Summary
Myoglobin (Mb) is highly concentrated in the myocytes of diving mammals such as whales and seals, in comparison with land animals, and its molecular evolution has played a crucial role in their deep-sea adaptation. We previously resurrected ancestral whale Mbs and demonstrated the evolutional strategies for higher solubility under macromolecular crowding conditions. Pinnipeds, such as seals and sea lions, are also expert diving mammals with Mb-rich muscles. In the present study, we resurrected ancestral pinniped Mbs and investigated their biochemical and structural properties. Comparisons between pinniped and whale Mbs revealed the common and distinctive strategies for the deep-sea adaptation. The overall evolution processes, gaining precipitant tolerance and improving thermodynamic stability, were commonly observed. However, the strategies for improving the folding stability differed, and the pinniped Mbs exploited the shielding of hydrophobic surfaces more effectively than the whale Mbs.
Subject areas: Biomolecules, Evolutionary biology, Evolutionary mechanisms
Graphical abstract

Highlights
-
•
Ancestral pinniped myoglobins (Mbs) were resurrected to analyze deep-sea adaptation
-
•
The structural and biochemical parameters of pinniped and whale Mbs were compared
-
•
Electrostatic repulsion, fold stabilization, and surface entropy increment were common
-
•
Hydrophobic patch shielding was detected as a unique strategy of pinniped Mbs
Biomolecules; Evolutionary biology; Evolutionary mechanisms.
Introduction
The thermodynamic stability and solubility of native proteins are closely related with their in vivo kinetics and physiological functions in living cells. These properties of extant proteins have evolved to be optimal for or not harmful to the survival of organisms. One of the best examples is myoglobin (Mb), which functions in O2 storage in muscle tissues for aerobic exercise. Mb is highly concentrated in the tissues of deep-diving animals, whereas its concentration is significantly lower in land animals (Dasmeh and Kepp, 2012; Davis, 2014; Davis and Kanatous, 1999; Helbo and Fago, 2012; Isogai et al., 2018; McGowen et al., 2014; McIntyre et al., 2002; McLellan, 1984; Nery et al., 2013; Noren and Williams, 2000; Samuel et al., 2015; Shire et al., 1975; Snyder, 1983; Velten et al., 2013; Wright and Davis, 2006). Thus, the diving capacity of mammals is considered to be correlated with the Mb concentration in their myocytes.
Amino acid sequences of ancestral Mbs were predicted to have less positive ZMb values than the extant diving mammals (Berenbrink, 2021; Dasmeh et al., 2013; Isogai et al., 2018; Mirceta et al., 2013; Romero-Herrera et al., 1978). Wilkinson and Harrison (1991) investigated the relationship between amino acid composition of natural proteins and their liability to form inclusion body and showed their average charge magnitude was the most significant parameter determining the inclusion body formation (Wilkinson and Harrison, 1991). Kuroda et al. (2016) have indicated repulsive Coulomb interactions were dominant in ensuring high solubility, based on molecular dynamics (MD) simulation of artificial polypeptides (Kuroda et al., 2016). Thus, the large positive charges of diving mammal Mbs are considered to cause electrostatic repulsion among the Mb molecules, to prevent their precipitation, and to maintain the high protein concentration (Kuroda et al., 2016; Mirceta et al., 2013; Wilkinson and Harrison, 1991). However, a simple increase of the protein solubility with an increase in the ZMb value has not been observed under normal buffer conditions (Chan et al., 2013; Hojgaard et al., 2016; Kramer et al., 2012; Shaw et al., 2001).
We previously resurrected ancestral whale Mbs and confirmed that ZMb increased during their deep-sea adaptation (Isogai et al., 2018). The Mbs of the common ancestor between toothed and baleen whales (assumed to be a relative of Basilosaurus) and the further common ancestor between whale and land animals (assumed to be a relative of Pakicetus) were synthesized. Their structures and biochemical properties, including the stabilities and solubilities of the molecules, were investigated and compared with those of the extant sperm whale Mb. Unexpectedly, the protein solubility in dilute buffer (log S0) decreased, rather than increased, along with the ZMb increase. On the other hand, the precipitant tolerance or solubility robustness against a macromolecular precipitant (β) significantly improved with ZMb during the evolution.
The adaptation processes of mammal Mbs are thought to be generally common among the diving animals, which independently evolved and returned to the sea (Isogai et al., 2018; Mirceta et al., 2013). Pinnipeds, such as seals, sea lions, and walruses, are also expert diving mammals comparable with whales and have Mb-rich muscles (Berenbrink, 2021; Dasmeh et al., 2013; Kooyman, 1989; Samuel et al., 2015). The leading theory, based on paleontology and molecular biology, is that they evolved from a terrestrial Mustelid-like form (Berta, 2018). The common ancestor between seals and sea lions is assumed to be an extinct genus of a large pinniped, such as Enaliarctos mealsi (Figure 1), whose fossils have been recovered from late Oligocene or early Miocene (ca. 24–22 million years ago) strata in North America (Berta et al., 1989; Poust and Boessenecker, 2018). The further common ancestor between pinnipeds and Mustelidae is assumed to be a walking seal, such as Puijila darwini, which lived during the late Oligocene and early Miocene epoch (ca. 24–21 million years ago) (Flynn et al., 2005; Rybczynski et al., 2009).
Figure 1.
Molecular phylogenic tree of Mbs from pinnipeds, land animal relatives, and inferred ancestors
The presented tree is a part of the entire tree consisting of Hbs, Mbs, and other globins. The positions of aMbSp and aMbSe are indicated on the corresponding nodes. The illustrations of animals are shown for extant polar bear and elephant seal and for the fossil species Enaliarctos mealsi and Puijila darwini. The horizonal scale bar indicates relative evolutionary distance of 0.02. The positions of the fossil species are not identified on the tree nodes. Credit of the illustrations belongs to Satoshi Kawasaki. All rights reserved, used with permission.
The theory of a common strategy for the diving adaptation of Mbs among mammals, however, has not been tested by biochemical experiments yet (Berenbrink, 2021; Dasmeh et al., 2013; Isogai et al., 2018; Mirceta et al., 2013). A comparison of the evolutional process of pinniped Mbs with that of whales would clarify the commonalities and differences in the strategies. In the present study, we resurrected ancestral pinniped Mbs, aMbSe and aMbSp, which are assumed to be from the relatives of the fossil species Enaliarctos mealsi and Puijila darwini, respectively. We investigated their structural and biochemical properties and compared them with those of an extant elephant seal Mb (esMb) and ancestral whale Mbs.
Results
Resurrection of ancestral pinniped Mbs
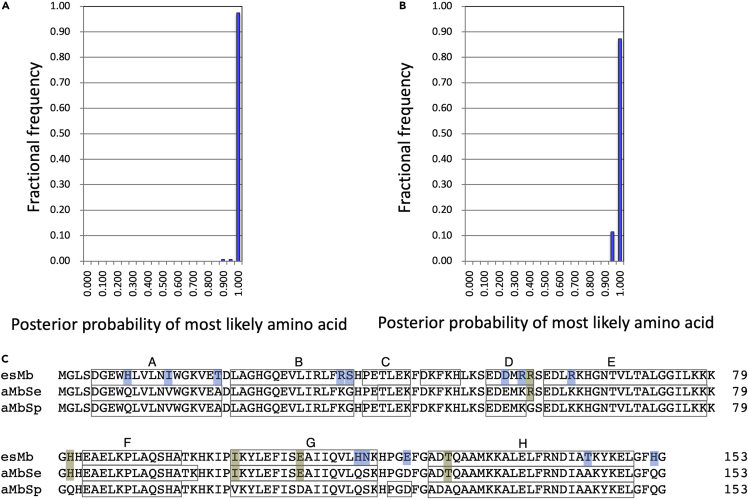
The amino acid sequences of the ancestral pinniped Mbs, aMbSe and aMbSp, were inferred based on the molecular phylogeny of Mbs, Hbs, and other closely related globins known to date (Isogai et al., 2018) and compared with that of the extant esMb, as shown in Figure 2. The aMbSe is an ancestral Mb from the common ancestors of seals and sea lions and expected to be related to the early pinniped Enaliarctos mealsi (Figure 1) (Berta et al., 1989; Poust and Boessenecker, 2018). The aMbSp is from a further common ancestor of pinnipeds and Mustelidae, which is assumed to be a relative of the quadruped land/semiaquatic animal Puijila darwini (Flynn et al., 2005; Rybczynski et al., 2009). Note that the species, to which the ancestral Mbs belonged, might not be identified to be the fossil species. The residues in the ancestral proteins were successfully deduced with the minimum 0.944 posterior probability (Figure 2B).
Figure 2.
Posterior probability distribution and inferred amino acid sequences of ancestral Mbs
(A and B) Posterior probability distributions for the sites of aMbSp (A) and aMbSe (B) sequences. Horizontal and vertical axes show posterior probability bins and fractional frequency of inferred sites, respectively.
(C) Amino acid alignment of the inferred esMb, aMbSe, and aMbSp sequences. Single-amino-acid substitutions are highlighted in blue, and residues conserved in two of the three sequences are highlighted in light brown. Letters above the boxed sequences indicate canonical helices A-H.
Five residue replacements, G57R, Q81H, V101I, D109E, and A127T, were deduced to have occurred during the evolution from aMbSp to aMbSe (Figure 2). Furthermore, 13 residue replacements, Q8H, V13I, A19T, K34R, G35S, E54D, K56R, K62R, Q116H, S117N, D122E, A144T, and Q152H, occurred during the evolution from aMbSe to extant esMb (elephant seal). Consequently, a total of 18 residue replacements were deduced during the evolution from a terrestrial ancestral seal to the existing elephant seal. The relative molecular masses (Mr), formal net charges, and formal pI values were calculated from the deduced amino acid sequences, as shown in Table 1.
Table 1.
Molecular properties of ancestral and elephant seal Mbs based on their sequences and X-ray crystallographic structures
| Mb | Mra | Formal net chargesb | Formal pIc | ZMbd (pH 7.0) | pId | ΔGsolve kcal/mol | dEeff/dRgf kcal/mol/nm | ΔΔGmutg kcal/mol |
|---|---|---|---|---|---|---|---|---|
| aMbSp | 17,125.5 | +9 | 8.95 | +1.92 | 7.65 | −1745.5 ± 20.2 | 1875.8 ± 125.0 | – |
| aMbSe | 17,291.7 | +11 | 9.30 | +3.06 | 8.03 | −1685.0 ± 33.1 | 780.0 ± 46.3 | −14.0 |
| esMb | 17,533.9 | +14 | 9.34 | +4.50 | 8.24 | −1661.0 ± 14.2 | 484.1 ± 62.6 | −5.9 |
Relative molecular masses of the apo-proteins, which were calculated from the amino acid sequences.
Formal net charges were calculated based on the amino acid sequences, assuming that the charges of the positively charged residues, Lys, Arg, and His, and the negatively charged residues, Asp and Glu, are +1 and −1, respectively.
Formal isoelectric points were calculated based on the amino acid sequences, according to Skoog and Wichman (1986) (Skoog and Wichman, 1986).
Total net charges (ZMb) at pH 7.0 and isoelectric points (pI) were taken from the pH dependence of ZMb (Figure S1), which was calculated based on the X-ray crystallographic structures according to Spassov and Yan (2008) (Spassov and Yan, 2008).
The solvation free energies were calculated based on the X-ray structures and the MD simulations, according to Sumi et al. (2015) (Sumi et al., 2015).
See Solvation free energy calculation in STAR Methods.
The mutational energy changes (ΔΔGmut) were calculated based on the X-ray structures, according to Spassov and Yan (2013) (Spassov and Yan, 2013). The values in each line are ΔΔGmut for the residue replacements from the proximate ancestral Mb, i.e., the value of aMbSe is for the sum of the replacements on aMbSp. The ΔΔGmut for each residue replacement and the itemized energy terms are shown in Figure S2.
The two ancestral and extant elephant seal Mbs were synthesized in the holo-forms and purified to homogeneity (see STAR Methods). The purification by size-exclusion chromatography and the subsequent small-angle X-ray scattering (SAXS) analysis indicated that all of the Mb samples are essentially monomeric within a broad range of Mb concentrations (see below). The atomic structures of the synthesized aMbSp, aMbSe, and esMb were determined by X-ray crystallography to 1.5, 2.9, and 1.9 Å resolutions, respectively (Figure 3 and Table S2). The main chain structures were well conserved among the ancestral and extant Mbs, as in those of the whale Mbs (Isogai et al., 2018).
Figure 3.
Residue replacements of seal myoglobin during the evolution from the terrestrial animal to elephant seal
(A) The replaced residues are shown on the crystal structures of aMbSp (PDB code 7DDS), aMbSe (7DDT), and esMb (7DDU) from left to right. The structures are superposed to each other. The canonical helices A-H are indicated on the structure of aMbSp.
(B and C) (B) Close-up views of the superposed aMbSp (light brown) and aMbSe (light blue) structures around the residue sites V101I and A127T, which have been replaced from aMbSp to aMbSe. (C) Close-up views of the superposed aMbSe (light blue) to esMb (blue) structures around the residue sites V13I, A19T, K34R, K56R, K62R, and A144T, which have been replaced from aMbSe to esMb. The electrostatic interactions/hydrogen bonds and cavity filling positions are indicated with yellow dotted lines and red circles, respectively.
MD simulations for 60 ns were executed for each of the pinniped Mbs based on the crystal structures, in order to derive several parameters related to their structural dynamics. The solvent-accessible surface areas (ASA), net surface charges (ZMb), isoelectric points (pI), SFEs (ΔGsolv), effective-energy changes involved in structural fluctuations (dEeff/dRg), and mutational energy changes (ΔΔGmut) were calculated, based on the crystal structures and the trajectories of the MD simulations (Table 1). For dEeff/dRg, the energy landscape Eeff is provided by the effective energy for conformation i from the MD trajectories, Eeff, defined as the sum of the intramolecular energy and the solvation free energy (SFE) . Thus, dEeff/dRg represents the wideness of the basins on an energy landscape Eeff, as a function of the radius of gyration Rg. The ΔΔGmut for each residue substitution and the contribution of the thermodynamic energy terms to the mutational energy changes are shown in Figure S2.
Biochemical characterization of ancestral and extant pinniped Mbs
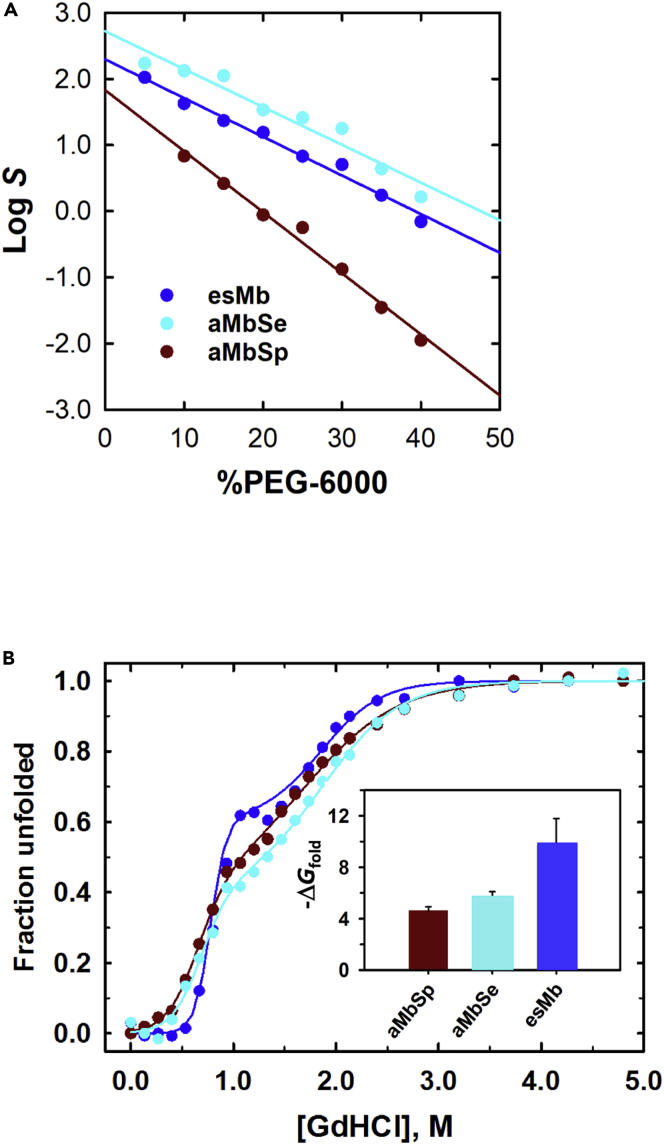
The solubilities of the synthesized Mbs were examined for the precipitant tolerance with polyethylene glycol (PEG) (Isogai et al., 2018; Kramer et al., 2012). The solubility dependence on the PEG-6000 concentration in the protein solution was measured at room temperature, as shown in Figure 4A. The relationship between protein solubility (S) and precipitant concentration ([precipitant]) can be described by a linear equation: log S = log S0 + β[precipitant], where log S0 is the y-intercept of the plot and β is the slope (Kramer et al., 2012). S0 and β represent the solubility in dilute solution without precipitant and the precipitant tolerance, respectively (Table S3).
Figure 4.
Solubility and stability of ancestral and extant seal Mbs
The values of aMbSp, aMbSe, and esMb are plotted in brown, sky blue, and blue, respectively.
(A) Solubility dependence of holo-forms of ancestral and elephant seal Mbs on the PEG-6000 concentration. Log solubility values in mg/mL (S) are plotted against the precipitant concentration.
(B) Chemical denaturation profiles of apoMbs. The unfolded fractions estimated by the CD signal intensity at 222 nm are plotted against the Gd-HCl concentration. The inset shows the ΔGfold (kcal mol−1) of the proteins.
Log S0 increased during the evolution from aMbSp to aMbSe, but slightly decreased from aMbSe to esMb, indicating that the solubility in a dilute buffer solution was increased during the early phase of evolution but decreased during the last phase (Table S3). On the other hand, the precipitant tolerance (β) significantly increased during the evolution from aMbSp to aMbSe and remained almost unchanged from aMbSe to esMb, which was similar to the whale Mb evolution (Isogai et al., 2018).
Chemical denaturation experiments of the apoMbs with guanidine hydrochloride (GdHCl) were performed to determine their thermodynamic stabilities, as shown in Figure 4B. The denaturation profiles were analyzed by assuming the three-state transition (Barrick and Baldwin, 1993; Isogai, 2006; Isogai et al., 2000), and the thermodynamic parameters were estimated for the folding reactions (Table S4 and Figure S3). The folding free energy from the intermediate state to the native state (ΔG1) and the total free energy from the unfolded to the native state (ΔGfold) slightly decreased from aMbSp to aMbSe and significantly decreased from aMbSe to esMb. These results indicated that the folding stability mainly improved during the late phase of the evolution.
We performed SAXS experiments with the pinniped Mbs to monitor their intermolecular interactions. The Mb solutions at pH 6.8 ± 0.2 and concentrations from ∼38.0 to ∼1 mg/mL were irradiated with X-rays. The zero-angle X-ray scattering, I(0), in the SAXS profile (Figure S4) decreased as the Mb concentration increased. This indicates the ordering of the Mb molecules in solution by their mutual repulsion, which is quantified by the second virial coefficient (A2) with Equation (4) (Figure 5I); a more positive A2 represents stronger repulsion. A2 increased from aMbSp to aMbSe in the early phase of evolution, but decreased from aMbSe to esMb in the late phase. This is in contrast to the case of whale Mb, for which A2 increased in both phases.
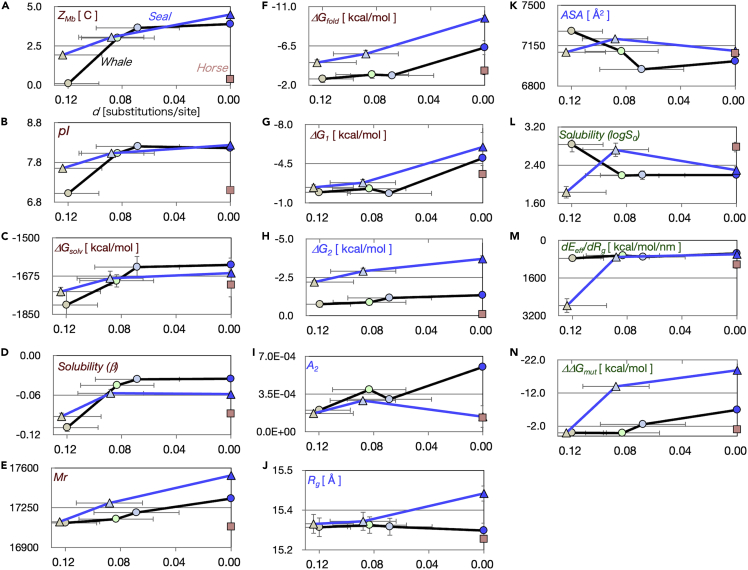
Figure 5.
Molecular parameters of ancestral and extant Mbs
The values of seal Mbs, aMbSp, aMbSe, and esMb, are indicated by light brown, light green, and blue triangles, respectively. Those of whale Mbs, aMbWp, aMbWb’, aMbWb, swMb, and hsMb (Isogai et al., 2018), are also shown in light brown, light green, light blue, blue, and salmon squares, respectively. In all plates, the horizontal axis is the evolutionary distance (d) of each Mb sequence from the extant swMb or esMb and the data points are those of the most ancient to extant Mbs from left to right. The labels and units (except for dimensionless numbers) of the vertical axes are shown inset. (A) Positive net charge (ZMb); (B) isoelectric point (pI); (C) solvation free energy (ΔGsolv); (D) solubility slope against precipitant (β); (E) relative molecular mass (Mr); (F) folding free energy (ΔGfold); (G) folding free energy of the folded state relative to the intermediate state (ΔG1); (H) folding free energy of the intermediate state relative to the unfolded state (ΔG2); (I) second virial coefficient (A2); (J) radius of gyration (Rg); (K) molecular surface area (ASA); (L) log of solubility (log S0); (M) effective-energy changes involved in structural fluctuation (dEeff/dRg); (N) mutational folding energy changes (ΔΔGmut).
Analyzing the molecular parameters of ancestral and extant Mbs
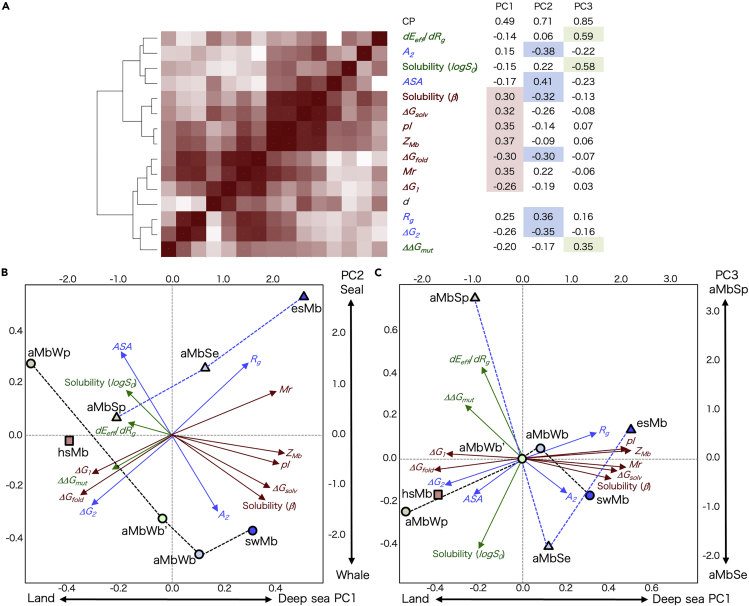
The molecular parameters of the ancestral and extant seal Mbs, as well as those of the whale Mbs (Isogai et al., 2018), are plotted against the evolutionary distance (d) in Figure 5. These parameters were subjected to a principal component analysis (PCA). All parameters except for d were applied, and the principal component (PC) axes were defined, where the cumulative proportion of variance was 85% to the third axis, as in Figure 6A.
Figure 6.
Principal component analyses of molecular parameters of ancestral and extant Mbs
(A) Clustering of the Mb molecular parameters (as shown in Figure 5) by the correlation coefficients. The heatmap indicates higher (red) to lower (white) absolute values of the correlation coefficients between parameters. The eigenvectors for principal components (PC1 - PC3) are shown on the right, where the CP row shows the cumulative proportion of variance. The parameters are colored red (major PC1 contributors), blue (major PC2 contributors), or green (major PC3 contributors). The dendrogram on the left indicates a hierarchical clustering of the parameters. The pinniped Mbs (aMbSp, aMbSe, and esMb), whale Mbs (aMbWp, aMbWb’, aMbWb, and swMb), and horse Mb (hsMb) are distributed (same notation as in Figure 5) over the biplots for (B) PC1 - PC2 and (C) PC1 - PC3. The loading of each parameter to the principal axes is indicated as overlaid arrows, colored as in panel (A). The scales on left and bottom of the plots are the scores of the proteins for corresponding principle axis, and those on right and top are the factor loadings of the variables.
The first principal component (PC1) clearly defined the evolutionary process by separating the Mbs of land/semiaquatic (aMbSp, aMbWp, and horse Mb [hsMb]), shallow-sea adapted (aMbSe, aMbWb, and aMbWb’), and deep-sea adapted (esMb and swMb) animals (Figure 6B). The eigenvectors indicted that ZMb, pI, Mr, ΔGsolv, β, ΔGfold, and ΔG1 were the major contributors to PC1. These parameters were further clustered into two groups by referring to the correlation between parameters (Figure 6A); ΔGfold, Mr, and ΔG1 showed higher correlations with the evolutionary distance, indicating that they constantly changed during the entire adaptation process, while ZMb, pI, and β and ΔGsolv mainly changed in the early phase (Figure 5).
The PC2 visibly separated the pinniped and whale Mbs and thus conceivably defines the differences in adaptation strategies between the major diving animal lineages (Figure 6B). The predominant contributing parameters for PC2 were A2, Rg, ASA, and ΔG2 (Figure 6B), while β and ΔGfold also significantly contributed to this axis. Although the meaning of PC3 was relatively ambiguous, this axis also appeared to define a specific strategy in the pinniped lineage, considering that aMbSp and aMbSe were distantly separated along this axis. The dEeff/dRg, log S0, and ΔΔGmut values were the major contributors for PC3 (Figure 6C). These pinniped-whale discriminating parameters were clustered into two groups through correlation: the first group consisted of dEeff/dRg, A2, log S0, and ASA and the second group contained Rg, ΔΔGmut, and ΔG2 (Figure 6A; see the major PC2 contributors colored by blue).
Distinctive strategies for pinniped and whale Mbs
Since Rg, dEeff/dRg, and ASA contributed to PC2 and PC3, this implied that the difference in the strategies between pinniped and whale Mbs would be relevant to the structures and dynamics of the molecules. Therefore, the MD trajectories were further analyzed by discriminating the conformations with lower (<15.35Å) and higher (>15.45 Å) Rg values (Figure 7A). The most remarkable contrast between pinniped and whale Mbs was found in the ASAs separately evaluated for hydrophobic (ASAhydrophobic) and hydrophilic (ASAhydrophilic) properties. As a general trend, ASAhydrophobic increased and ASAhydrophilic decreased during the process of diving adaptation (Figures 7C and 7D). However, these changes were rather mild for the pinniped Mbs (aMbSp, aMbSe, and esMb). Remarkably, the ASAhydrophilic values in the higher Rg conformation for the pinniped Mbs were nearly constant during the evolution or decreased and became smaller than the whale Mbs, indicating that the hydration effects on the softness and flexibility of the native conformations would be kept at a higher level than those of the whale Mbs (Figure 7D). This suggests that a relatively high ASAhydrophilic and low ASAhydrophobic are favorable for increasing the wideness of the basins on an energy landscape and thus might explain the decrease of the relatively high dEeff/dRg of aMbSp to a value comparable with those of other Mbs during the evolution from aMbSp to aMbSe (see Figure 5M).
Figure 7.
Rg, ASA, and intramolecular interactions in higher and lower Rg models of ancestral and extant Mbs
(A) The Rg values of the snapshot models in the MD trajectories are box-plotted for aMbWp, aMbSp, aMbWb’, aMbWb, aMbSe, swMb, esMb, and hsMb (marks are placed at median points, with the same notation as in Figure 5). The regions of lower (<15.35 Å) and higher (>15.45 Å) Rg are shown in light green and light blue, respectively. The values of (B) total; (C) hydrophobic; and (D) hydrophilic ASAs and the numbers of (E) hydrogen bonds; (F) ion pairs; and (G) van der Waals (vdW) contacts are separately box-plotted for lower (<15.35 Å, left, at start point of red arrow) and higher (>15.45 Å, right, at endpoint of red arrow) Rg models of aMbWp, aMbSp, aMbWb’, aMbWb, aMbSe, swMb, esMb, and hsMb, from left to right in each panel.
The observed stability of ASAhydrophilic might also be related to the improved solubility of aMbSe, regarding the high correlation between log S0 and dEeff/dRg (Figures 5L, 5M, and 6A). This high affinity for water protects the direct favorable interactions between Mbs and also enhances the softness and flexibility of the Mb molecules. This analysis also demonstrated that the intramolecular interactions increased during the diving adaptation process for both the pinniped and whale Mbs (Figures 7E–7G). Remarkably, the number of ion pairs and van der Waals (vdW) contacts in aMbSe increased in the early phase (from aMbSp to aMbSe) to a level comparable with that of swMb.
An inspection of the hydrophobic/hydrophilic surface areas on the lower and higher Rg conformations of esMb and swMb revealed that the hydrophobic patches in the higher Rg model of swMb were more significant than those in the esMb model (Figure 8). The shielding of the hydrophobic areas in the esMb model was mainly achieved with 8 hydrophilic residues, including the G57R, Q81H, and D109E substitutions from aMbSp to aMbSe, the A19T substitution from aMbSe to esMb, and K16 and E27, which interact with the substituted residues (Figure 3C). Thus, the hydrophilic residues (side chains) obtained during the diving adaptation process are used for shielding the hydrophobic surfaces in the higher Rg structures.
Figure 8.
Hydrophobic/hydrophilic ASA patches on swMb and esMb models
Hydrophobic (orange) and hydrophilic (light blue) surface patch distributions on typical snapshots of lower (bottom) and higher (top) Rg models of swMb and esMb. The Rg values are indicated for the models in front views on the left, and the rear views of the same models are on the right. Red ellipses indicate the remarkable hydrophobic patches, which are different between swMb and esMb, in the higher Rg conformations, and the residues contributing to these patches are indicated. The residues substituted during the diving adaptation process or those interacting with the substituted residues are shown in blue.
Discussion
The main experimental and computational data obtained here are summarized, along with those for the whale Mb evolution (Isogai et al., 2018), as shown in Figures 5 and 6. The isoelectric point (pI), net surface charges (ZMb), solubility slope or precipitant tolerance (β), and SFE (ΔGsolv) for both the pinniped and whale Mbs showed similar patterns of changes, which significantly increased during the early phase of the evolution and remained relatively constant during the late phase. It is well known that β depends primarily on attractive interactions between protein molecules caused by crowding agents, which could be explained by the Asakura-Oosawa (AO) theory (Asakura and Oosawa, 1954). According to the AO theory, the volumes of the protein and the crowding agent determine the magnitude of β (Atha and Ingham, 1981); therefore, in this study, none of the variations in the volumes of the Mbs and the crowding agent (PEG-6000) should contribute to the present variation of the β values in the Mb evolution. Alternatively, the correlation of β with ZMb can be explained by electrostatic repulsion between Mb molecules with the dielectric property of PEG solutions. The positively charged amino acids that were introduced during the evolution from aMbSp to aMbSe, namely, G57R and Q81H, did not participate in the remarkable intramolecular interactions in the crystal structure (Figure 3). This observation might support the major role of the increased ZMb in the intermolecular interactions.
In the previous study on whale Mbs, β and log S0 showed coherent movements during the evolution (Isogai et al., 2018). In contrast, the present study on the pinniped Mbs revealed that the behaviors of β and log S0 had deviated, and β was highly correlated with ZMb. However, the correlation between log S0 and ZMb was not clear, suggesting that the improvement of the precipitant tolerance (β) was rather important in the early phase of diving adaptation (Figures 5D and 5L). It is also possible that the tolerance was obtained not only for the crowding of cognate Mbs but also for other precipitant agents because a vast number of molecules interact with Mb in vivo (Fulton, 1982; Luby-Phelps, 2000).
In addition, we found a high correlation between β and ΔGsolv: the larger the ΔGsolv was, the larger the β was (Figures 5C, 5D, 6B, and 6C). In other words, the Mbs without preferred hydration are less apt to precipitate. The crowding effects could yield not only attractive interactions (the AO theory as noted earlier) but also repulsive interactions between proteins. The latter repulsive interactions are caused by energetically favorable interactions between the protein of interest and crowders (Bloustine et al., 2006). Thus, the observed sedimentation of Mb molecules resulted primarily from the entropic depletion effect by the AO theory and was modulated by the repulsive charge interactions between Mbs with ZMb and the effective repulsive interactions between Mbs by the Mb-PEG attraction. During the evolutionary stages, the increase in nonpolar interactions of Mb-PEG due to the increase in ASAhydrophobic (Figure 7C) and the decrease in the hydration of Mb due to the decrease in ASAhydrophilic (Figure 7D) would both be responsible for the improvement of the precipitant tolerance β, although they might reduce log S0. Therefore, the opposing strategies were needed to improve log S0 and β, respectively. In fact, the pinniped Mbs, which gained an improved log S0 during the evolution, had a smaller improvement of β than the whale Mbs, while the whale Mbs, which gave up the improvement of log S0, had a larger gain in β (Figure 5).
The folding stability ΔGfold and molecular mass Mr for both the seal and whale Mbs slightly increased in the early phase and significantly increased in the late phase (deep-sea adaptation) (Figures 5 and 6), possibly by the formation of hydrogen bonds and/or salt bridges (K34R, K56R, and K62R in the late phase) and van der Waals contacts (V101I and A127T in the early phase, V13I, A19T, and A144T in the late phase) near the protein surface by the replacements of small residues with larger ones (Figures 3 and S2). These parameters defined the common strategy in the late phase adaptations for pinniped and whale Mbs and are basically consistent with the previous conclusion obtained for whale Mbs (Isogai et al., 2018).
The present study quantitatively compared the diving adaptation strategies between pinniped and whale Mbs, and the PC analysis highlighted the contributions of the parameters dEeff/dRg, A2, log S0, ASA, Rg, ΔΔGmut, and ΔG2 to the difference. Our results indicated that the pinniped Mbs had evolved while reducing the effective-energy changes involved in their structural fluctuations (dEeff/dRg). The decrease of dEeff/dRg was slight for the whale Mb evolution. However, in pinniped Mbs, this parameter significantly decreased to a level comparable with that of the whale Mbs in the early phase of evolution (Figure 5M). The pattern of dEeff/dRg variations is similar to that of ΔΔGmut (Figure 5N).
Analyses of the MD trajectories of the Mbs revealed that the decrease in dEeff/dRg resulted from the relatively high retention of the hydrophilic surface area ASAhydrophilic in the pinniped Mbs under the high Rg conformation. The dEeff/dRg characterizes the wideness of basins on an energy landscape, Eeff against structural perturbation to increase Rg. Generally, an elastic motion or perturbation of a protein structure increases both Rg and the exposed hydrophobic surface area. Because exposed hydrophobic surfaces might cause molecular self-association, shielding of these surfaces should be one of the effective strategies for increasing molecular solubility. The residues substituted from aMbSp to esMb significantly contributed to shielding of the hydrophobic surfaces (Figures 3 and 8), and dEeff/dRg and the solubility log S0 are highly correlated (Figure 6A).
In the previous study of whale Mbs, the strategy for the late phase was fold stabilization, which was indicated by the decreased ΔGfold (Figure 5F) and the increased numbers of intramolecular interactions (Figures 7E–7G). This stabilization would also contribute to the prevention of hydrophobic surface exposure to the solvent and other molecules. The shielding of hydrophobic surfaces detected in the present study would be a strategy congruent with the fold stabilization to avoid molecular precipitation. This strategy was more extensively adopted in the pinniped Mbs.
The dEeff/dRg also demonstrated higher correlation with the second virial coefficient A2 (Figure 6A). In the previous study of whale Mb evolution, it was suggested that the increase in repulsive intermolecular interactions (higher A2) was originated from surface entropy increment (Isogai et al., 2018). A substitution of flexible surface amino acid, such as arginine, into smaller one, typically alanine, is one of the efficient strategies to induce protein crystallization (precipitation), known as the surface entropy reduction (Cooper et al., 2007; Derewenda, 2004; Derewenda and Vekilov, 2006). The amino acid substitutions in the opposite direction observed in the diving adaptation of Mbs are thought to suppress protein precipitation (He et al., 2021; Romero-Herrera et al., 1978). The dEeff/dRg decrease implies a wider conformational space around the native structure, within which conformational flexibility is expected to be high. The higher conformational flexibility has been also suggested to induce the strongly repulsive short-range forces between protein surfaces (Israelachvili and Wennerstroem, 1990). This might result in stabilization of the monomeric state by the surface entropy increment or the increase of conformational entropy against the polymeric states in the pinniped Mb evolution.
The major strategies for the diving adaptation of Mbs revealed in this and previous studies are schematically summarized in Figure 9 (Dasmeh et al., 2013; He et al., 2021; Isogai et al., 2018; Mirceta et al., 2013). In conclusion, a total of 18 residues were replaced on or near the protein surface of Mb, during the pinniped evolution from the terrestrial ancestor to the deep-diving extant seal. The time range of the early phase of evolution from the terrestrial (aMbSp) to the intermediate (aMbSe) is assumed to be ∼16 M years, from the middle Eocene to the early Miocene with five residue replacements, and that of the late phase from the intermediate to the elephant seal (esMb) is estimated to be approximately 20 M years from the early Miocene to the present, with 13 residue replacements. Over the entire evolution processes of both the pinniped and whale Mbs, the pI, ZMb, β, and ΔGsolv parameters showed similar changes, i.e., mainly in the early phase of the evolution. The fold stabilization was another common strategy adopted in the late phase of evolution, as represented by the ΔG1 and ΔGfold parameters. This was accomplished by increasing the intramolecular interactions in both the pinniped and whale Mbs. However, most probably, the prevention from sedimentation or self-association was extensively achieved by shielding the hydrophobic patches on the molecular surfaces of the pinniped Mbs, thus highlighting the differences between the two deep-sea adapted animals.
Figure 9.
Schemes for the diving adaptation strategies of Mbs
The four strategies of diving adaptation of Mb are shown with the changes (upward and downward arrows) of mainly contributing parameters.
Hence, the resurrection of pinniped Mbs, along with whale Mbs, was quite helpful not only to elucidate the common and essential molecular strategy but also to detect the applicable alternative strategies for diving adaptation. The detected strategies appeared to be general, rather than specific to Mb molecules, and would be applicable to a wide variety of proteins to improve their solubility.
Limitations of the study
In the present study, we have analyzed the various parameters and revealed their correlations during the Mb evolution for the deep-sea adaptation of marine mammals. As an apparent limitation, however, not every detected correlation has been experimentally tested and discussed in detail in the present study. For example, the correlation between β with ZMb could be further theoretically and experimentally explained by electrostatic repulsion between Mb molecules with the dielectric property of PEG solutions, which will be described elsewhere in detail. Thus, additional reports are required to complete a quantitative view of diving adaptation of Mbs in molecular crowding conditions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli strain BL21(DE3) | Takara Bio | Cat#9126 |
| Chemicals, peptides, and recombinant proteins | ||
| Isopropyl 1-Thio-β-D-galactopyranoside | Nacalai Tesque | Cat#06289-67; CAS: 367-93-1 |
| Polyethylene Glycol #6,000 | Nacalai Tesque | Cat#10200-25; CAS: 25322-68-3 |
| Profinity eXact pPAL RIC-Ready Expression Vector | Bio-Rad Laboratories | Cat#1563001 |
| Profinity eXact™ Purification Resin | Bio-Rad Laboratories | Cat#1563005 |
| Ammonium sulfate | Sigma-Aldrich | Cat#7783-20-2; EC: 231-984-1 |
| Tris(hydroxymethyl)aminomethane | FUJIFILM Wako Pure Chemical Corporation | Cat#207-06275; CAS: 77-86-1 |
| N-(2-Hydroxyethyl)piperazine-N'-2-ethanesulfonic Acid | Nacalai Tesque | Cat#02443-05; CAS: 7365-45-9 |
| PEG 1500 | Sigma-Aldrich | Cat#7783-20-2; EC: 231-984-1 |
| Sodium malonate dibasic monohydrate | Nacalai Tesque | Cat# 21108-02; CAS: 26522-85-0 |
| Boric acid | FUJIFILM Wako Pure Chemical Corporation | Cat# 021-02195; CAS: 10043-35-3 |
| Imidazole | Nacalai Tesque | Cat# 19028-64; CAS:288-32-4 |
| Tri-sodium citrate | FUJIFILM Wako Pure Chemical Corporation | Cat# 203-13605; CAS: 68-04-2 |
| Di-sodium tartrate | FUJIFILM Wako Pure Chemical Corporation | Cat#197-13975; CAS: 6106-24-7 |
| Methanol | FUJIFILM Wako Pure Chemical Corporation | Cat#057-00456; CAS: 64-17-5 |
| 2-Methyl-2,4-pentanediol | FUJIFILM Wako Pure Chemical Corporation | Cat# 137-08165; CAS: 107-41-5 |
| Deposited data | ||
| Ancestral myoglobin aMbSp of Puijila darwini relative (imidazol ligand) structure | This paper | PDB: 7DDR |
| Ancestral myoglobin aMbSp of Puijila darwini relative structure | This paper | PDB: 7DDS |
| Ancestral myoglobin aMbSe of Enaliarctos relative (imidazol ligand) structure | This paper | PDB: 7DDT |
| Elephant seal myoglobin esMb structure | This paper | PDB: 7DDU |
| Sperm whale myoglobin swMb structure | Isogai et al. (2018) | PDB: 5YCE |
| Horse heart myoglobin hsMb structure | Evans and Brayer, 1990 | PDB: 1YMB |
| Ancestral myoglobin aMbWp of Pakicetus relative structure | Isogai et al. (2018) | PDB: 5YCG |
| Ancestral myoglobin aMbWb of Basilosaurus relative structure | Isogai et al. (2018) | PDB: 5YCH |
| Ancestral whale myoglobin aMbWb' of Basilosaurus relative structure | Isogai et al. (2018) | PDB: 5YCI |
| Recombinant DNA | ||
| pPAL7-aMbSp | This paper | N/A |
| pPAL7-aMbSe | This paper | N/A |
| pPAL7-esMb | This paper | N/A |
| Software and algorithms | ||
| Nika | Ilavsky (2012) | https://usaxs.xray.aps.anl.gov/software/nika |
| IGOR Pro version 6.22A | Wavemetrics | https://www.wavemetrics.com |
| ClustalW | Thompson et al. (2002) | http://www.clustal.org/download/current/ |
| XCED | Katoh et al. (2002) | N/A |
| PAML | Yang (2007) | http://abacus.gene.ucl.ac.uk/software/paml.html |
| MOSFLM | Battye et al. (2011) | https://www.mrc-lmb.cam.ac.uk/mosflm/imosflm/ver740/downloads.html |
| PHENIX | Adams et al., (2010) | http://www.phenix-online.org/ |
| MOLREP | Vagin and Teplyakov (2010) | https://www.ccp4.ac.uk/ |
| COOT | Emsley et al. (2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | Williams et al. (2018) | http://molprobity.biochem.duke.edu/ |
| Discovery Studio 2020 | Biovia | https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ |
Resource availability
Lead contact
Further information and requests for resources and materials should be directed to and will be fulfilled by the Lead Contact, Yasuhiro Isogai (yisogai@pu-toyama.ac.jp) and Tsuyoshi Shirai (t_shirai@nagahama-i-bio.ac.jp).
Material availability
Materials are available upon reasonable request.
Data and code availability
The atomic models for the ancestral and extant seal Mbs have been deposited at PDB and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The present manuscript did not generate any custom software. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Methods details
Prediction of ancestral Mb sequences
The pinnipeds’ ancestral sequences were calculated based on the sequence alignment and molecular phylogeny previously used for the analyses of the whales’ ancestral Mbs (Isogai et al., 2018). Briefly, the amino acid sequences were retrieved from the Genbank, Refseq, and UniProt databases, aligned with ClustalW, and manually refined with the XCED program (Altschul et al., 1990; Kaminuma et al., 2010; Katoh et al., 2002; O'Leary et al., 2016; Thompson et al., 2002). The constructed alignment contained a total of 2,449 sequences, including 266 Mbs, 2,152 Hbs, and 31 other globins. The phylogenetic tree was inferred with the neighbor joining (NJ) method with the JTT matrix (Jones et al., 1992; Saitou and Nei, 1987).
The phylogeny and the alignment were subjected to a PAML analysis (Yang, 2007; Yang and Nielsen, 1998; Yang and Rannala, 2006) (Figures 1, S2B, and S2C). The program codeml with the JTT matrix for the substitution rate matrix was used. Because the phylogeny contained a wide variety of species/molecules and it suggested the constraints were largely varied over local trees, a model with the least prerequisite parameter was adapted. A uniform substitution rate over the sites was assumed by fixing the shape parameter α of the gamma distribution for substitution rates. No molecular clock was assumed, and tree topology was fixed to that of the initial tree. The distributions of the posterior probabilities of the sites in the ancestral sequences are shown in Figure 2 and Table S1. No site showed posterior probabilities below 0.9, and the lowest value was 0.944 for D109 of aMbSp.
For verification, a comparison of the ancestral sequences between the present and previous works was attempted. Although no complete sequence was available in the published data, the charge-altering substitutions, namely, G57R and Q81H from aMbSp to aMbSe and Q8H, Q116H, and Q152H from aMbSe to esMb (Figure 3), were totally consistent with the previous reconstruction (Mirceta et al., 2013). The sequence alignment and phylogeny data supporting the findings of this study are available from the corresponding author T.S. on request.
Protein synthesis and purification
The ancestral and extant elephant seal Mbs were synthesized from artificial genes encoding the entire protein sequences, which were predicted as described earlier and obtained from NCBI Reference Sequence: XP_034843514.1, respectively. The DNA sequences were optimized for Escherichia coli expression. The genes, cloned into a pUC vector, were purchased from FASMAC Co., Ltd., Japan. The cloning vector was replaced with the expression vector pPAL7 (Bio-Rad Laboratories, Inc.), which contained a Profinity eXact fusion-tag gene, according to the Bio-Rad instruction manual (Ruan et al., 2004). E. coli strain BL21 (DE3) was transformed with the vector DNA, and the recombinant proteins were synthesized by expression under the control of the T7 promoter in Terrific broth (TB) medium, in cells induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) and hemin at 30°C. The harvested cells were disrupted by intense sonication at 4°C, in buffer containing 0.2 M sodium phosphate (pH 7.0) and 0.5% β-mercaptoethanol.
The synthesized protein with the affinity tag was extracted from the cell lysate by repeated centrifugation steps and purified by affinity chromatography on Profinity eXact resin (Bio-Rad Laboratories, Inc.). The affinity-tagged Mb was digested on the column, and the targeted Mb with the native N-terminal sequence was eluted with buffer containing 0.1 M NaF and 0.1 M sodium phosphate (pH 7.0), after washing with 0.2 M sodium phosphate buffer (pH 7.0). The protein was finally purified by size-exclusion chromatography, in 50 mM HEPES-NaOH (pH 7.0) and 200 mM NaCl, on a Superdex 75 Increase 10/300 GL column (GE Healthcare). The resultant solution was concentrated with an Amicon Ultra-4 centrifugal unit, after dialysis against buffer suitable for the next experiment. The protein identities were verified by matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry. Their apo-forms were prepared from the holo-proteins by the acid-butanone method, as described previously (Wittenberg and Wittenberg, 1981). The protein concentrations of the met and apo-forms in an aqueous solution at pH 7 were determined spectrophotometrically, using ε409 = 157,000 M-1 cm-1 and ε280 = 15200 M-1 cm-1, respectively (Antonini and Brunori, 1971).
As shown in Figure 2B, the synthesized Mb sequences had N-terminal methionine, whereas naturally occurring Mbs from animal skeletal muscle lack the methionine residue. These sequence differences are possible to affect functional and chemical properties. However, these properties of recombinant Mbs were indistinguishable from those of corresponding natural Mbs (Dodson et al., 1988; Guillemette et al., 1991; Scott et al., 2000; Springer and Sligar, 1987). Thus, we assumed that the recombinant Mbs synthesized here have properties identical to those from the natural products.
Crystal structure analyses
The structures of the extant and ancestral pinniped Mbs were determined by X-ray crystallography. The crystals were grown by the hanging drop vapor diffusion method. The esMb crystals were obtained under conditions using 3.1 M ammonium sulfate and 5.5% (w/v) PEG3350 in a 0.5-mL reservoir and a hanging drop mixture of 0.5 - 1.0 μL of reservoir solution and 1 μL of protein solution in 50 mM Tris-HCl (pH 8.0) buffer, containing 1 - 2% (w/v) protein. The crystals of aMbSp were obtained under conditions with 25% (w/v) PEG 1500 in MIB buffer (20 mM sodium malonate dibasic monohydrate, 30 mM boric acid, and 30 mM imidazole, pH 5.0) (imidazole-ligand structure) or 1.6 M tri-sodium citrate for the reservoir. The crystals of aMbSe were obtained under conditions using 2.5 M ammonium sulfate, 80 mM di-sodium tartrate, and 4% (v/v) methanol in MIB buffer (20 mM sodium malonate dibasic monohydrate, 30 mM boric acid, and 35 mM imidazole, pH 9.0) for the reservoir. All crystals were grown at 18°C for a few weeks.
X-ray diffraction data were collected from loop-mounted crystals under cryogenic conditions, using an EIGER4M (Dectris) detector at BL26B2 or a Quantum315r (ADSC) CCD detector at BL38B1 in SPring-8 (Hyogo, Japan). The crystals were soaked for 10–30 s in the corresponding crystal growth buffer, containing 15% (v/v) 2-methyl-2,4-pentanediol (MPD) for cryoprotection. The MOSFLM program was used for processing the diffraction images (Battye et al., 2011).
The Phaser-MR application of PHENIX or MOLREP in the CCP4 suites were used to solve the crystal structures by the molecular replacement method (Adams et al., 2010; Vagin and Teplyakov, 2010), and the structures were refined by using COOT and the phenix.refine application of PHENIX (Adams et al., 2010; Emsley et al., 2004). The quality of the models was evaluated with the MolProbity program (Williams et al., 2018). The crystallographic parameters, data collection and refinement statistics, and PDB codes are summarized in Table S2. The atomic coordinates and structure factors of imidazole-ligated aMbSp, aMbSp, aMbSe, and esMb have been deposited in the Protein Data Bank, with the accession codes 7DDR, 7DDS, 7DDT, and 7DDU, respectively. The UCSF-CHIMERA was used for preparing molecular graphics (Pettersen et al., 2004).
Molecular dynamics simulations
The AMBER12 package with the ff99SB force field for proteins and an amber-compatible force field for O2-bound heme were used for the MD simulations of the Mbs (Arcon et al., 2015; Case et al., 2012; Hornak et al., 2006). The crystal structures of the Mbs determined in this study were used as the starting structures by modeling the ligand O2 molecules.
The tautomeric states of the His residues at neutral pH were inferred by using the protonate3D function in the Molecular Operating Environment software (Chemical Computing Group). The His residues were protonated as follows: His12, 48, 64, 82, 97, 113, 116, and 119 in HIE form (protonated at Nε2 atom); His24, 81 and 93 in HID form Nδ1 (protonated at Nδ1 atom); and His36 in positively charged HIP form (protonated at Nδ1 and Nε2 atoms). All other residues and both termini of the proteins were considered in their standard protonation state at pH 7. The net charges of Mb in the holo-form were +1, +1, +1, and −2, and those in the apo-form were +3, +3, +3, and 0, for aMbSp, aMbSe, esMb, and hsMb, respectively. Na+ or Cl− ions were added to neutralize the simulation system. The solvent was explicitly considered with a truncated octahedral box of a TIP3P water model, with periodic boundary conditions. The box size was set so that the minimal distance from the protein atoms to the box faces was 10 Å. The particle-mesh Ewald method was used to treat the long-range electrostatic interactions by exploiting the periodic boundary conditions (Darden et al., 1993). The solvated systems were energy-minimized and then heated to 298 K and equilibrated.
First, the energy minimization was performed for 1,000 cycles with the heavy atoms restrained with a force constant of 500 kcal mol-1 Å-2, followed by 2,500 cycles without any restraints. Then, the heating from 0 to 298 K was performed under the heat bath coupling algorithm for temperature control with a time constant of 1 ps, in a 20-ps MD calculation at constant volume with the Mb atoms restrained by a force constant of 10 kcal mol-1 Å-2 (Berendsen et al., 1984). The system was then subjected to an 80-ps MD calculation at constant temperature (298 K) controlled by Langevin dynamics with a collision frequency of 2 ps-1 and constant pressure (1 bar) controlled by isotropic position scaling with a relaxation time of 1 ps, with the same restraints on the Mb atoms. Finally, a 60-ns MD calculation with the same controls for constant temperature and constant pressure (the NTP ensemble) was performed after releasing all of the restraints, and the last 50-ns trajectory (snapshot coordinates of the system, sampled every 5 ps) was used for the data analyses. The tools in the AMBER package were used to calculate the radius of gyration (Rg), the RMSD (root mean square deviation) from the starting structure, the number of hydrogen bonds between Mb and water, and the conformational energy of Mb. The ASAs of the models in the trajectories were evaluated by an in-house program based on the Shrake-Rupley algorithm (Shrake and Rupley, 1973). The hydrophobic (ASAhydrophobic) and hydrophilic (ASAhydrophilic) ASAs were the sums of the areas belonging to carbon and hetero atoms, respectively.
SFE calculations
The reference-modified density functional theory (RMDFT) was applied to calculate the SFE for the Mbs in the same manner as our previous work (Isogai et al., 2018; Sumi et al., 2015, 2016, 2018). To calculate the site-density distribution functions of water around the Mbs that were used in the RMDFT, the three-dimensional reference-interaction-site-model (3D-RISM) integral equation with the Kovalenko-Hirata (KH) closure (Kovalenko and Hirata, 1999) was used. To prepare the site-site direct correlation functions for bulk water and for the reference hard-sphere fluid, the one-dimensional (1D) RISM integral equation with the KH closure (Kovalenko and Hirata, 1999) and the effective-density approximation (EDA) integral equation (Sumi and Sekino, 2008) were respectively applied.
For the 1D-RISM and EDA calculations, 0.00125 Å and 32,768 were used as the grid spacing and the number of grids, respectively. The number density of water and the temperature used in all the calculations were 0.033329 molecule/Å3 and 298 K, respectively. The 3D-RISM integral equation calculations were performed using a grid of 2563 points in an 80 Å3 cubic cell and accelerated by graphics processing units (GPUs) (Maruyama and Hirata, 2012). The SFE was determined by the following equation using 10,000 conformations of each Mb, which were provided by the MD simulations:
| (Equation 1) |
where is the SFE for conformation i, is the ensemble average over the conformations, is the Boltzmann constant, and is the temperature. In Equation (1), the first term is the simple average of and the second term yields the fluctuation effect on due to the conformation fluctuations.
Here, we defined the effective energy , where is the intramolecular energy for conformation i of the protein. It is noted that provides a Boltzmann distribution of the conformation i for a protein in water. To characterize the fluctuation of , we determined the derivative of as a function of the radius of gyration using least squares fitting to four blocks of 2,500 conformations and obtained their average as . A positive value of indicates the rigidity of the protein structure around stable basins, and thus, a smaller positive value of represents the greater flexibility of the protein structure around the basins.
PEG sedimentation analyses
The Mb solubility was measured in the presence of a precipitant PEG-6000. The purified holo-Mbs were first solubilized at 1.8∼4.6 mM in 10 mM HEPES-NaOH (pH6.8) and 10∼40% PEG-6000. The samples were equilibrated for 2 hours at a room temperature and were centrifuged at 20,000×g for 30 min to remove the precipitant. The visible absorption spectra of the supernatants were measured using a Nano Drop spectrometer, and the Mb concentrations were determined by using an ε409nm of 157000 M-1 cm-1 (Walenta, 1985). The Mb concentrations or solubilities (S) were plotted against the PEG concentration. The solubility dependence on the PEG concentration was analyzed by assuming
| (Equation 2) |
where S0 and β are the solubility in the absence of precipitant and the dependence of S on the precipitant concentration, respectively (Figure 4A) (Kramer et al., 2012). We have confirmed that β and Log S0 did not depend on the initial Mb concentrations used here because these values were determined by the solubilities in the presence of the precipitant at the concentrations, under which the S values were less than the initial Mb concentrations.
Small-angle X-ray scattering
The SAXS experiments were performed at the beamline BL-10C, in the Photon Factory (PF) of the High Energy Accelerator Research Organization (KEK), Tsukuba, Japan. The purified Mb solutions were dialyzed against a 2 mM HEPES–NaOH buffer solution (pH 6.8) at 4°C for one day. The dialyzed Mb solutions were concentrated to ∼3–5 mM and then centrifuged to remove the precipitates. The Mb solutions were diluted to the desired concentrations at a pH of 6.8 ± 0.2 and irradiated with an X-ray wavelength λ = 0.15 nm (camera length of 1 m) for 2 s in a cell with quartz windows, using a sample-flow system (∼14.5 μL/min) at 20 ± 0.1°C. X-ray intensities were recorded using a PILATUS3 2M detector (DECTRIS Ltd., Switzerland). A total of 30 images were collected for each condition, and the circular 1D averaging of the images was performed using the program Nika (Ilavsky, 2012).
The scattering parameter q = |q| = 4πsinθ/λ, where q is the scattering vector and 2θ is the X-ray scattering angle, which in this experiment was 0.01–0.6 Å-1. The scattering intensity was corrected by the intensity of the incident light and the transmittance of the X-rays. The absolute scattering intensity of the protein (I(q)) was determined as
| I(q) = [IS(q) – (1 – cpv)IB(q)]/f, | (Equation 3) |
where cp is the protein concentration (g/cm3), v is the specific volume of the solute (0.7425 cm3/g), and f is the correction factor to convert the observed intensity in arbitrary units to the absolute intensity in units of cm-1. The procedure was detailed in the study by Isogai et al., 2018.
The absolute scattering intensity of the protein (I(q)) was extrapolated to the absolute scattering intensity I(0) at q = 0. The I(0) value is related to the second virial coefficient A2 by
| I(0) = kMcp / (1 + 2A2Mcp), | (Equation 4) |
where M is the molecular weight of the protein and the k value is equal to v2(ρm – ρsolv)2/NA. NA is Avogadro's number, and ρm – ρsolv is the electron density difference between the protein and the solvent (typically 2.8 × 1010 cm-2) (Goldenberg and Argyle, 2014; Zimm, 1948).
Folding stability analyses
Thermodynamic stabilities of the synthesized Mbs in the apo-forms were measured by chemical denaturation experiments with guanidine hydrochloride (Gd-HCl). Each apoMb solubilized in 50 mM HEPES-NaOH (pH 7) at 5 μM protein was denatured by the addition of Gd-HCl, and the signal intensity of circular dichroism (CD) at 222 nm was monitored at 20°C and was plotted against Gd-HCl concentration. The denaturation profile was analyzed using a theoretical curve derived from the three-state transition model (Barrick and Baldwin, 1993), according to Isogai, (2006), Isogai et al. (2000). The theoretical curve was fitted to the observed denaturation data to obtain the thermodynamic parameters ΔG1, ΔG2, m1 and m2. The sum of ΔG1 and ΔG2 (ΔG1+2 =ΔG1+ ΔG2) gives the free energy change from the unfolded state to the folded state.
Data analyses
The PCA and clustering of the parameters were executed by using the R package (Team, 2007). All parameters except for evolutionary distance (d) were applied to the prcomp function, with scaling for the PCA. The parameters including d were subjected to clustering with the Ward method by referring to a matrix of 1- |Rij|, where Rij is the Pearson product-moment correlation coefficient between parameters i and j.
Acknowledgments
We thank Satoshi Kawasaki (https://paleontology.sakura.ne.jp/) for the kind permission to use his illustrations of animals. X-ray diffraction and SAXS experiments were conducted at SPring-8 and KEK, Japan, respectively, under the approval of the Photon Factory Program Advisory Committee: Proposal No. 2018G032 and No. 2020G077. This work was partly supported by Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology-Japan (JP17H01818, JP21K06503 and JP21H03547) and the Platform Project for Supporting Drug Discovery and Life Science Research (Basis of Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED (JP20am0101111j0004 support number 0792).
Author contributions
Y.I. and T. Shirai designed the research. K.T. and T. Shirai predicted the ancestral protein sequences. Y.I. synthesized proteins and performed biochemical analyses. S.N. and T. Shirai performed protein crystallization and 3D structure determination. Y.I., T. Sumi, K.T., and T. Shirai conducted theoretical calculations. H.I. and Y.I. performed SAXS measurements and analyses. Y.I. and T. Shirai wrote the main manuscript text. All authors reviewed the manuscript.
Declaration of interests
The authors declare that they have no financial and/or nonfinancial competing interests with the contents of this article.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102920.
Contributor Information
Yasuhiro Isogai, Email: yisogai@pu-toyama.ac.jp.
Tsuyoshi Shirai, Email: t_shirai@nagahama-i-bio.ac.jp.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antonini E., Brunori M. North-Holland); 1971. Hemoglobin and Myoglobin in Their Reactions with Ligands. [DOI] [Google Scholar]
- Arcon J.P., Rosi P., Petruk A.A., Marti M.A., Estrin D.A. Molecular mechanism of myoglobin autoxidation: insights from computer simulations. J. Phys. Chem. B. 2015;119:1802–1813. doi: 10.1021/jp5093948. [DOI] [PubMed] [Google Scholar]
- Asakura S., Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J. Chem. Phys. 1954;22:1255–1256. doi: 10.1063/1.1740347. [DOI] [Google Scholar]
- Atha D.H., Ingham K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981;256:12108–12117. doi: 10.1016/S0021-9258(18)43240-1. [DOI] [PubMed] [Google Scholar]
- Barrick D., Baldwin R.L. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993;32:3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbrink M. The role of myoglobin in the evolution of mammalian diving capacity - the August Krogh principle applied in molecular and evolutionary physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021;252:110843. doi: 10.1016/j.cbpa.2020.110843. [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- Berta A. Pinniped evolution. In: Würsig B., Thewissen J.G.M., Kovacs K.M., editors. Encyclopedia of Marine Mammals. Third Edition. Academic Press; 2018. pp. 712–722. [DOI] [Google Scholar]
- Berta A., Ray C.E., Wyss A.R. Skeleton of the oldest known pinniped, Enaliarctos mealsi. Science. 1989;244:60–62. doi: 10.1126/science.244.4900.60. [DOI] [PubMed] [Google Scholar]
- Bloustine J., Virmani T., Thurston G.M., Fraden S. Light scattering and phase behavior of lysozyme-poly(ethylene glycol) mixtures. Phys. Rev. Lett. 2006;96:087803. doi: 10.1103/PhysRevLett.96.087803. [DOI] [PubMed] [Google Scholar]
- Case D.A., Darden T.A., Cheatham T.E., Simmerling C.L., Wang J., Duke R.E., Luo R., Walker R.C., Zhang W., Merz K.M. University of California; 2012. AMBER 12.https://ambermd.org/doc12/Amber12.pdf [Google Scholar]
- Chan P., Curtis R.A., Warwicker J. Soluble expression of proteins correlates with a lack of positively-charged surface. Sci. Rep. 2013;3:3333. doi: 10.1038/srep03333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D.R., Boczek T., Grelewska K., Pinkowska M., Sikorska M., Zawadzki M., Derewenda Z. Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr. D Biol. Crystallogr. 2007;63:636–645. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Dasmeh P., Kepp K.P. Bridging the gap between chemistry, physiology, and evolution: quantifying the functionality of sperm whale myoglobin mutants. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2012;161:9–17. doi: 10.1016/j.cbpa.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Dasmeh P., Serohijos A.W., Kepp K.P., Shakhnovich E.I. Positively selected sites in cetacean myoglobins contribute to protein stability. PLoS Comput. Biol. 2013;9:e1002929. doi: 10.1371/journal.pcbi.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.W. A review of the multi-level adaptations for maximizing aerobic dive duration in marine mammals: from biochemistry to behavior. J. Comp. Physiol. B. 2014;184:23–53. doi: 10.1007/s00360-013-0782-z. [DOI] [PubMed] [Google Scholar]
- Davis R.W., Kanatous S.B. Convective oxygen transport and tissue oxygen consumption in Weddell seals during aerobic dives. J. Exp. Biol. 1999;202:1091–1113. doi: 10.1242/jeb.202.9.1091. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S., Vekilov P.G. Entropy and surface engineering in protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- Dodson G., Hubbard R.E., Oldfield T.J., Smerdon S.J., Wilkinson A.J. Apomyoglobin as a molecular recognition surface: expression, reconstitution and crystallization of recombinant porcine myoglobin in Escherichia coli. Protein Eng. 1988;2:233–237. doi: 10.1093/protein/2.3.233. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2004;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.V., Brayer G.D. High-resolution study of the three-dimensional structure of horse heart metmyoglobin. J. Mol. Biol. 1990;213:885–897. doi: 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- Flynn J.J., Finarelli J.A., Zehr S., Hsu J., Nedbal M.A. Molecular phylogeny of the carnivora (mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst. Biol. 2005;54:317–337. doi: 10.1080/10635150590923326. [DOI] [PubMed] [Google Scholar]
- Fulton A.B. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- Goldenberg D.P., Argyle B. Self crowding of globular proteins studied by small-angle x-ray scattering. Biophys. J. 2014;106:895–904. doi: 10.1016/j.bpj.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette J.G., Matsushima-Hibiya Y., Atkinson T., Smith M. Expression in Escherichia coli of a synthetic gene coding for horse heart myoglobin. Protein Eng. 1991;4:585–592. doi: 10.1093/protein/4.5.585. [DOI] [PubMed] [Google Scholar]
- He K., Eastman T.G., Czolacz H., Li S., Shinohara A., Kawada S.-i., Springer M.S., Berenbrink M., Campbell K.L. Myoglobin primary structure reveals multiple convergent transitions to semi-aquatic life in the world's smallest mammalian divers. Elife. 2021;10:e66797. doi: 10.7554/eLife.66797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbo S., Fago A. Functional properties of myoglobins from five whale species with different diving capacities. J. Exp. Biol. 2012;215:3403–3410. doi: 10.1242/jeb.073726. [DOI] [PubMed] [Google Scholar]
- Hojgaard C., Kofoed C., Espersen R., Johansson K.E., Villa M., Willemoes M., Lindorff-Larsen K., Teilum K., Winther J.R. A soluble, folded protein without charged amino acid residues. Biochemistry. 2016;55:3949–3956. doi: 10.1021/acs.biochem.6b00269. [DOI] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilavsky J. Nika: software for two-dimensional data reduction. J. Appl. Crystallogr. 2012;45:324–328. doi: 10.1107/S0021889812004037. [DOI] [Google Scholar]
- Isogai Y. Native protein sequences are designed to destabilize folding intermediates. Biochemistry. 2006;45:2488–2492. doi: 10.1021/bi0523714. [DOI] [PubMed] [Google Scholar]
- Isogai Y., Imamura H., Nakae S., Sumi T., Takahashi K.I., Nakagawa T., Tsuneshige A., Shirai T. Tracing whale myoglobin evolution by resurrecting ancient proteins. Sci. Rep. 2018;8:16883. doi: 10.1038/s41598-018-34984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Ishii A., Fujisawa T., Ota M., Nishikawa K. Redesign of artificial globins: effects of residue replacements at hydrophobic sites on the structural properties. Biochemistry. 2000;39:5683–5690. doi: 10.1021/bi992687+. [DOI] [PubMed] [Google Scholar]
- Israelachvili J.N., Wennerstroem H. Hydration or steric forces between amphiphilic surfaces? Langmuir. 1990;6:873–876. doi: 10.1021/la00094a028. [DOI] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaminuma E., Mashima J., Kodama Y., Gojobori T., Ogasawara O., Okubo K., Takagi T., Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooyman G.L. Springer; 1989. Diverse Divers : Physiology and Behavior. [DOI] [Google Scholar]
- Kovalenko A., Hirata F. Self-consistent description of a metal–water interface by the Kohn–Sham density functional theory and the three-dimensional reference interaction site model. J. Chem. Phys. 1999;110:10095–10112. doi: 10.1063/1.478883. [DOI] [Google Scholar]
- Kramer R.M., Shende V.R., Motl N., Pace C.N., Scholtz J.M. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys. J. 2012;102:1907–1915. doi: 10.1016/j.bpj.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y., Suenaga A., Sato Y., Kosuda S., Taiji M. All-atom molecular dynamics analysis of multi-peptide systems reproduces peptide solubility in line with experimental observations. Sci. Rep. 2016;6:19479. doi: 10.1038/srep19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Maruyama Y., Hirata F. Modified anderson method for accelerating 3D-RISM calculations using graphics processing unit. J. Chem. Theor. Comput. 2012;8:3015–3021. doi: 10.1021/ct300355r. [DOI] [PubMed] [Google Scholar]
- McGowen M.R., Gatesy J., Wildman D.E. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 2014;29:336–346. doi: 10.1016/j.tree.2014.04.001. [DOI] [PubMed] [Google Scholar]
- McIntyre I.W., Campbell K.L., MacArthur R.A. Body oxygen stores, aerobic dive limits and diving behaviour of the star-nosed mole (Condylura cristata) and comparisons with non-aquatic talpids. J. Exp. Biol. 2002;205:45–54. doi: 10.1242/jeb.205.1.45. [DOI] [PubMed] [Google Scholar]
- McLellan T. Molecular charge and electrophoretic mobility in cetacean myoglobins of known sequence. Biochem. Genet. 1984;22:181–200. doi: 10.1007/BF00499297. [DOI] [PubMed] [Google Scholar]
- Mirceta S., Signore A.V., Burns J.M., Cossins A.R., Campbell K.L., Berenbrink M. Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science. 2013;340:1234192. doi: 10.1126/science.1234192. [DOI] [PubMed] [Google Scholar]
- Nery M.F., Arroyo J.I., Opazo J.C. Accelerated evolutionary rate of the myoglobin gene in long-diving whales. J. Mol. Evol. 2013;76:380–387. doi: 10.1007/s00239-013-9572-1. [DOI] [PubMed] [Google Scholar]
- Noren S.R., Williams T.M. Body size and skeletal muscle myoglobin of cetaceans: adaptations for maximizing dive duration. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2000;126:181–191. doi: 10.1016/s1095-6433(00)00182-3. [DOI] [PubMed] [Google Scholar]
- O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poust A., Boessenecker R. Expanding the geographic and geochronologic range of early pinnipeds: new specimens of Enaliarctos from Northern California and Oregon. Acta Palaeontologica Pol. 2018;63:25–40. doi: 10.4202/app.00399.2017. [DOI] [Google Scholar]
- Romero-Herrera A.E., Lehmann H., Joysey K.A., Friday A.E. On the evolution of myoglobin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1978;283:61–163. doi: 10.1098/rstb.1978.0018. [DOI] [PubMed] [Google Scholar]
- Ruan B., Fisher K.E., Alexander P.A., Doroshko V., Bryan P.N. Engineering subtilisin into a fluoride-triggered processing protease useful for one-step protein purification. Biochemistry. 2004;43:14539–14546. doi: 10.1021/bi048177j. [DOI] [PubMed] [Google Scholar]
- Rybczynski N., Dawson M.R., Tedford R.H. A semi-aquatic Arctic mammalian carnivore from the Miocene epoch and origin of Pinnipedia. Nature. 2009;458:1021–1024. doi: 10.1038/nature07985. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Samuel P.P., Smith L.P., Phillips G.N., Jr., Olson J.S. Apoglobin stability is the major factor governing both cell-free and in vivo expression of holomyoglobin. J. Biol. Chem. 2015;290:23479–23495. doi: 10.1074/jbc.M115.672204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.E., Paster E.V., Olson J.S. The stabilities of mammalian apomyoglobins vary over a 600-fold range and can be enhanced by comparative mutagenesis. J. Biol. Chem. 2000;275:27129–27136. doi: 10.1074/jbc.M000452200. [DOI] [PubMed] [Google Scholar]
- Shaw K.L., Grimsley G.R., Yakovlev G.I., Makarov A.A., Pace C.N. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001;10:1206–1215. doi: 10.1110/ps.440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shire S.J., Hanania G.I., Gurd F.R. Electrostatic effects in myoglobin. Application of the modified Tanford-Kirkwood theory to myoglobins from horse, California grey whale, harbor seal, and California sea lion. Biochemistry. 1975;14:1352–1358. doi: 10.1021/bi00678a002. [DOI] [PubMed] [Google Scholar]
- Shrake A., Rupley J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Skoog B., Wichman A. Calculation of the isoelectric points of polypeptides from the amino acid composition. Trac Trends Anal. Chem. 1986;5:82–83. doi: 10.1016/0165-9936(86)80045-0. [DOI] [Google Scholar]
- Snyder G.K. Respiratory adaptations in diving mammals. Respir. Physiol. 1983;54:269–294. doi: 10.1016/0034-5687(83)90072-5. [DOI] [PubMed] [Google Scholar]
- Spassov V.Z., Yan L. A fast and accurate computational approach to protein ionization. Protein Sci. 2008;17:1955–1970. doi: 10.1110/ps.036335.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov V.Z., Yan L. pH-selective mutagenesis of protein-protein interfaces: in silico design of therapeutic antibodies with prolonged half-life. Proteins. 2013;81:704–714. doi: 10.1002/prot.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B.A., Sligar S.G. High-level expression of sperm whale myoglobin in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T., Maruyama Y., Mitsutake A., Koga K. A reference-modified density functional theory: an application to solvation free-energy calculations for a Lennard-Jones solution. J. Chem. Phys. 2016;144:224104. doi: 10.1063/1.4953191. [DOI] [PubMed] [Google Scholar]
- Sumi T., Maruyama Y., Mitsutake A., Mochizuki K., Koga K. Application of reference-modified density functional theory: temperature and pressure dependences of solvation free energy. J. Comput. Chem. 2018;39:202–217. doi: 10.1002/jcc.25101. [DOI] [PubMed] [Google Scholar]
- Sumi T., Mitsutake A., Maruyama Y. A solvation-free-energy functional: a reference-modified density functional formulation. J. Comput. Chem. 2015;36:1359–1369. doi: 10.1002/jcc.23942. [DOI] [PubMed] [Google Scholar]
- Sumi T., Sekino H. A self-consistent density-functional approach for homogeneous and inhomogeneous classical fluids. J. Phys. Soc. Jpn. 2008;77:034605. doi: 10.1143/jpsj.77.034605. [DOI] [Google Scholar]
- Team R.D.C. R Foundation for Statistical Computing; 2007. R : A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002;2:2 3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909052925. [DOI] [PubMed] [Google Scholar]
- Velten B.P., Dillaman R.M., Kinsey S.T., McLellan W.A., Pabst D.A. Novel locomotor muscle design in extreme deep-diving whales. J. Exp. Biol. 2013;216:1862–1871. doi: 10.1242/jeb.081323. [DOI] [PubMed] [Google Scholar]
- Walenta E. Small angle X-ray scattering. Acta Polymerica. 1985;36:296. doi: 10.1002/actp.1985.010360520. [DOI] [Google Scholar]
- Wilkinson D.L., Harrison R.G. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (N Y) 1991;9:443–448. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg J.B., Wittenberg B.A. Methods in Enzymology. Academic Press; 1981. [2] Preparation of myoglobins; pp. 29–42. [DOI] [Google Scholar]
- Wright T.J., Davis R.W. The effect of myoglobin concentration on aerobic dive limit in a Weddell seal. J. Exp. Biol. 2006;209:2576–2585. doi: 10.1242/jeb.02273. [DOI] [PubMed] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998;46:409–418. doi: 10.1007/PL00006320. [DOI] [PubMed] [Google Scholar]
- Yang Z., Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Zimm B.H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 1948;16:1093–1099. doi: 10.1063/1.1746738. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic models for the ancestral and extant seal Mbs have been deposited at PDB and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The present manuscript did not generate any custom software. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.