Abstract
HER2-positive (HER2+) breast cancer (BC) is a heterogenous and multifaceted disease, with interesting therapeutic implications. First, all intrinsic molecular subtypes can be identified in HER2+ tumors, with the HER2-enriched being the most frequent. Such subtypes do not differ much from their counterparts in HER2-negative disease, apart for the high expression of genes in/near the HER2 amplicon on chromosome 17. Intrinsic subtyping, along with the quantification of ERBB2 mRNA levels, is associated with higher rates of pathologic complete response across neoadjuvant trials of dual HER2 blockade and might help select patients for de-escalation and escalation treatment strategies. Secondly, HER2+ tumors have a broad range of DNA alterations. ERBB2 mutations and alterations in the PI3K/Akt/mTOR pathway are among the most frequent and might predict benefit from potent pan-HER, PI3K and mTOR inhibitors. Moreover, HER2+ tumors are usually infiltrated by lymphocytes. These tumor infiltrating-lymphocytes (TILs) predict response to neoadjuvant anti-HER2-based treatment and exert a prognostic role. PD-L1, detected in ∼42 % of HER2+ BC, might also be useful to define patients responding to novel anti-PD1/PD-L1 immunotherapies. New multiparametric clinicopathologic and genomic tools accounting for this complexity, such as HER2DX, are under development to define more tailored treatment approaches. Finally, HER2-targeted antibody-drug conjugates (ADC) such as trastuzumab deruxtecan might be active in tumors with low expression of HER2. Overall, there is a need to molecularly characterize and develop novel targeted therapies for HER2+ disease.
Keywords: HER2, Breast cancer, Intrinsic subtypes, HER2-enriched, PAM50, TILs
Highlights
-
•
Almost 50 % of HER2+ breast cancer (BC) are molecularly HER2-Enriched (HER2-E).
-
•
Most relevant mutations are found in ERBB2 (∼4 %) and PI3K/AKT/mTOR pathway (>30 %).
-
•
Tumor infiltrating lymphocytes are frequent, predictive and prognostic in HER2+ BC.
-
•
HER2 heterogeneity and HER2 low status are gaining therapeutic relevance.
-
•
New treatments need to consider HER2+ molecular and microenvironmental complexity.
1. Introduction
HER2-positive (HER2+) breast cancer (BC) represents 10–20 % of all breast tumors [1] and are characterized by the amplification of the ERBB2/HER2 gene and/or overexpression of its related kinase receptor protein [2]. This confers a more aggressive behavior, leading to a reduced disease-free survival (DFS) and overall survival (OS) [2,3]. However, the introduction of the humanized anti-HER2 monoclonal antibody trastuzumab in clinical practice and the subsequent development of several other anti-HER2 targeted agents (i.e. lapatinib, pertuzumab and T-DM1), have lowered the risk for relapse in the early stage disease and/or significantly improved survival in the metastatic setting [4]. Despite therapeutic improvements, around 4–23 % of patients with localized disease still experience relapse after (neo)adjuvant anti-HER2-based regimens and become metastatic [5,6]. When systemic, the disease remains incurable, though median survival rates have improved consistently over the years, now reaching a median of ∼57 months [7].
HER2+ BC is clinically defined, according to the ASCO/CAP guidelines [8], when a complete and intense circumferential membrane staining for the HER2 protein in >10 % of tumor cells (3+ score) is found at immunohistochemistry (IHC) and/or the HER2 gene (ERBB2) is amplified at in situ immunofluorescence (ISH) techniques, with an HER2/CEP17 ratio ≥2.0 and an average HER2 gene copy number≥4.0 signals/cell.This definition is based on the one adopted for HER2+ clinical trials evaluating trastuzumab, pertuzumab and T-DM1. However, under the curtain of clinical (c)HER2+ disease lies a more complex and heterogenous disease that needs to be appropriately dissected, to develop and validate novel and more effective diagnostic and therapeutic approaches.
1.1. Clinically HER2+ disease based on hormone receptor status
Breast tumors are usually classified in 4 IHC subtypes (Luminal A-like, Luminal B-like, HER2-positive and triple-negative) that mostly overlap with four different corresponding molecular subtypes (Luminal A, Luminal B, HER2-Enriched [HER2-E] and Basal-like, respectively) [9,10]. The IHC-surrogate classification of cHER2+ disease endorsed by the St. Gallen Expert Consensus divides cHER2+ tumors into two. More precisely, a Luminal B-like subtype that features estrogen (ER) and/or progesterone receptor (PgR) expression (roughly 40–50 % of cHER2+ tumors) [11], and an HER2-E-like, which does not express both hormone receptors (HR) [9].
The IHC-based categorization of HR-positive (+) vs. HR-negative cHER2+ disease is undoubtedly useful in the clinical setting to indicate the need for endocrine therapy (ET). Nonetheless, HR status within cHER2+ BC is relevant from other points of view. For example, several meta-analyses regrouping neoadjuvant trials in cHER2+ disease have demonstrated a strong association between HR-negative status and pathologic complete response (pCR) [[12], [13], [14]] and better outcomes in HR-negative tumors compared with HR + disease [14]. Moreover, albeit pCR was found to be associated with more favorable survival outcome compared to non-pCR, the strength of the association was more evident in the HR-negative disease than in the HR+ [12].
According to several studies, cHER2+ tumors also differ in main clinicopathological features and natural history of disease depending on HR expression. More specifically, cHER2+/HR-negative tumors are more likely to present with high histologic grade and higher tumor stage, less likely to first relapse in bone and more likely to recur in brain, than cHER2+/HR + disease. Moreover, patients with cHER2+/HR-negative tumors vs. cHER2+/HR + seem to be at increased risk of early death (i.e. first 5 years of follow-up) [15]. An exploratory analysis from the ALTTO adjuvant phase III trial of CT combined with trastuzumab, lapatinib or both, compared survival outcomes periods 0–5 years versus >5 years based on HR status. Patients with HR + tumors had better DFS, distant disease-free survival (DDFS) and OS for years 0–5 compared to HR-negative disease. Nevertheless, survival outcomes at 8 years were similar between the two groups [16].
Regarding the benefit to trastuzumab, the joint analysis of two large adjuvant randomized controlled trials (RCT), the NCCTG N9831 and the NSABP B-31, comparing different chemotherapy (CT) regimens ± trastuzumab in cHER2+ BC, revealed that the 10-year DFS and OS rates were similar between HR-negative vs. HR + tumors [17]. Concordant results were observed also in the adjuvant trastuzumab pivotal trial HERA [18]. Yet, the levels of ER protein might help to identify a subgroup of patients with cHER2+/HR + disease who might not benefit much from trastuzumab. In fact, a retrospective population-based study on 872 HR + cHER2+ BC patients treated with adjuvant CT ± trastuzumab, revealed a clear benefit from its addition, in the overall population; however, the effect on BC specific survival (BCSS) in tumors expressing ER in >30 % of cells was not statistically significant (p = 0.26). Furthermore, adjuvant trastuzumab did not improve both Relapse-Free Survival (RFS) and BCSS in tumors with ER overexpressed in >50 % of cells, as opposite to the significant effect observed in case of ER staining in ≤50 % of tumor cells [19]. A possible explanation might reside in a higher dependency on the ER rather than HER2 pathway for tumor growth and survival, which might limit the therapeutic efficacy of agents specifically blocking the HER2 downstream signaling pathway as their main mechanism of action.
Although these findings will require further validation, they do suggest that a subgroup of HR + tumors with high luminal features and hormone-dependency might not benefit much from (neo)adjuvant trastuzumab.
1.2. The intrinsic subtypes within cHER2+ disease
The four BC molecular subtypes, also called intrinsic subtypes, have shown different outcome patterns and response to therapy [[20], [21], [22]]. Clinically HER2+ BC has been considered a single tumor entity for a long time; however, in 2012, The Cancer Genome Atlas (TCGA) project demonstrated that not all cHER2+ BC had the same genomic profile. More precisely, only 50 % of cHER2+ tumors were HER2-E, while the other half were Luminal A or B, with high expression of typical luminal genes such as ESR1, GATA3 and BCL2, among others [23]. More recent studies have revealed the presence of cHER2+ Basal-like tumors, especially in HR-negative disease [[24], [25], [26], [27], [28], [29], [30]]. Among the different subtypes within cHER2+ disease, the HER2-E is characterized by the highest levels of ERBB2 mRNA, phosphorylated (p)HER2, total HER2 protein, pEGFR and EGFR protein, suggesting that this group has the highest activation of the HER2 signaling pathway [[23], [24], [25]].
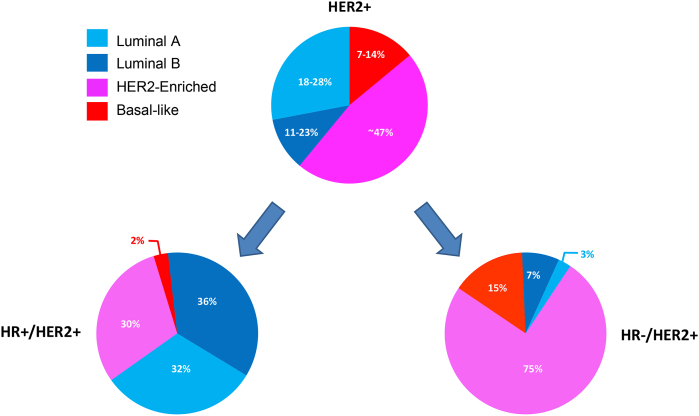
In summary, cHER2+ tumors comprises all of the 4 BCE intrinsic subtypes, with the HER2-E being the most frequent (∼47 %), followed by Luminal B (∼18–28 %), Luminal A (11–23 %) and Basal-like (7–14 %) [10,24]. Yet, the distribution seems to be heavily influenced by HR status, with HER2-E subtype representing 30 % of molecular subtypes within HR+/cHER2+ BC and 75 % in HR-negative/cHER2+ tumors [10,[26], [27], [28], [29], [30]] (Fig. 1). Although related, the concordance rate between the pathology-based subtypes and the intrinsic subtypes is moderate (67.4 %; kappa statistic: 0.50) [28].
Fig. 1.
Intrinsic subtype distribution in cHER2+ tumors.Legend. HR: hormone receptors; +: positive; -: negative. Number of samples: 3390 HR+/HER2+ tumors and 2567 HR-/HER2+ tumors.
An important question to address was how different an intrinsic subtype is when it is cHER2+ versus cHER2-negative. The answer is that a given subtype within cHER2+ disease is largely undistinguishable, biologically speaking, from its cHER2-negative counterpart, except for the high expression of genes in, or near, the HER2 amplicon on chromosome 17 in cHER2+ disease [24,31]. Furthermore, HER2 status is an independent prognostic factor beyond clinical-pathological variables, but such prognostic relevance disappears when intrinsic subtypes are considered [24].
1.3. Intrinsic subtype, ERBB2 mRNA and treatment response
Results from 6 randomized phase III, 5 randomized phase II, 3 single arm phase II, 2 non-randomized neoadjuvant clinical trial and 1 retrospective observational study evaluating several anti-HER2-based treatment regimens, +/− CT, concordantly suggested the association between the HER2-E subtype and pCR [[25], [26], [27], [28], [29], [30],[32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]]. A subsequent trial-level joint analysis of such studies confirmed a strong association between the HER2-E molecular subtype with pCR (pooled odds ratio [OR]: 3.50, p < 0.001), also beyond HR status (p < 0.001) and CT use (p < 0.001) [13]. These results suggest that intrinsic subtypes might identify a subgroup of cHER2+ tumors that benefit the most from anti-HER2 therapies, irrespective of CT and HR status.
Furthermore, 3 of those trials reported ERBB2 mRNA-high tumors achieving more frequently a pCR, compared to mRNA-low tumors (41.9–79.3 % vs. 25.5–51.72 % pCR rates) [26,39,43], albeit with different methodologies and cut-offs for defining high vs. low levels. Another secondary analysis from the GeparQuattro neoadjuvant trial evaluating anthracyline/taxane-based CT + trastuzumab, showed that the response to the combination correlated to ERBB2 mRNA levels [44]. Finally, high ERBB2 mRNA levels were found associated with better tumor responses and progression-free survival (PFS) in the metastatic setting and higher rates of pCR in the neoadjuvant setting, when treated with the antibody-drug conjugate (ADC) ado-trastuzumab-emtansine (T-DM1) [45].
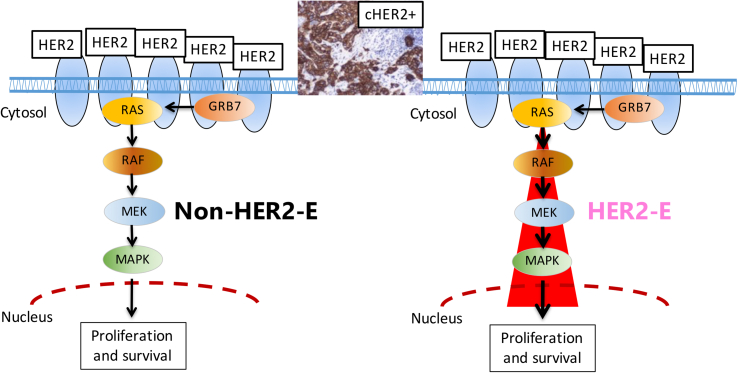
It is interesting to note that the combination of the HER2-E subtype with ERBB2 mRNA expression levels might better predict response to anti-HER2-based treatments than each variable alone. In fact, in a combined retrospective analysis of the TBCRC006 and PAMELA CT-free neoadjuvant trials of double anti-HER2 blockade (lapatinib + trastuzumab ± ET) in cHER2+ BC, our group evaluated intrinsic subtypes and ERBB2 mRNA levels of expression, dichotomizing the latter as low or high. As expected, pCR was more likely to occur in HER2-E tumors compared to non-HER2-E (35.1 % vs. 9.9 %; p < 0.001), but also in tumors with ERBB2 mRNA high compared to low levels (36.1 % vs. 8.2 %; p < 0.001). Notably, HER2-E/ERBB2 mRNA high BC showed pCR rates higher than the rest (adjusted OR: 6.0; p < 0.001), suggesting that combining HER2-E subtype and ERBB2 mRNA levels better identifies anti-HER2 sensitivity than each variable alone [46]. The most likely explanation is that the HER2-E subtype is more indicative of the activation of the HER2 signaling pathway than the expression of the HER2 protein or mRNA, which are more representative of the presence of the therapeutic target (Fig. 2). This might open up the possibility of de-escalating neoadjuvant systemic therapy to avoid CT, at least for a small proportion of patients carrying cHER2+ BC and with high “molecular addiction” to HER2 (i.e. HER2-E/ERBB2 mRNA high) and low-risk clinicopathological prognostic parameters (e.g. node negative, small primary tumor). Ultimately, more data are needed to draw definitive conclusions.
Fig. 2.
HER2-dependent pathway for proliferation and survival in HER2+ breast cancer.Legend. cHER2+: clinically HER2-positive; HER2-E: HER2-Enriched. The red cone visually suggests a higher activation of the HER2-related pathway in cHER2+/HER2-E tumors, compared to cHER2+/non-HER2-E.
Differently, no evidence of an added predictive value for the ERBB2 mRNA levels beyond molecular subtypes has been evaluated in the context of CT + anti-HER2 therapy, so far.
Interestingly, an ongoing single arm phase II trial in cHER2+ BC cases with high molecular HER2 addiction as previously defined and clinical low risk (clinically node negative, unifocal primary lesion of ≤2 cm), is evaluating the omission of surgery and sentinel lymph-node dissection, in case of achievement of a radiologic complete response at magnetic resonance, following neoadjuvant therapy with trastuzumab, pertuzumab and paclitaxel (ELPIS trial, NCT04301375). Results might be important to prove for the first time that in cHER2+ early stage BC, a combination of molecular and clinical tumor characteristics might be useful to de-escalate the therapeutic strategy, this time focusing on surgery, rather than systemic treatments. Recruitment is currently ongoing.
1.4. Intrinsic subtypes and survival outcome
Retrospective analyses from (neo)adjuvant trials evaluated the association of the intrinsic molecular subtypes with survival outcomes within the cHER2+ disease. By using archived tumor blocks of cHER2+ disease from the NSABP B-31 adjuvant trial, 47 % samples were classified as HER2-E. Similar DFS benefit was observed for trastuzumab in HER2-E and non-HER2-E tumors [47]. Differently, a retrospective analysis from the N9831 trial showed that HER2-E and Luminal tumors derived benefit from adjuvant trastuzumab in terms of relapse-free survival (RFS), while Basal-like did not [48]. In addition, another analysis from the adjuvant pertuzumab APHINITY trial showed that the Luminal A subtype was associated with strikingly better outcomes, mostly when compared to patients with a Basal-like subtype. However, no significant interaction was observed between intrinsic subtypes and treatment [49].
As opposite to previous evidence, a novel molecular analysis from the phase III Short-HER study of 1 year vs 9 weeks of adjuvant trastuzumab demonstrated that the HER2-E subtype was associated with worse metastasis-free survival (MFS) compared to the other intrinsic subtypes taken together, both in the short and standard arm [50]. Within the NeoALTTO trial there was no association between the HER2-E subtype and event-free survival (EFS), although the authors accounted for an overall low statistical power to perform this analysis [33]. The NSABP B-41 trial demonstrated that OS was increased in patients who achieved pCR, with no significant differences in pCR according to treatment arm. With respect to intrinsic subtyping, HER2-E patients on trastuzumab-based arms had higher pCR than those on the lapatinib arm, though no differences in terms of EFS and OS were observed [30]. Finally, albeit demonstrating better pCR rates for the HER2-E subtype compared to the others, a post-hoc analysis from the CALGB 40601 trial showed that the Luminal A subtype was favored over the HER2-E in terms of invasive DFS (iDFS). Moreover, within HR + disease, the Luminal A subtype was associated to a longer iDFS compared to those with Luminal B or HER2-E tumors [51].
1.5. Molecular alterations: focus on ERBB2, PIK3CA and PTEN
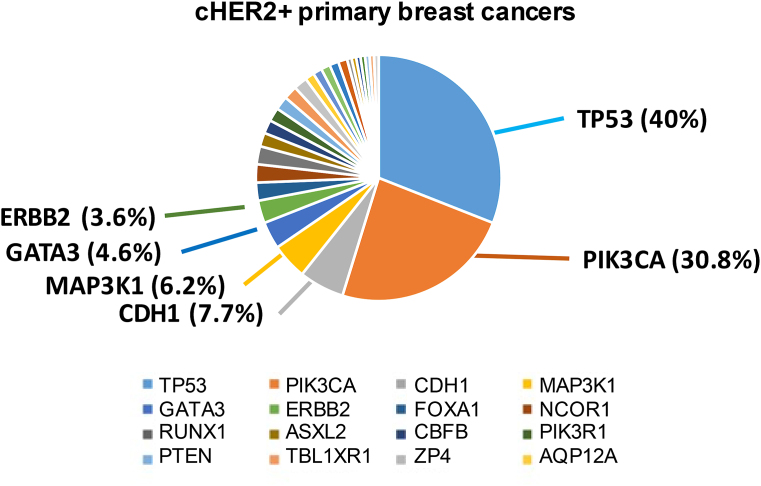
Recent studies have addressed the molecular complexity that characterize cHER2+ and HER2-E tumors. Analysis of somatic mutations from the TCGA showed that, other than ERBB2 amplification, cHER2+ primary tumors were characterized by a broad range of infrequent mutations (<10 %), including in ERBB2 (3.6 %). However, the most frequent mutations were observed in PIK3CA (∼31 % cases) and in TP53 (40 % cases) (Fig. 3).
Fig. 3.
Mutational spectrum of cHER2+ primary tumors.Legend. cHER2+: clinically HER2-positive.
Similarly, the HER2-E subtype, showed frequent TP53 (72 %) and PIK3CA mutations (39 %), as well as other mutated genes (i.e. PTEN, PIK3R1), albeit at significantly lower frequencies (usually <10 %) and ERBB2 amplification in 80 % cases [23,52].
ERBB2 mutations are infrequent with respect to amplifications; however, they can exert an oncogenic effect by constitutively activating HER2 tyrosine kinase activity or by increasing HER2 dimerization with other member of the EGFR family. These mutations have been found to cluster in the tyrosine kinase and extra-cellular domains of the HER2 protein and might be counterbalanced by irreversible TKI inhibitors like neratinib [53]. The most frequent activating mutations identified are the G309A, D769H, D769Y, V777L, P780ins, V842I, del755–759 and R896C, while the L755S, despite being not activating, seem to induce lapatinib resistance [53]. These mutations were mostly characterized in HER2-non-amplified tumors. Interestingly, in a recent study, the mutational landscape of ERBB2 was also assessed in HER2 amplification-positive patients. The study showed a higher ERBB2 mutation frequency in HER2-amplified tumors than in HER2-negative (19.5 % vs. 4.8 %; p < 0.001) [54]. One-hundred-ninety-four ERBB2 variants were predicted as driver mutations and 192 variants were predicted as passenger mutations. The top three frequency mutations were represented by the already known V777L, L755S and D769Y and were all predicted as driver mutations [54]. These rare mutations, at least in part, seem to be induced by HER2-targeted therapy and ET, and prevails in metastatic tumors, compared to primary ones [[54], [55], [56]]. Furthermore, in case of concomitant HER2 mutations and amplification, resistance to trastuzumab has been detected [54]. Conversely, in this cHER2+ subset, the tyrosin kinase inhibitor (TKI) neratinib showed promising activity [57], such as the TKI pyrotinib in BC with HER2 mutations without amplification [58]. However, randomized studies are needed to validate these preliminary findings.
PIK3CA, which codifies for the phosphatidil-inositol-3-kinase (PI3K), is on the top of a cascade that positively modulates cell survival, proliferation and metabolism. The pathway is negatively regulated by PTEN, which codifies for a protein/lipid phosphatase [59]. Usually, oncogenic PTEN and PIK3CA mutations do not occur together [23,60]. In cHER2+ tumors PIK3CA mutations have been shown to be associated with trastuzumab resistance in preclinical models [[61], [62], [63]]. Therefore, PIK3CA and PTEN potential role as prognostic and predictive biomarkers of response to treatments was further investigated in cHER2+ BC.
Retrospective studies on tumor samples deriving from adjuvant trastuzumab trials found PIK3CA mutations in ∼25 % cHER2+ BC, but failed to find an association with trastuzumab benefit [47,64]. At the same time a prognostically favorable effect was observed, albeit this effect disappeared after 3 years [64]. On the other hand, PIK3CA mutational status might be relevant in the neoadjuvant setting. Indeed, a patient-level pooled-analysis involving 5 neoadjuvant RCT investigating trastuzumab-/lapatinib-based treatments proved that PIK3CA-mutant/cHER2+ tumors had significantly lower pCR rates compared to wild-type tumors (16.2 % vs. 29.6 %; p < 0.001). However, such results seemed to be mostly driven by the HR+/cHER2+ cohort, for which the pCR rates differences were significantly more pronounced (7.6 % in PIK3CA mutant vs. 24.2 % in the wild-type group; p < 0.001) than in the HR-negative/cHER2+ cohort (p = 0.125) [65]. Mutational status did not affect the outcome in terms of DFS and OS in the overall population examined, though. However, HR+/PIK3CA-mutant patients seemed to have significantly worse DFS (p = 0.050) [65].
A possible explanation for PIK3CA influence on response to anti-HER2 agents might lie in the fact that the rates of PIK3CA mutation seem to be inversely proportional to ERBB2 mRNA levels (raw data obtained from a TCGA-derived dataset of 94 cHER2+ patients).
Interestingly, in advanced setting, PIK3CA mutational status proved to be a relevant prognostic marker for patients treated with pertuzumab + trastuzumab + docetaxel, in a biomarker analysis of the first-line CLEOPATRA trial. More specifically, longer median PFS was observed for patients whose tumors expressed wild-type versus mutated PIK3CA in the pertuzumab groups (21.8 v 12.5 months), as well as in the control group (13.8 v 8.6 months). However, pertuzumab showed a PFS benefit independently from PIK3CA mutational status, making it not suitable for selecting patients that might benefit from this drug [66]. Similar results were observed in a post-hoc biomarker analysis of the EMILIA and TH3RESA phase III RCT, where T-DM1 proved its effectiveness in both cHER2+ PIK3CA-mutant and wild-type patients over the therapeutic standard of second and subsequent lines [47,67]. Interestingly, PIK3CA-mutant patients appeared to better respond to lapatinib + capecitabine in the control arm of the EMILIA trial, both in terms of median PFS (mutant vs. wild type: 4.3 vs. 6.4 months) and OS (17.3 vs. 27.8 months) [67].
Concerning PTEN, retrospective analyses from the NeoALTTO and N9831 trials respectively showed no correlation between low levels of PTEN and pCR [75] and that the benefit from adjuvant trastuzumab was not affected by PTEN levels [76]. On the contrary, a retrospective analysis from the neoadjuvant TBCRC006 trial, where double blockade therapy with lapatinib + trastuzumab was administered without CT, showed that high levels of PTEN were associated to higher rates of pCR, compared to low PTEN levels (32 % vs. 9 %; p = 0.04). Moreover, in case of a combination of PTEN loss/low expression and PIK3CA mutation, pCR was achieved significantly less than in case of wild-type PIK3CA and high PTEN expression levels (4 % vs. 39 %; p = 0.006) [77].
In advanced setting, low PTEN levels were associated with better PFS outcome but, surprisingly, with worse OS in the CLEOPATRA trial. However, as observed with PIK3CA mutational status, a consistent PFS benefit from pertuzumab was shown independently from the expression levels of any biomarker [66]. Within the EMILIA, T-DM1 was associated with longer median PFS and OS compared to lapatinib + capecitabine, regardless of PTEN expression. Anyway, in case of low/absent PTEN, the PFS benefit obtained with T-DM1 appeared to be more pronounced than in case of normal/high PTEN expression. This relationship was not observed for OS [67].
Two recent phase III trials, BOLERO-1 and BOLERO-3 evaluated the addition of the mTOR inhibitor everolimus to trastuzumab and CT (paclitaxel or vinorelbine, respectively) in cHER2+ advanced BC. The serine/threonin kinase mTOR is a downstream effector of the PI3K pathway. A joint analysis on 377 samples from both trials was performed to evaluate genomic alterations. The overall genetic landscape was similar between the 2 trials. Nevertheless, PTEN was altered in 1.5 % of the BOLERO-1 samples, compared to the 8 % in BOLERO-3. Differently from pertuzumab and T-DM1, a statistically significant PFS benefit from the addition of everolimus was only observed in patients with PIK3CA mutations, PTEN loss or hyperactive PI3K pathway (namely, low PTEN expression/mutation and/or known PIK3CA and/or other activating mutations in the downstream pathway) [60]. These findings suggest that PIK3CA and/or PTEN mutational status might be considered a predictive biomarker for everolimus efficacy in this BC subtype. However, the clinical development of this drug in cHER2+ BC in first-line setting has been somewhat stopped by the introduction of pertuzumab-based regimens in first-line and T-DM1 in second-line, given the outstanding performances observed in their pivotal trials [[68], [69], [70]]. Its role might be revalued in more advanced treatment lines, in patients with hyperactivation of the PIK3CA pathway, albeit trials designed to prospectively assess the efficacy and safety of this drug in patients with alterations in the PI3K pathway are needed to confirm this hypothesis.
1.6. TILs and immune system
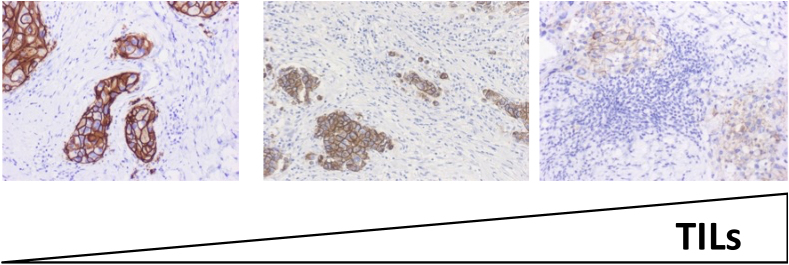
A constantly growing body of evidence strongly suggests that cancer can induce an immune response by producing new antigens and modulate such immune response through the induction of microenvironmental changes and interplay with immune cells and immune response (cancer immunoediting) [71,72]. Being this process heavily related to T cells activity, tumor-infiltrating lymphocytes (TILs) have been claimed to be a morphological representation of the complex interplay between cancer and immune system [73]. Interestingly, there are several evidences reporting a previously unacknowledged interaction between anticancer treatments and immune system. For example, CT and radiotherapy seem to elicit an immune system activation against cancer cells [74], while trastuzumab anticancer activity seems to heavily rely also on antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity, rather than merely blocking the HER2 pathway. Such activity might be in turn modulated by the innate or adaptive cancer immune status [[75], [76], [77]]. Therefore, the biological complexity of cHER2+ BC not only lies in its molecular heterogeneous profile, but also in its microenvironmental features. Indeed, cHER2+ tumors are characterized by differential levels of TILs, which seems to play a role in modulating the responsiveness to CT and anti-HER2 treatments. In fact, it has been reported that in early stage, around 50 % of cHER2+ BC present with ≥10 % TILs, around 20 % shows ≥20 % and about 10 % ≥ 40 % TILs (Fig. 4) [[78], [79], [80]].
Fig. 4.
Representative sections of TILs in early-stage HER2+ breast tumors.Legend. TILs: tumor-infiltrating lymphocytes. TILs levels increase from left to right.
A recent study showed that cHER2+ HER2-E and Basal-like subtypes were also characterized by higher levels of PD-L1, compared to the Luminal A and B [80]. PD-L1 is a transmembrane protein that is implied in suppressing the adaptive immune response by interacting with the PD1 on the surface of T cells, reducing the proliferation of antigen-specific T-cells in lymph nodes, while simultaneously reducing apoptosis in immunosuppressive T-regulatory (Treg) cells [81]. Recent studies have observed the presence of PD-L1 in ∼42 % of cHER2+ BC, with different detecting assays [82,83]. PD-L1 levels correlate with response to anti-PD-L1 immune-checkpoint inhibitors (ICI), as observed in the phase III trial IMPASSION130 in advanced triple negative breast cancer (TNBC), as well as in other trials in different solid tumors [81,84]. In cHER2+ disease, the phase I/II trial PANACEA showed that only PD-L1+ tumors responded to the combination of trastuzumab with the anti-PD1 pembrolizumab, in a group of trastuzumab-pretreated metastatic cHER2+ BC patients [82]. Similarly, the randomized phase II KATE-2 showed a trend for a PFS benefit in pretreated HER2+ PDL1+ tumors with T-DM1 and the anti-PD-L1 atezolizumab, with no benefit for the overall population enrolled, or in the PD-L1-negative subset [83].
Up to now, the role of TILs as prognostic/predictive factor has been more extensively studied than PD-L1 in several clinical trials involving cHER2+ BC. In the adjuvant setting, contrasting results have been reported so far, based on retrospective analyses from adjuvant trastuzumab trials [85,86]. Conversely, more interesting results have come from neoadjuvant anti-HER2-based trials. A retrospective analysis of the NeoALTTO from Salgado et al. showed that levels of TILs greater than 5 % were associated with higher pCR rates independent of treatment group (p = 0.01). Moreover, every 1 % increase in TILs was associated with a 3 % decrease in the rate of an event (p = 0.002) across all treatment groups [78]. In the CHER-LOB trial, both stromal (Str)TILs and intratumoral (It-)TILs were significantly associated with pCR, also independently from ER status [29]. However, continuous TILs levels significantly predicted pCR only for patients in lapatinib-containing arms, while no statistically significant effect was observed in the CT + trastuzumab group. In a dichotomous comparison between low-TILs BC and high-TILs BC, with a cut-off for TILs levels of 60 %, pCR rates were 64.7 % for high-TILs BC and 25 % for low-TILs BC patients, respectively (p < 0.001). Of note, molecular intrinsic subtypes outperformed TILs evaluation in predicting pCR at the multivariate analysis [29].
Opposite results were observed in the NeoSphere trial, where both TILs and PD-L1 did not associate with pCR [35]. Conversely, TILs as continuous variable and high vs low TILs were significantly linked to pCR in the HER2+ cohort of the neoadjuvant GeparSixto trial, where an anthracycline/taxane-based CT regimen with trastuzumab + lapatinib ± carboplatin was tested [87].
A retrospective analysis on 175 patients with primary cHER2+ BC receiving neoadjuvant CT ± trastuzumab failed to demonstrate a correlation between pCR rates and basal TILs levels. However, TILs levels decreased during treatment in 78 % of the patients and TILs’ variation was strongly associated with pCR (p < 0.001). Furthermore, post-neoadjuvant TILs were strongly associated with post-neoadjuvant tumor residue parameters and were higher in tumors with aggressive characteristics (p < 0.001), high nodal involvement (p = 0.054) and large nodal metastases (p = 0.001). In the population of patients that failed to achieve pCR, levels of TILs in residual tumor higher than 25 % were an independent poor prognostic factor at the multivariate analysis [88].
In a secondary analysis of the PAMELA trial, the rates of pCR in high-level TILs BC vs. non-high-TILs BC were 58.3 % and 27.2 %, respectively (p = 0.024). However, basal TILs lost their association with pCR at the multivariable analysis. Interestingly, the higher the TILs at day 15 (D15), the higher the odds of achieving a pCR and the correlation between D15 TILs and pCR was retained at the multivariate analysis. This study also proposed a mathematic model based on tumor cellularity and TILs at D15 (the CelTIL) to predict benefit from anti-HER2 therapy. The CelTIL was significantly associated with pCR in univariate and multivariate analysis either as a continuous variable or using a pre-specified cut point [89].
Notably, there are accumulating data on the capability of TILs or specific TILs subpopulations (e.g. CD8+) to predict clinical benefit of ICI in solid tumors like melanoma [[90], [91], [92]]. Whether this might be the case for cHER2+ BC is yet to be assessed. Moreover, the prognostic impact of specific immune cell infiltration (e.g. monocytes, B lymphocytes, T lymphocytes) is still a matter of debate. In this context, a more a favorable outcome was observed in cHER2+ BC with lymphocyte infiltration and expression of lymphocyte-related genes [93]. Other groups discovered several immune-system-related metagenes regrouping specific clusters of genes that seem to be representative of different tumor-infiltrating immune cell subpopulations (e.g. the IgG metagene related to B lymphocytes, the LCK metagene related to T lymphocytes, the HCK metagene related to macrophages and monocyte/myeloid lineage), with different prognostic implications according to HR status [94,95]. More recently, a cluster of immune-related genes contained in a bigger six-metagene signature (138 genes), was found to be associated with higher pCR rates (p = 0.019) following neoadjuvant CT and better prognosis in cHER2+/HR-negative BC (p = 0.026). Immunity metagene expression was also associated with the presence of tumor-infiltrating lymphocytes (TILs) in the same study [96]. Overall, there is an accumulating evidence supporting the prognostic and predictive roles of immune gene signatures, especially in primary BC [97]. However, the published signatures are numerous and partially overlapping. Their prognostic and/or predictive role has been tested in different therapeutic contexts, as well. Therefore, a better harmonization and characterization of these signatures is required to properly understand their clinical utility and applicability [97]. Furthermore, whether the prognostic and predictive information captured by gene-expression-based signatures can be predicted by the assessment of cheaper and easy-to-detect immunologic peripheral blood markers is also yet to be determined. At present, it is also unknown the capability of immune metagenes to predict any benefit from novel immunotherapies.
Importantly, multiple evidences show that the HER2-E intrinsic BC subtype is associated with higher levels of TILs infiltration, compared to the others [29,75,89]. A possible explanation might reside in the fact that HER2-E subtype is frequently characterized by the lack of co-amplification of the 17q12 chromosomal region, which contains genes encoding chemokines, and is located proximal to the HER2 gene amplicon [71]. Such lack of co-amplification, which as opposite is relatively frequent in cHER2+ Luminal tumors, results in higher levels of T-cells infiltration [98]. Additionally, several data confirm a link between APOBEC-mediated mutagenesis and the acquisition of subclonal mutations, leading to genomic instability, potential neoantigens expression and subsequently more immune tumor infiltration, as well as better response to ICI [[99], [100], [101]]. This might be particularly relevant for cHER2+ BC, considering that APOBEC genetic signature is frequently associated to the HER2-E subtype [22,102].
1.7. Beyond HER2 overexpression: HER2-low breast tumors and HER2 heterogeneity
Based on IHC assays, current ASCO/CAP guidelines recommend to classify tumors characterized by an HER2 IHC score of 1+ or 2+ (with the latter accompanied by an ISH negative result) as HER2-negative BC, for which there is no clinical recommendation for anti-HER2 targeted agents. However, 2 of the pivotal trials that demonstrated the efficacy of trastuzumab-based adjuvant therapies were found to contain a cohort of patients without amplification/overexpression of HER2 on tissue submitted for central testing. These so-called HER2-low cohorts appeared to benefit from the addition of trastuzumab, retrospectively [103], but the phase III RCT NSABP B-47 failed to demonstrate prospectively this benefit [103]. Similarly, the 3-year results of the KRISTINE trial demonstrated that replacing systemic CT with T-DM1 in a pertuzumab-containing neoadjuvant regimen leads to a greater risk of relapse, mostly locoregional. Interestingly, tumors from these early-relapsing patients had lower HER2 expression (2+ vs. 3+) and HER2 2+/3+ heterogeneity (focal vs. heterogeneous), compared to those from other patients in the same treatment arm [104]. However, recent phase I trials of potent ADC trastuzumab deruxtecan and trastuzumab duocarmazine showed impressive response rates and median PFS in heavily pretreated metastatic HER2-low BC [105,106], leading to their further clinical development, along with other promising anti-HER2 ADC (e.g. MEDI4276 and XMT-1522) [107].
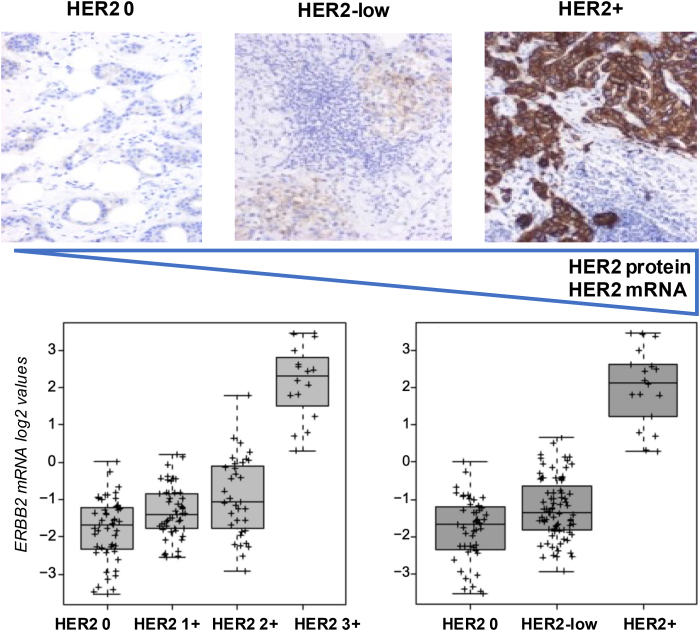
Our group conducted a large retrospective study on more than 3500 patients and comprehensively dissected HER2-low clinicopathological and molecular features [108]. Overall, we observed that HER2-low disease within HER2-negative BC is frequent (65 % of HR+/HER2-negative tumors and 37 % of TNBC) and mostly characterized by Luminal A/B tumors. Nevertheless, TNBC/HER2-low showed higher levels of Basal-like tumors, compared to HR+/HER2-low. However, while intrinsic subtypes’ distribution, Basal- and proliferation-related genes levels of expression were higher in TNBC, with no differences between HER2 0 and HER2-low tumors, Luminal genes, as well as ERBB2 and its companion gene GRB7 were significantly more expressed in HR+/HER2-low than in both HR+/HER2 0 and TNBC, irrespectively of HER2 status. These findings were also accompanied by a higher prevalence of Luminal A/B subtypes in HR+/HER2-low compared to the rest [108]. Interestingly, the higher the HER2 IHC score, the higher the ERBB2 mRNA levels [108]. This finding was also confirmed in a following study were we showed a significant positive correlation between ERBB2 mRNA levels and HER2 protein levels (Spearman Correlation = 0.531, p < 0.001) [109]. Finally, we found no survival differences between HER2-low vs 0 tumors, however the large biological heterogeneity observed, confirmed by a following study based on TCGA data, might have potential therapeutic implications [108,110]. An exemplification of the correlation between HER2 protein and mRNA levels is reported in Fig. 5.
Fig. 5.
HER2 protein levels detected by immunohistochemistry and ERBB2 mRNA levels according to HER2 IHC status.Legend. Representative pathological images of cHER2+ and cHER2-negative tumors courtesy of the Hospital Clinic of Barcelona Pathology Department and ERBB2 mRNA expression levels (log2 values) across HER2 IHC categories, as observed in 146 metastatic samples analyzed at the Translational Genomics and Targeted Therapies in Solid Tumors laboratory at IDIBAPS. HER2 protein and mRNA levels increase from left to right. HER2+: HER2-positive; IHC immunohistochemistry.
Notably, about 1–34 % of breast tumors are characterized by heterogeneity in HER2 expression levels, which is responsible for poor response to anti-HER2-based regimens and worse prognosis, compared to cHER2+ BC [108,111]. This feature is more common in tumors with an HER2 equivocal status [111]. A recent single arm phase II trial of neoadjuvant T-DM1+pertuzumab in centrally confirmed cHER2+ BC, tested tumor tissues for HER2 intratumor heterogeneity (ITH-HER2), defined as the presence in at least one tumor area of either 1) HER2 positivity by ISH in >5 % and <50 % of tumor cells or 2) an area of tumor that tested HER2 negative. ITH-HER2 was more frequent in ER + tumors compared to ER-negative (81 % vs 19 %) and no pCR was observed among cases classified as heterogeneous. A significant association between ITH-HER2 and pCR stratified by ER status (p < 0.0001) was observed. This study suggests that, if further validated, a routine ITH-HER2 assessment might be an additional biomarker for selecting patients for (de)escalated therapeutic approaches [112].
1.8. HER2DX
The increasingly complex and rich molecular and clinicopathological heterogeneity that characterizes cHER2+ BC, is negatively counterbalanced by a substantial absence of clinically validated biomarkers other than HER2 and HR status. In order to help predict treatment benefit and better personalize the therapeutic approaches, we and others developed a prognostic assay that integrates multiple data types for predicting survival outcome in patients with newly diagnosed HER2-positive breast cancer. This so-called HER2DX combined prognostic model, was built on retrospective clinicopathological and genomic data from patients who participated in the previously mentioned Short-HER phase 3 trial. The final prognostic model was then evaluated in an independent combined dataset of patients with early-stage cHER2+ BC treated with different neoadjuvant and adjuvant anti-HER2-based combinations from four other studies with available DFS data [113]. The model included tumor dimension, nodal status, StrTILs, PAM50 subtypes, and expression of 13 genes [113].
HER2DX score as a continuous variable was significantly associated with DFS and distant metastasis-free survival (DMFS) but not with pCR. The model could also stratify patients in low-, intermediate- and high-risk categories. Study results suggested that a substantial proportion of early-stage cHER2+ BC might not need additional treatments (e.g. pertuzumab, neratinib, T-DM1) other than CT + trastuzumab (and ET in HR + tumors) [113].
2. Conclusions
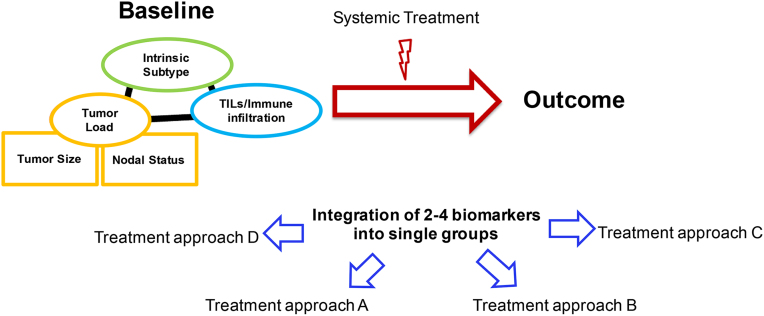
cHER2+ BC is a complex and multifaceted disease. Molecular profiling, gene expression patterns, clinicopathological tumor features and microenvironmental features altogether provide a faceted portrait of a nosological entity that no longer we should consider as one. Importantly, the integration of multiple biomarkers, also depending on the clinical setting, might lead towards different therapeutic strategies, depending on the presence/absence/value of several tumor/microenvironmental features or scores/categories identified by their combination (Fig. 6). In fact, in the era of personalized medicine [114], a multiple biomarker-driven diagnostic and therapeutic approach is progressively replacing the old “one-size-fit-all” formula, and single-biomarker-driven approaches, when possible.
Fig. 6.
Personalized medicine in cHER2+ breast cancer.Legend. TILs: tumor-infiltrating lymphocytes.
For example, the multiparametric score HER2DX might be useful in the (neo)adjuvant setting to select candidates for (de)escalated therapeutic approaches, while the identification of HER2-E/ERBB2 mRNA-high cHER2+ tumors might be useful to select candidates for CT-free anti-HER2-based regimes, in early and advanced setting [25,45,113]. Similarly, the detection of TILs and PD-L1 might be crucial to identify candidates for ICI with anti-HER2 combinations [82,83]. Alterations of the PIK3CA/Akt/mTOR pathway might play a role in selecting patients that might gain benefit with PI3K-, Akt- and mTOR-inhibitors [60,66,67]; similarly, ERBB2 mutations might be overcome with potent anti-HER family TKI such as neratinib and pyrotinib [57,58]. Finally, the novel category of HER2-low tumors seems to benefit from new potent anti-HER2 ADC, such as trastuzumab deruxtecan [105,106]. All these strategies are worthy of further investigation and need to be tested in prospective clinical trials.
The final goal will be to avoid or reduce treatment toxicity where unnecessary, without compromising patient's outcome at the same time, or correctly use escalated regimens to gain the maximum benefit at the cost of higher toxicity only in the most appropriate candidates. These tools might also help to correctly allocate financial resources, in a context where novel targeted agents are reaching unsustainable costs and their use is substantially unfeasible for numerous countries all over the world [[115], [116], [117]].
Author contributions
The authors equally contributed in the conception, drafting, revision and final approval of the manuscript.
Fundings
This article did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
No new datasets were generated or analyzed for this report.
Declaration of competing interest
F.S. declares no conflict of interest. A.P. declares an immediate family member being employed by Novartis; fees for Non-CME Services Received Directly from Commercial Interest or their Agents (e.g. speakers' bureaus); personal honoraria from Pfizer, Novartis, Roche, MSD Oncology, Lilly and Daiichi Sankyo; travel, accommodations and expenses paid by Daiichi Sankyo; research funding from PUMA, Nanostring Technologies, Roche and Novartis; consulting/speaker/advisory role for NanoString Technologies, Amgen, Roche, Novartis, Pfizer, PUMA, Seattle Genetics, AstraZeneca, Guardant Health, Foundation Medicine and Bristol-Myers Squibb; patent PCT/EP2016/080056: HER2 AS A PREDICTOR OF RESPONSE TO DUAL HER2 BLOCKADE IN THE ABSENCE OF CYTOTOXIC THERAPY and HER2DX (filed); founder of Reveal Genomics.
Acknowledgements
We thank Dr. Pedro L. Fernández Ruiz, from the Department of Pathology of Hospital Clinic of Barcelona, for the anatomopathological images shown in Fig. 4, Fig. 5.
References
- 1.Cronin K.A., Harlan L.C., Dodd K.W., Abrams J.S., Ballard-Barbash R. Population-based estimate of the prevalence of HER-2 positive breast cancer tumors for early stage patients in the US. Canc Invest. 2010;28:963–968. doi: 10.3109/07357907.2010.496759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 3.Seshadri R., Firgaira F.A., Horsfall D.J., McCaul K., Setlur V., Kitchen P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The South Australian Breast Cancer Study Group. J Clin Oncol. 1993;11:1936–1942. doi: 10.1200/JCO.1993.11.10.1936. [DOI] [PubMed] [Google Scholar]
- 4.Loibl S., Gianni L. HER2-positive breast cancer. Lancet. 2017;389:2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 5.von Minckwitz G., Procter M., de Azambuja E., Zardavas D., Benyunes M., Viale G. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377:122–131. doi: 10.1056/NEJMoa1703643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Minckwitz G., Huang C.-S., Mano M.S., Loibl S., Mamounas E.P., Untch M. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 7.Swain S.M., Miles D., Kim S.-B., Im Y.-H., Im S.-A., Semiglazov V. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA): end-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:519–530. doi: 10.1016/S1470-2045(19)30863-0. [DOI] [PubMed] [Google Scholar]
- 8.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline focused update. J Clin Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 9.Curigliano G., Burstein H.J., Winer E.P., Gnant M., Dubsky P., Loibl S. De-escalating and escalating treatments for early-stage breast cancer: the st. Gallen international Expert Consensus conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2017;28:1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cejalvo J.M., Martínez de Dueñas E., Galván P., García-Recio S., Burgués Gasión O., Paré L. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Canc Res. 2017;77:2213–2221. doi: 10.1158/0008-5472.CAN-16-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schettini F., Buono G., Cardalesi C., Desideri I., De Placido S., Del Mastro L. Hormone Receptor/Human Epidermal Growth Factor Receptor 2-positive breast cancer: where we are now and where we are going. Canc Treat Rev. 2016;46:20–26. doi: 10.1016/j.ctrv.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 13.Schettini F., Pascual T., Conte B., Chic N., Brasó-Maristany F., Galván P. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: a systematic review and meta-analysis. Canc Treat Rev. 2020;84:101965. doi: 10.1016/j.ctrv.2020.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Minckwitz G., Untch M., Blohmer J.-U., Costa S.D., Eidtmann H., Fasching P.A. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 15.Vaz-Luis I., Ottesen R.A., Hughes M.E., Marcom P.K., Moy B., Rugo H.S. Impact of hormone receptor status on patterns of recurrence and clinical outcomes among patients with human epidermal growth factor-2-positive breast cancer in the National Comprehensive Cancer Network: a prospective cohort study. Breast Cancer Res. 2012;14:R129. doi: 10.1186/bcr3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambertini M., Campbell C., Gelber R., Viale G., McCullough A., Hilbers F. Dissecting the effect of hormone receptor status in patients with HER2-positive early breast cancer: exploratory analysis from the ALTTO (BIG 2-06) randomized clinical trial. Breast Canc Res Treat. 2019;177:103–114. doi: 10.1007/s10549-019-05284-y. [DOI] [PubMed] [Google Scholar]
- 17.Perez E.A., Romond E.H., Suman V.J., Jeong J.-H., Sledge G., Geyer C.E. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32:3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Untch M., Gelber R.D., Jackisch C., Procter M., Baselga J., Bell R. Estimating the magnitude of trastuzumab effects within patient subgroups in the HERA trial. Ann Oncol. 2008;19:1090–1096. doi: 10.1093/annonc/mdn005. [DOI] [PubMed] [Google Scholar]
- 19.Vici P., Pizzuti L., Sperduti I., Frassoldati A., Natoli C., Gamucci T. “Triple positive” early breast cancer: an observational multicenter retrospective analysis of outcome. Oncotarget. 2016;7:17932–17944. doi: 10.18632/oncotarget.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 21.Prat A., Ellis M.J., Perou C.M. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2011;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prat A., Pineda E., Adamo B., Galván P., Fernández A., Gaba L. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–S35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat A., Carey L.A., Adamo B., Vidal M., Tabernero J., Cortés J. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prat A., Pascual T., De Angelis C., Gutierrez C., Llombart-Cussac A., Wang T. HER2-enriched subtype and ERBB2 expression in HER2-positive breast cancer treated with dual HER2 blockade. J Natl Cancer Inst. 2019 doi: 10.1093/jnci/djz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carey L.A., Berry D.A., Cirrincione C.T., Barry W.T., Pitcher B.N., Harris L.N. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llombart-Cussac A., Cortés J., Paré L., Galván P., Bermejo B., Martínez N. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18:545–554. doi: 10.1016/S1470-2045(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 28.Prat A., Bianchini G., Thomas M., Belousov A., Cheang M.C.U., Koehler A. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Canc Res. 2014;20:511–521. doi: 10.1158/1078-0432.CCR-13-0239. [DOI] [PubMed] [Google Scholar]
- 29.Dieci M.V., Prat A., Tagliafico E., Paré L., Ficarra G., Bisagni G. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Ann Oncol. 2016;27:1867–1873. doi: 10.1093/annonc/mdw262. [DOI] [PubMed] [Google Scholar]
- 30.Swain S.M., Tang G., Lucas P.C., Robidoux A., Goerlitz D., Harris B.T. Intrinsic subtypes of HER2-positive breast cancer and their associations with pathologic complete response (pCR) and outcomes: findings from NSABP B-41, a randomized neoadjuvant trial. J Clin Orthod. 2018;36 doi: 10.1200/JCO.2018.36.15_suppl.580. 580–580. [DOI] [Google Scholar]
- 31.Prat A., Chaudhury A., Solovieff N., Paré L., Martinez D., Chic N. Correlative biomarker analysis of intrinsic subtypes and efficacy across the MONALEESA phase III studies. J Clin Oncol. 2021:JCO2002977. doi: 10.1200/JCO.20.02977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernas S., Petit A., Climent F., Paré L., Perez-Martin J., Ventura L. PAM50 subtypes in baseline and residual tumors following neoadjuvant trastuzumab-based chemotherapy in HER2-positive breast cancer: a consecutive-series from a single institution. Front Oncol. 2019;9 doi: 10.3389/fonc.2019.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fumagalli D., Venet D., Ignatiadis M., Azim H.A., Maetens M., Rothé F. RNA sequencing to predict response to neoadjuvant anti-HER2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial. JAMA Oncol. 2017;3:227–234. doi: 10.1001/jamaoncol.2016.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bianchini G., Parker J.S., Carey L.A., Perou C.M., Sica L., Prat A. Research-based PAM50 predicts risk of relapse in residual disease after anti-HER2 therapies. Ann Oncol. 2018;29 viii58–86. [Google Scholar]
- 35.Bianchini G., Pusztai L., Pienkowski T., Im Y.-H., Bianchi G.V., Tseng L.-M. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann Oncol. 2015;26:2429–2436. doi: 10.1093/annonc/mdv395. [DOI] [PubMed] [Google Scholar]
- 36.Ignatiadis M., Van den Eynden G., Roberto S., Fornili M., Bareche Y., Desmedt C. Tumor-infiltrating lymphocytes in patients receiving trastuzumab/pertuzumab-based chemotherapy: a TRYPHAENA substudy. J Natl Cancer Inst. 2019;111:69–77. doi: 10.1093/jnci/djy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes F.A., Espina V., Liotta L.A., Nagarwala Y.M., Danso M., McIntyre K.J. Pathologic complete response after preoperative anti-HER2 therapy correlates with alterations in PTEN, FOXO, phosphorylated Stat5, and autophagy protein signaling. BMC Res Notes. 2013;6:507. doi: 10.1186/1756-0500-6-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swain S.M., Ewer M.S., Viale G., Delaloge S., Ferrero J.-M., Verrill M. Pertuzumab, trastuzumab, and standard anthracycline- and taxane-based chemotherapy for the neoadjuvant treatment of patients with HER2-positive localized breast cancer (BERENICE): a phase II, open-label, multicenter, multinational cardiac safety study. Ann Oncol. 2018;29:646–653. doi: 10.1093/annonc/mdx773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gavilá J., Oliveira M., Pascual T., Perez-Garcia J., Gonzàlez X., Canes J. Safety, activity, and molecular heterogeneity following neoadjuvant non-pegylated liposomal doxorubicin, paclitaxel, trastuzumab, and pertuzumab in HER2-positive breast cancer (Opti-HER HEART): an open-label, single-group, multicenter, phase 2 trial. BMC Med. 2019;17:8. doi: 10.1186/s12916-018-1233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prat A., Slamon D., Hurvitz S., Press M., Lewis Phillips G., Lopez Valverde V. Abstract PD3-06: association of intrinsic subtypes with pathological complete response (pCR) in the KRISTINE neoadjuvant phase 3 clinical trial in HER2-positive early breast cancer (EBC) Canc Res. 2018;78 doi: 10.1158/1538-7445.SABCS17-PD3-06. PD3-06. [DOI] [Google Scholar]
- 41.Guarneri V., Dieci M.V., Bisagni G., Frassoldati A., Bianchi G.V., De Salvo G.L. De-escalated therapy for HR+/HER2+ breast cancer patients with Ki67 response after 2 weeks letrozole: results of the PerELISA neoadjuvant study. Ann Oncol. 2019;30:921–926. doi: 10.1093/annonc/mdz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimawi M.F., Mayer I.A., Forero A., Nanda R., Goetz M.P., Rodriguez A.A. Multicenter phase II study of neoadjuvant lapatinib and trastuzumab with hormonal therapy and without chemotherapy in patients with human epidermal growth factor receptor 2-overexpressing breast cancer: TBCRC 006. J Clin Oncol. 2013;31:1726–1731. doi: 10.1200/JCO.2012.44.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gianni L., Pienkowski T., Im Y.-H., Roman L., Tseng L.-M., Liu M.-C. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 44.Denkert C., Huober J., Loibl S., Prinzler J., Kronenwett R., Darb-Esfahani S. HER2 and ESR1 mRNA expression levels and response to neoadjuvant trastuzumab plus chemotherapy in patients with primary breast cancer. Breast Canc Res. 2013;15:R11. doi: 10.1186/bcr3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griguolo G., Brasó-Maristany F., González-Farré B., Pascual T., Chic N., Saurí T. ERBB2 mRNA expression and response to ado-trastuzumab emtansine (T-DM1) in HER2-positive breast cancer. Cancers. 2020;12 doi: 10.3390/cancers12071902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prat A., De Angelis C., Pascual T., Gutierrez C., Llombart-Cussac A., Wang T. HER2-enriched subtype and ERBB2 mRNA as predictors of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer: a combined analysis of TBCRC006/023 and PAMELA trials. J Clin Orthod. 2018;36 doi: 10.1200/JCO.2018.36.15_suppl.509. 509–509. [DOI] [Google Scholar]
- 47.Pogue-Geile K.L., Kim C., Jeong J.-H., Tanaka N., Bandos H., Gavin P.G. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105:1782–1788. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez E.A., Ballman K.V., Mashadi-Hossein A., Tenner K.S., Kachergus J.M., Norton N. Intrinsic subtype and therapeutic response among HER2-positive breaty st tumors from the NCCTG (alliance) N9831 trial. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krop I.E., Paulson J., Campbell C., Kiermaier A.C., Andre F., Fumagalli D. Genomic correlates of response to adjuvant trastuzumab (H) and pertuzumab (P) in HER2+ breast cancer (BC): biomarker analysis of the APHINITY trial. J Clin Orthod. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.1012. 1012–1012. [DOI] [Google Scholar]
- 50.Conte P.F., Griguolo G., Dieci M.V., Bisagni G., Brandes A.A., Frassoldati A. PAM50 HER2-enriched subtype as an independent prognostic factor in early-stage HER2+ breast cancer following adjuvant chemotherapy plus trastuzumab in the ShortHER trial. J Clin Oncol. 2019;37:544. [Google Scholar]
- 51.Krop I., Hillman D., Polley M.-Y., Tanioka M., Parker J., Huebner L. Abstract GS3-02: invasive disease-free survival and gene expression signatures in CALGB (Alliance) 40601, a randomized phase III neoadjuvant trial of dual HER2-targeting with lapatinib added to chemotherapy plus trastuzumab. Canc Res. 2018;78 doi: 10.1158/1538-7445.SABCS17-GS3-02. GS3-02. [DOI] [Google Scholar]
- 52.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Canc Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bose R., Kavuri S.M., Searleman A.C., Shen W., Shen D., Koboldt D.C. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Canc Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi Z., Rong G., Guan Y., Li J., Chang L., Li H. Molecular landscape and efficacy of HER2-targeted therapy in patients with HER2-mutated metastatic breast cancer. NPJ Breast Cancer. 2020;6:59. doi: 10.1038/s41523-020-00201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth L.M., Piha-Paul S.A., Won H.H., Schram A.M., Saura C., Loi S. Efficacy and determinants of response to HER kinase inhibition in HER2-mutant metastatic breast cancer. Canc Discov. 2020;10:198–213. doi: 10.1158/2159-8290.CD-19-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croessmann S., Formisano L., Kinch L.N., Gonzalez-Ericsson P.I., Sudhan D.R., Nagy R.J. Combined blockade of activating ERBB2 mutations and ER results in synthetic lethality of ER+/HER2 mutant breast cancer. Clin Canc Res. 2019;25:277. doi: 10.1158/1078-0432.CCR-18-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cocco E., Javier Carmona F., Razavi P., Won H.H., Cai Y., Rossi V. Neratinib is effective in breast tumors bearing both amplification and mutation of ERBB2 (HER2) Sci Signal. 2018;11 doi: 10.1126/scisignal.aat9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma F., Li Q., Chen S., Zhu W., Fan Y., Wang J. Phase I study and biomarker analysis of pyrotinib, a novel irreversible pan-ErbB receptor tyrosine kinase inhibitor, in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2017;35:3105–3112. doi: 10.1200/JCO.2016.69.6179. [DOI] [PubMed] [Google Scholar]
- 59.Carnero A., Blanco-Aparicio C., Renner O., Link W., Leal J.F.M. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 60.André F., Hurvitz S., Fasolo A., Tseng L.-M., Jerusalem G., Wilks S. Molecular alterations and everolimus efficacy in human epidermal growth factor receptor 2-overexpressing metastatic breast cancers: combined exploratory biomarker analysis from BOLERO-1 and BOLERO-3. J Clin Oncol. 2016;34:2115–2124. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
- 61.Berns K., Horlings H.M., Hennessy B.T., Madiredjo M., Hijmans E.M., Beelen K. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Canc Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 62.Junttila T.T., Akita R.W., Parsons K., Fields C., Lewis Phillips G.D., Friedman L.S. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Canc Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 63.Serra V., Markman B., Scaltriti M., Eichhorn P.J.A., Valero V., Guzman M. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Canc Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 64.Loi S., Michiels S., Lambrechts D., Fumagalli D., Claes B., Kellokumpu-Lehtinen P.-L. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105:960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loibl S., Majewski I., Guarneri V., Nekljudova V., Holmes E., Bria E. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016;27:1519–1525. doi: 10.1093/annonc/mdw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baselga J., Cortés J., Im S.-A., Clark E., Ross G., Kiermaier A. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32:3753–3761. doi: 10.1200/JCO.2013.54.5384. [DOI] [PubMed] [Google Scholar]
- 67.Baselga J., Lewis Phillips G.D., Verma S., Ro J., Huober J., Guardino A.E. Relationship between tumor biomarkers and efficacy in EMILIA, a phase III study of trastuzumab emtansine in HER2-positive metastatic breast cancer. Clin Canc Res. 2016;22:3755–3763. doi: 10.1158/1078-0432.CCR-15-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baselga J., Cortés J., Kim S.-B., Im S.-A., Hegg R., Im Y.-H. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swain S.M., Kim S.-B., Cortés J., Ro J., Semiglazov V., Campone M. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2013;14:461–471. doi: 10.1016/S1470-2045(13)70130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verma S., Miles D., Gianni L., Krop I.E., Welslau M., Baselga J. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations (∗) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu-Trantien C., Willard-Gallo K. Tumor-infiltrating follicular helper T cells: the new kids on the block. OncoImmunology. 2013;2 doi: 10.4161/onci.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zitvogel L., Kepp O., Senovilla L., Menger L., Chaput N., Kroemer G. Immunogenic tumor cell death for optimal anticancer therapy: the calreticulin exposure pathway. Clin Canc Res. 2010;16:3100–3104. doi: 10.1158/1078-0432.CCR-09-2891. [DOI] [PubMed] [Google Scholar]
- 75.Pruneri G., Bonizzi G., Vingiani A. Biomarkers for the identification of recurrence in human epidermal growth factor receptor 2-positive breast cancer patients. Curr Opin Oncol. 2016;28:476–483. doi: 10.1097/CCO.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 76.Bianchini G., Gianni L. The immune system and response to HER2-targeted treatment in breast cancer. Lancet Oncol. 2014;15:58–68. doi: 10.1016/S1470-2045(13)70477-7. [DOI] [PubMed] [Google Scholar]
- 77.Andre F., Dieci M.V., Dubsky P., Sotiriou C., Curigliano G., Denkert C. Molecular pathways: involvement of immune pathways in the therapeutic response and outcome in breast cancer. Clin Canc Res. 2013;19:28–33. doi: 10.1158/1078-0432.CCR-11-2701. [DOI] [PubMed] [Google Scholar]
- 78.Salgado R., Denkert C., Campbell C., Savas P., Nuciforo P., Nucifero P. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the NeoALTTO trial. JAMA Oncol. 2015;1:448–454. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Denkert C., von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 80.Barroso-Sousa R., Barry W.T., Guo H., Dillon D., Tan Y.B., Fuhrman K. The immune profile of small HER2-positive breast cancers: a secondary analysis from the APT trial. Ann Oncol. 2019;30:575–581. doi: 10.1093/annonc/mdz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Canc Therapeut. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 82.Loi S., Giobbie-Hurder A., Gombos A., Bachelot T., Hui R., Curigliano G. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial. Lancet Oncol. 2019;20:371–382. doi: 10.1016/S1470-2045(18)30812-X. [DOI] [PubMed] [Google Scholar]
- 83.Emens L.A., Esteva F.J., Beresford M., Saura C., De Laurentiis M., Kim S.-B. Trastuzumab emtansine plus atezolizumab versus trastuzumab emtansine plus placebo in previously treated, HER2-positive advanced breast cancer (KATE2): a phase 2, multicentre, randomised, double-blind trial. Lancet Oncol. 2020;21:1283–1295. doi: 10.1016/S1470-2045(20)30465-4. [DOI] [PubMed] [Google Scholar]
- 84.Schmid P., Adams S., Rugo H.S., Schneeweiss A., Barrios C.H., Iwata H. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 85.Loi S., Michiels S., Salgado R., Sirtaine N., Jose V., Fumagalli D. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 86.Perez E.A., Ballman K.V., Tenner K.S., Thompson E.A., Badve S.S., Bailey H. Association of stromal tumor-infiltrating lymphocytes with recurrence-free survival in the N9831 adjuvant trial in patients with early-stage HER2-positive breast cancer. JAMA Oncol. 2016;2:56–64. doi: 10.1001/jamaoncol.2015.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Denkert C., von Minckwitz G., Brase J.C., Sinn B.V., Gade S., Kronenwett R. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol. 2015;33:983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 88.Hamy A.-S., Pierga J.-Y., Sabaila A., Laas E., Bonsang-Kitzis H., Laurent C. Stromal lymphocyte infiltration after neoadjuvant chemotherapy is associated with aggressive residual disease and lower disease-free survival in HER2-positive breast cancer. Ann Oncol. 2017;28:2233–2240. doi: 10.1093/annonc/mdx309. [DOI] [PubMed] [Google Scholar]
- 89.Nuciforo P., Pascual T., Cortés J., Llombart-Cussac A., Fasani R., Paré L. A predictive model of pathologic response based on tumor cellularity and tumor-infiltrating lymphocytes (CelTIL) in HER2-positive breast cancer treated with chemo-free dual HER2 blockade. Ann Oncol. 2018;29:170–177. doi: 10.1093/annonc/mdx647. [DOI] [PubMed] [Google Scholar]
- 90.Linette G.P., Carreno B.M. Tumor infiltrating lymphocytes in the checkpoint inhibitor era. Curr Hematol Malig Rep. 2019;14:286–291. doi: 10.1007/s11899-019-00523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klein S., Mauch C., Brinker K., Noh K.-W., Knez S., Büttner R. Tumor infiltrating lymphocyte clusters are associated with response to immune checkpoint inhibition in BRAF V600 E/K mutated malignant melanomas. Sci Rep. 2021;11:1834. doi: 10.1038/s41598-021-81330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bai R., Lv Z., Xu D., Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomarker Research. 2020;8:34. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alexe G., Dalgin G.S., Scanfeld D., Tamayo P., Mesirov J.P., DeLisi C. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Canc Res. 2007;67:10669–10676. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 94.Rody A., Holtrich U., Pusztai L., Liedtke C., Gaetje R., Ruckhaeberle E. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wirapati P., Sotiriou C., Kunkel S., Farmer P., Pradervand S., Haibe-Kains B. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamy A.-S., Bonsang-Kitzis H., Lae M., Moarii M., Sadacca B., Pinheiro A. A stromal immune module correlated with the response to neoadjuvant chemotherapy, prognosis and lymphocyte infiltration in HER2-positive breast carcinoma is inversely correlated with hormonal pathways. PloS One. 2016;11 doi: 10.1371/journal.pone.0167397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bedognetti D., Hendrickx W., Marincola F.M., Miller L.D. Prognostic and predictive immune gene signatures in breast cancer. Curr Opin Oncol. 2015;27:433–444. doi: 10.1097/CCO.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 98.Gil Del Alcazar C.R., Huh S.J., Ekram M.B., Trinh A., Liu L.L., Beca F. Immune escape in breast cancer during in situ to invasive carcinoma transition. Canc Discov. 2017;7:1098–1115. doi: 10.1158/2159-8290.CD-17-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lefebvre C., Bachelot T., Filleron T., Pedrero M., Campone M., Soria J.-C. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smid M., Rodríguez-González F.G., Sieuwerts A.M., Salgado R., Smissen WJCP-V der, Vlugt-Daane M van der. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang S., Jia M., He Z., Liu X.-S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018;37:3924–3936. doi: 10.1038/s41388-018-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanu N., Cerone M.A., Goh G., Zalmas L.-P., Bartkova J., Dietzen M. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016;17:185. doi: 10.1186/s13059-016-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fehrenbacher L., Cecchini R.S., Geyer C.E., Rastogi P., Costantino J.P., Atkins J.N. NSABP B-47/NRG Oncology phase III randomized trial comparing adjuvant chemotherapy with or without trastuzumab in high-risk invasive breast cancer negative for HER2 by FISH and with IHC 1+ or 2. J Clin Oncol. 2020;38:444–453. doi: 10.1200/JCO.19.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hurvitz S.A., Martin M., Jung K.H., Huang C.-S., Harbeck N., Valero V. Neoadjuvant trastuzumab emtansine and pertuzumab in human epidermal growth factor receptor 2-positive breast cancer: three-year outcomes from the phase III KRISTINE study. J Clin Oncol. 2019;37:2206–2216. doi: 10.1200/JCO.19.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iwata H., Tamura K., Doi T., Tsurutani J., Modi S., Park H. Trastuzumab deruxtecan (DS-8201a) in subjects with HER2-expressing solid tumors: long-term results of a large phase 1 study with multiple expansion cohorts. J Clin Orthod. 2018;36 doi: 10.1200/JCO.2018.36.15_suppl.2501. 2501–2501. [DOI] [Google Scholar]
- 106.Banerji U., van Herpen C.M.L., Saura C., Thistlethwaite F., Lord S., Moreno V. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019;20:1124–1135. doi: 10.1016/S1470-2045(19)30328-6. [DOI] [PubMed] [Google Scholar]
- 107.Rinnerthaler G., Gampenrieder S.P., Greil R. HER2 directed antibody-drug-conjugates beyond T-DM1 in breast cancer. Int J Mol Sci. 2019;20:1115. doi: 10.3390/ijms20051115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schettini F., Chic N., Brasó-Maristany F., Paré L., Pascual T., Conte B. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer. 2021;7:1. doi: 10.1038/s41523-020-00208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brasó-Maristany F., Paré L., Chic N., Martínez-Sáez O., Pascual T., Mallafré-Larrosa M. Gene expression profiles of breast cancer metastasis according to organ site. Mol Oncol. 2021 doi: 10.1002/1878-0261.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Agostinetto E., Rediti M., Fimereli D., Debien V., Piccart M., Aftimos P. HER2-Low breast cancer: molecular characteristics and prognosis. Cancers. 2021;13:2824. doi: 10.3390/cancers13112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marchiò C., Annaratone L., Marques A., Casorzo L., Berrino E., Sapino A. Evolving concepts in HER2 evaluation in breast cancer: heterogeneity, HER2-low carcinomas and beyond. Semin Canc Biol. 2020 doi: 10.1016/j.semcancer.2020.02.016. [DOI] [PubMed] [Google Scholar]
- 112.Metzger Filho O., Viale G., Trippa L., Li T., Yardley D.A., Mayer I.A. HER2 heterogeneity as a predictor of response to neoadjuvant T-DM1 plus pertuzumab: results from a prospective clinical trial. J Clin Orthod. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.502. 502–502. [DOI] [Google Scholar]
- 113.Prat A., Guarneri V., Paré L., Griguolo G., Pascual T., Dieci M.V. A multivariable prognostic score to guide systemic therapy in early-stage HER2-positive breast cancer: a retrospective study with an external evaluation. Lancet Oncol. 2020;21:1455–1464. doi: 10.1016/S1470-2045(20)30450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buono G., Schettini F., Perri F., Arpino G., Bianco R., Criscitiello C. The impact of translational research in breast cancer care: can we improve the therapeutic scenario? Anticancer Agents Med Chem. 2018;18:832–836. doi: 10.2174/1871520617666171103105247. [DOI] [PubMed] [Google Scholar]
- 115.Nelson R. Targeted therapies offer promise, but are they affordable? Medscape n. https://www.medscape.com/viewarticle/810147 d. accessed.
- 116.Barrios C.H., Reinert T., Werutsky G. Access to high-cost drugs for advanced breast cancer in Latin America, particularly trastuzumab. Ecancermedicalscience. 2019;13 doi: 10.3332/ecancer.2019.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nixon N., Verma S. American Society of Clinical Oncology Educational Book; 2016. A value-based approach to treatment of HER2-positive breast cancer: examining the evidence. e56–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new datasets were generated or analyzed for this report.