Abstract
The emergence and rapid spread of resistant bacteria has become a serious public health concern worldwide. Delayed antimicrobial therapy significantly increases mortality in high-risk infections with a particularly strong association with septic shock. Therefore, antimicrobial agents are often injudiciously used without any evidence-based microbiological confirmation. Antimicrobial consumption is strongly linked to the emergence and dissemination of antimicrobial-resistant bacteria strains in several epidemiological studies. According to CDC's recent publication, an estimated 30% of outpatient oral antimicrobial prescriptions may have been inappropriate. A compact and rapid pathogen identification (ID) and antimicrobial susceptibility testing (AST) can assist to address both the unnecessary use and overuse of antimicrobials, and therefore effectively reduce antimicrobial resistance. The overall goal of these AST protocols is to deliver a molecular diagnostic platform that is capable of profiling the antimicrobial susceptibility of causative pathogens in hours, not days. The presented AST utilizes an electrochemical sensor to quantify the microbial changes of 16S rRNA after exposure to various antimicrobial conditions either from clinical isolates or directly from unprocessed clinical specimens such as urine and blood. These protocols can be performed by our robotic lab automation systems or manual benchtop assays with associated reagent kits, AST stripwells and sensor chips.
-
•
A rapid, quantifiable antimicrobial efficacy profiling comparable to traditional AST reporting.
-
•
Customized antimicrobials and dilution ranges tailored to unique specifications for research and development.
-
•
Direct antimicrobial susceptibility of viable pathogen from whole blood, urine, or subculture.
Keywords: Phenotypic microbiological response to antimicrobial conditions, Molecular quantification of species-specific 16S rRNA content, Direct-from-specimen antimicrobial susceptibility testing
Graphical abstract
Specifications table
| Subject Area: | Immunology and Microbiology |
| More specific subject area: | Antimicrobial susceptibility testing |
| Method name: | Molecular analysis of microbial responses to antimicrobial exposure |
| Name and reference of original method: | Chen CH, Lu, Y, Sin MLY, Mach KE, Zhang DD, Gau V, Liao JC, Wong PK. Rapid antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal Chem. 2010; 82(3): 1012. Mach KE, Mohan R, Baron EJ, Shih M-C, Gau V, Wong PK, Liao JC. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J Urol. 2011; 185(1): 148–153. Altobelli E, Mohan R, Mach KE, Sin MLY, Anikst V, Buscarini M, Wong PK, Gau V, Banaei N, Liao JC. Integrated biosensor assay for rapid uropathogen identification and phenotypic antimicrobial susceptibility testing. Eur Urol Focus. 2017; 3(2–3): 293–299. doi:10.1016/j.euf.2015.12.010 Liu D, Gau V, Tomasek M, Chen J, Singh V, Memmer M, et al. Evaluation of an automated rRNA quantitation system for rapid AST in clinical lab diagnostics. J Mol Diagn. 2020; 11(22): Suppl 28. doi: https://doi.org/10.1016/S1525–1578(20)30513–4 |
| Resource availability: | All resources including reagent, consumables and materials necessary to reproduce the method are described in the Methods details. |
Method details
Method overview
This method utilizes an electrochemical-based sandwich hybridization test to report categorical antimicrobial susceptibility (R for resistant, I for intermediate, S for susceptible) directly from suspensions of non-fastidious Gram-negative organisms grown on solid media or directly from clinical whole blood and urine specimens [1]. After exposure to different antimicrobial conditions, viable bacteria are quantified according to the amount of 16S rRNA contained in each condition. The amperometric signal generated from each antimicrobial condition can be compared to that of the growth control to determine if the sample is susceptible, intermediate, or resistant to a given antimicrobial.
The panel of Gram-negative organisms for this method contains the following: Citrobacter freundii, Enterobacter cloacae, Escherichia coli, Klebsiella aerogenes, Klebsiella oxytoca, Klebsiella pneumoniae, Morganella morganii, Proteus mirabilis, Pseudomonas aeruginosa, and Serratia marcescens. The antimicrobials tested in this method include ciprofloxacin, gentamicin, and meropenem in stripwells containing antibiotic concentrations based on Clinical and Laboratory Standards breakpoints. The materials and assay conditions detailed below have been selected for the indicated species and antimicrobials.
Materials
-
-
0.45% saline, Cat. No. U159, Hardy Diagnostics, Santa Maria, CA
-
-
Mueller-Hinton II Broth, Cat. No. M5860, Teknova, Hollister, CA or Cat. No. RK1000-IDAST, GeneFluidics, Irwindale, CA
-
-
1 M NaOH, Cat. No. RK1000-IDAST, GeneFluidics, Irwindale, CA
-
-
1 M HCl, Cat. No. RK1000-IDAST, GeneFluidics, Irwindale, CA
-
-
0.75 U/mL Anti-Fluorescein-POD, Fab fragments in Blocker Casein in PBS (Enzyme), Cat. No. 11426346910, Roche, Basel, Switzerland or Cat. No. RK1000-IDAST, GeneFluidics, Irwindale, CA
-
-
Enhanced K-Blue® Substrate (TMB), Cat. No. 308177, Neogen, Lansing, MI or Cat. No. RK1000-IDAST, GeneFluidics, Irwindale, CA
-
-
AST sensor chip, Cat. No. SC1000-ASTGN, GeneFluidics, Irwindale CA
-
-
AST stripwell, Cat. No. SW-1000-CGM, GeneFluidics, Irwindale CA
-
-
Ciprofloxacin stripwell, Cat. No. SW-2000-CIP, GeneFluidics, Irwindale CA
-
-
Gentamicin stripwell, Cat. No. SW-2000-GEN, GeneFluidics, Irwindale CA
-
-
Meropenem stripwell, Cat. No. SW-2000-MEM, GeneFluidics, Irwindale CA
-
-
Grant DEN-1B densitometer, Grant Instruments, Cambridge, UK
-
-
16-channel electrochemical reader system, Cat. No. RS-1000–16 Helios, GeneFluidics, Irwindale, CA
Protocol for AST from clinical isolates (standard and one-drug)
The following protocol details the procedure for performing AST from clinical isolates grown on solid media using the AST stripwell or one-drug stripwell.
-
1.Prepare a 2-mL saline suspension of bacteria at 0.5–0.6 McFarland (MF) units by inoculating 2 mL of 0.45% saline with 5–6 colonies of bacteria grown for 18–24 h at 37 °C on a tryptic soy agar plate with 5% sheep's blood.
-
a.If the densitometer reading is greater than 0.5-0.6 MF, dilute the suspension with saline until the desired range is met.
-
b.If the densitometer reading is below the 0.5-0.6 MF range, add more colonies to the suspension until the desired range is met.
-
a.
-
2.Dilute the bacterial suspension to the appropriate starting concentration depending on the species tested.
-
a.If testing Escherichia coli or Klebsiella pneumoniae, add 120 µL of the suspension to 1960 µL of Mueller-Hinton II (MH) broth.
-
b.If testing Pseudomonas aeruginosa or Serratia marcescens, add 450 µL of the suspension to 1550 µL of MH broth.
-
c.If testing Citrobacter freundii, Enterobacter cloacae, Klebsiella aerogenes, Klebsiella oxytoca, Morganella morganii, or Proteus mirabilis, add 150 µL of the suspension to 1960 µL of MH broth.
-
a.
-
3.
Once the sample is at the correct starting concentration, inoculate the selected stripwell (AST or one-drug) by adding 100 µL of the diluted sample to each well of the stripwell.
-
4.Incubate the stripwell at 37 °C.
-
a.If using the AST stripwell, incubate for 2 h.
-
b.If using a one-drug stripwell, incubate for 3 h.
-
a.
-
5.
At the conclusion of the incubation period, begin lysing the stripwell by adding 36 µL of 1 M NaOH to each well, followed by a 5-min incubation at room temperature.
-
6.
Complete the lysing step by adding 24 µL of 1 M HCl to each well. Immediately add 15 µL of the lysed sample, or lysate, from each well to its corresponding sensors on two sensor chips.
-
7.
Incubate all sensor chips at 43 °C for 30 min to allow for hybridization.
-
8.
Following the hybridization period, wash one sensor chip with DI water and dry with pressurized air. Immediately add 10 µL of enzyme to all sensors on the chip, followed by a 5-min incubation at room temperature.
-
9.
Repeat step 8 for any remaining sensor chips.
-
10.
After the 5-min incubation period, wash and dry one chip. Immediately add 40 µL of TMB to all sensors on the chip. Wait 30 s, then read with the Helios reader.
-
11.
Repeat step 10 for any remaining sensor chips.
Protocol for AST directly from urine specimens
The procedure outlined below describes the AST assay to be performed directly from urine specimens using a one-drug stripwell.
-
1.
Obtain a urine sample of 4-mL volume collected in a BD Vacutainer C&S Preservative tube (REF BD364951).
-
2.
Perform the urine pelleting protocol as previously described to remove the urine matrix interference components [2,6].
-
3.
Add 4 mL of MH broth to the pellet as inoculum for antimicrobial exposure.
-
4.
Deliver 100 µL of the resuspended pellet to each well of the selected one-drug stripwell and incubate for 3 h at 37 °C.
-
5.
Perform steps 5–11 of the above protocol to complete the AST assay directly from urine specimens.
Protocol for AST directly from whole blood specimens
The procedure outlined below describes the AST assay to be performed directly from whole blood specimens using a one-drug stripwell.
-
1.
Obtain a whole blood sample of 2-mL volume collected in a BD Vacutainer Serum tube (REF BD368975).
-
2.
Perform the blood pelleting protocol similarly to the urine protocol to remove the blood matrix interference components.
-
3.
Add 3 mL of MH broth to the pellet to generate inoculum for the stripwell.
-
4.
Deliver 100 µL of the resuspended pellet to each well of the selected one-drug stripwell and incubate for 10 h at 37 °C for the target microbial load to be 1 CFU/mL or higher.
-
5.
Complete steps 5–11 of the above protocol to complete the AST assay directly from whole blood specimens.
Method validation
We performed the described protocols with various strains from the CDC AR Bank to validate the assay parameters detailed above. The microbiological response is quantified through an electrochemical-based molecular analysis sensor and scalable antimicrobial stripwell [3,7]. The electrochemical sensor assay provides an amperometric readout of the concentration of ribosomal RNA present in a sample. Capture and detector probes (probe pair) are designed to hybridize to group- (Fig. 1A) or species-specific (Fig. 1B) regions of the 16S rRNA molecule that are accessible to independent hybridization with oligonucleotide probes on each sensor. A universal sensor chip configuration as shown in Fig. 1A can be used if the causative pathogen has been confirmed to be Gram-negative. For polymicrobial or unknown causative pathogens, a species-specific sensor chip configuration such as the one in Fig. 1B can be used for species-specific microbiological response to antimicrobial conditions. Antimicrobial susceptibility is assessed through microdilutions with various antimicrobial conditions inside each well of the polystyrene stripwells. The AST stripwell in Fig. 1C is an example of a panel of three antimicrobial classes: ciprofloxacin (fluoroquinolone class), gentamicin (aminoglycoside class), and meropenem (carbapenem class). [4] As shown in Fig. 1D-F, stripwells can also be customized by drying antimicrobials in DI water with 0.1% Tween onto EIA/RIA 8-well strips (Corning, Corning, NY) at the following concentrations: 0.25, 0.5, 1, 2, 4, 8, 16 μg/mL or any antimicrobial spectrum needed. The first well of each stripwell is left without antimicrobial to be used as a growth control (GC). Antimicrobial stripwell reagents are dried onto flat-bottom medical grade polystyrene 8-well strips at selected concentrations such that resuspension of these reagents with 100 µL of bacterial inoculum results in the final working concentrations for each antimicrobial [5].
Fig. 1.
(A) Sensor configuration of the same probe pair (EB/KE1 for most clinically relevant Gram-negative strains including Enterobacterales and Pseudomonas aeruginosa. EB capture probe: 5′-GCACTTTATGAGGTCCGCTTGCTCT-3′, EB detector probe: 5′-TCAGAGTTCCCGAAGGCACCAATCCATC-3′, KE1 capture probe: 5′- TCAGAGTTCCCGAAGGCACCAATCCATC-3′, KE1 detector probe: 5′- TCTGGAAAGTTCTCTGGATGTCAAGAGT-3′) for measurement of 16S rRNA from the same species with different conditions such as antimicrobial responses. (B) Sensor configuration of different probe pairs for detection of 16S rRNA of a polymicrobial sample for species-specific susceptibility reporting. NC stands for Negative Control and PC stands for Positive Control. Additional probe pairs (EC, KE1, MM, PA, SM) can be added for a customized panel for Escherichia coli, Pseudomonas aeruginosa, Serratia marcescens, Morganella morganii, Klebsiella pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, Klebsiella aerogenes and Citrobacter freundii to quantify 16S rRNA content individually. (C-F) various configurations of antimicrobial stripwells to be used in the proposed study.
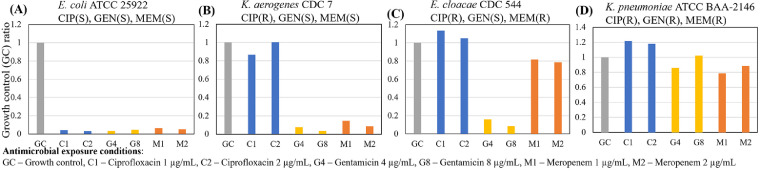
Validation of microbiological responses to antimicrobial exposure was performed with a panel of clinical isolates consisting of species of Gram-negative rods from the CDC AR bank and the American Type Culture Collection (ATCC) evaluated against ciprofloxacin, gentamicin, and meropenem as shown in Fig. 2. The microbiological response is plotted as electrochemical signal levels in nano amperes normalized to that of the growth control (GC) and are then translated into categorical susceptibility reporting (Susceptible, Intermediate, Resistant). Low GC ratios indicate inhibited microbial growth or a susceptible response to the antimicrobial conditions. High GC ratios suggest uninhibited growth or resistance to antimicrobials.
Fig. 2.
AST assay from clinical isolates using the AST stripwell in Fig. 1C. Microbiological response from each sensor-well combination for (A) ciprofloxacin, (B) gentamicin, and (C) meropenem AST stripwells with antimicrobial concentrations on or near breakpoints.
The goal of the validation study in Fig. 3 was to assess the ability to report susceptibility within a much shorter time frame; however, the susceptibility reporting based on a short exposure time may not be able to differentiate borderline intermediate/susceptible strains. Two ATCC QC strains (ATCC E. coli 25922 and ATCC K. pneumoniae BAA-2146) were used for antimicrobial exposure times of 30, 60, and 90 min as shown in Fig. 3. Susceptibility is evaluated based on the GC ratio across the antimicrobials spectrum tested. Additional GC wells were allocated to replace low antimicrobial conditions (0.25 µg/mL and 0.5 µg/mL) in this study.
Fig. 3.
Microbiological responses to antimicrobial exposure times using the one-drug stripwell.
The feasibility of the direct-from-blood AST was evaluated using the blood pellet as the inoculum for antimicrobial exposure. Higher contrived microbial loads (10, 100, and 1000 CFU/mL) were used in this proof-of-concept AST testing directly from whole blood, instead of positive blood cultures (Fig. 4).
Fig. 4.
Direct-from-blood antimicrobial efficacy profiling of pathogen (E. coli 77 susceptible to meropenem) with MEM one-drug stripwell.
Declaration of Competing Interest
Jade Chen, Michael Tomasek, Eliseo Nuñez, and Vincent Gau are employed by GeneFluidics. As a non-academic, commercial company, the employer and funder provided support in the form of salaries for authors [J.C., M.T., E.N., V.G.] but did not have any additional role in the protocol design, data collection, and decision to publish.
References
- 1.Liu D., Gau V., Tomasek M., Chen J., Singh V., Memmer M. Evaluation of an automated rRNA quantitation system for rapid AST in clinical lab diagnostics. J. Mol. Diagn., 2020;11(22) doi: 10.1016/S1525-1578(20)30513-4. Suppl 28. doi. [DOI] [Google Scholar]
- 2.Chen J., San S.S.S., Kung A., Tomasek M., Liu D., Rodgers W., Gau V. Direct-from-specimen microbial growth inhibition spectrums under antibiotic exposure and comparison to conventional antimicrobial susceptibility testing, bioRxiv. 2021. doi: 10.1101/2021.02.12.430910. [DOI] [PMC free article] [PubMed]
- 3.Chen J., Tomasek M., Cruz A., Faron M.L., Liu D., Rodgers W.H., Gau V. Feasibility and potential significance of rapid in vitro qualitative phenotypic antimicrobial susceptibility testing of gram-negative bacilli with the ProMax system. PLoS ONE. 2021;16(3) doi: 10.1371/journal.pone.0249203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamma P.D., Cosgrove S.E., Maragakis L.L. Combination therapy for treatment of infections with gram-negative bacteria. Clin. Microbiol. Rev., 2012;25:450–470. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altobelli E., Mohan R., Mach K.E., Sin M.L.Y., Anikst V., Buscarini M., Wong P.K., Gau V., Banaei N., Liao J.C. Integrated biosensor assay for rapid uropathogen identification and phenotypic antimicrobial susceptibility testing. Eur. Urol. Focus. 2017;3(2–3):293–299. doi: 10.1016/j.euf.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Tomasek M., Nuñez E., Gau V. Method for concentrating viable microorganisms for microbial load determination and eliminating uncertainty from matrix effects from urine and whole blood. MethodsX. 2021 doi: 10.1016/j.mex.2021.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Tomasek M., Gau V. Categorizing microbial growth inhibition through quantification of 16S rRNA growth marker with stripwells covering a spectrum of antimicrobial conditions. MethodsX. 2021 doi: 10.1016/j.mex.2021.101453. [DOI] [PMC free article] [PubMed] [Google Scholar]