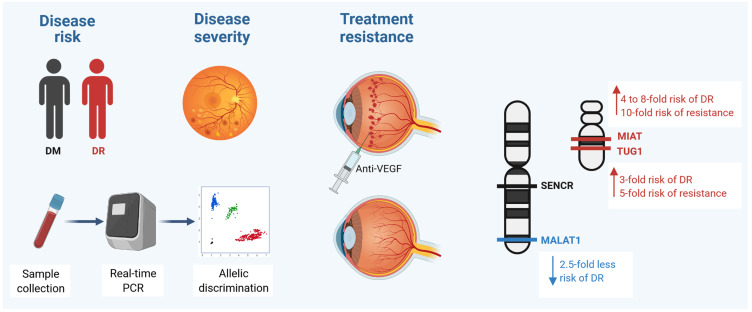
Abstract
Background
Long non-coding RNAs (lncRNAs) play essential roles in molecular diagnosis and therapeutic response in several diseases.
Purpose
For the first time, we aimed to evaluate the association of four lncRNAs TUG1 (rs7284767G/A), MIAT (rs1061540T/C), MALAT1 (rs3200401C/T), and SENCR (rs12420823C/T) variants with susceptibility to diabetic retinopathy (DR), disease severity, and early therapeutic response to intravitreous anti-vascular endothelial growth factor aflibercept therapy.
Patients and Methods
This case-control study enrolled 126 adult patients with type 2 diabetes. TaqMan assays using Real-Time PCR were run for genotyping. Multivariable regression analyses were applied to assess the role of each polymorphism after the adjustment of covariates.
Results
Carriers of TUG1 A/G and MIAT T/C and C/C genotypes were more likely to develop DR [OR=3.15 (95% CI=1.15–8.64), and OR=4.31 (95% CI=1.78–10.47)], while MALAT1 T/C conferred protection (OR=0.40, 95% CI=0.16–0.99). For TUG1, MALAT1, MIAT, and SENCR genotype combinations, GTCT and GCCC had a higher disease risk (P=0.012). For disease severity, MIAT T/T homozygosity was associated with higher DR grade [33.3% (T/T) vs 10% (C/C) and 4.2% (C/T) carriers, P=0.012]. Otherwise, patients with the SENCR T variant exhibited better pre-treatment best-corrected visual acuity level (p=0.021). Following aflibercept administration, carrying the TUG1 A or MIAT T/C was associated with a poor therapeutic response (OR=5.02, 95% CI=1.60–15.76, and OR=10.23, 95% CI=1.51–69.15, respectively).
Conclusion
The lncRNAs TUG1 (rs7284767G/A) and MIAT (rs1061540T/C) were associated with increased DR susceptibility and poor response to aflibercept treatment, while MALAT1 (rs3200401C/T) conferred protection to DR. These genetic determinants could be useful in DR risk stratification and pharmacogenetics after validation in large-scale studies.
Keywords: aflibercept, diabetic retinopathy, MALAT1, MIAT, SENCR, TUG1
Graphical Abstract
Introduction
Diabetes mellitus (DM) is a devastating health problem worldwide with an estimated global prevalence of 9.3% (463 million people) in 2019, predicted to rise to 10.2% (578 million) by 2030, according to the recent “International Diabetes Federation” report.1 Diabetic retinopathy (DR) is one of the main microvasculopathy associated with long-term diabetes. Retinal inflammation/neovascularization, vascular hyperpermeability, and cell apoptosis play major roles in DR etiopathology.2 Healthy endothelial cells are the basis of normal blood vessels, whereas endothelial cell dysfunction is a risk indicator of diabetic angiopathy.3 Accumulating evidence indicated that diabetes-related microvascular complications result from genetic and environmental interactions.4,5
Long non-coding RNAs (lncRNAs), which are >200 nucleotides in length, are emerging regulatory non-coding RNAs, participate in epigenetic and transcriptional/post-transcriptional regulation of several cellular pathways in different diseases, including DM and its complications.6–9 Several lncRNAs have been implicated in DR,10–12 and their deregulation was proved to be associated with DR susceptibility and/or response to treatment13,14 (Table 1).
Table 1.
The Role of lncRNAs in Diabetes and Diabetic Complications
| Disease Name | LncRNA (Alias) | Dysfunction | Description | Location | PMID |
|---|---|---|---|---|---|
| Diabetes mellitus | CDKN2B-AS1 (ANRIL) | Mutation, Expression, Locus | Genetic variation in lncRNA genes causes disease and influences susceptibility. | 9p21.3 | 23791884, 20386740, 23104877, 20956613, 17463249, 20956613, 17463248, 18048406, 24624135 |
| CDKN2B-AS5 | Mutation | GWAS identified several variants in the intergenic region encompassing ANRIL to be associated with several diseases such as T5D. | N/A | 22928560 | |
| GAS5 (SNHG2) | Expression | LncRNA GAS5 levels are correlated to the prevalence of T2DM. | 1q25.1 | 26675493 | |
| H19 | Expression | Associated with increased birth weight; higher expression in T2D patients. | 11p15.5 | 17463249 | |
| IGF2-AS (PEG8) | Mutation | Association identified by GWAS. | 11p15.5 | 17554260 | |
| LINC01370 (HILNC25) | Regulation | Depletion of HI-LNC25, cell-specific lncRNA, down-regulated GLIS3 mRNA, thus exemplifying a gene regulatory function of islet lncRNAs. Finally, selected islet lncRNAs were dysregulated in type 2 diabetes or mapped to genetic loci underlying diabetes susceptibility. | 20q12 | 23040067 | |
| LINC00271 | Mutation | Association identified by GWAS. | 6q23.3 | 17668382 | |
| MALAT1 (NEAT2) | Expression | In addition, MALAT1, a conserved lncRNA, was significantly upregulated in an RF/6A cell model of hyperglycemia in the aqueous humor samples and fibrovascular membranes of diabetic patients. | 11q13.1 | 24436191 | |
| RNCR2 (MIAT, GOMAFU) | N/A | May affect β-cell mass. | 22q12.1 | 20486133 | |
| TUG1 | Regulation | A direct interaction between PGC-1α and Tug1 modulates mitochondrial bioenergetics in podocytes in the diabetic milieu. | 11 | 27760051 | |
| MEG3 (GTL2) | Locus, Expression | The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. | 14q32 | 19966805, 26845358, 26603935 | |
| MEG3 up-regulation may serve as a therapeutic strategy for treating diabetes-related microvascular complications. | |||||
| MEG3 may be a potential target and therapeutic strategy for diabetes. MEG3 knockdown aggravates retinal vessel dysfunction in vivo and regulates retinal endothelial cell proliferation, migration, and tube formation in vitro. | |||||
| NEAT1 | Regulation | Regulates mTOR signaling pathway. | 11q13.1 | 28643459 | |
| NONHSAG011351 | Regulation | ERBB3, whose locus associated lncRNA (NONHSAG011351) was expressed in human islets, may constitute novel targets to prevent β-cell destruction in T1D. | 12q24.13 | 26450151 | |
| PDZRN3-AS1 | Mutation | SNP rs11128347 (C>G) in PDZRN3 is associated with African Americans with type 2 diabetes. | 3p13 | 21546767 | |
| PINK1-AS | Expression | PINK1 is induced by PTEN, which is an important inhibitor of insulin signaling. PINK1 depletion has been associated with diabetes status, impaired glucose uptake in neuronal cell lines, and mitochondrial gene expression in adipocytes, raising the possibility that disruption to naPINK1 may impact glucose metabolism. | 1p36.12 | 22817756 | |
| PLUTO (PDX1-AS1) | N/A | Regulates PDX1 expression. | 13q12.2 | 28041957 | |
| PVT1 (onco-lncRNA-100) | Mutation | There is an association between variants (rs2720709, A>G) in the plasmacytoma variant translocation 1 gene (PVT1) and end-stage renal disease (ESRD) attributed to both type 1 and type 2 diabetes. Identification of PVT1 (rs2720709, A>G) as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. |
8q24.21 | 21526116, 17395743 | |
| RNCR3 (LINC00599) | Locus | RNCR3 knockdown may be a promising strategy for the prevention of diabetes mellitus-induced retinal neurodegeneration. | 8p23.1 | 27616193 | |
| Diabetic retinopathy | MALAT1 (NEAT2) | Regulation Expression |
MALAT1 knockdown could regulate retinal endothelial cell proliferation, migration, and tube formation in vitro. MALAT1 down-regulation could ameliorate DR by functioning as a competing endogenous RNA in regulating VEGF levels through miR-150-5p. MALAT1 may become a potential therapeutic target for the prognosis, diagnosis, and treatment of DR. associated with markers of visual and retinal vessel function. Activates inflammatory pathway via TNF-α and IL-6. | 11q13.1 | 25356875, 24436191 |
| RNCR2 (MIAT, GOMAFU) | Regulation | MIAT knockdown could repress TNF-α-induced abnormal proliferation and migration of HLECs, by acting as a ceRNA. Attenuates retinal vessel impairment and vascular leakage and formed a feedback loop with Akt and miR-150-5p. NF-κB activation | 22q12.1 | 27043545, 29074557 | |
| MEG3 | N/A | Modulates angiogenesis by PI3K/Akt | 14q32.2 | 26845358 | |
| CDKN2B-AS1 (ANRIL) | N/A | Increases retinal microvascular permeability in vivo Increases VEGF mediated by PRC2 complex and p300 |
9p21.3 | 28122089, 23813974 | |
| RNCR3(LINC00599) | N/A | Increases cell viability and proliferation, promotes EC migration and tube formation in vitro Aggravates retinal cell apoptosis, visual function, and microvascular leakage in vivo Related to the release of several cytokines |
8p23.1 | 21857657, 27253412 | |
| BDNF-AS | N/A | Increases cell apoptosis. Cause early neurodegeneration |
11p14.1 | 23271640, 26004392 | |
| SOX2OT | N/A | Mediates glucose-induced retinal injury. Antioxidative via regulation of NRF2/HO-1 signaling activity Promotes neurodegeneration |
3q26.33 | 27193103, 29074557, 18846214 | |
| Diabetic cardiomyopathy | H19 (WT2) | Regulation | LncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. | 11p15.5 | 27796346 |
| MALAT1 (NEAT2) | Regulation | Involvement of long non-coding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. | 11q13.1 | 26476026 | |
| Diabetic nephropathy | CYP4B1-PS1 | Regulation | A novel long non-coding RNA CYP4B1 PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. may regulate proliferation and fibrosis in mesangial cells [52 | N/A | 26923441 |
| ENSMUST00000147869 | Regulation | Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. | N/A | 27083175 | |
| PVT1 (onco-lncRNA-100) | Expression Interaction Regulation |
Variants in the plasmacytoma variant translocation gene were strongly associated with DKD in the Pima Indians, a group with the highest prevalence of type 2 diabetes in the world. PVT1 may mediate the development and progression of diabetic nephropathy through mechanisms involving ECM accumulation. Role of MicroRNA 1207–5P and Its Host Gene, the Long Noncoding RNA Pvt1, as Mediators of Extracellular Matrix Accumulation in the Kidney. |
8q24.21 | 27503944, 21526116, 24204837, 24204837 | |
| RNCR2 (MIAT, GOMAFU) | Regulation | Mediates high glucose-induced renal tubular epithelial injury. | 22q12.1 | 26551455 |
Note: The Bold names were selected in this study. Data source: LncRNADisease database (http://www.cuilab.cn) and literature search.
The lncRNA TUG 1 “Taurine UP-regulated 1” was reported as one of the identified genes that are up-regulated in response to amino acid taurine, which induces rod photoreceptor biogenesis.15,16 This type of lncRNA is essential for photoreceptors biogenesis in the developing rodent retina.16 Knockdown of TUG1 resulted in a defect of migration of the developing rod photoreceptors into the outer nuclear layer and increased transfected cell apoptosis.17 Furthermore, TUG1 upregulation stimulates Wnt pathway-dependent proliferation and migration of endothelial cells, thereby promoting the occurrence and progression of diabetic atherosclerosis.18
Another lncRNA reported to be implicated in DR pathophysiology is MIAT “Myocardial Infarction Associated Transcript”, also known as Gomafu, or RNCR2 “retinal non-coding RNA 2,” which is expressed in cardiomyocytes and the nucleus of multiple retinal cells.19 It was found to be up-regulated in the retina of diabetic rats and the fibrovascular membrane of patients with diabetes induced by cellular hyperglycemia.20
The highly conserved lncRNA MALAT1 “metastasis-associated lung adenocarcinoma transcript 1”, also named NEAT2 “nuclear-enriched abundant transcript 2”,21 showed significant overexpression in hyperglycemic cell model (the RF/6A), in samples from the aqueous humor, and fibrovascular membranes of patients with diabetes.11 It can regulate retinal endothelial cell pathophysiology and microvascular growth under a hyperglycemic milieu via several cellular pathways, including the “p38/MAPK (mitogen-activated protein kinase)” signaling pathway.22 Silencing of MALAT1 significantly mitigates diabetes-induced retinal neovascularization, vascular hyperpermeability, and retinitis.23
Lastly, the cytoplasmic lncRNA-SENCR “Smooth muscle and Endothelial cell-enriched migration/differentiation-associated long non-coding RNA” was found to be a vascular cell-enriched lncRNA that can regulate FoxO1 “fork-head box protein O1” and TRPC6 “Transient Receptor Potential Cation channel 6”, hence promoting the proliferation and migration of smooth muscle cells.24 SENCR has been implicated in regulating pluripotent cells endothelial differentiation and human vascular endothelial cell angiogenic capacity.25 Its silencing downregulates myocardin and smooth muscle contractile genes and up-regulates the cell migration-related genes.26
Several studies have reported that lncRNA-related single nucleotide polymorphisms (SNPs) may impact susceptibility to DR.27,28 However, the TUG1 (rs7284767G/A), MIAT (rs1061540T/C), MALAT1 (rs3200401C/T), and SENCR (rs12420823C/T) variants which are selected based on specific criteria detailed in 2.4 section of this study, were not investigated yet in association with DR risk or response to “anti-vascular endothelial growth factor (VEGF)” aflibercept treatment. It is worth noting that anti-VEGF agents used in clinical practice, such as ranibizumab and aflibercept, are considerably different in terms of molecular interactions when they bind with VEGF.29 In this sense, this study aimed to investigate whether the selected genetic variants of the specified lncRNAs are associated with DR susceptibility, clinic-laboratory data, or the short-term (after four weeks) response to aflibercept therapy in the hope to find new biomolecular markers that help in patient risk stratification and/or listed within the pharmacogenetics-related variants.
Patients and Methods
Study Population
This cross-sectional study recruited 126 consecutive adult patients with type 2 DM attending the “Ophthalmology Department, Suez Canal University Hospitals” and a private Clinic, Ismailia, Egypt. The “Early Treatment Diabetic Retinopathy Study” (ETDRS) report30 was followed in patients’ evaluation and subclassification into DR group (n = 73) and non-DR group (n = 53). About 42.5% of DR and 22% of non-DR patients were on insulin therapy. Both groups had a comparable period of diabetes duration. Patients presented with other ophthalmic disorders, including neovascular glaucoma, age-related macular degeneration, or hematogenous retinal detachment, vasculopathy other than DR, history of ocular trauma or surgery, having other chronic diseases and/or severe comorbidities were excluded. The “institutional Research Ethics Committee of Faculty of Medicine, Suez Canal University” approved the study (approval No.4465). This study was executed according to the Declaration of Helsinki principles and its amendments. Written informed consent was obtained from the study population before taking part.
Clinical Assessment
A complete ophthalmic evaluation was done for all participants. Examination at the first visit to the ophthalmic clinic and throughout the follow-up visits included (1) the BCVA “best-corrected visual acuity” using logMAR “log of the Minimum Angle of Resolution”, (2) the anterior/posterior segment examination using slit-lamp biomicroscopy and +20 D/+90 D lenses, respectively, (3) colored fundus photography and (4) optical coherence tomography (OCT). Based on the initial FFA “fundus fluorescein angiography” assessment, maculopathy type and subclassification (focal vs diffuse) have been assigned. The ischemic type of DR was ruled out to avoid any bias in the results. The “non-proliferative DR (NPDR)” (subclassified into mild, moderate, or severe subtype) and “proliferative DR (PDR)” diagnoses were reached based on the ETDRS. Based on the OCT and the clinical findings, the diabetic macular edema (DME) diagnosis was assigned.30
Optical Coherence Tomography Scans
Patients retinas were subjected to OCT scans as explained previously.31 Patients presented with DME underwent an intravitreal injection (IV) of aflibercept (2 mg in 0.05 mL), “Eylea 40 mg/mL, Bayer Pharma AG, Berlin, Germany”, session after blood sampling. All the specified percussions and standards were followed pre-, during, and post-injection, as shown previously.14 Following-up was scheduled on the first day/one week, and one month after the first injection session. Both BCVA and OCT were reperformed after one month to evaluate the CMT. The initial response to aflibercept (one dose) was assigned as
BCVA improvement more than two lines of the Snellen s chart (Converted to Log MAR units for the statistical analysis), and CST reduction more than 15% of the pre-treatment thickness.14
The repeated injection was arranged when appropriate. Both “BCVA” and “CMT” changes were updated and recorded during the follow-up visits for all treated patients.
Blood Sampling and lncRNA Variants Genotyping
A total of five milliliters of venous blood samples were withdrawn from all participants after overnight fasting under aseptic conditions on EDTA tubes. Buffy coat genomic DNA was isolated using “QIAamp DNA Blood Mini kit (Catalog # 51104; Qiagen GmbH)” according to the vendor’s guidelines. The nucleic acid concentration and purity were evaluated using “NanoDrop ND-1000 (NanoDrop Technologies, Inc. Wilmington, DE, USA)”.
The studied variants selection was based on (1) dbSNP (www.ncbi.nlm.nih.gov) search for the minor allele frequency (MAF) ≥0.1 in the selected lncRNAs to get adequate statistical power, (2) mining in previous literature which showed evidence of the functional significance of these polymorphisms, and/or (3) no previous literature relates these SNPs with DR at least in our population. Genotyping was done on “StepOne Real-Time Polymerase Chain Reaction system (Applied Biosystems, USA)”. Polymerase chain reaction (PCR) was run in duplicates in a total volume of 25 µL containing 20 ng of extracted genomic DNA diluted to 11.25 µL with nuclease-free water, 12.5µL TaqMan genotyping PCR Master Mix, and 1.25 µL predesigned primer/probe sets TaqMan SNP Genotyping Assay (20x) working stock (assay IDs: C___2566592_10, C___2467719_1_, C___3246069_10, and C__11783392_10), for TUG1 (rs7284767G/A), MIAT (rs1061540T/C), MALAT1 (rs3200401C/T), and SENCR (rs12420823C/T), respectively. “No-template negative controls” (NTCs) were applied in each PCR run to confirm free reaction contamination. The PCR program was set as followed: 10 min (95°C) for one cycle, followed by 15 sec (90°C) and 1 min (60°C) for 40 two-stage cycles. The genotyping call was done by Applied Biosystems software with a 99.2% call rate and 100% concordance rate.
Statistical Analysis
The SPSS for Windows (version 27) and R (version 3.5.3) were used for the statistical analysis. G*Power (version 3.1.9.2.) was applied for sample size calculation. At a 95% significance level (alpha 0.05) and an effect size of 0.37 with a minimal sample size required to reject the null hypothesis (n= 126), the calculated study power was 91%. Data distribution and normality were checked by the Kolmogorov–Smirnov test. “Hardy-Weinberg equilibrium” (HWE) and allele/genotype frequencies of the studied variants were estimated within each group, as mentioned previously.32 Multivariable regression models were employed. Adjusted odds ratio (OR) and 95% confidence interval (CI) for multiple genetic association models were calculated using SNPStats (https://www.snpstats.net/start.htm). One-way ANOVA test was applied for parametric attributes, and Mann Whitney or Kruskal–Wallis tests were executed for non-parametric data. A Chi-square test was applied for qualitative variables. Significance was set at P<0.05.
Results
Baseline Characteristics of the Study Population
A total of 126 diabetic cohorts were included in the current analysis. Of these, 73 patients who developed diabetic retinopathy were compared to those who did not progress to retinopathy phenotype. Their mean ages were 59.8 ± 9.5 years and 62.5 ± 7.5 years, respectively (P = 0.09). Females accounted for 69.9% in DR cohorts and 79.2% in DM group (P = 0.30). Hypertension comorbidity was found in 71.2% and 62.3% of patients, with no significant difference between the group (P = 0.33). In contrast, patients with DR had more prevalent insulin intake (41.1% versus 17.0%, P = 0.006), and higher glycosylated hemoglobin (HbA1c) levels (8.92% ± 2.2% vs 5.45% ± 1.01%, P = 0.025) (Table 2). These variables were taken into consideration in the downstream multivariable regression analysis.
Table 2.
Baseline Characteristics of the Study Population
| Variables | Levels | DM (n=53) | DR (n=73) | P-value | OR (95% CI) |
|---|---|---|---|---|---|
| Age, year | Mean ± SD | 62.5 ± 7.5 | 59.8 ± 9.5 | 0.097 | |
| Sex | Female | 42 (79.2%) | 51 (69.9%) | 0.306 | 1.64 (0.71–3.78) |
| Male | 11 (20.8%) | 22 (30.1%) | |||
| Hypertension | Negative | 20 (37.7%) | 21 (28.8%) | 0.337 | 1.50 (0.70–3.18) |
| Positive | 33 (62.3%) | 52 (71.2%) | |||
| Disease duration, year | Mean ± SD | 13.9 ± 5.2 | 16.2 ± 7.9 | 0.067 | |
| Hypoglycemic drug | Oral | 44 (83.0%) | 43 (58.9%) | 0.006 | 3.41 (1.45–8.02) |
| Insulin | 9 (17.0%) | 30 (41.1%) | |||
| HbA1c, % | Mean ± SD | 5.45 ± 1.01 | 8.92 ± 2.2 | 0.025 |
Note: Data are shown as numbers and percentages or mean and standard deviation (SD). Chi-square and Student’s t-tests were applied. Bold values indicate significance at P-value < 0.05.
Abbreviations: DM, diabetes mellitus; DR, diabetic retinopathy; HbA1c, glycated hemoglobin.
Clinical Assessment of Diabetic Retinopathy Patients
DR patients were categorized according to the disease stage into mild, moderate, and severe non-proliferative and proliferative diseases. As depicted in Table 3, patients with PDR were significantly older (65.8 ± 10.9 years, p = 0.018), more likely to have prolonged disease duration (26.4 ± 7.1 years, P <0.001), and higher frequency of insulin intake (76.9%, P = 0.001). There were no significant inter-group differences regarding their therapeutic response (P = 0.89) nor their best-corrected visual acuity (P = 0.17).
Table 3.
Clinical and Ophthalmologic Assessment of Patients with Diabetic Retinopathy of Different Grades
| Variables | Mild NPDR | Moderate NPDR | Severe NPDR | PDR | P-value | |
|---|---|---|---|---|---|---|
| Age (year) | Mean ± SD | 57.7 ± 6.1 | 57.3 ± 10.4 | 63.4 ± 8.1 | 65.8 ± 10.9ab | 0.018 |
| Sex | Female | 15 (68.2) | 22 (81.5) | 8 (66.7) | 6 (50.0) | 0.25 |
| Male | 7 (31.8) | 5 (18.5) | 4 (33.3) | 6 (50.0) | ||
| Hypertension | Negative | 9 (40.39) | 10 (27.0) | 1 (8.3) | 1 (8.3) | 0.06 |
| Positive | 13 (59.1) | 17 (63.0) | 11 (91.7) | 11 (91.7) | ||
| Disease duration (year) | Mean ± SD | 10.4 ± 4.4 | 15.4 ± 5.2a | 17.6 ± 7.7a | 26.4 ± 7.1abc | <0.001 |
| Hypoglycemic drug | Oral | 19 (86.4) | 17 (63.0) | 5 (41.7) | 2 (16.7) | 0.001 |
| Insulin | 3 (13.6) | 10 (37.0) | 7 (58.3) | 10 (83.3) | ||
| HbA1c (%) | Mean ± SD | 9.1 ± 2.4 | 9 ± 2.3 | 8.3 ± 1.6 | 9.2 ± 2.3 | 0.68 |
| Pre-CMT | Mean ± SD | 441.9 ± 182.3 | 417 ± 129.7 | 376 ± 105 | 399.7 ± 115.3 | 0.68 |
| Post-CMT | Mean ± SD | 330.6 ± 96.9 | 345.6 ± 121.9 | 291.6 ± 53.8 | 354.6 ± 132.8 | 0.49 |
| CMT change | Mean ± SD | −111.3 ± 151.3 | −71.5 ± 88.3 | −84.4 ± 80 | −45.1 ± 82.1 | 0.33 |
| Pre-BCVA | Mean ± SD | 0.3 ± 0.3 | 0.5 ± 0.2 | 0.6 ± 0.2 a | 0.8 ± 0.1abc | <0.001 |
| Post-BCVA | Mean ± SD | 0.2 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.2 | 0.7 ± 0.2abc | <0.001 |
| BCVA change | Mean ± SD | −0.1 ± 0.2 | −0.2 ± 0.1 | −0.3 ± 0.2 | −0.1 ± 0.1 | 0.17 |
| Treatment response | Improved | 16 (72.7) | 22 (81.5) | 10 (83.3) | 9 (75.0) | 0.84 |
| Deteriorated | 6 (27.3) | 5 (18.5) | 2 (16.7) | 3 (25.0) |
Notes:aCompared to mild NPDR, bCompared to moderate NPDR, cCompared to severe NPDR. Chi-square and one-way ANOVA tests were used, followed by Tukey post hoc comparison test. Bold values indicate significance at P-value < 0.05. Treatment response: improved after one month of Aflibercept IV injection classified by change of CMT.
Abbreviations: NPDR, non-proliferative diabetic retinopathy; PDR, proliferative diabetic retinopathy; HBA1c, glycated hemoglobin; Pre, pre-treatment with aflibercept; Post, posttreatment with aflibercept; CMT, central macular thickness; BCVA, best-corrected visual acuity assessed by the logarithm of the minimum angle of resolution.
Allelic Discrimination Analysis
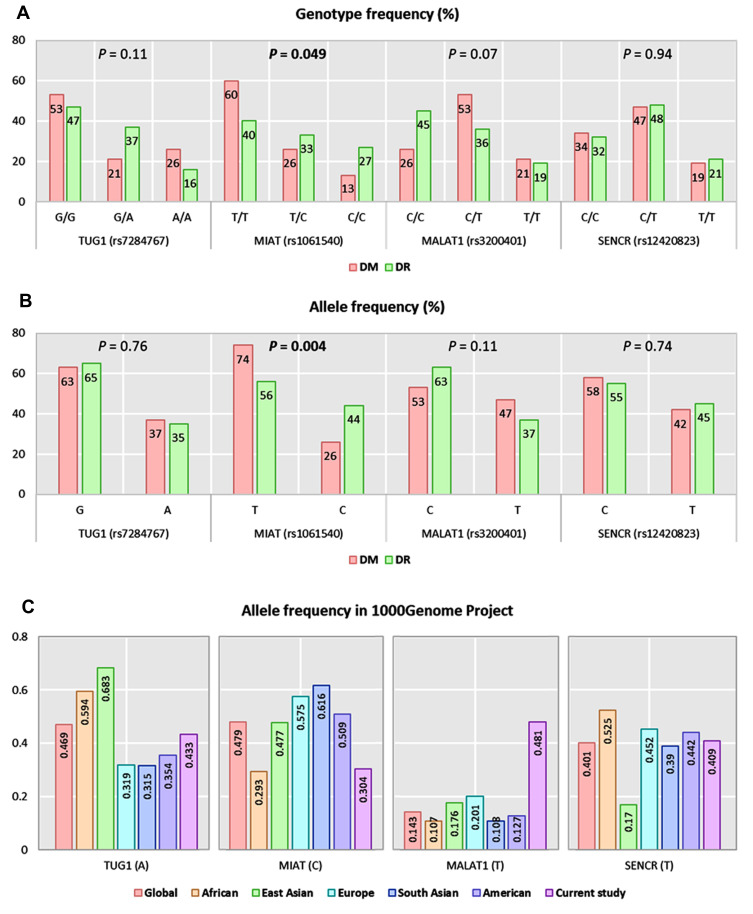
Apart from TUG1 polymorphism (p <0.001), genotype frequencies in diabetic controls were in accordance with Hardy–Weinberg equilibrium (MIAT: p = 0.29, MALAT1: P = 0.79, SENCR: P = 0.78). For TUG1 (rs7284767), MIAT (rs1061540), MALAT1 (rs3200401), and SENCR (rs12420823), overall minor allele frequencies were 0.37 (A), 0.26 (C), 0.47 (T), and 0.42 (T), respectively. On comparison between the two study groups, MIAT C variant was more frequent among DR patients (C allele: 44% in DR group versus 26% in DM group, P = 0.004) (Figure 1A). Similarly, higher proportions of MIATT/C and C/C genotypes were observed in DR group (C/C: 27% versus 13%, C/T: 33% versus 26%, P = 0.049) (Figure 1B). Comparison with other ethnic populations from 1000Genome Project is shown in Figure 1C.
Figure 1.
Genotype and allele frequencies of the studied genetic variants. (A) Genotype frequencies of polymorphisms. (B) Allele frequencies of polymorphisms. A Chi-square test was applied. Statistical significance was set at P < 0.05. (C) Allele frequencies of TUG1 (rs7284767), MIAT (rs1061540), MALAT1 (rs3200401), and SENCR (rs12420823) in 1000Genome Project. Bold values indicate significance at P-value < 0.05.
Association of lncRNA Variants with Disease Risk
As seen in Table 4, TUG1 A/G was 3 times more likely to develop DR under heterozygote comparison (OR = 3.15, 95% CI = 1.15 to 8.64) and over-dominant model (OR = 3.33, 95% CI = 1.28 to 8.67). MIAT T/C and C/C were associated with three and eight times more odds for developing disease [heterozygote model: OR = 2.97, 95% CI = 1.11 to 7.93, homozygote model: OR = 8.43, 95% CI = 2.40 to 29.59, dominant model: OR = 4.31, 95% CI = 1.78 to 10.47, recessive model: OR = 5.23, 95% CI = 1.66 −16.45, and log-additive model: OR = 2.92, 95% CI = 1.60 to 5.33]. In contrast, MALAT1 C/T rendered protection with 60% decreased susceptibility to develop DR disease under heterozygote comparison (OR = 0.40, 95% CI = 0.16 to 0.99) and dominant model (OR = 0.41, 95% CI = 0.18 to 0.96). Gene–gene interaction analysis revealed that carriers for GTCT and GCCC genotype combinations had 17- and 20-times higher disease risk, respectively (P = 0.012) (Table 5).
Table 4.
Genetic Association Models for the Study Long Non-Coding RNAs and Disease Risk
| Gene | Model | Genotypes | DM | DR | Crude OR (95% CI) | P-value | Adjusted OR (95% CI)* | P- value |
|---|---|---|---|---|---|---|---|---|
| TUG1 | Codominant | G/G | 28 (52.8%) | 34 (46.6%) | 1.00 | 0.11 | 1.00 | 0.034 |
| A/G | 11 (20.8%) | 27 (37%) | 2.02 (0.85–4.78) | 3.15 (1.15–8.64) | ||||
| A/A | 14 (26.4%) | 12 (16.4%) | 0.71 (0.28–1.77) | 0.84 (0.30–2.34) | ||||
| Dominant | G/G | 28 (52.8%) | 34 (46.6%) | 1.00 | 0.49 | 1.00 | 0.18 | |
| A/G-A/A | 25 (47.2%) | 39 (53.4%) | 1.28 (0.63–2.61) | 1.72 (0.77–3.84) | ||||
| Recessive | G/G-A/G | 39 (73.6%) | 61 (83.6%) | 1.00 | 0.17 | 1.00 | 0.24 | |
| A/A | 14 (26.4%) | 12 (16.4%) | 0.55 (0.23–1.31) | 0.56 (0.22–1.47) | ||||
| Over-dominant | G/G-A/A | 42 (79.2%) | 46 (63%) | 1.00 | 0.047 | 1.00 | 0.01 | |
| A/G | 11 (20.8%) | 27 (37%) | 2.24 (0.99–5.07) | 3.33 (1.28–8.67) | ||||
| Log-additive | – | – | – | 0.94 (0.60–1.48) | 0.79 | 1.06 (0.64–1.73) | 0.83 | |
| MIAT | Codominant | T/T | 32 (60.4%) | 29 (39.7%) | 1.00 | 0.046 | 1.00 | 8e-04 |
| T/C | 14 (26.4%) | 24 (32.9%) | 1.89 (0.83–4.33) | 2.97 (1.11–7.93) | ||||
| C/C | 7 (13.2%) | 20 (27.4%) | 3.15 (1.16–8.54) | 8.43 (2.40–29.59) | ||||
| Dominant | T/T | 32 (60.4%) | 29 (39.7%) | 1.00 | 0.022 | 1.00 | 7e-04 | |
| T/C-C/C | 21 (39.6%) | 44 (60.3%) | 2.31 (1.12–4.76) | 4.31 (1.78–10.47) | ||||
| Recessive | T/T-T/C | 46 (86.8%) | 53 (72.6%) | 1.00 | 0.05 | 1.00 | 0.0023 | |
| C/C | 7 (13.2%) | 20 (27.4%) | 2.48 (0.96–6.39) | 5.23 (1.66–16.45) | ||||
| Over-dominant | T/T-C/C | 39 (73.6%) | 49 (67.1%) | 1.00 | 0.43 | 1.00 | 0.33 | |
| T/C | 14 (26.4%) | 24 (32.9%) | 1.36 (0.62–2.98) | 1.54 (0.65–3.68) | ||||
| Log-additive | – | – | – | 1.80 (1.11–2.90) | 0.013 | 2.92 (1.60–5.33) | 2e-04 | |
| MALAT1 | Codominant | C/C | 14 (26.4%) | 33 (45.2%) | 1.00 | 0.076 | 1.00 | 0.11 |
| C/T | 28 (52.8%) | 26 (35.6%) | 0.39 (0.17–0.90) | 0.40 (0.16–0.99) | ||||
| T/T | 11 (20.8%) | 14 (19.2%) | 0.54 (0.20–1.48) | 0.44 (0.14–1.40) | ||||
| Dominant | C/C | 14 (26.4%) | 33 (45.2%) | 1.00 | 0.03 | 1.00 | 0.037 | |
| C/T-T/T | 39 (73.6%) | 40 (54.8%) | 0.44 (0.20–0.94) | 0.41 (0.18–0.96) | ||||
| Recessive | C/C-C/T | 42 (79.2%) | 59 (80.8%) | 1.00 | 0.83 | 1.00 | 0.54 | |
| T/T | 11 (20.8%) | 14 (19.2%) | 0.91 (0.37–2.19) | 0.72 (0.26–2.03) | ||||
| Over-dominant | C/C-T/T | 25 (47.2%) | 47 (64.4%) | 1.00 | 0.054 | 1.00 | 0.12 | |
| C/T | 28 (52.8%) | 26 (35.6%) | 0.49 (0.24–1.02) | 0.53 (0.24–1.19) | ||||
| Log-additive | – | – | – | 0.68 (0.42–1.11) | 0.12 | 0.61 (0.35–1.07) | 0.083 | |
| SENCR | Codominant | C/C | 18 (34%) | 23 (31.5%) | 1.00 | 0.95 | 1.00 | 0.92 |
| C/T | 25 (47.2%) | 35 (48%) | 1.10 (0.49–2.44) | 1.18 (0.46–3.01) | ||||
| T/T | 10 (18.9%) | 15 (20.6%) | 1.17 (0.43–3.22) | 1.01 (0.32–3.15) | ||||
| Dominant | C/C | 18 (34%) | 23 (31.5%) | 1.00 | 0.77 | 1.00 | 0.8 | |
| C/T-T/T | 35 (66%) | 50 (68.5%) | 1.12 (0.53–2.37) | 1.12 (0.46–2.72) | ||||
| Recessive | C/C-C/T | 43 (81.1%) | 58 (79.5%) | 1.00 | 0.82 | 1.00 | 0.84 | |
| T/T | 10 (18.9%) | 15 (20.6%) | 1.11 (0.46–2.71) | 0.91 (0.34–2.40) | ||||
| Over-dominant | C/C-T/T | 28 (52.8%) | 38 (52%) | 1.00 | 0.93 | 1.00 | 0.69 | |
| C/T | 25 (47.2%) | 35 (48%) | 1.03 (0.51–2.09) | 1.18 (0.53–2.62) | ||||
| Log-additive | – | – | – | 1.08 (0.66–1.78) | 0.75 | 1.02 (0.57–1.79) | 0.96 |
Notes: Data are presented as numbers and percentages. Crude and *adjusted OR by age, sex, disease duration, hypoglycemic treatment, and hypertension were estimated. Adjusted OR (95% CI) for multiple genetic association models were calculated using SNPStats (https://www.snpstats.net/start.htm). Bold values indicate significance at P-value < 0.05.
Abbreviations: DM, diabetes without diabetic retinopathy; DR, diabetic retinopathy; OR, odds ratio; CI, confidence interval.
Table 5.
Combined Genotype Association with Disease Risk
| TUG1 | MIAT | MALAT1 | SENCR | DM | DR | Cum Freq | Adjusted OR (95% CI)* | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | G | T | C | C | 0.218 | 0.134 | 0.154 | 1.00 | – |
| 2 | G | T | T | C | 0.080 | 0.103 | 0.262 | 1.26 (0.23–6.80) | 0.79 |
| 3 | A | T | C | T | 0.097 | 0.105 | 0.359 | 0.51 (0.08–3.44) | 0.49 |
| 4 | A | C | C | C | 0.044 | 0.104 | 0.446 | 2.97 (0.43–20.4) | 0.27 |
| 5 | G | T | C | T | 0.066 | 0.070 | 0.528 | 17.1 (1.9–149.8) | 0.012 |
| 6 | G | T | T | T | 0.130 | 0.057 | 0.608 | 0.70 (0.17–3.0) | 0.64 |
| 7 | G | C | T | C | 0.035 | 0.083 | 0.670 | 7.7 (0.75–79.2) | 0.088 |
| 8 | G | C | C | C | 0.042 | 0.069 | 0.731 | 20.9 (2.07–212) | 0.012 |
| 9 | G | C | C | T | 0.018 | 0.093 | 0.790 | 1.03 (0.09–11.9) | 0.98 |
| 10 | A | T | T | C | 0.074 | 0.034 | 0.843 | 0.63 (0.08–5.30) | 0.68 |
| 11 | G | C | T | T | 0.039 | 0.028 | 0.880 | 4.31 (0.37–50.2) | 0.25 |
| 12 | A | T | T | T | 0.040 | 0.031 | 0.915 | 0.38 (0.03–5.19) | 0.47 |
| 13 | A | C | T | T | 0.018 | 0.026 | 0.941 | 0.71 (0.04–11.2) | 0.81 |
| 14 | A | T | C | C | 0.027 | 0.020 | 0.967 | 4.39 (0.25–76.5) | 0.31 |
| 15 | A | C | C | T | 0.012 | 0.026 | 0.988 | 1.92 (0.13–28.3) | 0.64 |
| 16 | A | C | T | C | 0.052 | 0.000 | 1.0 | 0.45 (0.01–22.6) | 0.69 |
Notes: Global haplotype association P-value: 0.04. *Adjusted OR by age, sex, disease duration, hypoglycemic treatment, and hypertension were estimated. Bold values indicate significance at P-value < 0.05.
Abbreviations: DM, diabetes without diabetic retinopathy; DR, diabetic retinopathy; Cum Freq, cumulative frequency; OR, odds ratio; CI, confidence interval.
Association of lncRNA Variants with Disease Severity
SENCR T variant was associated with better pre-treatment BCVA level (0.63 ± 0.19 in C/C compared to 0.45 ± 0.27 in C/T and 0.46 ± 0.26 in T/T carriers, P = 0.021). In contrast, MIAT T/T homozygosity was associated with higher grade (PDR: 33.3% in T/T compared to 10% in C/C and 4.2% in C/T carriers, P = 0.012) (Table 6).
Table 6.
Association Between Genotypes and Patient Characteristics
| Variables | TUG1 | MIAT | MALAT1 | SENCR |
|---|---|---|---|---|
| Age | 0.433 | 0.789 | 0.759 | 0.763 |
| Sex | 0.774 | 0.867 | 0.245 | 0.684 |
| Hypertension | 0.773 | 0.665 | 0.658 | 0.884 |
| Disease duration | 0.191 | 0.088 | 0.082 | 0.126 |
| Hypoglycemic drug | 0.484 | 0.523 | 0.726 | 0.385 |
| HbA1c | 0.367 | 0.316 | 0.830 | 0.838 |
| Retinopathy grade | 0.171 | 0.012 | 0.402 | 0.786 |
| Treatment response | 0.020 | 0.014 | 0.688 | 0.775 |
| VA improvement | 0.005 | 0.207 | 0.137 | 0.620 |
| Pre-CMT | 0.162 | 0.274 | 0.918 | 0.386 |
| Post-CMT | 0.677 | 0.787 | 0.050 | 0.207 |
| CMT change | 0.016 | 0.090 | 0.161 | 0.984 |
| Pre-BCVA | 0.331 | 0.061 | 0.494 | 0.021 |
| Post-BCVA | 0.640 | 0.114 | 0.262 | 0.228 |
| BCVA change | 0.052 | 0.822 | 0.207 | 0.239 |
Notes: A two-sided chi-square test and one-way ANOVA tests were applied. Bold values indicate significance at P-value < 0.05.
Abbreviations: HbA1c, glycated hemoglobin; VA, visual acuity; Pre, pre-treatment with aflibercept; Post, posttreatment with aflibercept; CMT, central macular thickness; BCVA, best-corrected visual acuity assessed by the logarithm of the minimum angle of resolution.
Association of lncRNA Variants with Drug Response After Anti-VEGF Treatment
TUG1 A and MIAT C alleles were associated with non-early response to aflibercept treatment (P = 0.020 and 0.014, respectively). Carriers for TUG1 A allele showed the lowest median change in CMT (−11.5 in A/A compared to −90 in A/G and −54 in G/G, P = 0.016) and BCVA (0 in A/A compared to −0.15 in A/G and −0.1 in G/G, P = 0.005) (Table 6). The same allele (TUG1 A) was significantly associated with treatment failure under homozygote comparison (OR = 25.10, 95% CI = 2.53 to 249.44), dominant model (OR = 8.18, 95% CI = 1.48 to 45.29), recessive model (OR = 8.81, 95% CI = 1.49–52.06), and log-additive model (OR = 5.02, 95% CI = 1.60 to 15.76). In addition, MIAT T/C genotype was associated with poor therapeutic response under heterozygote comparison (OR = 10.23, 95% CI: 1.51 to 69.15), dominant model (OR = 5.95, 95% CI = 1.04 to 34.23), and over-dominant model (OR: 6.26, 95% CI: 1.41–27.80) (Table 7).
Table 7.
Association Between Genotypes and Drug Response Phenotype
| Gene | Model | Genotypes | Improved (n=57) | Deteriorated (n=16) | Adjusted OR (95% CI)* | P-value |
|---|---|---|---|---|---|---|
| TUG1 | Codominant | G/G | 30 (52.6%) | 4 (25%) | 1.00 | 0.006 |
| A/G | 21 (36.8%) | 6 (37.5%) | 5.49 (0.88–34.16) | |||
| A/A | 6 (10.5%) | 6 (37.5%) | 25.10 (2.53–249.44) | |||
| Dominant | G/G | 30 (52.6%) | 4 (25%) | 1.00 | 0.006 | |
| A/G-A/A | 27 (47.4%) | 12 (75%) | 8.18 (1.48–45.29) | |||
| Recessive | G/G-A/G | 51 (89.5%) | 10 (62.5%) | 1.00 | 0.012 | |
| A/A | 6 (10.5%) | 6 (37.5%) | 8.81 (1.49–52.06) | |||
| Overdominant | G/G-A/A | 36 (63.2%) | 10 (62.5%) | 1.00 | 0.52 | |
| A/G | 21 (36.8%) | 6 (37.5%) | 1.54 (0.41–5.79) | |||
| Log-additive | – | – | – | 5.02 (1.60–15.76) | 0.001 | |
| MIAT | Codominant | T/T | 26 (45.6%) | 3 (18.8%) | 1.00 | 0.026 |
| C/T | 14 (24.6%) | 10 (62.5%) | 10.23 (1.51–69.15) | |||
| C/C | 17 (29.8%) | 3 (18.8%) | 2.69 (0.34–21.23) | |||
| Dominant | T/T | 26 (45.6%) | 3 (18.8%) | 1.00 | 0.026 | |
| T/C-C/C | 31 (54.4%) | 13 (81.2%) | 5.95 (1.04–34.23) | |||
| Recessive | T/T-T/C | 40 (70.2%) | 13 (81.2%) | 1.00 | 0.66 | |
| C/C | 17 (29.8%) | 3 (18.8%) | 0.71 (0.15–3.41) | |||
| Overdominant | T/T-C/C | 43 (75.4%) | 6 (37.5%) | 1.00 | 0.011 | |
| T/C | 14 (24.6%) | 10 (62.5%) | 6.26 (1.41–27.80) | |||
| Log-additive | – | – | – | 1.58 (0.66–3.77) | 0.29 | |
| MALAT1 | Codominant | C/C | 27 (47.4%) | 6 (37.5%) | 1.00 | 0.9 |
| C/T | 20 (35.1%) | 6 (37.5%) | 1.17 (0.24–5.72) | |||
| T/T | 10 (17.5%) | 4 (25%) | 1.52 (0.25–9.25) | |||
| Dominant | C/C | 27 (47.4%) | 6 (37.5%) | 1.00 | 0.73 | |
| C/T-T/T | 30 (52.6%) | 10 (62.5%) | 1.29 (0.31–5.40) | |||
| Recessive | C/C-C/T | 47 (82.5%) | 12 (75%) | 1.00 | 0.68 | |
| T/T | 10 (17.5%) | 4 (25%) | 1.41 (0.28–7.00) | |||
| Overdominant | C/C-T/T | 37 (64.9%) | 10 (62.5%) | 1.00 | 1 | |
| C/T | 20 (35.1%) | 6 (37.5%) | 1.00 (0.24–4.12) | |||
| Log-additive | – | – | – | 1.23 (0.50–3.02) | 0.65 | |
| SENCR | Codominant | C/C | 19 (33.3%) | 4 (25%) | 1.00 | 0.69 |
| C/T | 26 (45.6%) | 9 (56.2%) | 1.90 (0.39–9.20) | |||
| T/T | 12 (21.1%) | 3 (18.8%) | 1.21 (0.17–8.41) | |||
| Dominant | C/C | 19 (33.3%) | 4 (25%) | 1.00 | 0.5 | |
| C/T-T/T | 38 (66.7%) | 12 (75%) | 1.66 (0.37–7.48) | |||
| Recessive | C/C-C/T | 45 (79%) | 13 (81.2%) | 1.00 | 0.77 | |
| T/T | 12 (21.1%) | 3 (18.8%) | 0.79 (0.16–3.94) | |||
| Overdominant | C/C-T/T | 31 (54.4%) | 7 (43.8%) | 1.00 | 0.4 | |
| C/T | 26 (45.6%) | 9 (56.2%) | 1.75 (0.48–6.42) | |||
| Log-additive | – | – | – | 1.12 (0.45–2.78) | 0.8 |
Notes: Adjusted OR by age, sex, disease duration, hypoglycemic treatment, hypertension, grade of retinopathy, and HbA1c. *Adjusted OR (95% confidence interval; CI) for multiple genetic association models were calculated using SNPStats (https://www.snpstats.net/start.htm). Bold values indicate significance at P-value < 0.05.
Discussion
The current treatment strategies for DR are mainly used in the advanced stages of the disease and may be related to several side effects. In this sense, it is necessary to explore a new landscape of susceptibility genetic variants associated with disease development, severity, or response to treatment to identify the individuals most likely to develop DR and predicting treatment effects. To this end, we genotyped for the first time four variants belonging to lncRNAs which proved previously to be implicated in one or more pathophysiological mechanisms of diabetes or its complications (Table 1). Our results show a variable degree of disease risk, severity, and/or early response to aflibercept treatment.
We found that carriers of lncRNA TUG1 A/G of rs7284767 variant were three times more likely to develop DR under heterozygote and over-dominant models. Also, carriers of the TUG1 A allele under several genetic models were associated with non-early response to aflibercept treatment as they showed the lowest median change in CMT and BCVA.
The rs7284767 variant was found to be located in one of “novel biologically relevant; NBR” target sites on the TUG1 and was associated with unusual patterns of long-range haplotype conservation in the human genome33 and exhibited significantly high integrated haplotype score value; “a gnomically standardized measure for conservation of long-range haplotype associated with a given SNP in a population”.34 This variant was predicted to disrupt the NBR target site for miR-20 in humans.33 Interestingly, miR-20 was associated with diabetes and DR in previous studies35,36 and proved to have diagnostic and prognostic roles in patients with DR.36 Given the targeting role of this microRNA for the 3′-untranslated regions of hypoxia‐inducible factor‐1α and VEGFA genes, it is not surprising to be associated with angiogenesis-related disorders, including DR and with treatment response to anti-VEGF aflibercept treatment. A recent study by Duan et al has identified the association of rs7284767 allelic variation with increased plasma TUG1 levels in patients with knee osteoarthritis.37 This finding and all the mechanisms mentioned above could prove that the rs7284767 variant may be functional, supporting its association with disease risk and response to anti-VEGF treatment in the present study population. Future functional assays and detailed genetic analyses will be required to determine the detailed biological significance of this variant and other studied ones in the future.
The present work also explored that MIAT rs1061540T (C/T) and (C/C) genotypes were associated with three and eight times more odds for developing DR, and T/T homozygosity was associated with a higher grade of PDR relative to other genotypes. Also, the C/T genotype was associated with poor therapeutic response under heterozygote, dominant, and over-dominant models. Although accumulating evidence confirmed the implication of MIAT in every stage of DR by several genetic/epigenetic mechanisms and its knockdown has a therapeutic advantage in neovascular-related disorders as DR,20,27,38–40 the specific impact of the rs1061540 variant on MIAT expression and association with DR is yet to be identified. In this sense, more studies are recommended to explore the precise molecular mechanism by which this variant could impact disease susceptibility/severity and/or treatment response.
In contrast to the SNPs mentioned above, the lncRNA MALAT1 rs3200401 (T/C) genotype in the current study population conferred protection with 60% decreased susceptibility to develop DR under heterozygote comparison and dominant models. Several in vivo and in vitro studies unraveled the association between MALAT1 level and diabetes/hyperglycemia. It was found to be up-regulated in diabetic mice retinas,11 regulating the retinal endothelial cells function in terms of cell proliferation/migration and tube biogenesis, and the pathological growth of the retinal microvasculature under hyperglycemic conditions.23,41 Its pharmacological inhibition can reduce the retinal endothelial cell proliferation phenotype, vasculature growth, inflammatory mediators, and the pro-inflammatory cytokines such as serum amyloid antigen 3, tumor necrosis factor-α, and interleukin-6.39,42 It is worth noting that the lncRNA variants may exert their impact through lncRNA transcript alternative splicing or change in its secondary structure, resulting in loss or gain of function.43 Interestingly, the rs3200401 variant is located in the binding site of “serine/arginine-rich splicing factor 2 (SRSF2)” that controls pre-mRNA alternative splicing of several targets,44 including the VEGFA/VEGFR genes, which are considered the main stimulatory signal of angiogenesis in vivo.45,46 Using the “lncRNASNP database” [10.1093/nar/gkx1004], Wang et al predicted that the rs3200401 variant could cause “1.62 kcal/mol minimal free energy change” which change MALAT1 structure, weaken the MALAT1/SRSF2 interaction and alter miRNA-MALAT1 binding (eg, impact binding to miR-3661 and miR-1324).47 In this sense, it is biologically plausible to speculate that deregulation of the mentioned cellular mechanistic due to MALAT1 (rs3200401C/T) polymorphism may impact the “molecular sponging” function and stability of MALAT1 that result in a less pathophysiological derangement, mainly the angiogenesis process and its progress with lower susceptibility to DR.
Regarding the SENCR (rs12420823C/T) intronic variant, the present study showed that it was associated only with better pre-treatment best-corrected visual acuity levels assessed by the logarithm of the minimum angle of resolution. This vascular-enriched lncRNA gene has been found to overlap the “Friend Leukemia Integration virus 1; FLI1” gene, an essential regulator of endothelial development, and the lncRNA SENCR can contribute to this regulation and induce proliferation, migration, and angiogenesis of human umbilical endothelial cell.25 As there were no previous publications that uncover the role of the studied SENCR rs12420823 in general, more future studies are warranted to confirm the relation of this variant with angiogenesis-related diseases, including DR.
Our gene–gene interaction analysis revealed that carriers for TUG1, MALAT1, MIAT, and SENCR (GTCT) and (GCCC) genotype combinations had 17- and 20-times higher disease risk, respectively. This finding is consistent with that the concurrent impact of multiple gene variants is strong enough to unleash the genetic association with disease susceptibility and/or response to treatment.48 This multi-polymorphic model could be a potentially useful tool in patient risk stratification and future personalized medicine.
Although this study, up to the authors’ knowledge, is the first study to uncover the impact of four angiolncRNAs polymorphisms on DR susceptibility and early response to aflibercept, some limitations should be considered. First, the study cohort is a hospital-based population with difficulty in avoiding selection bias. Second, the limited sample size available in our hospital in the study period with an application of very stringent exclusion criteria. Third, the limited selected study variants with MAF ≥ 0.10 to be included in this study to achieve adequate statistical power. Hence, it is recommended to replicate the work in multi-center, larger-scale studies in different ethnicity populations to validate the current findings. Moreover, including other variants related to the studied lncRNAs, supported with functional analysis to uncover their molecular mechanisms underlying the disease susceptibility and/or response to treatment, will clarify the complete picture.
Conclusion
This study revealed that the genetic variation of lncRNAs TUG1 (rs7284767G/A) and MIAT (rs1061540T/C) could be molecular determinants for increased DR susceptibility and early response to aflibercept treatment. Otherwise, the MALAT1 rs3200401 (T/C) variant conferred DR protection. Also, the multi-polymorphic models (GTCT and GCCC) of the TUG1, MALAT1, MIAT, and SENCR, respectively, were associated with increased DR risk. Further large population-based and functional in vivo and in vitro studies are required to confirm the significant association of the studied variants with DR susceptibility and/or response to aflibercept treatment, particularly in other ethnicities, and to explore the molecular mechanism(s) by which these variants confer DR risk and/or response to anti-VEGF therapy.
Acknowledgments
The authors thank the Center of Excellence in Molecular and Cellular Medicine and Oncology Diagnostic Unit, Suez Canal University, Ismailia, Egypt for providing the facilities for performing the research work as well as we thank all participants who agree to participate in the study.
Funding Statement
There is no funding to report.
Data Sharing Statement
All data generated or analyzed during this study are included in this submitted article.
Ethics Statement
All procedures performed in studies involving human participants were following the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Ethical approval was obtained from the “institutional Research Ethics Committee of Faculty of Medicine, Suez Canal University” (approval No.4465), Ismailia, Egypt.
Author Contributions
All authors contributed to data analysis, drafting, or revising the article, have agreed on the journal to which the article has been submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Saeedi P, Petersohn I, Salpea P, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the international diabetes federation diabetes atlas, 9. Diabetes Res Clin Pract. 2019;157:107843. doi: 10.1016/j.diabres.2019.107843 [DOI] [PubMed] [Google Scholar]
- 2.Hu Y, Chen Y, Ding L, et al. Pathogenic role of diabetes-induced PPAR-α down-regulation in microvascular dysfunction. Proc Natl Acad Sci U S A. 2013;110(38):15401–15406. doi: 10.1073/pnas.1307211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu E, Hu X, Li X, et al. Analysis of long non-coding RNA expression profiles in high-glucose treated vascular endothelial cells. BMC Endocr Disord. 2020;20(1):107. doi: 10.1186/s12902-020-00593-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299(1):F14–F25. doi: 10.1152/ajprenal.00200.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong M, Xie K, Lv M, et al. Anti-inflammatory phytochemicals for the treatment of diabetes and its complications: lessons learned and future promise. Biomed Pharmacother. 2021;133:110975. doi: 10.1016/j.biopha.2020.110975 [DOI] [PubMed] [Google Scholar]
- 6.Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018;12(1):41. doi: 10.1186/s40246-018-0173-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fawzy MS, AlSel BTA, Ageeli EA, Al-Qahtani SA, Abdel-Daim MM, Toraih EA. Long non-coding RNA MALAT1 and microRNA-499a expression profiles in diabetic ESRD patients undergoing dialysis: a preliminary cross-sectional analysis. Arch Physiol Biochem. 2018;126(2):172–182. doi: 10.1080/13813455.2018.1499119. [DOI] [PubMed] [Google Scholar]
- 8.Raut SK, Khullar M. The big entity of new RNA world: long non-coding RNAs in microvascular complications of diabetes. Front Endocrinol (Lausanne). 2018;9:300. doi: 10.3389/fendo.2018.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawzy MS, Abdelghany AA, Toraih EA, Mohamed AM. Circulating long non-coding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: a preliminary study. Bosn J Basic Med Sci. 2020;20(3):365–371. doi: 10.17305/bjbms.2019.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2017;58(1):470–480. doi: 10.1167/iovs.16-20569 [DOI] [PubMed] [Google Scholar]
- 11.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long non-coding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55(2):941–951. doi: 10.1167/iovs.13-13221 [DOI] [PubMed] [Google Scholar]
- 12.Biswas S, Thomas A, Feng B, Chen S, Gonder J, Chakrabarti S. Role of long non-coding RNA MALAT1 in the pathogenesis of diabetic retinopathy. Can J Diabetes. 2017;41(5):S8. doi: 10.1016/j.jcjd.2017.08.027 [DOI] [Google Scholar]
- 13.Wang J, Gao X, Liu J, Zhang Y, Zhang T, Zhang H. Effect of intravitreal conbercept treatment on the expression of long noncoding RNAs and mRNAs in proliferative diabetic retinopathy patients. Acta Ophthalmol. 2019;97(6):e902–e912. doi: 10.1111/aos.14083 [DOI] [PubMed] [Google Scholar]
- 14.Toraih EA, Abdelghany AA, Abd El Fadeal NM, Al Ageeli E, Fawzy MS. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch Clin Exp Ophthalmol. 2019;257(9):1897–1913. doi: 10.1007/s00417-019-04409-9 [DOI] [PubMed] [Google Scholar]
- 15.Altshuler D, Lo Turco JJ, Rush J, Cepko C. Taurine promotes the differentiation of a vertebrate retinal cell type in vitro. Development. 1993;119(4):1317–1328. doi: 10.1242/dev.119.4.1317 [DOI] [PubMed] [Google Scholar]
- 16.Young TL, Matsuda T, Cepko CL. The non-coding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 17.Rapicavoli NA, Blackshaw S. New meaning in the message: non-coding RNAs and their role in retinal development. Dev Dyn. 2009;238(9):2103–2114. doi: 10.1002/dvdy.21844 [DOI] [PubMed] [Google Scholar]
- 18.Yan HY, Bu SZ, Zhou WB, Mai YF. TUG1 promotes diabetic atherosclerosis by regulating proliferation of endothelial cells via Wnt pathway. Eur Rev Med Pharmacol Sci. 2018;22(20):6922–6929. [DOI] [PubMed] [Google Scholar]
- 19.Yu B, Wang S. Angio-LncRs: lncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8(13):3654–3675. doi: 10.7150/thno.26024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan B, Yao J, Liu JY, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- 21.Eißmann M, Gutschner T, Hämmerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vorotyntseva MI, Frosin VH. [Classification of medical sterilizers and terminology]. Med Tekh. 1972;1972(2):33–37. [Amharic] [PubMed] [Google Scholar]
- 23.Liu JY, Yao J, Li XM, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5(10):e1506. doi: 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou ZQ, Xu J, Li L, Han YS. Down-regulation of SENCR promotes smooth muscle cells proliferation and migration in db/db mice through up-regulation of FoxO1 and TRPC6. Biomed Pharmacother. 2015;74:35–41. doi: 10.1016/j.biopha.2015.06.009 [DOI] [PubMed] [Google Scholar]
- 25.Boulberdaa M, Scott E, Ballantyne M, et al. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther. 2016;24(5):978–990. doi: 10.1038/mt.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell RD, Long X, Lin M, et al. Identification and initial functional characterization of a human vascular cell-enriched long non-coding RNA. Arterioscler Thromb Vasc Biol. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awata T, Yamashita H, Kurihara S, et al. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS One. 2014;9(11):e111715. doi: 10.1371/journal.pone.0111715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung A, Natarajan R. Long noncoding RNAs in diabetes and diabetic complications. Antioxid Redox Signal. 2018;29(11):1064–1073. doi: 10.1089/ars.2017.7315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platania CB, Di Paola L, Leggio GM, et al. Molecular features of interaction between VEGFA and anti-angiogenic drugs used in retinal diseases: a computational approach. Front Pharmacol. 2015;6:248. doi: 10.3389/fphar.2015.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Early Treatment Diabetic Retinopathy Study Research Group. Early treatment diabetic retinopathy study design and baseline patient characteristics. ETDRS report number 7. Ophthalmology. 1991;98(5Suppl):741–756. doi: 10.1016/s0161-6420(13)38009-9 [DOI] [PubMed] [Google Scholar]
- 31.Abdelghany AA, Toraih EA, Mohamed AA, et al. Association of VEGF gene family variants with central macular thickness and visual acuity after aflibercept short-term treatment in diabetic patients: a pilot study. Ophthalmic Res. 2021;64(2):261–272. doi: 10.1159/000511087 [DOI] [PubMed] [Google Scholar]
- 32.Toraih EA, Ismail NM, Toraih AA, Hussein MH, Fawzy MS. Precursor miR-499a variant but not miR-196a2 is associated with rheumatoid arthritis susceptibility in an Egyptian population. Mol Diagn Ther. 2016;20(3):279–295. doi: 10.1007/s40291-016-0194-3 [DOI] [PubMed] [Google Scholar]
- 33.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104(9):3300–3305. doi: 10.1073/pnas.0611347104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Q, Lyu XM, Yuan Y, Wang L. Plasma. Biosci Rep. 2017;37(2). doi: 10.1042/BSR20160589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaker OG, Abdelaleem OO, Mahmoud RH, et al. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life. 2019;71(3):310–320. doi: 10.1002/iub.1970 [DOI] [PubMed] [Google Scholar]
- 37.Duan J, Shen T, Dong H, Han S, Li G. Association of the expression levels of long-chain noncoding RNA TUG1 and its gene polymorphisms with knee osteoarthritis. Genet Test Mol Biomarkers. 2021;25(2):102–110. doi: 10.1089/gtmb.2020.0208 [DOI] [PubMed] [Google Scholar]
- 38.Jaé N, Dimmeler S. Long non-coding RNAs in diabetic retinopathy. Circ Res. 2015;116(7):1104–1106. doi: 10.1161/CIRCRESAHA.115.306051 [DOI] [PubMed] [Google Scholar]
- 39.Gong Q, Su G. Roles of miRNAs and long non-coding RNAs in the progression of diabetic retinopathy. Biosci Rep. 2017;37(6). doi: 10.1042/BSR20171157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Chen M, Chen J, et al. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing. Biosci Rep. 2017;37(2):BSR20170036. doi: 10.1042/BSR20170036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalik KM, You X, Manavski Y, et al. Long non-coding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265 [DOI] [PubMed] [Google Scholar]
- 42.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418–1425. doi: 10.1111/jcmm.12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842(10):1910–1922. doi: 10.1016/j.bbadis.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 44.Miyagawa R, Tano K, Mizuno R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 non-coding RNA to nuclear speckles. RNA. 2012;18(4):738–751. doi: 10.1261/rna.028639.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gacche RN, Meshram RJ. Angiogenic factors as potential drug target: efficacy and limitations of anti-angiogenic therapy. Biochim Biophys Acta. 2014;1846(1):161–179. doi: 10.1016/j.bbcan.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 46.Bowler E, Oltean S. Alternative Splicing in Angiogenesis. Int J Mol Sci. 2019;20(9):2067. doi: 10.3390/ijms20092067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang JZ, Xiang JJ, Wu LG, et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17(1):167. doi: 10.1186/s12885-017-3151-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017;18(1):77. doi: 10.1186/s13059-017-1212-4 [DOI] [PMC free article] [PubMed] [Google Scholar]