Abstract
Heat shock factor 1, HSF1, is one of several family members that recognize repeated nGAAn sequences associated with the heat shock element of heat shock and other genes. This transactivator is activated from a monomeric to trimeric form by oxidative, thermal and other stressors. Various studies show that HSF1 levels increase with cancer and decrease with aging and neurodegenerative disorders. It has a role in development as well as infections and inflammation.
HSF1 is regulated by post-translational modifications and interactions with other proteins such as HSBP-1. Given its central importance in stress responsivity, various methods have been developed to identify HSF1 and its interacting partners. To date, multiple studies use conventional immunoprecipitation of HSF1 with commercially available antibodies which work well in cell lines but not whole tissue extracts. To remedy this shortfall, we developed a technique to retrieve activated HSF1 with an oligonucleotide link to a magnetic bead. The method captures HSF1 using a DNA sequence specific for HSF1 binding sites on promoter of heat shock genes. Confirmation of tissue derived HSF1 is identified using antibody against HSF1. The magnetic beads conjugated with DNA sequence specific to HSF1 binding was capable of yielding a reproducible band of high signal intensity with low background after native gel electrophoresis and ECL. Thus, the trimeric form of HSF1 can be isolated from tissue with magnetic beads conjugated with a short DNA sequence specific to HSF1 binding. This new method to identify HSF1 is economic, easy, and reproducible and does not require specialized equipment. It overcomes limitations of HSF1 tissue extraction by conventional immunoprecipitation, thus allowing for new approaches to understand HSF1 function in animal and human tissue.
-
•
HSF1 is a transcription factor that homotrimerize and binds to a conserved regulatory site, the heat shock element (HSE), consists of repeats of pentameric sequence ‘5-nGAAn-3’ present in the promoters of inducible heat shock protein genes.
-
•
This protocol allows isolation of trimeric forms of HSF1 from tissue lysate using magnetic beads conjugated with a short DNA sequence with specific binding to HSF1.
-
•
This method is easy, economic and does not require unique instrumentation.
Keywords: Heat shock factor1 (HSF1), Heat shock proteins, Immunoprecipitation, HSF1-DNA binding
Graphical Abstract
Specifications table
| Subject Area | Biochemistry, Genetics and Molecular Biology |
| More specific subject area | Biochemistry |
| Method name | Immunoprecipitation methods to pull down HSF1 from liver tissue extract. |
| Name and reference of original method | Kim, M.Y., et al., A DNA immunoprecipitation assay used in quantitative detection of in vitro DNA-protein complex binding. Anal Biochem, 2013. 441(2): p. 147-51. |
| Resource availability | NA |
Method details
Introduction
Immunoprecipitation (IP) is a key tool with multi-ordered steps for characterizing protein and interaction of proteins. Purified protein can be eluted and analyzed by various assays such as western blotting and mass spectroscopy [1,2]. This assay can be challenging, especially when low abundance proteins are sought. But the specific binding of these low abundance proteins to short DNA sequences can be utilized to overcome this challenge [3]. In this respect, a transcription factor HSF1 is a relatively low expressed protein as a master regulator of heat shock response. It is part of a feedback loop in which HSF1 controls expression of HSP70 [4] which in turn negatively regulates HSF1 action at the promoter region of heat shock genes. During unfavorable thermal conditions, the amount of unfolded proteins exceeds the capacity of heat shock proteins to refold and protect misfolded and damaged proteins (HSP40, HSP70 and HSP90). Under these circumstances, HSP70 is titrated away from HSF1, releasing HSF1 to self-assemble into a trimer unit with DNA binding capacity. As a transactivator to the heat shock genes, HSF1 drives HSP70 gene expression until enough HSP70 protein is produced and deactivates HSF1 in lieu of its association with damaged proteins [5,6]. Recent findings have shown that transcription level of the Hsp70 gene is significantly altered in cells from older organisms and is linked to impaired binding of HSF1 to the HSE present on heat shock genes promoter [7,8]. By studying alterations in HSF1 interaction with HSP70 in older organisms, we can gain insight into a key facet of aging-the reduced ability to respond to environmental stress. Thus, altered HSF1 and HSP70 interaction with aging may provide a good model for aging at the molecular level [9]. To study these changes, we initiated HSF1 immunoprecipitation in lysates from mouse liver tissue but found that the assay did not work as described by commercial vendors. To confirm this problem, we successfully immunoprecipitated HSF1 from cell lysate prepared from heat shocked HeLaS3 and SHY5Y cells but not from liver tissue lysates. To overcome this problem, we developed a novel method for pulling down HSF1 from tissue lysate by using a DNA sequence specific for HSF1 binding.
Materials
Buffers
-
1.
IP lysis buffer: 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol. # add protease inhibitor cocktail before use.
-
2.
2X Binding Buffer: 20 mM Tris pH = 7.5,100 mm NaCl, 2mM NaEDTA, 10% Glycerol,
-
3.
Sample buffer: 62.5 mM Tris−HCl pH 6.8, 10% (v/v) glycerol, 5% (w/v) SDS, 0.05% (w/v) bromophenol blue, 5% (v/v) β−mercaptoethanol.
-
4.
Non-reducing Sample buffer: sample buffer without β−mercaptoethanol.
-
5.
Running buffer: 25 mM Tris−HCl pH 8.3, 190 mM glycine, 0.1% (w/v) SDS.
-
6.
Transfer buffer: 25 mM Tris−HCl pH 8.3, 192 mM glycine, 20% (v/v) methanol.
-
7.
TBS buffer: 20 mM Tris−HCl pH 7.5, 150 mM NaCl.
-
8.
Washing buffer: TBS buffer containing 0.1% (v/v) Tween-20 detergent
-
9.
Blocking: washing buffer containing 5% (w/v) BSA.
-
10.
Stripping buffer: 62.5 mM Tris−HCl pH 6.7, 2% (w/v) SDS, 100 mM β−mercaptoethanol.
Reagents
Magnetic beads from BcMag Quick Oligo-DNA conjugation kit, HSF1 antibody (Proteintech, Catalog number 51034-AP), Clarity western ECL substarte, Goat anti rabbit HRP conjugated secondary antibody, Pierce Rapid gold BCA protein assay kit (Thermo scientific).
Conjugation of magnetic beads to oligo-DNA sequence
Materials required
Magnetic separator
BcMag Quick Oligo-DNA conjugation kit (Bioclone Inc) was used for the conjugation of Oligo-DNA to magnetic beads.
Oligo-DNA preparation
-
1.
The oligo-DNA must be modified with amino group at either 5’or 3’ end (Commercial oligo synthesis company can provide such service). In this protocol, we modified our oligo-DNA (CTAGAAGCTTCTAGAAGCTTCTAG) at 5’end. 10–15 extra nucleotides were added to our target DNA sequence between the terminal amino group and DNA sequence to overcome steric hindrance you design your target oligo–DNA sequence.
-
2.
Oligo-DNA was annealed and resuspend the in 1X suspension buffer at concentrations of 5–10 µg/ul.
Coupling
-
1.
Weigh 20 mg magnetic beads and transfer to a centrifuge tube.
-
2.
Add 200 µl oligo-DNA solution (5–10 µg oligo DNA/ul) and mix very well by pipetting.
-
3.
Incubate the beads overnight at 50 °C with gentle shaking.
-
4.
Place the tube on magnetic separator for 1–3 min. Remove the supernatant while the tube remains on the separator (Optional: Aspirate 2–5 µl supernatant, transfer to a new centrifuge tube and label the tube as C2).
-
5.
Wash the beads three times with 1 ml of washing buffer at room temperature and twice with double distilled H2O at 650C.
-
6.
Resuspend the beads at 5 mg/ml in PBS buffer containing 0.2% NaN3 and store at 40C.
Coupling efficiency calculation
-
1.
Measure OD at A260 Coupling Efficiency (%) = [(C1-C2)/C1] x 100% C1: A260 Pre-Coupling oligo DNA Solution x dilution factor; C2: A260 post-Coupling oligo DNA Solution.
-
2.
Or Run DNA on agarose gel to compare amount of oligo-DNA from C1 and C2.
Immunoprecipitation method
-
1.
Dissect the liver tissue with clean tools and as quickly as possible. Try to do this on ice to prevent protein degradation by proteases.
-
2.
Place the tissue in round-bottom micro centrifuge tubes and immerse in liquid nitrogen to snap freeze. Store samples at -80°C for later use or keep on ice for immediate homogenization.
-
3.
For a ∼10 mg piece of tissue, add ∼300 μL IP lysis buffer rapidly to the tube and homogenize with a bullet blender for 3-min at speed 8 using zirconium oxide beads.
-
4.
Centrifuge at 14,000 rpm for 20-min at 4 °C. Transfer the supernatant to a new tube. If necessary, lysate can be stored at -80°C. Quantify the protein concentration using Pierce Rapid gold BCA protein assay kit (Thermo scientific).
-
5.
Transfer 250 μL of magnetic beads slurry prepared in coupling step in a clean tube. Place the tube on magnetic stand for 30-s. Carefully remove the buffer once solution is clear.
# Briefly vortex the tube to resuspend the magnetic beads.
-
6.
Add 1mg of liver tissue lysate diluted in 1X binding buffer in a tube with magnetic beads (step 5).
-
7.
Incubate with rotation for 3–4 h at room temperature.
-
8.
Place the tube on magnetic stand for 30-s and discard the clear solution.
-
9.
Wash the beads twice with 1X Wash buffer (0.1% tween-20).
-
10.
Add 40 µl of 3X sample buffer without beta-mercaptoethanol.
-
11.
Heat the sample at 90 °C for 5-min.
-
12.
Centrifuge the tubes at 14,000 rpm for 5-min.
-
13.
Transfer the supernatant to a new tube.
-
14.
Separate load 35–40 µl of sample on a 10–12% SDS–polyacrylamide gel in running buffer following the manufacturer's instructions of the electrophoresis apparatus.
-
15.
Transfer proteins to PVDF membrane using iblot following the manufacturer's instructions of the transfer apparatus.
-
16.
Block non-specific binding on the membrane by incubation in blocking buffer at room temperature for 1hr under gentle agitation.
-
17.
Incubate the membrane with rabbit polyclonal antibody against HSF1 (Proteintech, Catalog number 51034-AP) at 4°C overnight under gentle agitation.
-
18.
Wash the membrane in washing buffer for 8 min under gentle agitation. Repeat wash three times.
-
19.
Incubate blots with anti-Rabbit Horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h under gentle agitation.
-
20.
Wash the membrane in washing buffer for 8-min under gentle agitation. Repeat wash three times.
-
21.
Detect protein-antibody reactions with chemiluminescent detection reagent following the manufacturer's instructions. Acquire images with an automated image acquisition system.
-
22.
To check the interaction partner of HSF1 such as HSP70, remove primary and secondary antibodies from the membrane and re-probe it with the primary antibody against HSP70.
-
23.
First, wash the membrane with washing buffer for 8-min under gentle agitation. Repeat this washing twice.
-
24.
Incubate membrane in Restore stripping buffer (Thermo Scientific) at room temperature for 15- minutes under gentle agitation.
-
25.
Wash the membrane with washing buffer twice for 8-min under gentle agitation.
-
26.
Block the membrane using blocking buffer for 1hr at room temperature.
-
27.
Incubate the membrane with antibody against target protein (HSP70).
-
28.
Repeat step 18–21 for the detection of the protein.
A workflow of representation of 5’ modified DNA sequence binding to magnetic bead for identification of HSF1 protein (Fig. 1)
Fig. 1.
HSF1 Immunoprecipitation.
Method validation
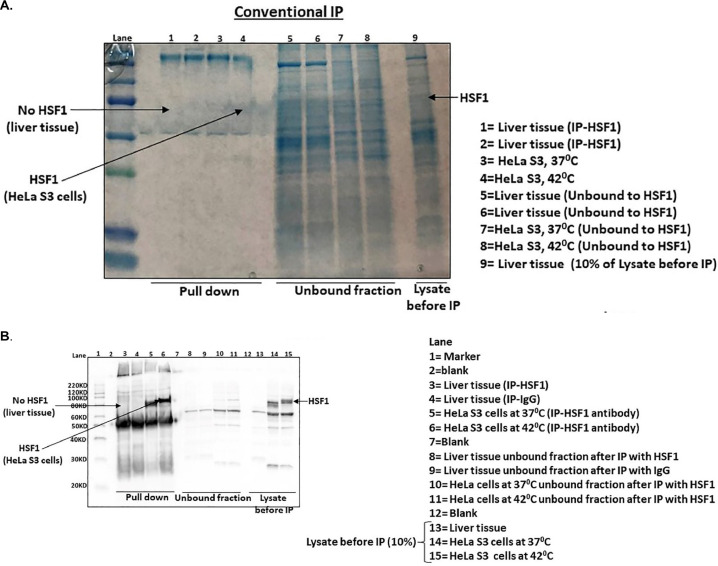
In the present study, we first performed immunoprecipitation using HSF1 antibody (Proteintech) to pull down activated HSF1 from HeLa S3 cells (control and heat shocked) and liver tissue lysates. In this conventional immunoprecipitation method, we first pre-cleared the cell/ tissue lysate with washed protein G plus/ Protein A agarose beads by incubating with gentle rocking at 4°C for 1 h. To the pre-cleared cell/tissue lysate, HSF1 antibody was added and incubated with gentle rocking overnight at 4°C. Samples were micro centrifuged for 30 s at 4°C and pellet was washed five times with 500 µl of wash buffer (PBS with 1% NP40). Pellet was resuspended in 20µl 3X SDS sample buffer, vortexed, then micro centrifuged for 30 s. Samples were heated to 95°C for 5-min and micro centrifuged for 1 min at 14,000 rpm. Samples were loaded on SDS PAGE and analyzed by western blotting. The conventional immunoprecipitation method of HSF1 pull down with commercially available antibodies worked well in HeLa S3 cells but it did not worked with liver tissue lysate (Fig 2B). To overcome this problem, we developed another technique (Described in method details) to retrieve the activated HSF1 from liver tissue lysate with an oligonucleotide specific to HSF1 binding at the promoter region of heat shock genes. Using this novel method, we could successfully pull down the activated HSF1 from liver tissue lysate (Fig 3B). We also loaded the HSF1 pulldown from heat- shocked cells and liver tissue samples on same gel along with two negative controls (Fig 3C). The first negative control represents beads incubated with buffer only while the second negative control represents liver protein samples incubated with beads only which resulted in no HSF1 pulldown.
Fig. 2.
Conventional IP method (A). Coomassie blue-stained SDS-PAGE analyses of HSF1 pulled down by IP using anti-HSF1 antibody from HeLa S3 and liver tissue lysates. (B). Western blot analysis of conventional HSF1 immunoprecipitation using protein HSF1 antibody demonstrates effective pull down of HSF1 from HeLa S3 but not Liver tissue.
Fig. 3.
New Assay (A). Coomassie blue-stained SDS-PAGE analyses of HSF1 pulled down by IP using magnetic bead conjugated HSE sequence from liver tissue lysates. (B). Western blotting of HSF1 pulled down using magnetic beads conjugated with a short DNA sequence (HSE) with specific binding to HSF1, clearly demonstrates successful pulldown of activated HSF1 from liver tissue lysates. (C) Western blot of HSF1 pull down using magnetic beads conjugated to short DNA from heat-shocked cells (HeLas3 and SHSY5Y) and from liver tissues with two negative controls.
HSF1 from tissue lysate using magnetic beads conjugate with a short DNA sequence with specific binding to HSF1.
The development of this easy and economic assay that can pull down activated HSF1 from liver tissue lysates assay is capable of measuring the other interacting partners of HSF1 such as HSP70, HSP40 and HSP90.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.DeCaprio J., Kohl T.O. Immunoprecipitation. Cold Spring Harb. Protoc. 2017;2017(12) doi: 10.1101/pdb.prot098640. p. pdb prot098640. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino J.S., Gershlick D.C., Dell’Angelica E.C. Immunoprecipitation. Curr. Protoc. Cell Biol. 2016;71 doi: 10.1002/cpcb.3. [DOI] [PubMed] [Google Scholar]
- 3.Kim M.Y. A DNA immunoprecipitation assay used in quantitative detection of in vitro DNA-protein complex binding. Anal. Biochem. 2013;441(2):147–151. doi: 10.1016/j.ab.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Trivedi R., Jurivich D.A. A molecular perspective on age-dependent changes to the heat shock axis. Exp. Gerontol. 2020;137 doi: 10.1016/j.exger.2020.110969. [DOI] [PubMed] [Google Scholar]
- 5.Krakowiak J. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. Elife. 2018;7 doi: 10.7554/eLife.31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kmiecik S.W., Breton L.Le, Mayer M.P. Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. EMBO J. 2020;39(14) doi: 10.15252/embj.2019104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heydari A.R. Expression of heat shock protein 70 is altered by age and diet at the level of transcription. Mol. Cell Biol. 1993;13(5):2909–2918. doi: 10.1128/mcb.13.5.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife. 2016;5 doi: 10.7554/eLife.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattan S.I., Eskildsen-Helmond Y.E., Beedholm R. Molecular mechanisms of anti-aging hormetic effects of mild heat stress on human cells. Nonlinearity Biol. Toxicol. Med. 2004;2(2):105–116. doi: 10.1080/15401420490464376. [DOI] [PMC free article] [PubMed] [Google Scholar]