Abstract
Background
Several SARS-CoV-2 lineages with spike receptor binding domain (RBD) N501Y mutation have spread globally. We evaluated the impact of N501Y on neutralizing activity of COVID-19 convalescent sera and on anti-RBD IgG assays.
Methods
The susceptibility to neutralization by COVID-19 patients’ convalescent sera from Hong Kong were compared between two SARS-CoV-2 isolates (B117-1/B117-2) from the α variant with N501Y and 4 non-N501Y isolates. The effect of N501Y on antibody binding was assessed. The performance of commercially-available IgG assays was determined for patients infected with N501Y variants.
Findings
The microneutralization antibody (MN) titers of convalescent sera from 9 recovered COVID-19 patients against B117-1 (geometric mean titer[GMT],80; 95% CI, 47–136) were similar to those against the non-N501Y viruses. However, MN titer of these serum against B117-2 (GMT, 20; 95% CI, 11–36) was statistically significantly reduced when compared with non-N501Y viruses (P < 0.01; one-way ANOVA). The difference between B117-1 and B117-2 was confirmed by testing 60 additional convalescent sera. B117-1 and B117-2 differ by only 3 amino acids (nsp2-S512Y, nsp13-K460R, spike-A1056V). Enzyme immunoassay using 272 convalescent sera showed reduced binding of anti-RBD IgG to N501Y or N501Y-E484K-K417N when compared with that of wild-type RBD (mean difference: 0.1116 and 0.5613, respectively; one-way ANOVA). Of 7 anti-N-IgG positive sera from patients infected with N501Y variants (collected 9-14 days post symptom onset), 6 (85.7%) tested negative for a commercially-available anti-S1-IgG assay.
Interpretation
We highlighted the importance of using a panel of viruses within the same lineage to determine the impact of virus variants on neutralization. Furthermore, clinicians should be aware of the potential reduced sensitivity of anti-RBD IgG assays.
Keywords: SARS-CoV-2 N501Y variant B.1.1.7, B.1.351, Neutralizing antibody Spike protein receptor binding domain, VOC
Research in context.
Evidence before this study
SARS-CoV-2 variants with spike protein N501Y mutation are of particular concern as they are associated with increased transmissibility, higher mortality, or reduced susceptibility to neutralization by antibody induced after natural infection or COVID-19 vaccination. We searched PubMed without language restrictions on 5th April 2021 for articles using the terms “COVID-19” or “SARS-CoV-2” and the terms “N501Y”, “variant”, or “antibody assay”. Most of the studies compared the susceptibility of different lineages using a single virus isolate from each lineage.
Added value of this study
We showed that there was a significant difference in the neutralizing antibody titer between two alpha variant (B.1.1.7 lineage) viruses we tested. There was no amino acid difference in the receptor binding domain between these two virus isolates. Instead, the amino acid differences occurred in the S2 of the spike protein, nsp2 and nsp13 (helicase). Furthermore, sera from patients infected with N501Y variant (either B.1.1.7 or B.1.351 lineage) are often negative for anti-RBD IgG even when the anti-nucleocapsid IgG is positive. This is unlike sera from patients infected with non-N501Y lineages, for whom the anti-RBD IgG is often positive when the anti-N IgG is positive.
Implication of all the available evidence
Our results highlight the importance of testing different viruses within the same lineage when assessing the susceptibility to neutralization for SARS-CoV-2 variants. As mutations outside the RBD can have a great impact on susceptibility to neutralization, it is important to monitor for mutations that emerge outside the RBD. Clinicians and clinical microbiologists should also be alerted to the possible reduced sensitivity of currently available anti-RBD IgG assays which are designed based on non-N501Y virus.
Alt-text: Unlabelled box
1. Introduction
SARS-CoV-2 was first detected in humans during a cluster of pneumonia in China in December 2019 [1]. Efficient person-to-person transmission was demonstrated by the high attack rate in a familial outbreak [2]. Early SARS-CoV-2 virus isolates in December 2019 and January 2020 were already genetically diverse. In the first two months after the discovery of SARS-CoV-2, different genetic clades have emerged, including the V, S and L clade according to the GISAID nomenclature [3]. The first major mutation was the spike D614G, which first emerged in February 2020 and then dominated the world. Spike D614G affects the transmissibility of the virus [4], and SARS-CoV-2 with D614G mutation has been found in reinfection cases [5,6].
SARS-CoV-2 enters cells via the attachment of the spike protein receptor binding domain (RBD) to the host receptor angiotensin-converting enzyme 2 (ACE2) [7], although other host factors, such as heparan sulphate, has been shown to play a role in virus entry [8]. In addition to cell surface ACE2, the interaction between spike protein and soluble ACE2 has been shown to facilitate endocytosis [9]. The spike protein N-terminal domain, RBD and S2 are targets of neutralizing antibodies, with the RBD being the most immunogenic part [[10], [11], [12]]. Anti-RBD antibody has a high correlation with neutralizing antibody titers [13,14]. COVID-19 vaccines are designed to elicit antibody against the spike protein. Hence, SARS-CoV-2 variants with mutations in the RBD, especially those at positions that interacts with ACE2 [15,16], is of particular concern because these variants may escape natural infection or vaccine-induced humoral immunity.
Since November 2020, several variants bearing mutations in the spike RBD have rapidly spread in the United Kingdom (B.1.1.7; α variant), South Africa (B.1.351; β variant) and Brazil (P.1; γ variant). All three variants contain spike N501Y mutation. The B.1.351 and P.1 variants also contain mutation at the spike amino acid position 484 (E484K) and at position 417 (K417N for B.1.351; K417T for P.1). These mutations have led to heightened concern as epidemiological studies suggest that they are more transmissible [17,18], and a higher viral load is found in patients with the B.1.1.7 variant [19]. Furthermore, these variants may jeopardize vaccine efficacy [20]. These variants have also caused reinfections [21].
Although some studies have suggested that the convalescent sera neutralizing antibody titer is lower for these variants than for wild type, these studies were conducted in Europe or USA. Since there are geographical differences in the lineages in different parts of the world, results from Europe or USA patients may not represent those from others. Furthermore, the number of patients’ serum specimens in these studies are relatively small. In the current study, we compared the neutralizing antibody titers against different lineages of viruses with convalescent serum from patients initially infected with different lineages. Furthermore, we assessed the difference in antibody against wild type and N501Y mutant RBD with a large serum panel consisting of > 250 patients.
2. Methods
2.1. Study setting and clinical specimens
Archived posterior oropharyngeal saliva, nasopharyngeal swab and serum specimens from hospitalized COVID-19 patients in Hong Kong were retrieved for viral genome sequencing, viral culture or the determination of antibody titers. The SARS-CoV-2 virus isolates used for the live virus microneutralization antibody (MN) assay were isolated from clinical specimens between March 2020 and January 2021, while the serum specimens used in the antibody assays were collected between July 2020 and January 2021.
2.2. Ethics statement
This study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 13–372 and UW 20–292), the Hong Kong Polytechnic University (approval no. RSA20021), the Kowloon West Cluster REC (KW/EX-20–038[144-26]), and the Kowloon Central/Kowloon East Cluster REC (KC/KE-20–0140/ER-1). Since archived specimens were used, written informed consent was waived.
2.3. Next generation sequencing of clinical specimens
Library preparation, nanopore sequencing and bioinformatic analysis were performed as we described previously [3]. Briefly, nanopore sequencing was performed following the Nanopore protocol - PCR tiling of COVID-19 (Version: PTC_9096_v109_revH_06Feb2020) according to the manufacturer's instructions with modifications (Oxford Nanopore Technologies). Briefly, extracted RNA was first reverse transcribed to cDNA using SuperScriptTM IV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA; Cat#18090200). PCR amplification was then performed using the hCoV-2019/nCoV-2019 Version 3 Amplicon Set [Integrated DNA Technologies (IDT), Coralville, IA, USA; Cat#10006788]. End preparation and native barcode ligation were performed according to the PCR tiling of COVID-19 virus protocol (EXP-NBD196, Oxford Nanopore Technologies). Barcoded and pooled libraries were then ligated to sequencing adapter and sequenced with the Oxford Nanopore MinION device using R9.4.1 flow cells for 24–48 h. For bioinformatics analysis, the recommended ARTIC bioinformatics workflow was used and minor modifications were applied as described previously [3] (Supplementary Method). The sequences of the SARS-CoV-2 isolates used in the MN assay have been deposited into the GISAID database (Supplementary Table S1).
2.4. Viral culture
Viral culture was performed using wild-type VeroE6 (ATCC Cat#CRL-1586; RRID:CVCL_0574) or TMPRSS2-expressing VeroE6 cells (JCRB Cat#JCRB1819; RRID:CVCL_YQ49) in a biosafety level-3 facility [22,23]. Briefly, cells were seeded with 1 mL of minimum essential medium (MEM) (Thermo Fisher Scientific, Gibco; Cat#11095) at 2 × 105 cells/mL in culture tubes and incubated at 37°C in a carbon dioxide incubator for 1–2 days until confluence for inoculation. Each tube was inoculated with 0.2 mL of specimen and was incubated in a slanted position so that the inoculum covered the monolayer for 60 min at 37°C. Then 1 mL of MEM was added and incubated in a roller apparatus at a speed 12 to 15 revolutions per hour. Virus-induced cytopathic effect was examined daily for up to 7 days. The cultures with more than 50% virus-induced cytopathic effect were expanded to large volume and the 50% tissue culture infective dose (TCID50) was determined.
2.5. Commercial IgG assays against N protein and spike protein S1 subunit
Anti-nucleocapsid (N) IgG was determined using Abbott SARS-CoV-2 IgG assay (Abbott, Abbott Park, Illinois, U.S.A). Anti-S1 IgG was determined using the Euroimmun anti-SARS-CoV-2 ELISA (IgG) (Euroimmun, Lubeck, Germany). The Euroimmun anti-S1 IgG assay is a semi-quantitative assay by calculating the ratio of the extinction coefficient of patient's serum over the extinction coefficient of the calibrator.
2.6. Live virus MN assay
Live virus MN assay was performed as we described previously [23,24]. The MN antibody titer was the highest dilution with 50% inhibition of the cytopathic effect, and an MN antibody titer of ≥20 was considered positive. All dilutions were performed in duplicates.
2.7. Expression and purification of RBD
Recombinant RBD (amino acid residues 306-543) of SARS-CoV-2 spike protein from the reference sequence Wuhan-Hu-1 (GenBank ID YP_009724390.1) (wild type) or with the mutations N501Y or N501Y-E484K-K417N were expressed and purified as we described previously with modifications [25]. Briefly, RBD gene sequences were codon-optimized for baculovirus expression and cloned into pFast dual baculovirus expression vector (Thermo Fisher Scientific, Gibco; Cat#10712024). The constructs were fused with an N-terminal gp67 signal peptide and C-terminal 6ⅹHis tag. The plasmid containing the RBD gene was transformed to DH10Bac to generate a recombinant bacmid DNA, which was used to transfect the Sf9 cells (ATCC Cat#CRL-1711; RRID:CVCL_0549) using Cellfectin II (Thermo Fisher Scientific, Gibco; Cat#10362100). After 72 h, the culture supernatant which contained the baculovirus was used to infect ExpiSf9 cell suspension culture (Thermo Fisher Scientific, Gibco; Cat#A35243) at a multiplicity of infection of 1 to 10. Infected ExpiSf9 cells were incubated at 27.5 °C with shaking at 125 r.p.m. for 96 h for protein expression. The supernatant was collected and then concentrated using a 10 kDa MW cutoff Labscale TFF System (Millipore). The RBD protein was purified by Ni-NTA purification system and size exclusion chromatography. The concentration of purified RBD was determined by using the Bradford Assay Kit (Bio-Rad; Cat#5000002) according to the manufacturer's instructions. The purity of recombinant RBD mutants were verified by western blotting.
2.8. Anti-RBD assay for wild type, N501Y, and N501Y-E484K RBD
An in-house enzyme immunoassay coated with either wild type, N501Y, or N501Y-E484K RBD was used to determine the impact of N501Y on RBD binding. Briefly, 96-well Nunc MaxiSorp™ flat-bottom immunoplates (Thermo Fisher Scientific, Invitrogen, Denmark; Cat#44-2404) were coated with 100 μL/well (0.1 μg/well) of His-tagged SARS-CoV-2 spike RBD with the wild type, N501Y or N501Y-E484K-K417N RBD in 0.05 M NaHCO3 (pH 9.6) overnight at 4°C and then followed by incubation with a blocking reagent. After blocking, 100 μL heat-inactivated serum samples at 1:100 dilution or human monoclonal antibody against SARS-CoV-2 RBD was added to the wells and incubated at room temperature for 1 h. For normalization, mouse monoclonal antibody against His-tag (ABclonal, ABclonal, Inc., Woburn, MA, USA; Cat# AE003; RRID:AB_2728734) was diluted in a series of two-fold dilution from 1:12,000 to 1:6,144,000. The attached human and mouse antibodies were detected using horseradish-peroxidase-conjugated goat anti-human IgG (Cat#A18811; RRID AB_2535588) and anti-mouse IgG antibody (Cat#31430; RRID AB_228307), respectively (Thermo Fisher Scientific, Invitrogen, Waltham, MA, USA). The reaction was developed by adding diluted 3,3’,5,5’-tetramethylbenzidine single solution (Thermo Fisher Scientific, Invitrogen; Cat#002023) and stopped with 0.3 N H2SO4. The optical density (OD) was read at 450 and 620 nm.
For each run, we included two positive samples as positive control, and an archived anonymous sample from 2018 as negative control. For OD values greater than 4, a value of 4 is assigned. Furthermore, we have compared the binding of WT, N501Y and N501Y-E484K RBD with a SARS-CoV-2 human antibody which was produced as previously described with modifications [26]. Briefly, SARS-CoV-2 RBD-specific memory B cells were sorted by multi-laser AriaII sorter (BD Biosciences, New Jersey, USA) from SARS-CoV-2 infected individuals. The IgG heavy and light chain variable regions were amplified independently by nested PCR. Full length IgG1 was expressed by co-transfecting HEK-293T cells (ATCC Cat#CRL-3216; RRID:CVCL_0063) with equal amounts of paired heavy and light chain plasmids based on the backbone of the pCI-neo mammalian expression vector (Promega; Cat#E1841). Culture media were harvested six days after transfection and purified using protein A agarose (Thermo Fisher Scientific; Cat#89931). The concentration of protein was determined by using the Bradford Assay Kit (Bio-Rad; Cat#5000002) according to the manufacturer's instructions.
2.9. Statistical analysis
Statistical analysis was performed using SPSS 26.0 (IBM SPSS Statistics; RRID:SCR_019096) and GraphPad PRISM 9.1.1 (GraphPad Software, San Diego CA, USA. RRID:SCR_002798). For the purpose of statistical analysis, an MN titer of <20 was considered as 10. The comparison of log-transformed MN titer was performed using one-way ANOVA with Dunnett's multiple comparisons test. B117-1 or B117-2 were used as the common control because our aim was to assess whether the viruses in the B.1.1.7 lineage are significantly different from other strains. We selected 3 samples from each of the 3 different waves in Hong Kong so that the results are not biased towards one single lineage. The comparison of anti-RBD IgG levels was determined using one-way ANOVA with Tukey's multiple comparisons test. The EC50 of monoclonal anti-RBD IgG was determined with the 5-parameter dose-response curve.
2.10. Role of funding source
The funding source had no role in the study design, data collection, data analysis, interpretation, or writing of the manuscript.
3. Results
3.1. Identification of patients with N501Y variants at RBD
Since January 2020 till April 2021, we have sequenced a total of 858 specimens. We have identified 14 patients infected with B.1.1.7 lineage with N501Y alone, 4 patients infected with B.1.351 lineage with N501Y, E484K and K417N, and 1 patient infected with P.3 lineage with N501Y and E484K. All patients were imported cases.
3.2. Microneutralization antibody titers
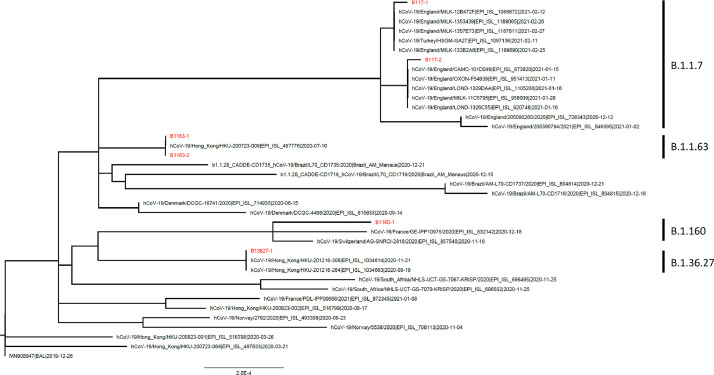
We have performed MN assay using a total of 9 serum specimens and 6 viruses. The 9 serum specimens were collected from Hong Kong patients, including 3 from second wave (March-April, 2020); 3 from third wave (August 2020); and 3 from fourth wave (November-December 2020) (Tables 1 and Supplementary S2). The median age was 55 years (range 28 to 65 years), and their sera were collected at median of 30 days post-symptom onset (range 18–47 days). For the 6 viruses, 3 were isolated from patients returning from England, including 2 viruses in the B.1.1.7 lineage and 1 virus in the B.1.160 lineage (a non-N501Y lineage); 3 other viruses were isolated from patients who acquired the infection in Hong Kong, including 2 viruses in the B.1.1.63 lineage collected in July 2020, and 1 virus in the B.1.36.27 lineage collected in December 2020 (Fig. 1 and Supplementary Table S1).
Table 1.
Details of serum specimens used for the MN assay.
| Case no. | Month of diagnosisa | Virus lineage/variant | Days after symptom onset | Anti-N IgG | Anti-RBD IgG |
|---|---|---|---|---|---|
| N1 | March | N/A | 32 | Positive | Positive |
| N2b | April | N/A | 18 | Positive | Positive |
| N3c | April | N/A | 31 | Positive | Positive |
| N4 | August | B.1.1.63 | 29 | Positive | Positive |
| N5c | August | B.1.2 | 29 | Positive | Positive |
| N6d | August | N/A | 20 | Positive | Positive |
| N7 | December | B.1.36.27 | 31 | Positive | Positive |
| N8 | December | N/A | 33 | Positive | Positive |
| N9 | November | B.1.36.27 | 47 | Positive | Positive |
N/A: Not available because sequencing not performed.
All diagnosed in 2020.
Patient is a returning traveler from Japan.
Patient is a returning traveler from USA.
Patient is a returning traveler from India.
Fig. 1.
Phylogenetic tree showing the 6 different virus isolates used in the microneutralization assay in this study.
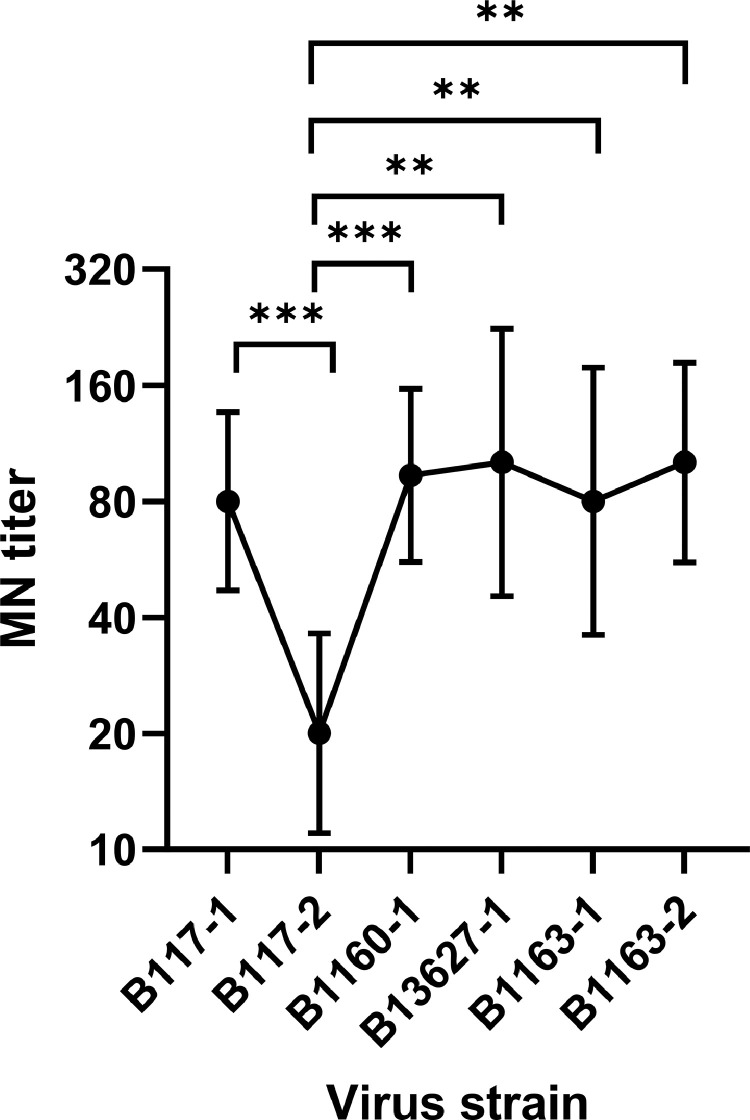
There was no statistically significant difference in MN titer between B117-1 virus and viruses in non-N501Y lineages (Fig. 2). However, for B117-2 virus, there was a statistically significant decrease in MN titer when compared with all non-N501Y lineages. The MN titer of B117-1 (geometric mean titer [GMT], 80; 95% confidence interval [CI], 47–136) were similar to those of non-N501Y viruses, while the MN titer of B117-2 (GMT, 20; 95% CI, 11–36) was statistically significantly reduced when compared with non-N501Y viruses (P < 0.01; repeated measures one-way ANOVA with Dunnett's multiple comparisons test).
Fig. 2.
Comparison of microneutralization titer between viruses from B.1.1.7 lineages (B117-1 and B117-2) and those from non-B.1.1.7 lineages (B1160-1, B13627-1, B1163-1, B1163-2). Data represent the geometric mean of convalescent serum specimens from 9 patients. Error bar indicates 95% confidence interval. One-way ANOVA with Dunnett's multiple comparison test was used for statistical analysis, with B117-1 as the reference group. **, P < 0.01; ***, P < 0.001.
To confirm the difference in the MN titer between the B117-1 and B117-2, we performed the MN assay against B117-1 and B117-2 for an additional 60 serum specimens collected. The MN titer against B117-1 was greater than B117-2 for 93.3% (56/60) of patients (Table 2).
Table 2.
Comparison of neutralization of B117-1 and B117-2 in serum specimens from 60 convalescent COVID-19 patients.
| Difference in microneutralization antibody titer between B117-1 and B117-2 | No. of patients (%) (n=60) |
|---|---|
| No difference | 4 (6.7) |
| B117-1 > B-117-2 | |
| ≥2-fold | 56 (93.3) |
| ≥4-fold | 35 (58.3) |
| ≥8-fold | 9 (15) |
Since there was a large difference in susceptibility to neutralization between the two B.1.1.7 lineage viruses, we compared the whole genome sequences of the two virus culture isolates. There were 3 amino acid differences between the two viruses, including nsp2 amino acid position 512 (B117-1: Y; B117-2: S), nsp13 amino acid position 460 (B117-1: K; B117-2: R) and spike amino acid position 1056 (B117-1: A; B117-2: V). To rule out the possibility of mutations that arose during virus culture, we have also compared the sequence from clinical specimen and from clinical culture isolate but there was no difference. In the GISAID database (as of 25th May 2021), nsp13 K460R was present in 23.7% (156257 of 658890) of sequences within the B.1.1.7 lineage (Supplementary Table S3), while spike A1056V and nsp2 S512Y mutation was present in 0.0127% (84/660147) and 0.0006% (4/657159) of sequences, respectively.
3.3. Comparison of anti-RBD IgG against wild type, N501Y, and N501Y-E484K RBD
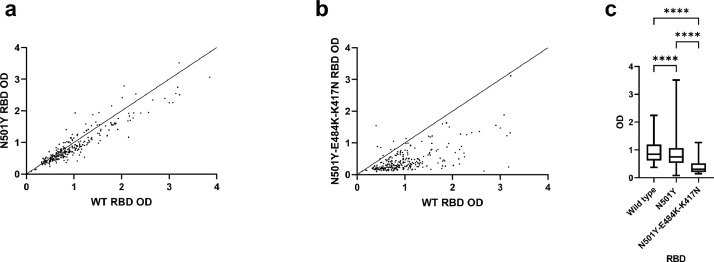
Although the neutralizing antibody titers are mainly affected by mutations in the RBD, mutations outside the RBD may also affect the neutralizing antibody titer [27]. To eliminate the effect from other mutations, we compared the levels of anti-RBD antibodies using recombinant RBD with or without N501Y mutation for 272 recovered COVID-19 patients’ sera. The normalized OD values for N501Y RBD (mean difference from wild type RBD, 0.1116; standard error [SE] of difference, 0.01405) and N501Y-E484K-K417N RBD (mean difference from wild type RBD, 0.5613, SE of difference, 0.02773) were statistically significantly lower than those from wild type RBD (P < 0.0001, repeated measures one-way ANOVA with Tukey's multiple comparisons test) (Fig. 3). Furthermore, the normalized OD values for N501Y-E484K-K417N RBD was significantly lower than that of N501Y RBD (mean difference, 0.4497; SE of difference, 0.02648). We have also tested a human anti-RBD monoclonal IgG, but there was no significant difference in the OD between the wild type, N501Y and N501Y-E484K-K417N RBD (Supplementary Fig. S1).
Fig. 3.
Comparison of anti-RBD IgG for serum optical density values of 272 convalescent COVID-19 patients between wild type, N501Y, and N501Y-E484K-K417N RBD mutant. (a) WT vs N501Y RBD; (b) WT vs N501Y-E484K-K417N RBD. For (a) and (b), Each dot represents the OD value of a single serum, and the line of identity is shown. (c) Box-whisker plot of the OD values, indicating the 5th, 25th, 50th, 75th and 95th percentiles . One-way ANOVA with Tukey's multiple comparisons test was used for statistical analysis. ****, P < 0.0001.
3.4. Performance of commercially-available antibody assays for patients infected with N501Y variants
We assessed the anti-N and anti-spike S1 subunit (containing the RBD) for 7 patients infected with N501Y variant, including 5 patients with B.1.1.7 lineage and 2 patients with B.1.351 lineage. The serum specimens were collected between 9 and 14 days post symptom onset for symptomatic patients. All 7 patients tested positive for anti-N IgG, while only 1 of 7 patients (14.3%) tested anti-S1 IgG positive (Table 3). As controls, we have randomly tested 99 anti-N IgG positive serum specimens from patients infected with non-N501Y viruses, that were collected between 9 and 14 days after symptom onset. Anti-S1 IgG was tested positive in 50% (49/98) of patients.
Table 3.
Details of serum specimens from patients infected with SARS-CoV-2 with N501Y mutation.
| Case no. | Month of diagnosisa | Virus lineage/variant | Days after symptom onset or hospital admission | Anti-N IgG | Anti-RBD IgG |
|---|---|---|---|---|---|
| SA1 | December | B.1.351 | 9 | Positive | Negative |
| SA2 | February | B.1.351 | 1b | Positive | Negative |
| UK1 | December | B.1.1.7 | 16b | Positive | Positive |
| UK2 | December | B.1.1.7 | 10 | Positive | Negative |
| UK3 | December | B.1.1.7 | 9 | Positive | Negative |
| UK4 | December | B.1.1.7 | 14 | Positive | Negative |
| UK5 | December | B.1.1.7 | 13b | Positive | Negative |
All diagnosed in 2020.
Days after hospital admission (patient was asymptomatic).
3.5. Correlation between commercially-available antibody assays and neutralizing antibody titer against B.1.1.7 and non-B.1.1.7 lineage viruses
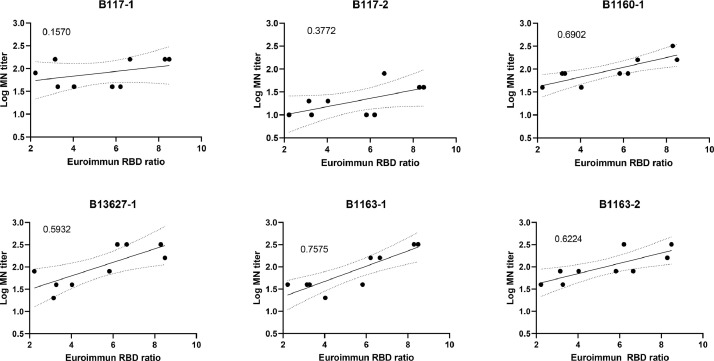
For the serum specimens from the 9 patients infected with non-B.1.1.7 lineage virus, we performed the Euroimmun anti-S1 IgG assay. The correlation between the Euroimmun ratio and log MN titers was better for non-B.1.1.7 lineage viruses than those of B.1.1.7 lineage viruses (0.59–0.76 for non-B.1.1.7 lineage viruses; 0.16 and 0.37 for B.1.1.7 lineage viruses) (Fig. 4).
Fig. 4.
Correlation between the ratio of Euroimmun anti-S1 IgG assay and the log MN titer of sera from 9 convalescent COVID-19 patients against different viruses. The ratio of Euroimmun anti-S1 IgG assay is calculated by dividing extinction coefficient of patient's serum over the extinction coefficient of the calibrator. The number in the upper left hand corner of each group represents the r2 value, and was calculated using the simple linear regression. The solid line indicates the best fit line, while the dotted lines indicate the 95% confidence band.
4. Discussions
Three SARS-CoV-2 variants with spike N501Y mutation, including B.1.1.7, B.1.351 and P.1, have been classified as “variants of concern” because of increased transmissibility, disease severity, or reduced susceptibility to neutralization by natural infection or vaccine-induced antibodies [28]. This study assessed the impact of N501Y variants on the neutralizing activity of convalescent sera from COVID-19 patients, and on anti-RBD immunoassays. Although one of the B.1.1.7 lineage virus (B117-1) did not exhibit a statistically significant difference with other non-B.1.1.7 lineage viruses, another B.1.1.7 lineage virus (B117-2) with 3 amino acid difference was much more resistant to neutralizing activity in convalescent sera, with ≥ 4-fold reduction in MN titer for most serum specimens tested. The binding of antibody in convalescent sera were statistically significantly lower for N501Y RBD than for wild type RBD. Most early serum specimens from patients infected with B.1.1.7 or B.1.351 lineage viruses tested positive with antibody assay against the N protein but negative against the S1 subunit of the spike protein.
Most previous studies assessing the impact of N501Y variants on the antibody titer used only a single virus isolate in the variant lineage for the determination of neutralizing antibody titer [29,30]. There are conflicting results from these studies, with some showing slight reduction of antibody titers for B.1.1.7 lineage virus [31,32], while others showed no difference [33]. In the current study, we have shown a large difference between the two B.1.1.7 lineage viruses, although there was no difference in the RBD amino acid sequences of these two isolates. Therefore, it is important to select multiple viruses to ensure the generalizability of the results. The difference between these two strains were located at nsp2 (S512Y), nsp13 (K460R), and the spike protein (A1056V). A1056V is located in the S2 subunit, which is required for the fusion of host and viral membrane. It has been suggested S2 can be a target of the neutralizing antibody for SARS-CoV, MERS-CoV and SARS-CoV-2 [34,35]. Indeed, neutralizing antibodies targeting the S2 region have been found [36]. D796H mutation in the S2 region has been proposed to reduce neutralization susceptibility by convalescent plasma and is found in the B.1.1.318 [37,38]. D614G causes conformation change of the spike protein, and therefore mutations in the S2 subunit may affect neutralization by allosteric mechanism [39]. Nsp13 is a helicase and a potent interferon antagonist, while the function of nsp2 is currently unknown. It remains to be determined which of these mutations is responsible for the difference in MN titers.
Our current study showed that N501Y variants may affect the time-to-seropositivity for anti-RBD IgG with the commercial assay, as 6 of the 7 anti-N IgG positive specimens tested negative for anti-RBD IgG. One study showed that the anti-N IgG assay by Abbott had earlier seroconversion than the Euroimmun anti-S1 IgG assay, although the seropositive percentage was above 35% for samples collected 9-10 days after symptom onset and 70% for samples collected on days 13-14 [40]. Our previous study showed that anti-N and anti-RBD IgG appear near the same time [13]. Therefore, anti-RBD IgG induced by infection with N501Y variant may not bind well to wild type RBD. These results suggest that anti-RBD IgG immunoassay should be modified for a better detection of anti-RBD antibodies. Furthermore, there is a possibility that second generation vaccines containing variant spike RBD may have reduced efficacy against non-N501Y strains.
Our study is specifically designed so that our findings are generalizable. Therefore, we have specifically chosen serum specimens from patients who were infected at different time periods, ranging from March to December 2020 (Table 1). Throughout this period, Hong Kong has experienced 4 major waves of COVID-19, and the virus lineages in each period varies [41]. Furthermore, to ensure that the virus culture isolates do not have mutations that arises during in vitro cell passage, we have sequenced the clinical specimen and the virus culture isolates.
There are several limitations in this study. First, we tested viruses from the B.1.1.7 lineage. As increasing number of variants are identified, testing on other variants are required. Second, the serum specimens were collected from patients in Hong Kong, although we have also selected travelers from other places. A global effort is required to understand how these variants affect the susceptibility of convalescent serum from different countries which may have been infected with unique lineages. Third, the number of patients infected with SARS-CoV-2 variants was small, and the anti-S1 antibody testing was performed with only a single commercial assay due to limited volume of the serum specimens. A larger cohort would be required to verify whether the variants will affect the performance of different serological assays.
Increasing number of variants with mutations at the spike RBD have been identified. The results from neutralization assays and anti-RBD antibody assays play a key role in the risk assessment of these variants. We demonstrated the large difference in neutralization antibody titer within the same variant lineage. Differences outside the RBD play an important role in neutralizing antibody titer. Therefore, it would be important to determine antigenic drift not only on variants with mutations in the RBD, but also with mutations outside RBD in the spike protein, or even outside the spike protein.
Contributors
LL and KKWT had roles in study design, data collection, data analysis, data interpretation, literature search and writing of the manuscript. DCL, ART, YSY, MYWK, WKT, OTYT, LLYL, VCC, and IFNH had roles in recruitment, data collection, and/or clinical management. AWHC, RRZ, WMC, JDI, HWT, LLC, JPC, HY, TCI, RWSP, GKHS and BWYM had roles in performing the experiments, data collection, data analysis, and/or data interpretation. KHC and KYY had roles in study design, data interpretation and writing of the manuscript. LL and KKWT verified the underlying data. All authors interpreted the data, revised the manuscript critically for important intellectual content and approved the final version of the manuscript.
Data sharing
Data are available upon reasonable request from the corresponding author.
Funding
Richard and Carol Yu, Michael Tong, and the Government Consultancy Service (see acknowledgments for full list).
Declaration of Competing Interest
All authors declare no conflict of interest.
Acknowledgements
We would like to thank Professor Ling Chen for providing the human monoclonal antibody against SARS-CoV-2 RBD. We gratefully acknowledge the originating and submitting laboratories who contributed sequences to Global Initiative on Sharing All Influenza Data (GISAID) (Supplementary Table S4). This work was supported by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance, Department of Health, Hong Kong SAR Government; and donations of Richard Yu and Carol Yu, May Tam Mak Mei Yin, the Shaw Foundation Hong Kong, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Jessie and George Ho Charitable Foundation, Kai Chong Tong, and Tse Kam Ming Laurence.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103544.
Appendix. Supplementary materials
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To K.K., Chan W.M., Ip J.D., Chu A.W., Tam A.R., Liu R. Unique SARS-CoV-2 clusters causing a large COVID-19 outbreak in Hong Kong. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric R.S. Emergence of a highly fit SARS-CoV-2 variant. N Engl J Med. 2020;383:2684–2686. doi: 10.1056/NEJMcibr2032888. [DOI] [PubMed] [Google Scholar]
- 5.To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J.S., Kim S.Y., Kim T.S., Hong K.H., Ryoo N.H., Lee J. Evidence of severe acute respiratory syndrome Coronavirus 2 reinfection after recovery from mild Coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021;12:134. doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung M.L., Teng J.L.L., Jia L., Zhang C., Huang C., Cai J.P. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.053. 2212-28 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.028. 2332-47 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.035. 1821-35 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.037. 1024-42 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong C.H., Cai J.P., Dissanayake T.K., Chen L.L., Choi C.Y., Wong L.H. Improved detection of antibodies against SARS-CoV-2 by microsphere-based antibody assay. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21186595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26:2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021:372. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B. S-variant SARS-CoV-2 Lineage B1.1.7 Is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. 2021;223:1666–1670. doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahase E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 21.Zucman N., Uhel F., Descamps D., Roux D., Ricard J.D. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: a case report. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To K.K., Cheng V.C., Cai J.P., Chan K.H., Chen L.L., Wong L.H. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: a multicohort study. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30053-7. e111-e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To K.K., Hung I.F., Chan K.H., Yuan S., To W.K., Tsang D.N. Serum antibody profile of a patient with Coronavirus disease 2019 reinfection. Clin Infect Dis. 2021;72 doi: 10.1093/cid/ciaa1368. e659-e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovitz D., Chan J.F., Oh Y.J., Yuan S., Chu H., Yeung M.L. 2021. Res Sq. [Google Scholar]
- 26.Yan Q., He P., Huang X., Luo K., Zhang Y., Yi H. Germline IGHV3-53-encoded RBD-targeting neutralizing antibodies are commonly present in the antibody repertoires of COVID-19 patients. Emerg Microbes Infect. 2021;10:1097–1111. doi: 10.1080/22221751.2021.1925594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021 doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021;397:462. doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A. Neutralizing activity of BNT162b2-elicited serum. N Engl J Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.033. 2201-2211.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.03.002. 529-539. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou D., Tian X., Qi R., Peng C., Zhang W. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: implications for vaccination and antibody therapeutics. Glycobiology. 2021;31:69–80. doi: 10.1093/glycob/cwaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemp S.A., Collier D.A., Datir R.P., Ferreira I., Gayed S., Jahun A. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tegally H., Ramuth M., Amoaka D., Scheepers C., Wilkinson E., Giovanetti M. Genomic epidemiology of SARS-CoV-2 in Mauritius reveals a new wave of infections dominated by the B.1.1.318, a variant under investigation. medRxiv. 2021 doi: 10.1101/2021.06.16.21259017:2021.06.16.21259017. [DOI] [Google Scholar]
- 39.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.032. 739-751. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Elslande J., Decru B., Jonckheere S., Van Wijngaerden E., Houben E., Vandecandelaere P. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect. 2020;26 doi: 10.1016/j.cmi.2020.07.038. 1557.e1- 1557.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan W.M., Ip J.D., Chu A.W., Tse H., Tam A.R., Li X. Phylogenomic analysis of COVID-19 summer and winter outbreaks in Hong Kong: an observational study. Lancet Reg Health West Pac. 2021;10 doi: 10.1016/j.lanwpc.2021.100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.