Summary
Fruit softening indicated by firmness determines the texture, transportability, and shelf life of tomato products. However, the regulatory mechanism underlying firmness formation in tomato fruit is poorly understood. Here, we report the regulatory role of SlBES1, an essential component of brassinosteroid hormone signaling, in tomato fruit softening. We found that SlBES1 promotes fruit softening during tomato fruit ripening and postharvest storage. RNA-seq analysis suggested that PMEU1, which encodes a pectin methylesterase, might participate in SlBES1-mediated softening. Biochemical and immunofluorescence assays indicated that SlBES1 inhibited PMEU1-related pectin de-methylesterification. Further molecular and genetic evidence verified that SlBES1 directly binds to the E-box of PMEU1 to repress its expression, leading to fruits softening. Loss-of-function SlBES1 mutant generated by CRISPR-Cas9 showed firmer fruits and longer shelf life during postharvest storage without other quality alteration. Collectively, our results indicated the potential of manipulating SlBES1 to regulate firmness without negative consequence on visual and nutrition quality.
Subject areas: Biological sciences, Molecular biology, Plant biology, Plant development
Graphical abstract

Highlights
-
•
SlBES1 promotes tomato fruit softening without affecting nutritional quality
-
•
SlBES1 inhibits PMEU1-related fruit pectin de-methylesterification
-
•
SlBES1 represses PMEU1 expression through directly binding to the E-box
-
•
Knockout of SlBES1 by CRISPR-Cas9 enhances fruit firmness and extends shelf life
Biological sciences; Molecular biology; Plant biology; Plant development
Tomato (Solanum lycopersicum) is not only one of the most important vegetables worldwide but also a model system for fruit development. The softening process during fruit ripening indicated by firmness determines the overall palatability, transportability, and shelf life of tomatoes. Fruit firmness is determined by diverse factors, including cell wall structure (Seymour et al., 2013) and cuticle properties (Li et al., 2020). Among them, the cell wall and alterations of its structure are considered as the predominant factors, such as changes in the complex of microfibrils and polysaccharides, which are composed of hemicellulose, cellulose, pectin, and other structural proteins. Pectins, known as pectic polysaccharides, are a group of complex polymers containing homogalacturonan (HG), rhamnogalacturonan-I, and rhamnogalacturonan-II and located in the primary cell wall and middle lamella with a high amount (Senechal et al., 2014). A wide range of cell wall remodeling by pectin metabolic enzymes has been investigated to explore the genetic, molecular, and biochemical basis of fruit softening. A methyl ester group is removed from HG in a reaction catalyzed by pectin methylesterase (PME/PE, EC 3.1.1.11), and the de-methylesterified pectin forms “egg box” through a cross-link with Ca2+, strengthening the cell wall (Senechal et al., 2014; Silva-Sanzana et al., 2019). Genetic analysis also suggested that total pectin methylesterase (PME) activity alters cell wall strengthening and represses tomato fruit softening (Tieman and Handa, 1994). Inhibition of PME ubiquitously 1 (PMEU1), a PME family gene, reduces the total PME activity and accelerates tomato fruit softening (Phan et al., 2007). In addition, pectin degradation by other pectin metabolic enzymes such as pectate lyase (PL) and polygalacturonase (PG) also contribute to targeted control of fruit softening (Uluisik et al., 2016; Wang et al., 2019).
Regulation of pectin metabolic enzymes to control fruit softening by phytohormones, especially ethylene, has been reported. Upstream transcriptional factors (TFs), such as RIPENING INHIBITOR (RIN) and NON-RIPENING (NOR), modulate fruit softening in both ethylene-dependent and ethylene-independent manners (Klee and Giovannoni, 2011; Osorio et al., 2020); but they negatively affect visual, flavor, and nutritional quality of tomato fruits. Therefore, it is necessary to search for other factors including upstream TFs that can precisely control fruit softening without negative effects on other quality traits. Brassinosteroids (BRs) are well-characterized phytohormones that are critical for plant vegetative growth, development, and response to environmental stimulus, influencing many important agronomic traits (He et al., 2005; Yu et al., 2011; Nolan et al., 2020). Exogenous BR application enhanced lycopene accumulation in tomato fruit (Liu et al., 2014). BRs were also found to be actively synthesized and accumulated during tomato fruit ripening (Li et al., 2016), indicating their potential role in tomato fruit ripening. As ethylene is regarded as the master regulator of climacteric tomato fruit ripening, BRs are found responsible for various aspects of fruit ripening, including improvement of carotenoid accumulation and visual quality (Liu et al., 2014), as well as the increase in soluble sugar content during tomato fruit ripening (Li et al., 2016), in either ethylene-dependent or ethylene-independent manner. Genetic and molecular biology studies in Arabidopsis have revealed the core components of the BRs signaling pathway, which constitutes a complete signal pathway from cell surface BR receptor to downstream transcription factors that regulate the expression of BR response genes (Nolan et al., 2020). BRI1-EMS-SUPPRESSOR1 (BES1), a key basic helix-loop-helix TF in the BR signaling pathway, balances plant growth and environment stress tolerance via binding a conserved E-box (CANNTG) and BRRE-box (CGTGC/TG) elements of its target genes (Jiang et al., 2019). Although the mechanism of BES1, acting as a transcription factor in regulating plant growth and development, has been well elucidated in Arabidopsis, its function in fruit softening has remained elusive.
In the present study, the specific role of SlBES1 on softening was investigated in tomatoes. SlBES1-overexpressing lines exhibited enhanced fruit softening while SlBES1-silencing lines showed improved fruit firmness, indicating its positive role in softening. PMEU1, a gene from PME family, which inhibited fruit softening, was downregulated in fruits of SlBES1 RNAi lines. Further genetic analyses along with molecular biology assays (ChIP-qPCR, EMSA, and luciferase [LUC] reporter) reveal that SlBES1 directly binds to the E-box in the promoter of PMEU1 to inhibit its expression, leading to fruit softening. Interestingly, we found knockdown or knockout of SlBES1 in tomato extended shelf life without detrimental effects on visual and nutrition quality, implying SlBES1 will be potential in tomato breeding.
Results and discussion
Functional identification of SlBES1 in tomato
To elucidate the function of BES1 in fruit softening, we identified SlBES1, the homologous gene of AtBES1 in tomato (Figures S1A and S1B), and analyzed its expression pattern in BR-deficient and BR-insensitive tomato mutants. The expression level of SlBES1 was decreased in both BR-deficient mutant dx and BR-insensitive mutant cu-3 that harbors a mutation in the BR receptor gene BRI1 (Figure S1C). To explore the function of SlBES1 in tomato, we then generated SlBES1-overexpressing transgenic lines, SlBES1-OX-3 and SlBES1-OX-8, as well as SlBES1 RNAi lines, SlBES1-RNAi-8 and SlBES1-RNAi-9 (Figure 1A). In Arabidopsis, the gain-of-function mutant of AtBES1 (bes1-D) has constitutive BR responses including tolerance to BR biosynthesis inhibitors, propiconazole (Pcz) or brassinazole (BRZ), in hypocotyl elongation assays in the dark (Yin et al., 2002; Hartwig et al., 2012). The hypocotyl elongation of SlBES1-OX-3 and SlBES1-OX-8 was not affected by 0.5 μM Pcz in the dark, whereas that of SlBES1-RNAi-8 and SlBES1-RNAi-9 was more susceptible to inhibition (Figures S1D and S1E), which was in agreement with the phenotypes of Arabidopsis BES1 mutants. In addition, BES1/BZR1 repressed the transcription of BR biosynthetic genes, such as DWARF, CPD, and DWARF4, through feedback regulation (He et al., 2005; Yu et al., 2011). In the present study, expression levels of the tomato BR biosynthetic genes (SlDWARF, SlCPD, SlCYP724B2, and SlCYP90B3) were significantly downregulated in SlBES1-OX-3 and SlBES1-OX-8 but upregulated in SlBES1-RNAi-8 and SlBES1-RNAi-9 fruits (Figure S1F). These results indicate that SlBES1 confers conserved function as a transcription factor in BR signaling pathway between tomato and Arabidopsis.
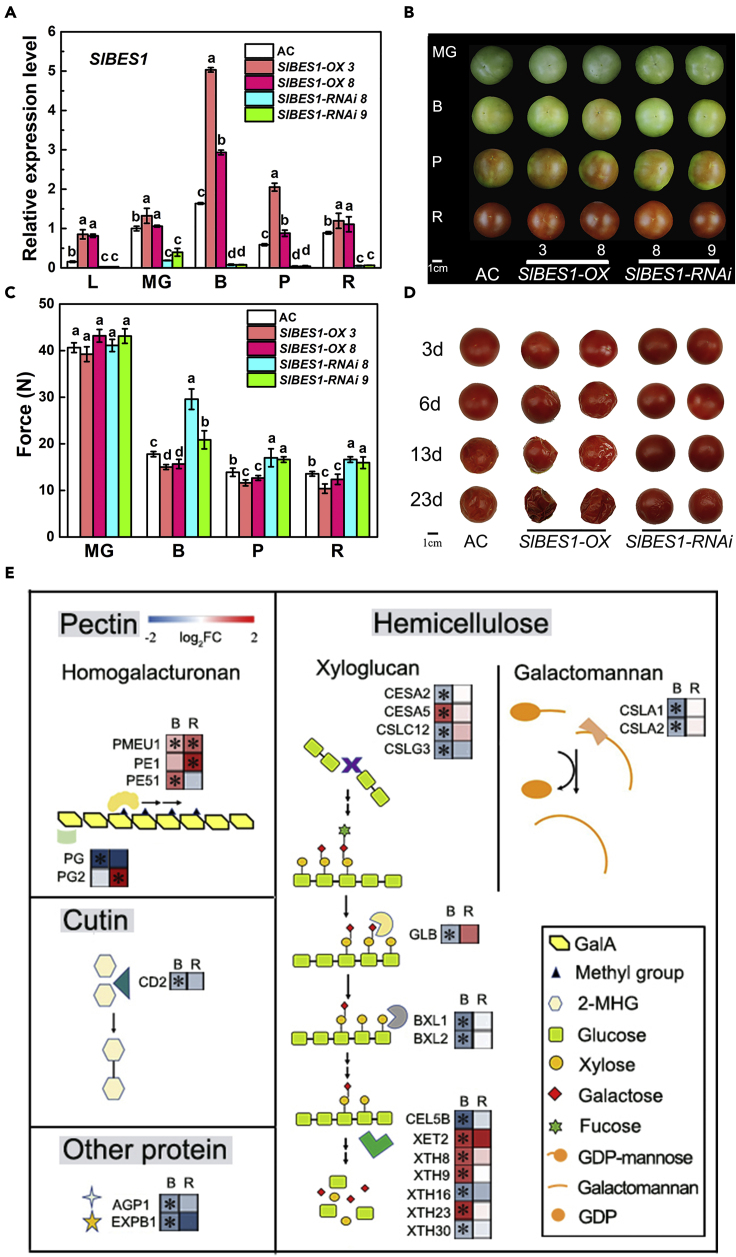
Figure 1.
Phenotypes of SlBES1-OX and SlBES1-RNAi tomato fruits
(A) Gene expression levels of SlBES1 in leaves and fruits of SlBES1-OX and SlBES1-RNAi plants. L, leaves. MG, mature green stage. B, breaker stage. P, pink stage. R, red ripe stage. Data shown represent the means ± SD of three biological replicates. Different letters indicate significant difference compared to AC (wild type) (one-way ANOVA with Tukey's test, p < 0.05).
(B) Phenotypes of AC (wild type), SlBES1-OX, and SlBES1-RNAi tomato fruits. Pictures were taken at four developmental stages of fruit. MG, mature green; B, breaker; P, pink; R, red ripe. Scale bar, 1 cm.
(C) Fruit firmness of AC (wild type), SlBES1-OX, and SlBES1-RNAi fruits at different development stages. MG, mature green. B, breaker. P, pink. R, red ripe. Data shown represent the means ± SD of twenty-four biological replicates from at least six independent fruits. Different letters indicate significant difference compared to AC (wild type) (one-way ANOVA with Tukey's test, p < 0.05).
(D) The shelf life of AC (wild type), SlBES1-OX, and SlBES1-RNAi fruits. Fruits were harvested at red ripe stage and stored at room temperature. The progression of fruit deterioration was recorded by time-lapse photography. Time after harvest is specified by days. The storage condition was 25°C and 35% relative humidity. Scale bar, 1 cm.
(E) RNA-seq results and related metabolic pathway of SlBES1-RNAi fruits at B and R stages. A heatmap showing the expression pattern of firmness-related genes in SlBES1-RNAi fruits and wild type. The values of fold change (log2) with or without significance (p < 0.05) were represented as colored blocks with or without asterisk, respectively. Each group contains three biological replicates. B, breaker stage. R, red ripe stage. Accession number, name, description, and the value of fold change of genes in the figure are listed in Table S1.
SlBES1 promotes tomato fruit softening without affecting nutritional quality
SlBES1 transgenic fruits were then used to investigate the role of BES1 in fruit development. As shown in Figure 1B, no significant difference in fruit appearance at different developmental stages was found between wild-type Ailsa Craig (AC) and transgenic tomato lines. However, SlBES1-OX lines and SlBES1-RNAi lines exhibited reduced and increased fruit firmness compared with AC, respectively (Figure 1C). The varying degrees of firmness correspond to the SlBES1 expression levels in different transgenic lines (Figure 1A), which demonstrated that SlBES1 negatively regulates tomato fruit firmness. Ethylene production in fruits of SlBES1 transgenic lines and wild type (AC) was similar at each development stage (Figure S2A), indicating that SlBES1-mediated fruit softening is via ethylene-independent manner and might be different with the action of formerly reported ripening-related TFs, RIN, and NOR (Osorio et al., 2020). Meanwhile, SlBES1 overexpressing or silencing (RNAi) did not affect nutritional qualities (Table S1), and no significant difference was observed in expression levels of genes related to carotenoid and ascorbic acid biosynthesis between SlBES1-RNAi lines and wild type (Table S2). The fruit weight and fruit number per plant were also not affected in SlBES1-OX or SlBES1-RNAi fruits. Softening due to reduction in firmness is usually essential in determining shelf life, representing postharvest fruit integrity. We found that SlBES1-OX fruits have a shorter shelf life, while SlBES1-RNAi fruits have a longer shelf life when compared with the wild type (Figure 1D). The SlBES1-OX lines have a similar phenotype as the loss of integrity in fruits with inhibited PME activity (Tieman and Handa, 1994). Furthermore, treatment of wild-type fruits with 3 μM 24-epibrassinolide (EBL), a bioactive BR, significantly promoted fruit softening (Figure S2B). Meanwhile, EBL treatment recovered fruit softening in the BR-deficient mutant dim (Figure S2C), indicating a positive role of BR in fruit softening.
To identify softening-related genes or cell wall metabolism processes that might participate in SlBES1-induced fruit softening, SlBES1-RNAi fruits were harvested at the breaker (B) and red ripe (R) stages for RNA-seq. Gene ontology (GO) analysis on RNA-seq data (Table S3) in the fruit development period category (p < 0.05) showed that the expression of 24 genes related to the metabolism of cell wall components, including genes involved in pectin metabolism (PMEs and PGs), xyloglucan metabolism (XTHs and CESAs), galactomannan metabolism (CSLAs), and cutin metabolism (CD), as well as other genes encoding cell wall-related proteins, was differentially regulated by SlBES1 (Figure 1E, Table S3). These genes participate in cell wall strengthening through remodeling, degrading or polymerizing cell wall composition as shown in Figure 1E. Among them, pectin degradation gene PG or its homologous genes in different species, such as EXP, CD, and XTHs, have been proved to affect softening-related texture change in fleshy fruit (Seymour et al., 2013; Wang et al., 2019), and the expression levels of XTHs were reported to be regulated by BR during hypocotyl elongation (Nolan et al., 2020). Although these genes have a relationship with softening or might be regulated by BR, the expression levels of most genes were not always influenced by SlBES1 at different stages of fruit development. The expression pattern of PG2 was oppositely regulated at different stages, whereas that of cutin, xyloglucan, and galactomannan metabolism genes, as well as genes encoding other cell wall-related proteins, such as CD, CESAs, XTHs, and EXPB1, was significantly altered at only one stage. However, the expression levels of PME genes, such as PMEU1 and PE1, were conspicuously upregulated at both stages of SlBES1-RNAi fruits. More than fifty PME family genes from tomato were identified with different gene expression patterns in various tissues by bioinformatics analysis (Wen et al., 2020). PMEU1 and PE1 have been initially reported as PME family genes in tomato fruits (Phan et al., 2007; Wen et al., 2013). PMEU1 was reported to negatively regulate tomato fruit softening by promoting cell wall strengthening (Phan et al., 2007), but PE1 did not function in cell wall strengthening of fruits (Wen et al., 2013). It was interesting that PMEU1 was the only one whose expression level was significantly increased at both B and R stages of SlBES1-RNAi fruits. This evidence suggested that PMEU1 and related pectin metabolic pathway might be involved in SlBES1-mediated softening.
Taken together, all the above results suggested that BR signaling pathway component SlBES1 positively modulates fruit softening, possibly through regulation of PMEU1-associated pectin metabolic pathways.
SlBES1 inhibited PMEU1-associated pectin metabolic pathway
Previous studies have established a negative role of PME in de-methylesterification to modulate fruit softening and maintain fruit integrity (Tieman and Handa, 1994; Phan et al., 2007). Our RNA-seq results indicated that PMEU1 and PE1 were upregulated in firmer SlBES1-RNAi fruits (Figure 1E, Table S3). As discussed above, SlBES1-OX fruits phenocopy the fruit with repressed PME activity. Based on these results, we hypothesize that SlBES1-mediated fruit softening might be caused by regulation of the PMEU1-associated pectin metabolic pathway.
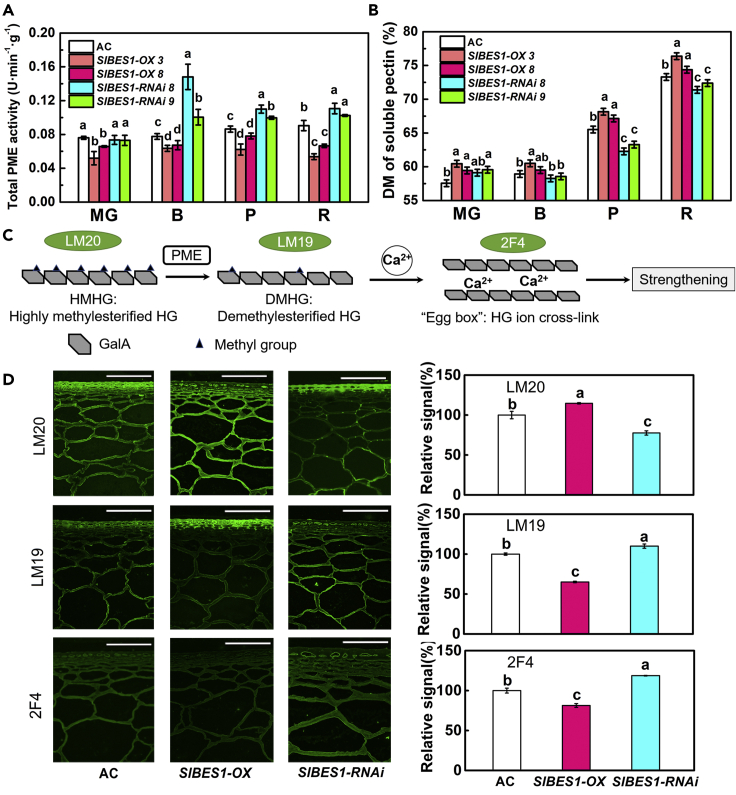
To test this hypothesis, we measured the change in the pectin metabolic pathway in SlBES1 transgenic fruits. The total PME activity was significantly reduced in SlBES1-OX-3 and SlBES1-OX-8 fruits, while increased in SlBES1-RNAi-8 and SlBES1-RNAi-9 fruits at both mature green (MG) and red ripe (R) stage when compared with the wild type (Figure 2A). PME catalyzes the de-methylesterification of pectin and then attenuates the degree of pectin methylesterification (DM) (Senechal et al., 2014; Figure 2C). With the decrease in total PME activity, the DM of soluble pectin was increased in SlBES1-OX-3 and SlBES1-OX-8 fruits, while decreased in SlBES1-RNAi-8 and SlBES1-RNAi-9 fruits at both B and R stage when compared with the wild type (Figure 2B). Additionally, the content and location of highly methylesterified HG, de-esterified HG, and “egg box” were detected through the immunofluorescence of LM20, LM19, and 2F4, respectively (Figures 2C and 2D).The results showed that the signal of methylesterified HG was stronger in SlBES1-OX and weaker in SlBES1-RNAi fruits (Figure 2D), whereas the signal of de-esterified HG was weaker in SlBES1-OX and stronger in SlBES1-RNAi fruits in comparison with the wild type (Figure 2D), consistent with the total PME activity and DM in the related SlBES1 transgenic fruits. The “egg box” signal was significantly elevated in SlBES1-RNAi and lowered in SlBES1-OX fruits when compared with that in wild type fruits (Figure 2D). Given the positive role of the “egg box” in cell wall strengthening (Senechal et al., 2014; Silva-Sanzana et al., 2019), the immunolocalization results suggested that the decreased content of the “egg box” structure in SlBES1 fruits was the direct reason for SlBES1-induced softening. Calcofluor-white staining showed no major differences in cell size or patterning between transgenic lines (Figure S3A). The reduced activity of PME and decreased content of its products, as revealed by DM and immunolocalization experiments, support our hypothesis that PME-mediated pectin de-methylesterification was repressed by SlBES1.
Figure 2.
SlBES1 represses the pectin de-methylesterification
(A and B) Total pectin methylesterase (PME) activity (A) and degree of methylesterification (DM) of soluble pectin (B) in SlBES1-OX and SlBES1-RNAi fruits at different development stages. MG, mature green stage. B, breaker stage. P, pink stage. R, red ripe stage.
(C) Schematic representation of the relationship between pectin methylesterase (PME) and cell wall strengthening.
(D) Immunolocalization of highly methylesterified HG (homogalacturonan), de-esterified HG, and Ca2+ cross-linked HG in SlBES1-OX and SlBES1-RNAi fruits at red ripe stage. Monoclonal LM20 antibody probe recognizing highly methylesterified HG, LM19 probe recognizing de-esterified pectin, and 2F4 probe recognizing Ca2+ cross-linked HG were used to label tomato pericarp tissue. Representative sections of fruits from each of AC (wild type), SlBES1-OX-3, and SlBES1-RNAi-8 lines are presented. Scale bar represents 100 μm. The right graphs show the relative fluorescence signal of each antibody. Relative signals are calculated through software ImageJ. Values were normalized with respect to AC (wild type).
In (A and B) and (D), each data point represents means ± SD of three determinations. Different letters indicate significant difference between different groups (one-way ANOVA with Tukey's test, p < 0.05).
The expression pattern of the PME gene during the fruit softening process was further investigated. The expression level of PMEU1 was significantly decreased in SlBES1-OX and increased in SlBES1-RNAi fruits when compared with the wild type at both the B, P, and R stages (Figure 3A), which is consistent with the change of total PME activity and DM (Figures 2A and 2B). Interestingly, the expression level of PMEU1 was also downregulated in EBL-treated fruits, while EBL treatments offset the downregulated PMEU1 expression in BR-deficient mutant dim (Figures S3B and S3C), suggesting a role of BR in repressing PMEU1 expression. These results indicated that both PMEU1-associated pectin de-methylesterification and the expression level of PMEU1 were inhibited by SlBES1. Our data suggest that SlBES1 might repress the expression of PMEU1 to promote fruit softening.
Figure 3.
SlBES1 represses the transcriptional expression of PMEU1
(A) Relative expression levels of PMEU1 in SlBES1-OX and SlBES1-RNAi fruits at different development stages. MG, mature green stage. B, breaker stage. P, pink stage. R, red ripe stage. Each data point represents means ± SD of three determinations. Different letters indicate significant difference between different groups (one-way ANOVA with Tukey's test, p < 0.05).
(B) ChIP-qPCR assays showing that SlBES1 was associated with the locus of candidate genes, LoxA, PMEU1, and PE1. Chromatin of transgenic plants expressing 35Spro:SlBES1-myc (SlBES1-myc) was immunoprecipitated with anti-myc antibody, and the result of ACTIN7 served as control. The relative enrichment for ChIP signal was displayed as the percentage of total input DNA. Values are means ± SD of three biological replicates. Statistical analysis was performed with ANOVA. Bars with asterisks indicate significant difference (∗∗p < 0.01).
(C) ChIP-qPCR assay to detect the association between SlBES1 and the PMEU1 promoter. The upper graph, schematic diagram of PMEU1, indicating the amplicons used for ChIP-qPCR. Positions of E-box and BRRE-box (BR response element) are indicated. The below graph, ChIP-qPCR assays, shows that SlBES1 associated with PMEU1 locus in tomato fruits. The relative enrichment for SlBES1 at two PMEU1 promoter motifs, E-box and BRRE-box, was calculated against the ACTIN2 promoter. AC (wild type) without and with antibody were set as blank control and negative control, respectively. SlBES1-myc without antibody was also set as negative control. The mean value of two technical replicates was recorded for each biological replicate. Values are means ± SD of three biological replicates. Different letters indicate significant difference among groups for each locus (one-way ANOVA with Tukey's test, p < 0.05).
(D) Transient expression assays showing that SlBES1 represses PMEU1 expression. Representative images of N. benthamiana leaves were taken 48 hr after infiltration. The bottom panel indicates the infiltrated constructs.
(E and F) Luminescence intensity (E) and SlBES1 expression level (F) under different treatments as indicated in (D). Values are means ± SD of six biological replicates. Different letters indicate significant difference among groups for each locus (one-way ANOVA with Tukey's test, p < 0.05).
(G) Motif sequence of labeled, unlabeled competitor, and mutant competitor probes. Mutant competitor in which the 5′-CACTTG-3′ motif was replaced with 5′-AAAAAA-3′.
(H) DNA electrophoretic mobility shift assay (EMSA) showing that the binding of SlBES1-His to the E-box of PMEU1 promoter in vitro. FAM-labeled probes were incubated with SlBES1-His and the free and bound DNAs were separated on an acrylamide gel.
SlBES1-mediated direct transcriptional inhibition of PMEU1 confers to fruit softening
To test the hypothesis of possible inhibited expression of PMEU1 by SlBES1, we carried out further tests to prove the transcriptional regulation of SlBES1 on the expression of PMEU1. Chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) results showed a significantly higher enrichment for the promoters of PMEU1 than for PE1 (Figure 3B), consistent with a stronger effect of SlBES1 on the transcriptional level of PMEU1 than that of PE1 (Figure 1E, Table S3). The two binding sites, E-box and BRRE-box, were located at −465 bp and −289 bp of the PMEU1 promoter as predicted by the JASPAR database, respectively (http://jaspar.genereg.net/) (Figure 3C). The ChIP-qPCR results further indicated that enrichment of SlBES1 bound to the region containing E-box instead of the BRRE-box (Figure 3C), suggesting that SlBES1 might regulate PMEU1 expression by binding to the E-box.
To test if E-box is necessary for regulation of PMEU1 expression by SlBES1, we generated a PMEU1pro:LUC reporter, in which LUC was fused with the PMEU1 promoter. Co-expression of PMEU1pro:LUC with the 35Spro:SlBES1 construct led to a significantly reduced luminescence intensity, suggesting that SlBES1 represses the expression of PMEU1. In addition, when PMEU1pro:LUC was replaced by PMEU1proΔEbox:LUC, in which the E-box of the PMEU1 promoter was deleted, SlBES1-mediated repression of the PMEU1 expression was abolished, further verifying that E-box rather than BRRE-box was involved in transcriptional regulation of SlBES1 on PMEU1 (Figures 3D–3F).
We then conducted an electrophoretic mobility shift assay with purified SlBES1-His and a 20-bp DNA probe containing the E-box motif. SlBES1-His bound to the DNA probe, and this binding was successfully outcompeted by the unlabeled DNA probe but not by the DNA probe without E-box. These results suggested that SlBES1 directly binds to the PMEU1 promoter through E-box (Figures 3G and 3H). E-box was the binding site of SlBES1 to repress gene expression in tomatoes. BES1/BZR1 usually binds to E-box to activate gene expression and binds to BRRE to repress gene expression (Nolan et al., 2020). There are exceptions; a recent study also showed that OsBZR1 bound to E-box to inhibit downstream gene expression in rice (Fang et al., 2020).
Silencing PMEU1 caused softer fruits with lower total PME activity and exhibited a complete loss of fruit integrity and shorter shelf life (Figures 4A–D), which was in accordance with previous reports (Tieman and Handa, 1994; Phan et al., 2007). More importantly, knocking down PMEU1 in the SlBES1-RNAi background (TRV-PMEU1/SlBES1-RNAi) suppressed the SlBES1-RNAi phenotypes of higher firmness and the longer shelf life (Figures 4B and 4C). Fruit firmness, shelf life, and total PME activity of TRV-PMEU1/SlBES1-RNAi were restored to almost the same as wild type, suggesting that SlBES1 might modulate fruit softening by inhibition of PMEU1 (Figures 4B–4D). Taken together, these results demonstrated that SlBES1 directly binds to the E-box of PMEU1 to repress its expression, thereby promoting fruit softening.
Figure 4.
Effects of PMEU1 silencing in SlBES1-RNAi on tomato firmness and shelf life
(A) Relative expression levels of SlBES1 and PMEU1 in fruits of AC (control), virus-induced PMEU1 silencing lines (TRV-PMEU1), SlBES1-RNAi, and virus-induced PMEU1 silencing in SlBES1-RNAi (TRV-PMEU1/SlBES1-RNAi) at red ripe stage.
(B) Fruit firmness of control, TRV-PMEU1, SlBES1-RNAi, and TRV-PMEU1/SlBES1-RNAi at red ripe stage. Values are means ± SD of twenty-four biological replicates.
(C) The shelf life of control, TRV-PMEU1, SlBES1-RNAi, and TRV-PMEU1/SlBES1-RNAi. Fruits were harvested at the red ripe stage and stored at room temperature. The progression of fruit deterioration was recorded by time-lapse photography. Time after harvest is specified by days. The storage condition was 25°C and 35% relative humidity.
(D) Total PME activity of control, TRV-PMEU1, SlBES1-RNAi, and TRV-PMEU1/SlBES1-RNAi fruits at the red ripe stage.
(A), (C), and (D) Values are means ± SD of three biological replicates. Different letters indicate significant difference among groups (one-way ANOVA, p < 0.05, Tukey's test).
(E) Schematic representation of relationship between transcriptional regulation of SlBES1 on PMEU1 and fruit firmness.
When BRs are absent as shown, receptor BRI1 and co-receptor BAK1 are separated. Transcriptional factor SlBES1 with phosphorylated pattern could not inhibit PMEU1 expression. PMEU1 is formed and secreted into the cell wall. Then, PMEU1 de-esterified HMHG and DMHG; DMHG formed “Egg box” through cross-link with Ca2+. Egg box provides strengthening support to fruit firmness. When BRs are present, BRs bind to BRI1 and BAK1. A series of phosphorylation and dephosphorylation reactions are occurred, leading to the dephosphorylation of SlBES1. Dephosphorylated SlBES1 binds to the E-box of PMEU1 promoter and then represses the expression of PMEU1. Less PMEU1 is secreted into the cell wall, and pectin methylesterase activity is attenuated, so the contents of DMHG and Egg box are decreased which causing fruit softening.
BRI1, BRASSINOSTEROID INSENSITIVE 1; BAK1, BRI1-ASSOCIATED KINASE1; BES1, BRI1-EMS-SUPPRESSOR 1; PMEU1, PECTIN METHYLESTERASE UBIQUITOUSLY 1; HMHG, highly methylesterified HG; DMHG, demethylesterified HG.
Gene editing of SlBES1 by CRISPR-Cas9 enhances fruit firmness without negative effect on nutritional quality
The above results suggested that SlBES1 might be a potential target to breed tomatoes with longer shelf life and maintaining optimum flavor and nutrients. Then, we generated SlBES1-KO through CRISPR-Cas9-mediated genome editing the 5′-TAGTTGGTGATGAAAGAGGTGG-3′ in the second extron of SlBES1 antisense strand with pYLCRISPR/Cas9 (Ma et al., 2015). The independent SlBES1-KO mutant was obtained in AC background, which was the same background with SlBES1-OX and SlBES1-RNAi, with 1 bp deletion (Figure S4A). The expression level of SlBES1 was decreased by 80%, and the expression level of PMEU1 was significantly upregulated as a result of reduced mRNA level of SlBES1 in SlBES1-KO fruits at the red ripe stage (Figure S4B). As expected, the fruit of SlBES1-KO had higher firmness and longer shelf life (Figures S4C and S4D) with negligible effects on other agronomic or quality traits (Table S1), which is in accordance with the phenotypes of SlBES1-OX and SlBES1-RNAi (Figures 1C and 1D; Table S1). Pectin metabolic genes, such as PL or PG, have been used as genome editing targets to obtain fruits with longer shelf life (Uluisik et al., 2016; Wang et al., 2019). Genome editing of upstream TFs that regulate pectin metabolic enzymes might be an alternative way for extending shelf life. However, the reported upstream TFs, RIN or NOR, regulated pectin metabolic genes to provide firmer fruit, while inherited poor quality in the meantime (Osorio et al., 2020). Our results suggest that SlBES1, as an upstream TF that directly regulates PMEU1 and associated pectin de-methylesterification, could be a powerful target to be explored to extend shelf life without defect in other quality traits to tomato fruits (Figure S4D). A previous study suggested that CRISPR-editing of a hormone biosynthetic gene generated firmer fruit without unfavorable changes in quality (Li et al., 2020). Considering that diverse phytohormone signaling components generally play essential functions via subtle control, such as transcriptional regulation on the target genes (Jiang et al., 2019), it might be potential to achieve improved agronomical traits of crops by genome editing of core signaling components of phytohormones.
In summary, this study provides genetic, biochemical, and molecular biology evidence for SlBES1-mediated fruit softening in an ethylene-independent manner by modulation of the pectin metabolic pathway. At the transcriptional level, SlBES1 directly binds to the E-box of the PMEU1 promoter to repress PMEU1 expression and pectin de-methylesterification, thereby promoting fruit softening (Figure 4E). Fruits with silenced SlBES1 described in this study provide a promising solution to improve fruit firmness and extended shelf life without negative effects on visual and nutritional quality.
Limitations of the study
In this study, we find that SlBES1 represses PMEU1 expression and pectin de-methylesterification to promote fruit softening. Although this study provides a regulation mechanism of SlBES1 on firmness under BR, the relationship between softening and other important homologous TFs, BZR1 or BEHs, in BR signaling remains unknown.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LM19 | Plantprobes | Cat# LM19;RRID:AB_2734788 |
| LM20 | Plantprobes | Cat# LM20; RRID:AB_2734789 |
| 2F4 | Plantprobes | Cat# 2F4 |
| Alexa Fluor 488 goat anti-rabbit | Jackson ImmunoResearch | Cat# 111-545-003;RRID:AB_2338046 |
| Bacterial and virus strains | ||
| E.coli strain DH5α | N/A | N/A |
| A. tumefaciens GV3101 | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 24-epibrassinolide | Sigma-Aldrich | CAS#78821-43-9 |
| Propiconazole | Sigma-Aldrich | CAS#60207-90-1 |
| Ni-His Resin | Thermo Fisher | 88221 |
| pET28a-SlBES1-His | This paper | N/A |
| Critical commercial assays | ||
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| Gateway LR Clonase II Enzyme mix | Thermo Fisher | Cat. 11791020 |
| Deposited data | ||
| RNA-seq data | This paper | PRJNA635540 |
| Experimental models: Organisms/strains | ||
| Tomato: SlBES1-OX 3 | This paper | N/A |
| Tomato: SlBES1-OX 8 | This paper | N/A |
| Tomato: SlBES1-RNAi 8 | This paper | N/A |
| Tomato: SlBES1-RNAi 9 | This paper | N/A |
| Tomato: SlBES1-KO | This paper | N/A |
| Oligonucleotides | ||
| More than 10, see Table S4 | ||
| Recombinant DNA | ||
| Plasmid: Pro35S:SlBES1-myc | This paper | N/A |
| Plasmid: pHANNIBAL-SlBES1i | This paper | N/A |
| Plasmid: pYLCRISPR/Cas9- SlBES1 | This paper | N/A |
| Plasmid: pGWB35-PMEU1pro:LUC | This paper | N/A |
| Plasmid: pGWB35-PMEU1proΔE-box:LUC | This paper | N/A |
| Plasmid: SlBES1-1300 | This paper | N/A |
| Plasmid: pTRV2-PMEU1 | This paper | N/A |
| Software and algorithms | ||
| ImageJ | National Institutes of Health, USA | https://imagej.nih.gov/ij/ |
| MEGA X | N/A | https://www.megas |
| SPSS 19 | N/A | https://www.ibm.com/cn-zh/analytics/spss-statistics-software |
| Indigo software | Berthold Technologies | Combined with equipment |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Qiaomei Wang (qmwang@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Plant materials and growth conditions
All transgenic lines were constructed in tomato cultivars Ailsa Craig (AC). Cultivars Lycopersicon pimpinellifolium (Spim), Condine Red (CR), and Craigella are the parental line of cu-3, dim, and dx (Bishop et al., 1996; Scheer et al., 2003; Li et al., 2015), respectively. Plants were cultivated under a 16 h photoperiod (22/28°C, night/day). The number of tagged flowers at anthesis was limited to fewer than five per cluster. The fruit ripening stages, including mature green stage (MG), breaker stage (B), pink stage (P), and red ripe stage (R), were defined based on fruit color as described previously (Giovannoni, 2004). After firmness determination and immunofluorescence, three biological replicates for transgenic fruit pieces (each biological replicate consisting of at least three fruits) or fully expanded leaves from 4-week-old BR mutants were frozen with liquid nitrogen as samples, and then stored at -80°C for further tests.
Accession numbers
The accession numbers for the genes described in this report are as follows: SlBES1 (Solyc04g079980), PMEU1 (Solyc03g123630), PE1 (Solyc07g064170), SlDWARF (Solyc02g089160), SlCPD (Solyc06g051750), SlCYP724B2 (Solyc07g056160), SlCYP90B3 (Solyc02g085360), LOXA (Solyc08g014000), ACTIN7 (Solyc03g078400), ACTIN2 (Solyc11g005330).
Method details
Vector constructs and plant transformation
Vector constructs for transgenic were generated following standard molecular biology protocols. For SlBES1-OX plants, the full-length coding sequence of SlBES1 without termination codon was amplified via PCR and inserted into the pGWB17 vector using Gateway (Invitrogen) technology (Nakagawa et al., 2007) to get the Pro35S:SlBES1-myc construct. For SlBES1-RNAi construct, a fragment of SlBES1 with length of 305 bp was amplified and then inserted into intermediate vector pHANNIBAL in the positive orientation, thereby generating vector pHANNIBAL-SlBES1. The same fragment of SlBES1 was inserted into pHANNIBAL-SlBES1 in the reverse orientation, generating the vector pHANNIBAL-SlBES1i. The target inverted repeat sequences were obtained by Sac I and Spe I digestion of pHANNIBAL-SlBES1i. Eventually, these sequences were inserted into pBIN19 under the control of CaMV 35S promoter to generate pBIN19-SlBES1-RNAi.
The above constructs were introduced into tomato cultivars AC via Agrobacterium tumefaciens LBA4404-mediated transformation (Liu et al., 2019). For SlBES1-OX lines, thirteen T1 lines were obtained according to their resistance to Kanamycin on the screening for regenerated shoots. Among them, six lines showed an increased expression level of SlBES1 in leaves. Two lines, SlBES1-OX 3 and SlBES1-OX 8, showed the significantly enhanced expression level of SlBES1 in leaves and fruits. Homozygous T2 or T3 transgenic plants from these two lines were used for further researches. For SlBES1-RNAi lines, six T1 lines were selected according to their resistance to Kanamycin. Two of them, SlBES1-RNAi 8 and SlBES1-RNAi 9, have a decreased expression level of SlBES1 in fruits and homozygous T2 or T3 transgenic plants from these two lines were used for further researches.
For fruits of overexpressing lines, the expression level of SlBES1 in SlBES1-OX 3 was increased by 32%, 208%, 250%, and 35% at MG, B, P, and R stage, respectively. The expression level of SlBES1 in SlBES1-OX 8 was increased by 6%, 79%, 50%, and 25% at MG, B, P, and R stage, respectively (Figure 1A). Meanwhile, for fruits of silencing lines, the expression level of SlBES1 in SlBES1-RNAi 8 was down-regulated by 81%, 95%, 94%, and 94% at MG, B, P, and R stage, respectively. The expression level of SlBES1 in SlBES1-RNAi 9 was reduced by 60%, 95%, 93%, and 93%, at MG, B, P, and R stage, respectively (Figure 1A).
Generation of the CRISPR/Cas9 mutant
The guide RNA sequence CCACCTCTTTCATCACCAACTA for SlBES1 was cloned into pYLCRISPR/Cas9 (Ma et al., 2015), and the base C at 556bp was deleted. After inducing construct into AC by Agrobacterium-mediated transformation, sequences containing guide RNA target sites were amplified and sequenced to confirm mutations in targeted regions. Homozygous T2 transgenic plants were selected for further experiments.
Chemical treatment
AC and dim at mature green stage were treated with 24-epibrassinolide (EBL, Sigma, St. Louis, MO). Lanolin with or without 3 μM EBL were used to cover fruits totally and treated fruits were put in a phytotron (Qiushi environment company, Hangzhou, China) with 16 h photoperiod at 24 °C and 80 % relative humidity (Liu et al., 2014) and then collected at 1st, 3rd, 6th, and 9th day for further tests. Tomato seeds were sown in half-strength MS containing 0.5 μM propiconazole (Pcz, Sigma, St. Louis, MO). Plants were grown at 28 °C with total darkness for 6 d, then the lengths of seedlings hypocotyls and roots were measured through software ImageJ after photographed.
Firmness determination
Firmness was tested at the fruit equatorial region with a texture analyzer (TA-XT2i, Godalming, UK) and a 7.5mm probe with 1mms−1 penetration speed (Liu et al., 2018).
RNA extraction and relative quantitative PCR
For RNA extraction, 0.1 g of leaves or fruits was mixed with 1 mL RNAiso plus according to manufacturer’s instruction (Takara, Kusatsu, Japan), then RNA was reverse-transcribed into cDNA using PrimeScript RT reagent with gDNA Eraser (Takara, Kusatsu, Japan). TB Green (Takara, Kusatsu, Japan) was then used in Step One Real-Time PCR System (Applied biosystem, CA, USA) for relative quantitative PCR (qPCR). The gene-specific primers used were listed in Table S4.
RNA-Seq
AC and SlBES1-RNAi-8 fruits at breaker and red ripe stages were collected for total RNA extraction and Illumina MiSeq library was constructed as described by manufacturer’s instructions (Illumina, San Diego, CA, USA) and then sequenced with the Illumina Miseq platform.
ChIP-qPCR
Chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) for fruits was performed following previous reports (Liu et al., 2019). In brief, 3 g of 0.5 cm2 fruit pieces of SlBES1-OE-3 at mature green stage were collected and cross-linked using 1 % (v/v) formaldehyde under vacuum for 10 min and ground to powder in liquid nitrogen. Then, the chromatin complexes were isolated, sonicated with Biorupter plus (Diagenode, Belgium, Antwerp), and immunoprecipitated with 5μg Mouse monoclonal anti-c-myc antibody (clone 9E10, IgG, Roche, Basel, Switzerland). About 200∼300 bp of ChIP DNA and input DNA was recovered and dissolved in water for further ChIP-qPCR analysis. For ChIP-qPCR, primer pairs were used to analyze the ChIP DNA (Table S4). Each ChIP value was normalized to its respective input DNA value. The fold enrichment on each candidate gene or PMEU1 promoter was calculated against the ACTIN7 or ACTIN2, respectively. The mean value of two technical replicates was recorded for each biological replicate. The values of three independent biological replicates were collected and error bars represented the standard error from these tests.
Transient expression assay in tobacco leaves
Transient expression assay was performed in tobacco (Nicotiana benthamiana) (Liu et al., 2019). The PMEU1 promoter was cloned with KOD FX (Toyobo, Japan, Osaka) and inserted into PQB vector using DNA Ligation Kit (Takara, Kusatsu, Japan). Then the promoter was fused with the LUC reporter gene into the plant binary vector pGWB35 using Gateway cloning kit (Thermo, Boston, USA) to generate the PMEU1pro:LUC reporter construct. SlBES1-1300 was used as an effector construct. These constructs were transformed into Agrobacterium cell GV3101 with P19 protein and pSoup vector, and then the cells were incubated, harvested, and re-suspended in infiltration buffer (10 mM MES, 40 mM AS, and 10 mM MgCl2) to a final concentration of OD600= 1.0. Equal volumes of transformed cells with different combinations were mixed and then co-infiltrated into tobacco leaves with a needleless syringe. Infiltrated plants were placed at 28 °C for 48 h before imaging. NightOWL II LB983 Ultrasens backlit (Berthold Technologies, Bad Wildbad, Germany) with Indigo software was used to capture LUC expression image and to quantify LUC luminescence intensity. 100 mM luciferin was sprayed on leaves and incubated in dark for 5 min before detection. Six independent determinations were performed.
EMSA
The full-length coding region of SlBES1 was amplified and then inserted into pET28a vector. The recombinant protein, SlBES1-His were expressed in E.coli Rosetta cells at 28°C and purified to homogeneity with Ni-His Resin (Thermo, Boston, USA) after identified by SDS-PAGE. Oligonucleotide probes were synthesized and labeled with FAM at 3’ ends. EMSAs (electrophoretic mobility shift assay) was performed as previously described (Liu et al., 2019) with minor modification. In brief, the FAM-labeled binding box probe, probe without any label (competitor), and mutant probe (mutant competitor) were incubated with SlBES1-His proteins at room temperature for 20 min, respectively. Bound and free probes were separated via PAGE. Typhon FLA7000 (GE, Fairfield, USA) was used to capture the final image. Probes were listed in Table S4.
Determination of PME enzyme activity and degree of pectin methylesterification
Total PME activity and degree of pectin methylesterifcation (DM) were determined as previously reported (Kyomugasho et al., 2015). Alcohol-insoluble substances (AIS) from cell wall was first obtained and then soluble and insoluble pectin were extracted from AIS. 4 g of fresh tomato pericarp at different fruit stage was boiled in 25 mL 95 % ethanol for 20 min and then mixed for 60 s. The precipitate was left after centrifuged at 1500 g for 10 min. Then the precipitate was mixed with 25 mL 80 % ethanol and the mixture was centrifuged at 1500 g for 20 min to obtain precipitates. This step was repeated three times until the supernatant showed colorless. The crude cell wall pellet was dried under air steam and then suspended in DMSO: water (9:1, v/v, 20 mL/g). After stirring in room temperature for 24 h, the slurry was centrifuged at 1500 g for 20 min to remove DMSO. The pellet was washed repeatedly in 95 % ethanol and then was dried under air. The dried material was washed once with acetone. Then this cell wall extraction was freeze-dried and weighted. It was centrifuged at 10500 g for 25 min after suspending in sterilized distilled water for 1h and this step was repeated for once. Supernatant and precipitate in these two steps were collected, respectively. The supernatant was soluble pectin while the precipitate was insoluble pectin. The pH was adjusted to 6 with 0.1M NaOH to ensure total ion amount of carboxyl group.
For pectin methylesterase (PME, EC3.1.1.11) activity determination, titration with an automatic titration was used. Weighted crude protein from cell wall was assayed in determination solution, 2 % (w/v) citrus pectin (pH=7.0). The volume of 5 mM NaOH consumed for titration was recorded over 5 min. Total PME activity was calculated as the volume of consumed NaOH per min (U·min-1·g-1).
Immunofluorescence
Fresh tomato pericarp materials were fixed with FAA for at least 24h. Fixed tissue was dehydrated by serial incubations at 4°C in solution with an increasing concentration of ethanolfrom 10 % to 100 % and then submerged with LR White resin (Sigma-Aldrich, St. Louis, MO) to place into capsules. 1 μm sections were obtained from the embedding block and then placed on glass slides for immunofluorescence assays. LM19 and LM20 mouse monoclonal antibodies bind demethylated and hypermethylated HGs, respectively. Additionally, 2F4 mouse monoclonal antibodies are used to bind "egg box" epitopes formed of chain dimer of de-esterified HG linked with calcium ions (Silva-Sanzana et al., 2019). The non-specific binding sites were blocked by incubating the slide with the sample portion at room temperature for 30 min with 5% fat-free milk powder dissolved in 1 × PBS and washed once with 1×PBS. The primary antibody was diluted with blocking solution (5%, 1×PBS) at a ratio of 1: 5, and the solution was incubated for 90 min at room temperature. Then samples were washed three times with 1×PBS before secondary antibody incubation. The secondary antibody Alexa Fluor 488 goat anti-rabbit (Jackson ImmunoResearch, Pennsylvania, USA) was diluted with blocking solution (5%, 1×PBS) at a ratio of 1: 100 and incubated for 60 min at room temperature. Then 1× PBS solution was used to wash for three times. 0.25 mg/mL Calcofluor White (Sigma-Aldrich, St. Louis, MO) dissolved in 1×PBS was added for 5 min to fix the cell wall. This sample was washed with 1×PBS twice and anti-fluorescence decay quencher Citifluor (Agar Scientific, Stansted, UK) was added before placing the coverslip. The images were observed with a NIKON ECLIPSE Ci-L upright microscope and an objective lens CFI 10∗/22. The immunolabels of different treatments were done at least three times, and the most representative batch of processed images was selected for display. The relative signals of images were calculated through software ImageJ.
Virus-inducing gene silencing
A coding region fragment of PMEU1 (1-300bp) was inserted into pTRV2 to generate pTRV2-PMEU1. pTRV1 and pTRV2-PMEU1 were induced into AC fruits following the previous protocol (Fu et al., 2005). Agrobacterium GV3101 containing pTRV1 or pTRV2-PMEU1 were harvested and resuspended in the Agrobacterium infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, 150 mM acetosyringone) to a final OD 600 of 1.0. After shaking for 4-6 h at 28°C, the mixture of Agrobacterium GV3101 cultures containing pTRV1 and pTRV2-PMEU1 in a 1:1 ratio was syringe-infiltrated into the carpopodium of fruit at MG stage. Tomato fruit infiltrated with empty TRV alone was used as the control.
Carotenoid and ascorbic acid analysis
For carotenoid analysis (Liu et al., 2018), 0.4 g fruit powder was added with 30 ml extracting solution (hexane:acetone:ethanol=1:1:1 by volume) and mixed at 150 r min−1 for 30 min. After adding with 15 ml double distilled water, the mixture was centrifuged at 1500 g for 10 min. The supernatant was concentrated by nitrogen and dissolved with 1.5 ml dissolution (tetrahydrofuran: acetonitrile: methanol=15:30:55 by volume) as a sample for analysis in HPLC (Shimadzu, Kyoto, Japan). 20 μl sample was injected into a C18 column (5 μm particle size, 4.6mm×250 mm, Elite analytical instruments Co., Ltd., Dalian, China). Mobile phase (methanol:acetonitrile=9:1, with 0.05% triethylamine) was set at a flow rate of 1.2 ml min−1. The absorbance of 475nm was detected via an SPD-M20A diode array detector. Authentic carotenoids (lycopene, β-carotene, and lutein; Sigma, St Louis, MO, USA) were chosen to calculate the amounts of carotenoids.
For ascorbic acid analysis (Liu et al., 2018), 0.4 g powder was mixed with 2.5 ml of 1% oxalate and centrifuged at 7000rpm for 10 min at 4°C. 20 μl sample from filtered supernatant was injected onto the same specification C18 column as carotenoid analysis described. 0.1% oxalic acid solution was chosen as the mobile phase with a flow rate of 1 ml min−1. The absorbance of 243 nm was detected via an SPD-M20A diode array detector. Authentic ascorbic acid was chosen as a standard chosen to calculate the amount of ascorbic acid.
Quantification and statistical analysis
Statistical analyses were performed using SPSS 19. Details of the statistical tests applied, including the statistical methods, number of replicates, mean and error bar details and significances, are indicated in the relevant figure legends. All replicates are biological, unless otherwise noted in the figure legend.
Acknowledgments
We thank Tomato Genetics Resource Center (University of California, Davis, CA) for providing tomato seeds used in our study. We also thank Professor Daqi Fu (China Agricultural University) and Professor Yaoguang Liu (South China Agriculture University) for kindly providing vectors for VIGS and pYLCRISPR/Cas9 plasmids, respectively. This research was supported by National Natural Science Foundation of China (Key Program, No.31830078), China Postdoctoral Science Foundation (No. 2019M662067), and Zhejiang Provincial Ten-thousand Program for Leading Talents of Science and Technology Innovation (2018R52026).
Author contributions
Q.W., C.L., and H.L. designed the research. H.L., M.Z., C.J., M.Q., F.M., S.H., Z.S., and D.L. performed the research. H.L. D.L., and Y.L. analyzed data. H.L., L.L., Q.W., Y.Y., and C.J. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102926.
Contributor Information
Yanhai Yin, Email: yin@iastate.edu.
Chuanyou Li, Email: cyli@genetics.ac.cn.
Qiaomei Wang, Email: qmwang@zju.edu.cn.
Supplemental information
Data and code availability
The RNA-seq data have been deposited at the NCBI Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra/) with the BioProject ID PRJNA635540 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bishop G.J., Harrison K., Jones J.D. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Ji Y., Hu J., Guo R., Sun S., Wang X. Strigolactones and Brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant. 2020;13:586–597. doi: 10.1016/j.molp.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Fu D.Q., Zhu B.Z., Zhu H.L., Jiang W.B., Luo Y.B. Virus-induced gene silencing in tomato fruit. Plant J. 2005;43:299–308. doi: 10.1111/j.1365-313X.2005.02441.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni J.J. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T., Corvalan C., Best N.B., Budka J.S., Zhu J.Y., Choe S., Schulz B. Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One. 2012;7:e36625. doi: 10.1371/journal.pone.0036625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Tang B., Xie Z., Nolan T., Ye H., Song G.Y., Walley J., Yin Y. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019;100:923–937. doi: 10.1111/tpj.14484. [DOI] [PubMed] [Google Scholar]
- Klee H.J., Giovannoni J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- Kyomugasho C., Christiaens S., Shpigelman A., Van Loey.A.M., Hendrickx M.E. FT-IR spectroscopy, a reliable method for routine analysis of the degree of methylesterification of pectin in different fruit- and vegetable-based matrices. Food Chem. 2015;176:82–90. doi: 10.1016/j.foodchem.2014.12.033. [DOI] [PubMed] [Google Scholar]
- Li X.J., Chen X.J., Guo X., Yin L.L., Ahammed G.J., Xu C.J., Chen K.S., Liu C.C., Xia X.J., Shi K. DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 2016;14:1021–1033. doi: 10.1111/pbi.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sun S., Wang H., Wang K., Yu H., Zhou Z., Xin P., Chu J., Zhao T., Wang H. FIS1 encodes a GA2-oxidase that regulates fruit firmness in tomato. Nat. Commun. 2020;11:5844. doi: 10.1038/s41467-020-19705-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Meng F., Miao H., Chen S., Yin T., Hu S., Shao Z., Liu Y., Gao L., Zhu C. Effects of postharvest methyl jasmonate treatment on main health-promoting components and volatile organic compounds in cherry tomato fruits. Food Chem. 2018;263:194–200. doi: 10.1016/j.foodchem.2018.04.124. [DOI] [PubMed] [Google Scholar]
- Liu L., Jia C., Zhang M., Chen D., Chen S., Guo R., Guo D., Wang Q. Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol. J. 2014;12:105–115. doi: 10.1111/pbi.12121. [DOI] [PubMed] [Google Scholar]
- Liu Y., Du M., Deng L., Shen J., Fang M., Chen Q., Lu Y., Wang Q., Li C., Zhai Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell. 2019;31:106–127. doi: 10.1105/tpc.18.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nolan T.M., Vukasinovic N., Liu D., Russinova E., Yin Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 2020;32:295–318. doi: 10.1105/tpc.19.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Carneiro R.T., Lytovchenko A., McQuinn R., Sorensen I., Vallarino J.G., Giovannoni J.J., Fernie A.R., Rose J.K.C. Genetic and metabolic effects of ripening mutations and vine detachment on tomato fruit quality. Plant Biotechnol. J. 2020;18:106–118. doi: 10.1111/pbi.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.D., Bo W., West G., Lycett G.W., Tucker G.A. Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol. 2007;144:1960–1967. doi: 10.1104/pp.107.096347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Pearce G., Ryan C.A. Generation of systemin signaling in tobacco by transformation with the tomato systemin receptor kinase gene. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10114–10117. doi: 10.1073/pnas.1432910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal F., Wattier C., Rusterucci C., Pelloux J. Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J. Exp. Bot. 2014;65:5125–5160. doi: 10.1093/jxb/eru272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour G.B., Ostergaard L., Chapman N.H., Knapp S., Martin C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013;64:219–241. doi: 10.1146/annurev-arplant-050312-120057. [DOI] [PubMed] [Google Scholar]
- Silva-Sanzana C., Celiz-Balboa J., Garzo E., Marcus S.E., Parra-Rojas J.P., Rojas B., Olmedo P., Rubilar M.A., Rios I., Chorbadjian R.A. Pectin methylesterases modulate plant homogalacturonan Status in Defenses against the Aphid Myzus persicae. Plant Cell. 2019;31:1913–1929. doi: 10.1105/tpc.19.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Handa A.K. Reduction in pectin methylesterase activity Modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol. 1994;106:429–436. doi: 10.1104/pp.106.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik S., Chapman N.H., Smith R., Poole M., Adams G., Gillis R.B., Besong T.M., Sheldon J., Stiegelmeyer S., Perez L. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 2016;34:950–952. doi: 10.1038/nbt.3602. [DOI] [PubMed] [Google Scholar]
- Wang D.D., Samsulrizal N.H., Yang C., Allcock N.S., Craigon J., Blanco-Ulate B., Ortega-Salazar I., Marcus S.E., Bagheri H.M., Perez-Fons L. Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 2019;179:544–557. doi: 10.1104/pp.18.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B., Strom A., Tasker A., West G., Tucker G.A. Effect of silencing the two major tomato fruit pectin methylesterase isoforms on cell wall pectin metabolism. BMC Plant Biol. 2013;15:1025–1032. doi: 10.1111/j.1438-8677.2012.00714.x. [DOI] [PubMed] [Google Scholar]
- Wen B., Zhang F., Wu X., Li H. Characterization of the tomato (solanum lycopersicum) pectin methylesterases: evolution, activity of isoforms and expression during fruit ripening. Front Plant Sci. 2020;11:238. doi: 10.3389/fpls.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited at the NCBI Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra/) with the BioProject ID PRJNA635540 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.