Abstract
Single molecule localization microscopy (SMLM) provided an unprecedented insight into the sub-nuclear organization of proteins and nucleic acids but apart from the nuclear envelope the role of the nuclear lipids in the functional organization of the cell nucleus was less studied. Nevertheless, nuclear lipids and specifically phosphatidylinositol phosphates (PIPs) play increasingly evident roles in gene expression. Therefore, here we provide the SMLM-based approach for the quantitative evaluation of the nuclear PIPs distribution while preserving the context of nuclear architecture. Specifically, on the example of phosphatidylinositol 4,5-bisphosphate (PIP2) we have:
-
•
Implemented and optimized the dual-color dSTORM imaging of nuclear PIP2.
-
•
Customized the Nearest Neighbor Distance analysis using ImageJ2 plug-in ThunderSTORM to quantitatively evaluate the spatial distribution of nuclear PIP2.
-
•
Developed an ImageJ2 tool for the visualization of the Nearest Neighbor Distance analysis results in cellulo.
Our customization of the dual-color dSTORM imaging and quantitative analysis provide a tool that is independent of but complementary to the biochemical and lipidomic analyses of the nuclear PIPs. Contrary to the biochemical and lipidomic analyses, the advantage of our analysis is that it preserves the spatial context of the nuclear PIP distribution.
Keywords: Immunofluorescence, Wide-field microscopy, Super-resolution microscopy, ImageJ, Nearest neighbor distance, Cell nucleus, Nuclear architecture, Nuclear speckles, SON, RNA polymerase II, Fibrillarin
Specifications table
| Subject Area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Super-resolution microscopy and quantitative imaged analysis for the nuclear lipid biology |
| Method name: | Q-DC-dSTORM: Quantitative dual-color dSTORM Two-color dSTORM imaging and nearest neighbor distance analysis in ThunderSTORM ImageJ plug-in of the nuclear phosphatidylinositol phosphate distribution |
| Name and reference of original method: | Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172-6. doi: 10.1002/anie.200802376. PMID: 18646237. van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011 Jun 16;6(7):991-1009. doi: 10.1038/nprot.2011.336. PMID: 21720313. ThunderSTORM: Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014 Aug 15;30(16):2389-90. doi: 10.1093/bioinformatics/btu202. Epub 2014 Apr 25. PMID: 24771516; PMCID: PMC4207427. ImageJ2: Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017 Nov 29;18(1):529. doi: 10.1186/s12859-017-1934-z. PMID: 29187165; PMCID: PMC5708080.a |
| Resource availability: | https://code.google.com/p/thunder-storm/ |
Nearest Neighbor Distance analysis: Malkusch S., Endesfelder U., Mondry J., Gelleri M., Verveer P.J., Heilemann M., Coordinate-based colocalization analysis of single-molecule localization microscopy data, Histochem. Cell Biol. 137 (1) (2012) 1–10.
Method details
Motivation
The eukaryotic nucleus has very dense interior comprised of viscoelastic polymer matrix. Therefore, it is difficult to visualize its individual components with classical diffraction-limited light microscopy. Super-resolution imaging methods provided a valuable insight into the sub-nuclear organization, however mostly focused on the proteins and nucleic acids while the role of the nuclear lipids has been neglected. However, there is growing evidence for the different roles of nuclear lipids and in particular nuclear phosphatidylinositol 4,5-bisphosphate (PIP2) in the establishment of the sub-nuclear organization and regulation of various nuclear processes [6], [7], [8], [9].
Super-resolution imaging methods allow to control in time the emission of fluorophores by either deterministic or stochastic approach. The deterministic approaches include stimulated emission depletion (STED) microscopy [10] and structured illumination microscopy (SIM) [11]. Stochastic methods include single-molecule localization microscopy (SMLM) approaches, such as photoactivation or fluorescence photoactivation localization microscopy (PALM or FPALM) [12,13], and stochastic optical reconstruction microscopy (STORM) [14] or direct STORM (dSTORM) [1], [2]. SMLM methods provide also a new paradigm for the quantitative analysis of the relative nanoscale organization characterized by the spatial relationships between molecules. Therefore, we provide here the detailed description of the indirect immunofluorescence labeling of nuclear PIP2 and other nuclear antigens, dual-color dSTORM image acquisition, reconstruction and quantitative evaluation of the nanoscale sub-nuclear distribution of PIP2 based on the ThunderSTORM [3], which is a plug-in for ImageJ2 [4]. The precise localization approach that preserves the sub-nuclear architecture and the detailed quantitative analysis of the nuclear PIP2 distribution will expand the set of standard biochemical and lipidomic analyses, which are powerful but in their nature cumulative and thus neglect the spatial context of the nuclear architecture. This experimental pipeline is applicable for the functional studies of the role of nuclear PIP2, other nuclear PIPs and lipids in the establishment of nuclear architecture and regulation of gene expression [5].
Labeling of the nuclear PIP2 and other nuclear antigens for the dual-color dSTORM
The U-2 OS cells were grown on the HellmanexTM III (Sigma cat. #Z805939) treated high-precision 12 mm 1.5H round coverslips (Paul Marienfeld GmbH & Co., cat. #0117520) placed in the 24-wells plate the day before processing and incubated in DMEM with 10% FBS at 37°C and 5% CO2.
HellmanexTM III treatment of the coverslips was performed according to the following procedure:
-
1.
Place the coverslips into the clean beaker filled with 2% HellmanexTM III in ddH2O
-
2.
Cover with aluminum foil and incubate over-night at 56°C
-
3.
Sonicate for 30 min in the heated HellmanexTM III solution
-
4.
Wash three times with ddH2O and sonicate again in ddH2O for 30 min
-
5.
Wash five times with ddH2O and store covered in ddH2O
For the immunofluorescence labeling of the endogenous, detergent resistant nuclear PIP2 and the nuclear speckle marker SON or RNAPII we used the typical protocol used for the fluorescence microscopy. This protocol consisted of the following steps and is generally applicable for other nuclear antigens:
-
1.
Aspirate the media and wash the cells twice with PBS
-
2.
Fix the cells in the original 24-wells plate with 2% PFA in PBS for 30 min at room temperature (RT)
-
3.
Permeabilize the cells in the original 24-wells plate with 0.1% Triton X-100 in PBS for 20 min at RT
-
4.
Wash the cells three times for 5 min with PBS
-
5.
Transfer the coverslips with the cells upside-down to block in the 30 µL of 5% BSA in PBS (filtered through the 0.2 µm syringe filter) for 30 min at RT in a humidified chamber.
-
6.
Transfer the coverslips with the cells upside down onto a parafilm placed in the humidified chamber with 30 µL of the primary antibodies diluted in 5% BSA in PBS (BSA in PBS was filtered through the 0.2 µm syringe filter and the diluted antibodies were spun down in a bench top centrifuge at 12.000g for 3 min to remove the aggregated antibodies and the supernatant was used). Incubate for 45 min at RT.
-
7.
Transfer the coverslips into the 24-wells plate with the cells facing up and wash three times for 5 min with PBS.
-
8.
Transfer the coverslips with the cells upside down onto a parafilm placed in a humidified chamber with 30 µL of the secondary antibodies diluted in 5% BSA in PBS (BSA in PBS was filtered through 0.2 µm syringe filter and the diluted antibodies were spun down in a bench top centrifuge at 12.000g for 3 min to remove the aggregated antibodies and the supernatant was used). Incubate for 30 min at RT.
-
9.
Transfer the coverslips into the 24-wells plate with the cells facing up and wash three times for 5 min with PBS.
-
10.
Aspirate PBS and post-fix with 2% PFA in PBS for 15 min to stabilize the interaction between antibodies and antigen.
-
11.
Eventually store in PBS in the fridge for short periods of time (over-night) until imaging.
In steps 6 and 8 we used the following primary antibodies at given final concentrations: anti-PI(4,5)P2 clone 2C11 (Z-P004; Echelon Biosci. Inc., USA) 5 µg/mL, anti-SON (ab121759; Abcam, UK) 1 µg/mL, anti-RNAPII CTD (ab210527; Abcam, UK) 3 µg/mL and the following secondary antibodies and concentrations: goat anti-mouse IgM (µ-chain) Alexa Fluor (AF)647 (115-605-075; Jackson ImmunoRes., UK) 10 µg/mL; goat anti-mouse IgM (µ-chain) AF555 (A24126; Jackson ImmunoRes., UK) 10 µg/mL, goat anti-rabbit IgG AF647 (A21245; Invitrogen) 10 µg/mL.
In addition we used fibrillarin (Fib) found in the nucleolar dense fibrillar component fused to the self-labeling SNAP tag [24] (Fib-SNAP) and stably exogenously over-expressed in the U-2 OS cell line [8]. We preferred to use the Fib-SNAP due to the lack of reliable primary antibodies for the immunofluorescence detection of endogenous Fib. The U-2 OS cells stably over-expressing exogenous Fib-SNAP were cultured as described above for the standard U-2 OS cells. We stained the Fib-SNAP with SNAP-Cell® 647-SiR (New England Biolabs, USA; cat. # S9102S) applied to the cell culture in the FluoroBrite DMEM media (A1896701 ThermoFischer) in the final concentration of 500 nM for 45 min at 37°C and 5% CO2. Then we aspirated the labeling media and washed the cells twice with PBS, added the fresh culture media and incubated the cells to remove the excessive dye at 37°C and 5% CO2. After 45 min incubation the cells were washed with PBS, fixed and processed for immunofluorescence staining of PIP2 as described above for the standard U-2 OS cells.
Image acquisition
Coverslips with the cells were mounted in the Chamlide chamber (Live Cell Instrument, Korea) and covered with the imaging buffer (see below). Single-molecule localizations (SMLs) were acquired by Zeiss Elyra PS.1 equipped with HR Diode 642-150 and HR DPSS 561-200 lasers, Alpha Plan-Apochromat 100x/1.46 oil DIC M27 Elyra objective and Andor EM CCD iXon DU 897 camera and Zeiss ZEN Black 2.1 SP3 software. AF647 and AF555 photo-switching was achieved by HiLo illumination and TIRF HP FOV setting with 100% power of 642nm and 561nm lasers and the signal was acquired via MBS 642 + EF LP 655 and MBS 561 + EF BP 570-620 / LP 750 filters, respectively. Exposure time was 40 ms and EM gain was 300 for both channels. We have tested the performance of AF647 and AF555 for dSTORM of nuclear PIP2 and RNAPII in two imaging buffers. The first buffer, called OxEA [15] included:
-
•
50 mM β-MercaptoEthylamine hydrochloride (MEA, Sigma-Aldrich)
-
•
3% (v/v) OxyFlour™ (Oxyrase Inc., Mansfield, Ohio, U.S.A.)
-
•
20% (v/v) of sodium DL-lactate solution (L1375, Sigma-Aldrich)
-
•
in PBS, pH adjusted to 8.5 with NaOH
The second, MEA buffer [1], [2], included:
-
•
50 mM β-Mercaptoethylamine hydrochloride (MEA, Sigma-Aldrich)
-
•
in PBS pH 7.4
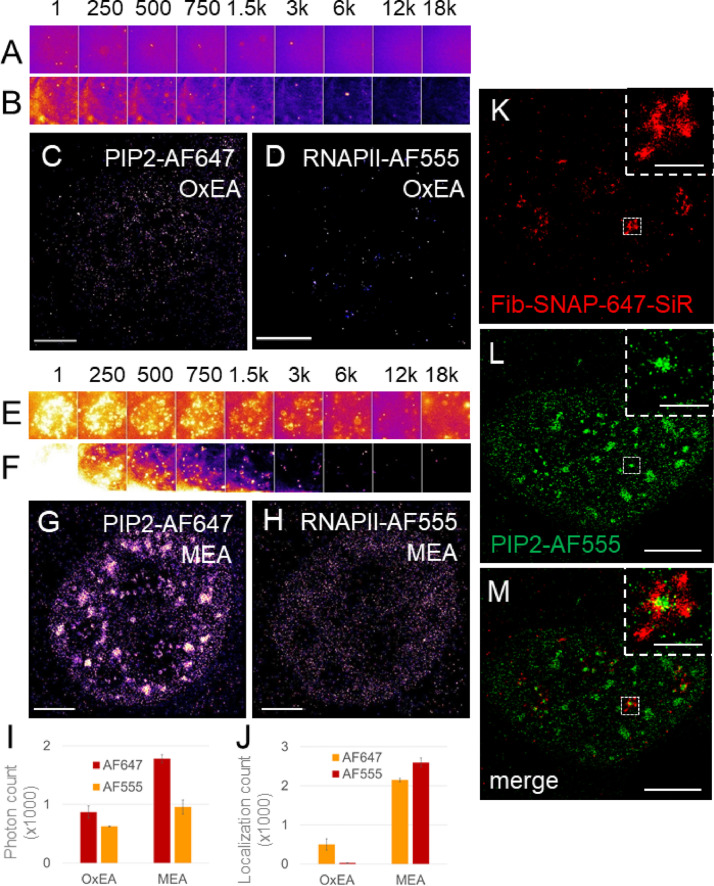
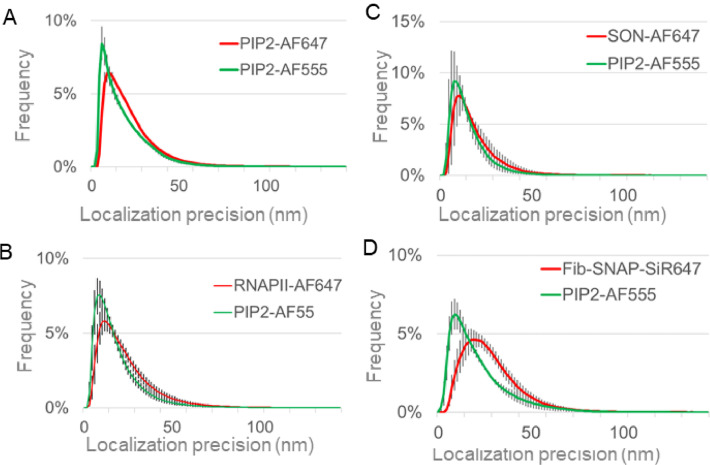
Two channels were recorded sequentially (Fig. 1A-H), with AF647 channel first. The 2D SMLs were calculated by Zeiss ZEN Black 2.1 SP3 software using x,y 2D Gauss fit with the point spread function (PSF) half width 177.9 nm. Peak mask size 9 pixels and peak intensity to noise factor of 6 were selected empirically and kept consistently. Due to the high density of the nuclear antigens, we used the method that accounted for the overlap in 2D with the max cluster size of 10. The calculated SMLs were rendered in ZEN software with 10 nm/pixel resolution and 1x PSF expansion factor. The possible drift was corrected using a model-based correction in ZEN and automatic number of segments was selected for the model calculation. Each drift-corrected localization file was then individually saved and the two channels (AF647 and AF555) corresponding to the same nucleus were then aligned in ZEN Channel Alignment using the affine fit setting. The localization coordinates and other image information, such as localization precision, photon count and background variation were exported as text files for further image analysis. Figure 1A-D documents the image acquisition in OxEA buffer and Fig. 1E-H in MEA buffer. We measured in MEA buffer the higher photon count (mean number of photons per localization) and higher number of localizations (Fig. 1I, J) for both, AF647 and AF555 conjugated to the secondary antibodies against the primary antibody that recognizes the detergent resistant nuclear PIP2 or RNAPII. We found that in our case the combination of AF647 and AF555 performs better in MEA buffer and therefore we used this buffer for the combination of these two fluorophores in our experiments. AF555 provided slightly better localization precision than AF647 (Fig. 2), and therefore for the dual-color dSTORM of nuclear PIP2-AF555 we combine it with AF647-conjugated secondary antibody against the primary antibodies that recognize other nuclear markers of interest, Fig. 5. The Fib-SNAP-647-SiR performed in the MEA buffer well (Fig. 1K-M) and we reached the localization precision 25.7±2.7nm (Fig. 2). Therefore, Fib-SNAP-647-SiR is suitable for the dual-color dSTORM studies of the nucleolar PIP2 and Fib.
Fig. 1.
Optimization of the dual-color dSTORM image acquisition. Nuclear PIP2 indirectly immunolabeled with Alexa Fluor (AF)647 and RNA polymerase II (RNAPII) with AF555 were imaged in OxEA (A-D) or MEA (E-H) buffer. Raw blinking frames of (A) PIP2-AF647 and (B) RNAPII-AF555 imaged in OxEA buffer. Reconstructed images of (C) PIP2-AF647 and (D) RNAPII-AF55 imaged in OxEA buffer (see Image reconstruction). Raw blinking frames of (E) PIP2-AF647 and (F) RNAPII-AF555 imaged in MEA buffer. Reconstructed images of (G) PIP2-AF647 and (H) RNAPII-AF555 imaged in MEA buffer using the Normalized Gaussian method (see Image reconstruction). (I) Mean number of photons per localization and (J) number of localizations collected from PIP2-AF647 and from RNAPII-AF555 in OxEA or MEA buffer. Cells stably expressing Fib-SNAP labeled with Cell SNAP SiR-647 (Fib-SNAP-SiR647) (K) and indirectly immunolabeled PIP2-AF555 (L) merged image (M) and zoom-in to the boxed nucleolar regions. Scale bars 5 µm; inset 1 µm.
Fig. 2.
Experimental single-molecule localization precisions. Plot of the measured localization precisions for PIP2-AF647 and PIP2-AF555 (A), RNAPII-AF647 and PIP2-AF555 (B), SON-AF647 and PIP2-AF555 (C) and Fib-SNAP-SiR647 and PIP2-AF555 (D).
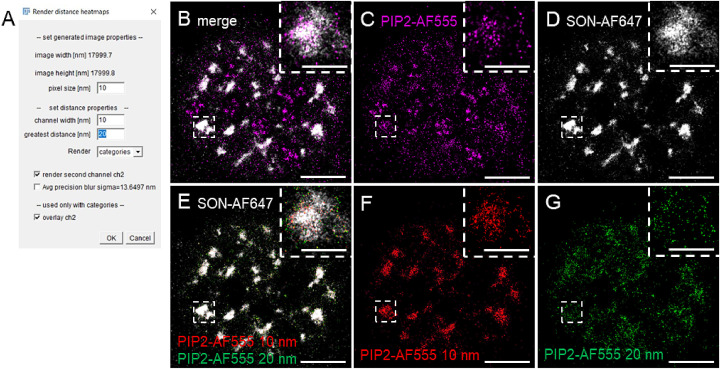
Fig. 5.
Color-coded in cellulo visualization of NND in ImageJ2. (A) Graphical user interface of the “Render distance heatmaps” macro which we wrote for the color-coding of the pixels in the “results” (in this example PIP2-AF555) image according to their NND to the “reference” (ground-truth) image (in this example SON-AF647). (B) Merged image of the original Normalized Gauss rendering of PIP2-AF555 in magenta (C) and SON-AF647 in gray (D). (E) Merge of the SON-AF647 in grey (the same as in D) and the pixels corresponding to the PIP2-AF555 at the NND of 10 nm (red, F) and 20 nm (green, G) to SON-AF647. Insets show zoom-in to the boxed regions. Scale bar = 5 µm; inset 1 µm.
Image reconstruction
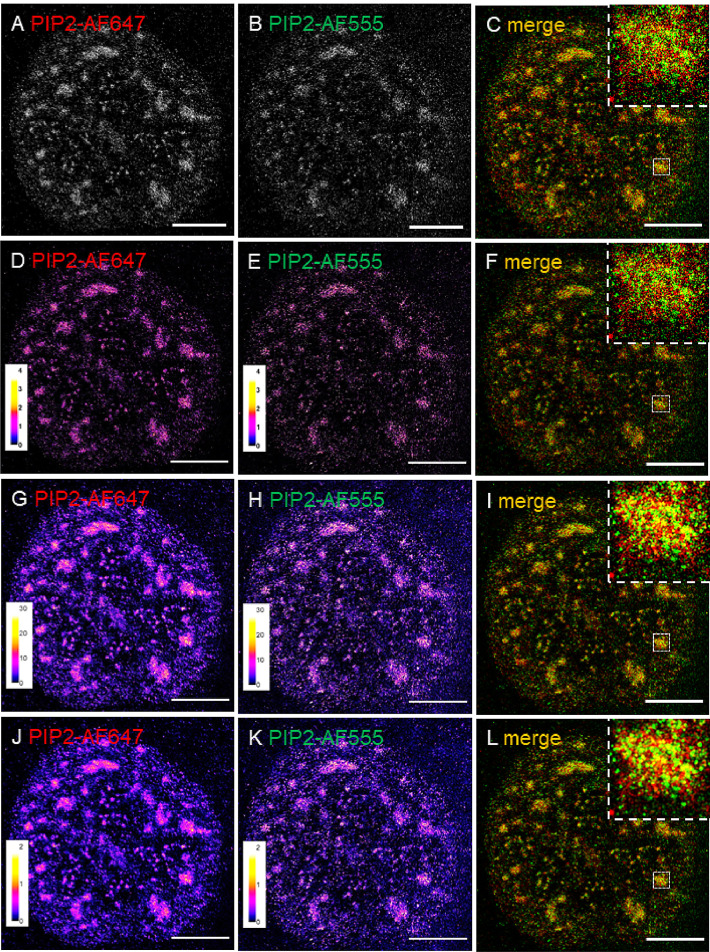
SML coordinates and other data were exported from ZEN software as txt files that contained in one file the information about both aligned channels. The image analysis software ThunderSTORM [3], which is an ImageJ plug-in [4], requires as input two csv files. First file represents “ground truth” and second file represents “result”. Then the Nearest Neighbor Distance (NND) and other parameters of the result channel will be calculated according to the ground truth channel (see further). Therefore it was first necessary to convert the txt files into the csv files and split the files according to the channels, then import the corresponding two channels (“marker”-AF647 as a ground truth and PIP2-AF555 as a result) into the ThunderSTORM, calculate the NND and save the results for further statistical evaluation. We developed an ImageJ macro to automate all these steps (Suppl. Material 1). This macro transforms in batch mode the common txt files containing the information of the two channels into the csv files that contain information about the individual channels, imports the data into ThunderSTORM and reconstructs the images. ThunderSTORM offers four options to render (visualize) the SML data [3]. First visualization is a simple scatter plot with the 0 (black pixels) or 1 (white pixels) coding of the individual pixels corresponding to the individual SMLs [16]. Fig. 3A and B shows the scatter plot of a representative PIP2-AF647 and PIP2-AF555 image reconstructions, resp. The magnification ratio is specified by the user and here we used the magnification factor of 10. Fig. 3C shows merged AF647-PIP2 and AF555-PIP2 scatter plots pseudo-colored in red and green, resp. This visualization however neglects the number of repeating SMLs in the same pixel (the information does not have a bit depth but is only binary, black for 0 or white for 1) and therefore lacks the information about the density of localizations. Second rendering method is Histogram, a two-dimensional histogram of molecular positions created with the bin size corresponding to the pixel size of the final super-resolution image [3,16]. Images in Fig. 3D and E were pseudo-colored in ImageJ with the Fire look-up table (LUT) to better visually illustrate that the image brightness increases with the molecule localizations accumulating at each bin (pixel). Fig. 3D and E show the histogram of a representative PIP2-AF647 and PIP2-AF555 image renderings, resp. and Fig. 3F shows merged PIP2-AF647 and PIP2-AF555 histograms that were pseudo-colored in red and green, resp. The images in Fig. 3D-F were rendered with lateral uncertainty set to 10 nm. Third method offered by ThunderSTORM for the rendering of SML data is based on the implementation of Average Shifted Histograms (ASH) [17]. In this rendering, the width of the histogram bin is given by the multiplication of the pixel size of the super-resolution image and the number of shifts in the lateral and axial directions. Both of these parameters can be independently specified and for the creation of images in the Fig. 3G-I we used magnification factor of 10 and lateral shifts factor of 2. The width of the histogram bin is in the Fig. 3G and H illustrated by the Fire LUT. The visual output of ASH rendering (Fig. 3G-I) is similar to the visualization by fourth rendering method that ThunderSTORM offers. However, the ASH is computationally faster [3]. Fourth rendering method by ThunderSTORM is Normalized Gaussian (Fig. 3J-L), which fits a normalized symmetric 2D function integrated over each molecule localization. The standard deviation of this normalized Gaussian fit can be individually specified by the localization uncertainty, which was set to 10 nm for the images in Fig. 3J-L. The visualized molecules accumulate in the final super-resolution image, which is illustrated by the Fire LUT in Fig. 3J and K. Although Normalized Gaussian is the most computationally demanding of the above mentioned four rendering methods that ThunderSTORM offers, we preferred it for the limited data sets, because it is the most precise rendering method.
Fig. 3.
Image reconstruction in ThunderSTORM. Nuclear PIP2 labeled with AF647 (A, D, G and J) and AF555 (B, E, H and K) was sequentially imaged in MEA buffer. SMLs were rendered using Scatter Plot (A-C), Histogram (D-F), Average Shifted Histogram (G-I) and Normalized Gaussian (J-L) methods. Images (A, B) show individual localizations as white pixels, other methods account for the accumulation of the localizations and therefore for the better visualization are pseudo-colored using “Fire” LUT and the intensities are indicated by the respective calibration bars (D, E, G, H, J and K). Merged images of PIP2-AF647 and PIP2-AF555 pseudo-colored in red and green, resp., rendered with the above indicated methods (C, F, I, L). Insets show zoom-in to the boxed regions. Scale bar = 5 µm; inset 1 µm.
Analysis of the spatial distribution of the nuclear PIP2
We characterized the spatial distribution of nuclear PIP2-AF555 with respect to the nuclear speckle marker SON-AF647 as a measure of the NNDs and then we compared the real NNDs with the randomized NNDs. Randomized data were created either by swapping x and y coordinates (Random Rotated, RR) or by generating randomly distributed (Random Generated, RG) SMLs. RR approach was based on the suggestion by Dunn et al. [19]. The authors suggested the randomization of the experimental data by rotating one channel of the dual-color image and comparing the spatial relationship between the two channels in the real and randomized data. RG SMLs were created using ThunderSTORM ImageJ2 plug-in, “Performance testing” option and “Generator of simulated data” function. Each RG ground-truth channel was created with respect to the number of frames, area and molecular density of the corresponding real ground-truth data. Molecular density for each RG data set was calculated from the real number of corresponding SMLs divided by the area of the real image divided by the real number of frames in the corresponding original image acquisition. Other parameters of the simulator were kept default. The advantage of RR approach is its simplicity and effectiveness. Its limitation, compared with computationally more extensive RG method, is that it preserves the original clustering of the SMLs. Therefore, RR can results in the “random” overlap of the RR SML clusters in channel 2 with the real clusters in the channel 1. In contrary, RG method creates randomly dispersed SMLs without their clustering. Therefore, RG method is more rigorous than RR method. These two randomization approaches were further discussed in [5]. We used these two randomization approaches to assess if the real mean NND and the fraction of neighbors at the distance smaller than mean NND differs from the RR and RG data.
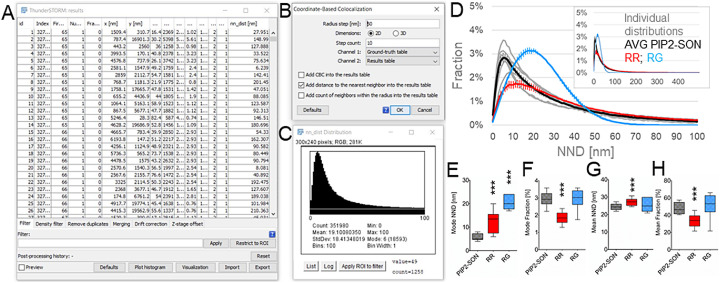
ThunderSTORM calculates the NNDs using one channel as a ground-truth, second channel as results channel and appends the quantitative output to the results table. We used the AF647-SON channel as a ground truth and calculated the NND of PIP2-AF555 to SON-AF647. After importing the ground-truth and results data the NND calculation is accessible in the ThundeSTORM menu as “colocalization”, CBC (coordinate-based co-localization) [18]. We selected the radius step 50 nm, according to the nature of our data, we used calculation in 2D (calculation in 3D is also available) and step count factor of 10. As “Channel 1” for the calculation was selected “Ground-truth table” and as “Channel 2” was selected “Results table” and together with the option “Add distance to the nearest neighbor into the results table”. The column “nn_dist [nm]” is then added to the Results table (Fig. 4A). Fig. 4B shows the user interface for the NND calculation and Fig. 4C shows a representative NND distributions. We used self-written macro that automates all above mentioned steps and at the end creates txt file with all resulting NNDs (Suppl. Material 2). For the quantitative evaluation, we imported the data from the txt file into Excel, calculated the sum of all SMLs and normalized the individual NND distributions by the number of SMLs. Then we calculated average normalized NND distributions (Fig. 4D). Although in total the NND was measured up to 500 nm, the vast majority of NND was in the range of tens of nm, and therefore we reported the NND distributions in the range of 0-100 nm. From the individual NND distributions we calculated the most frequent (mode) NND (Fig. 4E), fraction of the NND at the mode NND (Fig. 4F), mean NND (Fig. 4G) and fraction of the NND at the distances smaller than the mean NND (Fig. 4H). The mode NND of the RR and RG data were significantly higher as compared to the real NND (PIP2-SON, Fig. 4F). The fraction of the NND at the mode NND was significantly reduced for RR but not for RG data (Fig. 4G). This reflects the fact that for RR data the peak of the normalized NND distribution was lower as compared to the real data (PIP2-SON, Fig. 4H) but the tail of the normalized RR NND distribution was shifted to the higher values (Fig. 4H, red curve). In contrast, normalized RG NND distribution had the peak similar to the normalized real NND distribution but shifted to higher values (Fig. 4H, blue curve). We suggest to evaluate mode rather than the mean, because mode (which represents the peak or the most frequent value in the data set) better reflects the shift of the peaks of NND distributions that are not normally distributed but have long tails in large NND values. Therefore, the mean NNDs were higher than mode NNDs of all three (PIP2-SON, RR and RG) data sets, but only mean RR NND was slightly but significantly higher as compared to the real mean PIP2-SON NND. Similarly, fraction of the NND at the distances smaller than the mean NND was significantly reduced only in the case of RR. Taken together, for the quantitative evaluation of the spatial proximity between two probes we recommend comparison of the mode NNDs and the fraction of NNDs at the mode NND between real and randomly generated data sets. Our example of the NND analysis documents the specific proximity between nuclear PIP2 and the nuclear speckle marker SON.
Fig. 4.
Nearest Neighbor analysis in ThunderSTORM. (A) Results table containing SML coordinates (“x [nm]” and “y [nm]” columns) and Nearest Neighbor Distances (NND) (“nn_dist [nm]” column) of a representative dual-color dSTORM image of PIP2-AF555 to SON-AF647, which was calculated using the Coordinate-Based Colocalization (CBC) with the parameters indicated in (B). (C) Exemplar NND distribution on the interval 0-100 nm for the representative dual-color dSTORM image of PIP2-AF647 and SON-AF555. (D) Individual normalized NND distributions (Individual distributions, grey curves), average normalized PIP2-SON NND (AVG PIP2-SON, black curve), average normalized random rotated PIP2-SON NND (RR, red curve) and average normalized random generated PIP2-SON NND (RG, blue curve) distributions on the interval 0-100 nm and (inset) 0-500 nm. From the normalized PIP2-SON, RR and RG NND distributions were calculated the mode (most frequent) NND (E), fraction of the NND at the mode NND (F), mean NND (G) and fraction of neighbors at the distance smaller than the mean NND (H). *** P<0.001 (N=6; paired, one-tailed t test).
Visualization of the nanoscale sub-nuclear distribution of PIP2
For better understanding of the spatial organization of the cell nucleus and nuclear PIP2 distribution, it is useful to visualize the distribution of the NND within its spatial context. Therefore, we developed an ImageJ macro for the creation of visual in cellulo maps of the pixels color-coded according to NNDs (Suppl. Material 3). This macro prompts the user to load the csv file of the SML data for the channel in which the color-coded NND map will be created. If the corresponding “nn_dist” txt file of the NND of the marker of interest (in our example PIP2-AF555) to the reference marker (in our example SON-AF647) created by ThunderSTORM is located in the same folder it will be then loaded automatically, otherwise the user will be prompted to specify the location of “nn_dist” txt file. The user is then prompted to specify the pixel size in the graphical user interface (Fig. 5A), the “channel width”, which means the distance in [nm] that will be collectively visualized in one color and the “greatest distance” that will be visualized. The greatest distance value is estimated from the given NND distribution (Fig. 4D). The macro then creates several images. Resulting images include the stack of images in which each image has pixels corresponding to the marker of interest (in our example PIP2-AF555) (Fig. 5B and C) coded in individual colors according to their NND to the reference marker (in our example SON-AF647) (Fig. 5B and D). The other resulting images include first the rendering of the reference marker and second the merged stack of images with pixels corresponding to the marker of interest coded in individual colors according to their NND to the reference marker, and the overlay of these two images. We found that, at least in the case of PIP2-SON (Fig. 4D) –RNAPII or -Fib NNDs [5], the most informative are the distances up to 20-30 nm (“greatest distance” parameter) rendered in the 10 nm bins. Fig. 5E therefore shows the overlay of the reference marker SON-AF647 in grey, with the green and red pixels corresponding to the PIP2-SON NNDs of 10 nm (Fig. 5F) and 20 nm (Fig. 5G), resp. This tool is applicable for the color-coded in cellulo visualization of NNDs between any two markers.
Concluding remarks
We provided a detailed protocol for the immunofluorescence labeling of nuclear PIP2 and other nuclear antigens such as RNAPII or the nuclear speckle marker SON, and SNAP-labeling of Fib-SNAP, dual-color dSTORM image acquisition, reconstruction and - Q-DC-dSTORM analysis. We described in depth an approach for the quantitative evaluation of the nanoscale sub-nuclear distribution of PIP2 based on the ImageJ [4] plug-in ThunderSTORM [3]. We automated the majority of the steps, from the data import, through the image reconstruction, to the calculation of NND, by self-written ImageJ macros that allow for batch processing of the data. We recommend to extract two quantitative parameters from the normalized average NND distributions: the mode NND and the fraction of NND at the mode NND. The NND calculation by ThunderSTORM was previously applied to quantitatively describe at nanoscale the relationship between the transcription and replication sites in nucleoli [20]. Here we introduced the parameter fraction of NND at the mode NND, which represents the relative amount of the most frequent (mode) NNDs. Calculation of these two parameters is useful for the quantitative evaluation of the nanoscale organization of nuclear PIP2 and its relationship with other nuclear markers. This approach is applicable to other PIPs [5] and for the evaluation of the differences between various experimental conditions. Moreover, we developed ImageJ-based in cellulo visualization tool of the NNDs. Although the NND analysis provides the important information about the nanoscale subcellular organization, we also recommend to combine the distance-based analysis with other quantitative analyses suitable for the SMLM data. The powerful approach represents a method based on the Voronoi Tesselation, which allows for the quantitative evaluation of the co-distribution of two markers [21]. The technical aspects of the dSTORM with various conventional fluorophores are nicely reviewed for instance in van de Linde et al. [2] and various methods for the analysis of SMLM data were reviewed for example in Schermelleh et al. [22] or Jacquemet et al. [23]. In conclusion Q-DC-dSTORM is customized systematic approach for in-depth quantitative analysis of the dual-color dSTORM data of nuclear PIP2 and several nuclear markers. Q-DC-dSTORM represents easily implementable tool to study the nanoscale organization of nuclear PIP2 and other PIPs. This quantitative analysis approach is also applicable to other nuclear and cellular markers.
Declaration of Competing Interests
X The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge Imaging Methods Core Facility at BIOCEV, institution supported by the MEYS CR (Large RI Project LM2018129 Czech-BioImaging) and ERDF (project No. CZ.02.1.01/0.0/0.0/16_013/0001775) for their support. Microscopy was performed in the Laboratory of Confocal and Fluorescence Microscopy co-financed by the ERDF and the state budget of the Czech Republic, projects no. CZ.1.05/4.1.00/16.0347 and CZ.2.16/3.1.00/21515, and supported by the Czech-BioImaging large RI project LM2018129. This work was supported by the Grant Agency of the Czech Republic (Grant Nos. 19-05608S and 18-19714S); by the Czech Academy of Sciences (Grant No. JSPS-20-06); by the Institutional Research Concept of the Institute of Molecular Genetics (Grant No. RVO: 68378050); and by the MEYS CR (COST Inter-excellence internship LTC19048, LTC20024 and Action 15214 EuroCellnet) and BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University“ (CZ.1.05/1.1.00/02.0109).
We are grateful to Marie Olšinova and Aleš Benda for their initial help with microscopy, to Zdeněk Švindrych for the advice with ThunderSTORM, to Lenka Pišlová for her excellent administrative assistance, to Iva Jelínková for her assistance with the cell cultures, to Pavel Kříž for the lab management and to reviewers for helping us to improve our manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.mex.2021.101372.
Appendix. Supplementary materials
References
- 1.Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. PMID: 18646237. [DOI] [PubMed] [Google Scholar]
- 2.van de Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6(7):991–1009. doi: 10.1038/nprot.2011.336. Jun 16doi: 10.1038/nprot.2011.336. PMID: 21720313. [DOI] [PubMed] [Google Scholar]
- 3.Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30(16):2389–2390. doi: 10.1093/bioinformatics/btu202. Aug 15Epub 2014 Apr 25. PMID: 24771516; PMCID: PMC4207427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 2017;18(1):529. doi: 10.1186/s12859-017-1934-z. Nov 29PMID: 29187165; PMCID: PMC5708080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoboth P., Sztacho M., Šebesta O., Schätz M., Castano E., Hozák P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids. 2021;1866(5) doi: 10.1016/j.bbalip.2021.158890. [DOI] [PubMed] [Google Scholar]
- 6.Osborne S.L., Thomas C.L., Gschmeissner S., Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001;114(Pt 13):2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 7.Sobol M., Krausova A., Yildirim S., Kalasova I., Faberova V., Vrkoslav V., Hozak P. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018;131(8) doi: 10.1242/jcs.211094. [DOI] [PubMed] [Google Scholar]
- 8.Guillen-Chable F., Corona U.R., Pereira-Santana A., Bayona A., Rodriguez-Zapata L.C., Aquino C., Castano E. Fibrillarin ribonuclease activity is dependent on the GAR domain and modulated by phospholipids. Cells. 2020;9(5) doi: 10.3390/cells9051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sztacho M., Šalovská B., Červenka J., Balaban C., Hoboth P., Hozák P. Limited proteolysis-coupled mass spectrometry identifies phosphatidylinositol 4,5-bisphosphate effectors in human nuclear proteome. Cells. 2021;10(1):E68. doi: 10.3390/cells10010068. Jan 4PMID: 33406800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klar T.A., Jakobs S., Dyba M., Egner A., Hell S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U S A. 2000;97(15):8206–8210. doi: 10.1073/pnas.97.15.8206. Jul 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafsson M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000;198(Pt 2):82–87. doi: 10.1046/j.1365-2818.2000.00710.x. May. [DOI] [PubMed] [Google Scholar]
- 12.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. Sep 15. [DOI] [PubMed] [Google Scholar]
- 13.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahidiazar L., Agronskaia A.V., Broertjes J., van den Broek B., Jalink K. Optimizing imaging conditions for demanding multi-color super resolution localization microscopy. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0158884. Jul 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baddeley D., Cannell M.B., Soeller C. Visualization of localization microscopy data. Microsc. Microanal. 2010;16(1):64–72. doi: 10.1017/S143192760999122X. [DOI] [PubMed] [Google Scholar]
- 17.Scott D.W. Averaged shifted histograms: effective nonparametric density estimators in several dimensions. Ann. Stat. 1985;13(3):1024–1040. [Google Scholar]
- 18.Malkusch S., Endesfelder U., Mondry J., Gelleri M., Verveer P.J., Heilemann M. Coordinate-based colocalization analysis of single-molecule localization microscopy data. Histochem. Cell Biol. 2012;137(1):1–10. doi: 10.1007/s00418-011-0880-5. [DOI] [PubMed] [Google Scholar]
- 19.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300(4):C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smirnov E., Borkovec J., Kovacik L., Svidenska S., Schrofel A., Skalnikova M., Raska I. Separation of replication and transcription domains in nucleoli. J. Struct. Biol. 2014;188(3):259–266. doi: 10.1016/j.jsb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Levet F., Julien G., Galland R., Butler C., Beghin A., Chazeau A., Sibarita J.B. A tessellation-based colocalization analysis approach for single-molecule localization microscopy. Nat. Commun. 2019;10(1):2379. doi: 10.1038/s41467-019-10007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., Drummen G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019;21(1):72–84. doi: 10.1038/s41556-018-0251-8. Jan10.1038/s41556-018-0251-8. Epub 2019 Jan 2. PMID: 0602772. [DOI] [PubMed] [Google Scholar]
- 23.Jacquemet G., Carisey A.F., Hamidi H., Henriques R., Leterrier C. The cell biologist's guide to super-resolution microscopy. J. Cell Sci. 2020;133(11) doi: 10.1242/jcs.240713. Jun 11PMID: 32527967. [DOI] [PubMed] [Google Scholar]
- 24.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21(1):86–89. doi: 10.1038/nbt765. PMID: 12469133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.