCSVTCGNGIQVRIKPGSADKPKDQLDYINDIEKKICKMEKCS |
| H1 | HNMPNDPNRNVDENAKANNAVKNNNNEEPSDKHIEQYLKTIQNSLSTEWSPCSVTCGNGIQVRIKPGSANKPKDQLDYENDIEKKICKMEKCS |
| H50 | HNMPNDPNRNVDENANANNAVKNNNNEEPSDKHIEQYLKTIKNSLSTEWSPCSVTCGNGIQVRIKPGSAGKSKNELDYENDIEKKICKMEKCS |
| H234 | HNMPNDPNRNVDENANANSAVKNNNNEEPSDQHIEKYLKTIKNSLSTEWSPCSVTCGNGIQVRIKPGSANKPKDELDYANDIEKKICKMEKCS |
Titrations
-
1.
The linear range of the MSD platform in our work spans at least four logs (Fig. 1).Therefore, preparation of multiple dilutions for each sample is not necessary [1].
-
2.
Generate a pool of all samples to be tested and perform a titration to determine the optimal range (e.g., 1:100 vs 1:100,000).
-
3.
Subsequent testing of the individual samples can be done at a single dilution [1].
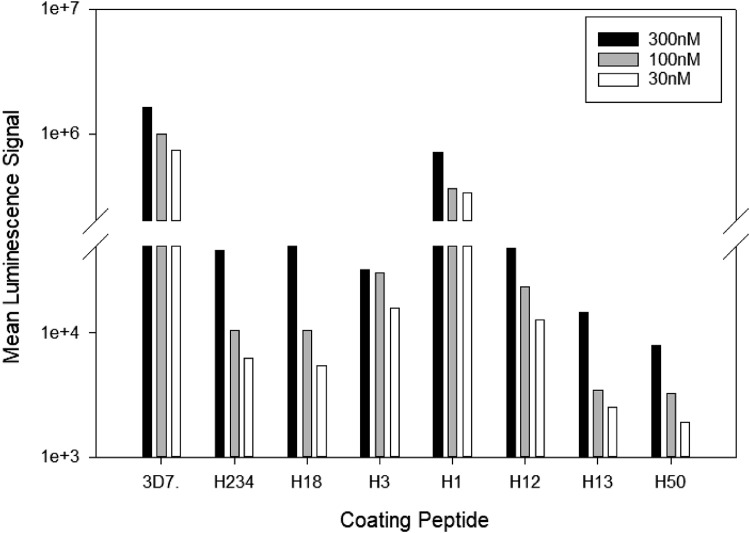
Fig. 1.
Establishing optimal coating concentration for the various peptides. Different coating concentrations (300 nM, 100 nM, 30 nM) of the various CSP C-terminal peptides were tested for reactivity with a human CSP-immune serum pool (1:3,000 dilution). The mean luminescence signal of the malaria-naïve serum pool (negative control) did not exceed the background (i.e., wells incubated with secondary antibody only (MLS < 1000 for all conditions)).
U-PLEX platform assay
-
1.
Designate a MSD Linker (1–10) to represent each biotinylated peptide. Our previous work has shown there is no significant difference in the efficiency of the linkers binding to biotinylated targets [1]. Each of the linkers can only bind to its unique spot in the assay well.
-
2.
Prepare tubes for each biotinylated peptide/linker mix. Combine 200 µl of biotinylated peptide and 300 µl of the assigned Linker in one tube. Vortex and incubate at room temperature (22-25°C) for 30 min.
-
3.
After 30 min, add 200 µl of Stop Solution to the biotinylated peptide/Linker mix. Vortex once combined. Incubate at room temperature (22-25°C) for 30 mins.
-
4.
Combine each of the ten Linker-coupled peptide solution at 600 µl each into one tube, resulting in a 6 ml final volume, and vortex. This will create the coating solution.
-
5.
Coat the U-PLEX plate: Add 50 µl of the coating solution to each well. Seal with an adhesive plate seal and incubate at room temperature (22-25°C) for 1 hour. Shake the plate on the Titramax plate shaker at 800 rpm during incubation.
-
6.
Wash plates three times with 150 µl /well of 1x MSD Wash Buffer. Pat gently on paper towel.
-
7.
Prepare sample dilutions: Dilute samples to desired concentration using Diluent 2.
-
8.
Add 50 µl of each sample in Diluent 2 mix to each well. Seal with an adhesive plate seal and incubate at room temperature (22-25°C) for 1 hour. Shake the plate on the Titramax plate shaker at 800 rpm during incubation.
-
9.
After incubating the samples, wash plates three times with 150 µl /well of 1x MSD Wash Buffer. Pat gently on paper towel.
-
10.
Prepare detection antibody solution: Dilute MSD SULFO-TAG goat-anti-human IgG (H+L) to 1 µg/ml using Diluent 3.
-
11.
Add 50 µl of detection antibody/ Diluent 3 mix to each well. Seal with an adhesive plate cover and incubate at room temperature (22-25°C) for 1 hour. Shake the plate on the Titramax plate shaker at 800 rpm during incubation.
-
12.
After incubating the plates with the detecting (secondary) antibody solution, wash plates three times with 150 µl /well of 1x MSD Wash Buffer. Pat gently on paper towel.
-
13.
Add 150 µl /well of 2x MSD Read Buffer to the plate.
-
14.
Immediately read plate(s) on MESO QuickPlex SQ 120 (MSD).
Data analysis
The quantitative analysis is performed using the DISCOVERY WORKBENCH software provided by MesoScale Discovery. The data are reported as mean luminescence signal (MLS).
Computational analysis
The MSD readout is typically log-transformed prior to statistical analysis, such as a univariate analysis. Although the MSD instrument appears to have a wide linear range, we have found that distributions of antibody responses in a given study typically appear normally distributed only when log-transformed, making the log-transformed data better suited for many types of analyses. If background (media only) is being subtracted, it should be done prior to log transformation.
Method application
Establishing the landscape of disease- or vaccine-specific serological responses is crucial for the identification of antigens and epitopes that can either serve as biomarkers or evaluation of vaccine antigens. This effort requires the application of a high-throughput and multiplex testing platform in order to efficiently process the workload, as well as maximize the information that can be gained from limited sample volumes. We previously compared the traditional ELISA with multiplex platforms [1,2] and determined that the MSD platform was superior for several reasons: ease of assay setup and performance, reproducibility, throughput, and the ability to simultaneously test closely related antigens without reduction of signal due to antigenic competition [1].
Multiplex serological platforms have a wide range of applications:
Vaccine development
Establishing serological profiles associated with protected vs. non-protected individuals and applying bioinformatics tools such as machine learning can identify antigens or epitopes associated with protection, or lack thereof. This, in turn, informs vaccine design to either focus the humoral immune response to these antigens/epitopes or to exclude them from a vaccine. Identifying a specific antigen or epitope(s) within antigens correlating with protective status constitutes an immune correlate or surrogate marker of protection and can advance understanding how vaccines or naturally acquired immunity works.
Sero-surveillance
Determination of prevalence of specific pathogens in a geographic region or resident population, and establishing the serological landscape associated with pathogen exposure identifies antigens that can serve as biomarkers of exposure. Monitoring the induction of antibodies to these biomarkers may even replace some more traditional surveillance tools [3], [4], [5]. Potential applications of sero-surveillance include for example: a) estimating the level of malaria transmission in populations, b) monitoring trends in transmission over extended periods of time, c) identifying individuals and populations with recent exposures (within several months), d) identifying focal areas or populations with ongoing transmission, e) screening for asymptomatic carriers, and f) identifying populations at high risk [3], [4], [5].
Identification of exposure biomarkers
Establishing pathogen-specific serological profiles is the basis for the identification of antibody specificities that can serve as diagnostic or prognostic tools for disease outcome [6].
Pathogen Cross-reactivity: Assessing the quality/quantity of pathogen-specific serological responses for breadth, i.e., ability of vaccine-induced antibodies to recognize polymorphic variants and different strains of the pathogen as a measure of vaccine coverage.
We developed the current protocol specifically for closely related antigens found in circulating field isolates and highlighted crucial steps for users to consider when adapting this protocol to other peptides or proteins. We also describe the computational approach for analyzing the dataset to enable users to not only integrate large datasets, but also as an approach to perform the data analysis that can assist in assessing the breadth of an immune response and potential correlation with protective status.
Selection of plate antigens
The current protocol can be adapted to any peptides/proteins. We have successfully employed this methodology for other applications, such as SARS-CoV-2, and mosquito saliva proteins for entomological surveillance (Bolton et al, manuscript in preparation). Vaccines targeting the main surface antigen CSP on the infectious malaria sporozoites can mediate sterile protection in malaria-naïve vaccinees [7] as well as in the field [8,9]. The leading malaria vaccine RTS,S/AS01 induces several immune parameters that are associated with protection [10]. While the role of CSP-specific antibodies, and, in particular, the role of antibodies binding to the central repeat region, in mediating protection is well established [10], [11], [12], the role of CSP C-terminal antibodies is less clear [13], [14], [15]. The current hypothesis is that the fine specificity of these C-terminus-specific antibodies is crucial for protective efficacy since this region of the CSP is essential for invading hepatocytes and, therefore, for successful infection of the liver and establishment of the disease [16]. Field data from a Phase III trial in African children and infants has demonstrated the importance of C-terminal responses in RTS,S/AS01E-mediated protection [13]. The variant sequences associated with protection identified in clinical field samples [14] represent the foundation for assay development outlined in this report. The genetic diversity of parasites in individuals immunized with the RTS,S/AS01 vaccine demonstrated that vaccination leads to improved protection against 3D7-matched parasites [14]. This implies that vaccine efficacy could be partially dependent on matching the C-term allele(s) to those prevalent among parasites in the geographic region of vaccine deployment. Recent work that employed the experimental protocol described here demonstrated that sera from RTS,S/AS01 vaccinees who were protected after a controlled human malaria infection (CHMI) had a significantly greater breadth of reactivity against these variant C-terminal peptides [2]. Furthermore, these data also supported the observations from Early et al [14], of the critical residues required for RTS,S/AS01 vaccine efficacy, and also suggested epitopes associated with greater breadth of C-terminal reactivity.
Optimizing coating concentration of peptides
Previous work establishing various panels with proteins ranging from 20-120 kD has demonstrated that the first step in assay adaption should be to determine the optimal coating concentration per well. The titration was done in half-log steps (300 nM, 100 nM, 30 nM) and demonstrated that 300 nM was the optimal coating concentration for these 93-mer peptides (Fig. 1). Each of the peptides is assigned to a specific MSD U-plex linker that can only bind to its unique spot in the assay well (see graphical abstract). After coupling the respective antigen/peptide with a linker, all peptides can be combined into a cocktail and the plates coated. Previously, we reported the equivalency of binding efficiency for the various MSD U-plex linkers, which is an important aspect of assay development. Therefore, differences in the signal strength solely reflect distinct reaction patterns of the antibodies [1,2] and not binding artifacts. For subsequent experiments we chose a coating concentration of 300 nM for each of the peptides.
Titration curves of the peptides to show the wide linear range of the assay
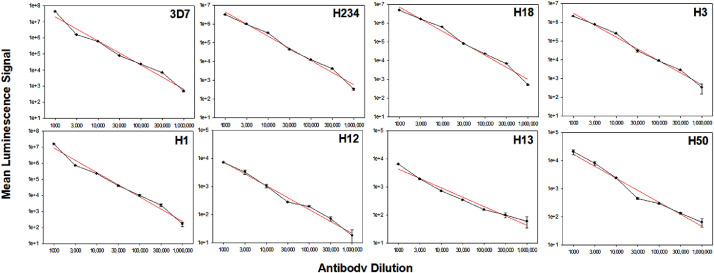
The linear range of an assay represents the portion of the dose-response curve where the luminescence signal of the samples is proportional to the specific antibody concentration in the sample. The wider the linear range of an assay, the lower the risk that samples with very different concentrations of specific antibodies fall outside the linear range and would have to be tested again or at multiple dilutions. Polyclonal human anti-CSP pooled serum (consisting of five donors having received a CSP-based vaccine) was titrated from 1:1000 to 1:1,000,000 for all eight CSP C-terminal peptides (Fig. 2). The results clearly demonstrate a linear response, i.e., proportional increase in mean luminescence signal with increasing antibody concentration over the entire range. The regression line for each plot had a R2 >0.97, hence confirming linearity.
Fig. 2.
Linear range of antibody response to peptides spans across four logs. Pooled human CSP-immune serum was tested at various dilutions for reactivity with the mutant peptides. The regression (red line) showed a fit with R2 >0.97 for all peptides. Data are expressed as mean luminescence signal of two independent experiments for each panel (error bars represent standard deviation). Note different ranges on Y-axis depending on the reactivity of the antibodies to the respective peptide. % CV was less than 5% for all tests. Plots arranged in decreasing sieve Hamming Distance (left to right, top to bottom) from 3D7. The signal with malaria-naïve serum (negative control) did not exceed MSL<780 at the 1:1,000 dilution.
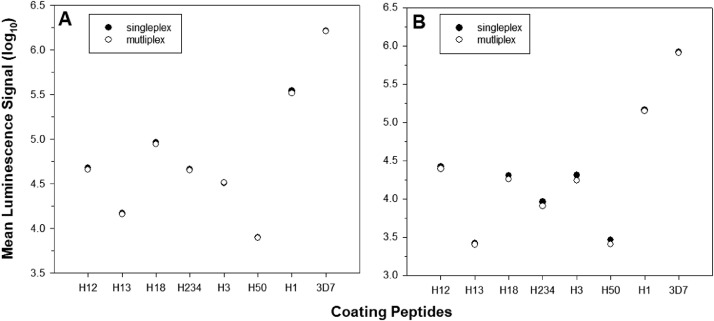
Antigens divergent in only few amino acids can be multiplexed without evidence of antigenic competition
The eight peptides tested (Table 1) diverged in only few amino acids raising the risk of antigenic competition, i.e., antibodies competing for binding to the closely related peptides. The presence of closely related antigens, and therefore the high level of crossreactivity can cause antibodies to bind not only to “their” peptide, but also others that are similar. Such antigenic competition would lead to an overall signal reduction when the antigens are multiplexed. To determine the extent of such competition, we tested each peptide alone or in the mixture with the other seven peptides (Fig. 3). In addition, we tested the optimal coating concentration (300 nM) where the antigen is well in excess and all binding sites on the respective spot for the linker saturated vs. a suboptimal concentration (100 nM) where the antigen is limited and may more readily show evidence of antigenic competition. The results did not indicate any reduction of signal when the peptides were tested multiplexed. Previously, we had tested a bead-based assay (Luminex) for this application and were not able to multiplex the various peptide-coated beads due to a significant reduction of signal (data not shown), further establishing the usefulness of the MSD platform for the assessment of closely related antigens in a multiplexed approach.
Fig. 3.
No evidence of antigenic competition. Plates were coated with (A) 300 nM (saturated concentration) or (B) 100 nM (limiting concentration) of peptides. Wells were either coated with only one specific peptide (single) or mixed as a cocktail (multiplex) and tested for reactivity with a human CSP-immune serum pool (1:3,000). The mean luminescence signal of the malaria-naïve serum pool (negative control) did not exceed the background (i.e., wells incubated with secondary antibody only (MLS < 1,000 for all conditions). Data expressed as mean luminescence signal (log-transformed).
Computational analysis
To measure the relative response of one antigen against a reference antigen, for example in the case of a strain variant and the wild-type antigen, the MLS readout of the antigen of interest () is divided by the MLS readout for the reference antigen () and then log-transformed (Eq. 1), reflecting the log of the fold change difference in the readout between the two antigens. We can express the ‘breadth’ of the antibody response across a series of antigen variants (…), relative to a reference antigen () as the median of relative response across those series of antigens, relative to the reference antigen (Eq. 2). For relative responses, the values are negative if the response to the reference antigen is higher than the response to the other antigens, as is typically the case with a vaccine strain and antigenic variants.
Declarations
Authors’ contributions
JB and EBL developed the protocol and JB performed the experiments. DEN and EMA provided the peptide sequences and participated in discussions. RSM, CRK, and EL provided critical reagents and scientific input. SC developed the computational approach and performed the data analysis. JB, EBL, and SC compiled the manuscript. All authors reviewed and edited the manuscript. The authors would like to thank Ms. Lisa Leitner for editorial assistance.
Availability of data and materials
The data will made available upon request by the corresponding author.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting the views of the Department of the Army or the Department of Defense. This paper has been approved for public release with unlimited distribution. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Funding
This study was funded by PATH's Malaria Vaccine Initiative and received additional support from the Military Infectious Disease Research Program (F041_20_WR_CS_OC).
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
References
- 1.Bolton J.S., Chaudhury S., Dutta S., Gregory S., Locke E., Pierson T. Comparison of ELISA with electro-chemiluminescence technology for the qualitative and quantitative assessment of serological responses to vaccination. Malar. J. 2020;19:159. doi: 10.1186/s12936-020-03225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.S. Chaudhury, R.S. MacGill, A.M. Early, J.S. Bolton, C.R. King, E. Locke, et al. Breadth of humoral immune responses to the C-terminus of the circumsporozoite protein is associated with protective efficacy induced by the RTS,S malaria vaccine. medRxiv. 2020:2020.11.15.20232033. [DOI] [PubMed]
- 3.Bousema T., Drakeley C., Gesase S., Hashim R., Magesa S., Mosha F. Identification of hot spots of malaria transmission for targeted malaria control. J. Infect. Dis. 2010;201:1764–1774. doi: 10.1086/652456. [DOI] [PubMed] [Google Scholar]
- 4.Cook J., Kleinschmidt I., Schwabe C., Nseng G., Bousema T., Corran P.H. Serological markers suggest heterogeneity of effectiveness of malaria control interventions on Bioko Island, equatorial Guinea. PLoS One. 2011;6:e25137. doi: 10.1371/journal.pone.0025137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corran P., Coleman P., Riley E., Drakeley C. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol. 2007;23:575–582. doi: 10.1016/j.pt.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Andagalu B., Lu P., Onyango I., Bergmann-Leitner E., Wasuna R., Odhiambo G. Evidence of interaction between humoral immunity and drug half-life in determining treatment outcome for artemisinin combination therapy in high transmission settings in western Kenya. bioRxiv. 2020 2020.08.12.248724. [Google Scholar]
- 7.Regules J.A., Cicatelli S.B., Bennett J.W., Paolino K.M., Twomey P.S., Moon J.E. fractional third and fourth dose of RTS,S/AS01 malaria candidate vaccine: a phase 2a controlled human malaria parasite infection and immunogenicity study. J. Infect. Dis. 2016;214:762–771. doi: 10.1093/infdis/jiw237. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J., Nussenzweig V., Nussenzweig R., Vekemans J., Leach A. From the circumsporozoite protein to the RTS, S/AS candidate vaccine. Human Vaccines. 2010;6:90–96. doi: 10.4161/hv.6.1.9677. [DOI] [PubMed] [Google Scholar]
- 9.Laurens M.B. RTS, S/AS01 vaccine (Mosquirix™): an overview. Human Vaccines Immunotherapeutics. 2019:1–10. doi: 10.1080/21645515.2019.1669415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White M.T., Bejon P., Olotu A., Griffin J.T., Riley E.M., Kester K.E. The relationship between RTS,S vaccine-induced antibodies, CD4(+) T cell responses and protection against Plasmodium falciparum infection. PLoS One. 2013;8:e61395. doi: 10.1371/journal.pone.0061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhury S., Regules J.A., Darko C.A., Dutta S., Wallqvist A., Waters N.C. Delayed fractional dose regimen of the RTS,S/AS01 malaria vaccine candidate enhances an IgG4 response that inhibits serum opsonophagocytosis. Sci Reports. 2017;7:7998. doi: 10.1038/s41598-017-08526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suscovich T.J., Fallon J.K., Das J., Demas A.R., Crain J., Linde C.H. Mapping functional humoral correlates of protection against malaria challenge following RTS,S/AS01 vaccination. Sci. Transl. Med. 2020:12. doi: 10.1126/scitranslmed.abb4757. [DOI] [PubMed] [Google Scholar]
- 13.Dobano C., Sanz H., Sorgho H., Dosoo D., Mpina M., Ubillos I. Concentration and avidity of antibodies to different circumsporozoite epitopes correlate with RTS,S/AS01E malaria vaccine efficacy. Nat. Commun. 2019;10:2174. doi: 10.1038/s41467-019-10195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neafsey D.E., Juraska M., Bedford T., Benkeser D., Valim C., Griggs A. Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. New Eng J Med. 2015;373:2025–2037. doi: 10.1056/NEJMoa1505819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scally S.W., Murugan R., Bosch A., Triller G., Costa G., Mordmuller B. Rare PfCSP C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J. Exp. Med. 2018;215:63–75. doi: 10.1084/jem.20170869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coppi A., Natarajan R., Pradel G., Bennett B.L., James E.R., Roggero M.A. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J. Exp. Med. 2011;208:341–356. doi: 10.1084/jem.20101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data will made available upon request by the corresponding author.