Abstract
Purpose:
To understand the prognostic value of The Cancer Genome Atlas (TCGA) for uveal melanoma metastasis, using a simplified 4-category classification, based on tumor DNA.
Methods:
A retrospective cohort study of 1001 eyes with uveal melanoma at a single center, categorized according to TCGA as Group A, B, C, or D (by fine-needle aspiration biopsy for DNA analysis), and treated with standard methods, was studied for melanoma-related metastasis at 5 and 10 years.
Results:
Of 1001 eyes with uveal melanoma, the TCGA categories included Group A (n = 486, 49%), B (n = 141, 14%), C (n = 260, 26%), and D (n = 114, 11%). By comparison, increasing category (A vs. B vs. C vs. D) was associated with features of older age at presentation (56.8 vs. 52.8 vs. 61.1 vs. 63.5 years, P < 0.001), less often visual acuity of 20/20–20/50 (80% vs. 67% vs. 70% vs. 65%, P = 0.001), tumor location further from the optic disc (P < 0.001) and foveola (P < 0.001), and greater median tumor basal diameter (10.0 vs. 13.0 vs. 14.0 vs. 16.0 mm, P < 0.001) and tumor thickness (3.5 vs. 5.2 vs. 6.0 vs. 7.1 mm, P < 0.001). The Kaplan–Meier (5-year/10-year) rate of metastasis was 4%/6% for Group A, 12%/20% for Group B, 33%/49% for Group C, and 60%/not available for Group D.
Conclusion:
A simplified 4-category classification of uveal melanoma using TCGA, based on tumor DNA, is highly predictive of risk for metastatic disease.
Keywords: Choroid, ciliary body, cytogenetics, genetics, melanoma, outcomes, TCGA, The Cancer Genome Atlas, uvea
The Cancer Genome Atlas (TCGA) is an international collaborative project designed by the National Cancer Institute’s Center for Cancer Genomics and the National Human Genome Research Institute for the investigation of human cancer-related mutations. This project explored molecular aberrations on various platforms including histologic features, chromosome copy numbers, genetic mutations, expression of RNA, DNA methylation status, proteins, biochemical pathways, and immune markers for 33 cancer types, including uveal melanoma.[1,2,3,4,5,6,7,8] In 2017, Robertson et al. published TCGA results of 80 eyes with uveal melanoma and identified four distinct molecular subsets, two associated with good prognosis (disomy 3 [D3]) and two associated with poor prognosis (monosomy 3 [M3]).[3] In 2018, Jager et al. commented on TCGA results for uveal melanoma in an editorial, indicating the enormity and organization of this project, and provided clarity to the 4 main prognostic categories of uveal melanoma as identified by TCGA.[4] They remarked that these clusters included Group A (D3, disomy 8), Group B (D3, 8q gain), Group C (M3, 8q gain possible), and Group D (M3, 8q gain multiple [isochromosome for 8q]).
In 2019, Vichitvejpaisal et al. applied the simplified 4-group classification of TCGA to a cohort of 658 patients with uveal melanoma at a single center and found the 5-year cumulative percentage of distant metastasis (odds ratio, OR) at 4% (OR 1.0) for Group A, 20% (OR 3.5) for Group B, 33% (OR 11.4) for Group C, and 63% (OR 26.4) for Group D.[5] Mazloumi et al. later documented that TCGA classification provided superior accuracy to the American Joint Committee on Cancer (AJCC) 8th ed.ition categories, subcategories, and stages for 5-year prediction of metastasis of uveal melanoma.[7] Herein, we explore a larger cohort of 1001 cases with a longer follow-up period using 5-year and 10-year Kaplan–Meier analyses to further validate TCGA in the prediction of uveal melanoma metastasis.
Methods
The medical records of the Ocular Oncology Service at the Wills Eye Hospital, Thomas Jefferson University, Philadelphia, Pennsylvania, USA, were retrospectively reviewed for patients with the clinical diagnosis of uveal melanoma between February 4, 2004 and June 2, 2020, who underwent genetic evaluation and assessment for The Cancer Genome Atlas (TCGA) classification. This study was approved by the Institutional Review Board of Wills Eye Hospital, adhering to the tenets of the Declaration of Helsinki, and complied with the Health Insurance Portability and Accountability Act. Informed consent was obtained from all patients.
All patients were examined by a trained ocular oncologist (CLS, SEL, JAS) for clinical confirmation of diagnosis of uveal melanoma based on indirect ophthalmoscopy with large detailed fundus drawings and imaging. Ophthalmic imaging included fundus photography with wide-angle imaging, fundus autofluorescence, ultrasonography, optical coherence tomography (OCT), fluorescein angiography, indocyanine green angiography, and OCT angiography, as needed for documentation at the first examination and subsequent examinations. Those patients who had undergone genetic testing by fine-needle aspiration biopsy or open biopsy with results on the status of chromosomes 3 and 8 were classified according to TCGA as A, B, C, or D and included in this study.
Data were recorded at each examination and documented in patient’s chart. The demographic data included age (years), sex (male, female), race (Caucasian, African American, Hispanic, Asian, others/unknown), affected eye (right, left), and visual acuity (20/20–20/50, 20/60–20/200, 20/400–no light perception [NLP]).
The tumor features at presentation included tumor location with distance from the optic disc (millimeters [mm]), distance from the foveola (mm), largest tumor basal diameter (mm), tumor thickness (mm), tumor epicenter (choroid, ciliary body, iris), anterior margin of the tumor (macula, macula to the equator, equator to ora serrata, ciliary body, iris), and posterior margin of the tumor (macula, macula to the equator, equator to ora serrata, ciliary body, iris).
Samples for genetic testing were obtained by fine-needle aspiration biopsy (FNAB), performed in the operating room at the time of uveal melanoma treatment, as described in the literature.[5] The samples were stored in Hanks’ balanced salt solution (Gibco, Life Technologies, Grand Island, NY) at 4 degrees Celsius, and DNA analysis was performed using a DNA Micro Kit (QIAGEN, Valencia, CA).[5]
Primary outcomes included the rate and mean time to melanoma-related metastasis and death. Information on metastasis was gathered through history from the patient and correspondence from physicians. Information on death was gathered through history from the family and correspondence from physicians and family. Metastasis was further stratified based on the rate of metastasis to the liver, lung, and other systemic locations. Kaplan–Meier analysis at 1 year, 2 years, 5 years, and 10 years for each outcome of melanoma-related metastasis and death as well as metastasis to the liver, lung, and other systemic locations was stratified by TCGA classification (Group A vs. Group B vs. Group C vs. Group D). Additionally, Cox regression analyses to assess competing risks were performed but did not differ significantly from Kaplan–Meier analysis in this population.
Statistical analysis was performed using SAS Software Suite (version 9.4; SAS Institute). Continuous variables were expressed as mean (median, range). The one-sample Shapiro–Wilk test was used to assess the normality of distribution. A comparison between groups was performed using the one-way ANOVA test for continuous variables with normal distribution and the Kruskal–Wallis test for continuous variables without normal distribution. A comparison of categorical variables was performed using the likelihood ratio Chi-square test and Fisher’s exact test when indicated. Binary logistic regression analysis was performed to identify factors potentially predictive of metastasis and death, which could act as confounders because of their strong correlation with the TCGA classification. Variables found to be significant in univariate analysis at a level of P < 0.05 (age, largest tumor basal diameter, tumor thickness, distance from the optic disc, distance from the foveola, location of the tumor epicenter, anterior margin of the tumor, and posterior margin of the tumor) were entered into multivariate multiple regression models using the stepwise Wald method, which further excluded variables non-contributory to the fit of the model (P > 0.05). Odds ratios and 95% confidence intervals were generated for both univariate and multivariate regression models. Kaplan–Meier analysis was performed for metastasis (liver metastasis, lung metastasis, any metastasis) and death from uveal melanoma. A P value < 0.05 was considered statistically significant for the results of multivariate multiple regression and Kaplan–Meier analysis.
Results
There were 1001 consecutive eyes with uveal melanoma in 999 patients that were sampled for DNA analysis of chromosomes 3 and 8 at the time of tumor management at the Ocular Oncology Service at the Wills Eye Hospital at Thomas Jefferson University, Philadelphia, Pennsylvania USA, over a 22-year period. Patients with no genetic testing and/or those with no follow-up information were not included in this analysis.
Of all 1001 eyes with uveal melanoma, TCGA categories included Group A (n = 486, 49%), B (n = 141, 14%), C (n = 260, 26%), and D (n = 114, 11%). Demographic features are listed in Table 1. By comparison, increasing category (A vs. B vs. C vs. D) was associated with older age at presentation (56.8 vs. 52.8 vs. 61.1 vs. 63.5 years, P < 0.001) and less often visual acuity of 20/20–20/50 (80% vs. 67% vs. 70% vs. 65%, P = 0.001). There was no difference in sex, race, or affected eye.
Table 1.
Ten-Year Outcomes of Uveal Melanoma Based on The Cancer Genome Atlas (TCGA) Classification in 1001 Cases. Patient demographics
| Patient Demographics | TCGA Class | Total Population | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| A (n=486) [n (%)] | B (n=141) [n (%)] | C (n=260) [n (%)] | D (n=114) [n (%)] | P | (n=1001) [n (%)] | |
| Age | ||||||
| Mean (years) (median, range) | 56.8 (58.0, 10.0 - 90.0) | 52.8 (54.0, 13.0 - 83.0) | 61.1 (62.5, 12.0 - 88.0) | 63.5 (64.0, 28.0 - 94.0) | <0.001 | 58.1 (59.0, 10.0 - 94.0) |
| Sex | ||||||
| Male | 266 (55) | 72 (51) | 121 (47) | 57 (50) | 0.194 | 516 (52) |
| Female | 220 (45) | 69 (49) | 139 (53) | 57 (50) | 485 (48) | |
| Race | ||||||
| Caucasian | 472 (97) | 130 (92) | 251 (97) | 112 (98) | 0.088 | 965 (96) |
| African American | 0 (0) | 2 (1) | 0 (0) | 0 (0) | 2 (<1) | |
| Hispanic | 10 (2) | 3 (2) | 5 (2) | 2 (2) | 20 (2) | |
| Asian | 1 (<1) | 3 (2) | 1 (<1) | 0 (0) | 5 (<1) | |
| Other/unknown | 3 (1) | 3 (2) | 3 (1) | 0 (0) | 9 (1) | |
| Affected eye | ||||||
| Right | 249 (51) | 77 (55) | 143 (55) | 60 (53) | 0.760 | 529 (53) |
| Left | 237 (49) | 64 (45) | 117 (45) | 54 (47) | 472 (47) | |
| Visual Acuity | ||||||
| 20/20 - 20/50 | 389 (80) | 94 (67) | 182 (70) | 74 (65) | 0.001 | 739 (74) |
| 20/60 - 20/200 | 55 (11) | 29 (21) | 53 (20) | 28 (25) | 165 (16) | |
| 20/400 - NLP | 42 (9) | 18 (13) | 25 (10) | 12 (11) | 97 (10) | |
Bold values indicate significant P. TCGA=The Cancer Genome Atlas; NLP=No light perception
Tumor characteristics are listed in Table 2. By comparison, increasing category (A vs. B vs. C vs. D) was associated with tumor location further from the optic disc (P < 0.001) and foveola (P < 0.001), increasing tumor basal diameter (10.5 mm vs. 12.7 mm vs. 13.6 mm vs. 15.3 mm, P < 0.001) and tumor thickness (4.4 mm vs. 6.2 mm vs. 6.7 mm vs. 7.6 mm, P < 0.001), greater frequency of the anterior margin involving the ciliary body (13% vs. 28% vs. 32% vs. 41%, P < 0.001), and less frequency of the posterior margin in the macula (63% vs. 56% vs. 48% vs. 55%, P < 0.001).
Table 2.
Ten-Year Outcomes of Uveal Melanoma Based on The Cancer Genome Atlas (TCGA) Classification in 1001 Cases. Tumor features
| Tumor Features | TCGA Class | Total Population | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| A (n=486) [n (%)] | B (n=141) [n (%)] | C (n=260) [n (%)] | D (n=114) [n (%)] | P | (n=1001) [n (%)] | |
| Distance to optic disc (mm) [mean (median, range)] | 3.9 (3.0, 0.0 - 20.0) | 4.7 (4.0, 0.0 - 18.0) | 5.5 (5.0, 0.0 - 17.0) | 4.9 (5.0, 0.0 - 18.0) | <0.001 | 4.5 (3.5, 0.0 - 20.0) |
| Distance to foveola (mm) [mean (median, range)] | 3.6 (2.3, 0.0 - 18.4) | 4.1 (3.3, 0.0 - 15.0) | 5.4 (4.0, 0.0 - 18.0) | 4.7 (3.0, 0.0 - 17.0) | <0.001 | 4.3 (3.0, 0.0 - 18.4) |
| Largest basal diameter (mm) [mean (median, range)] | 10.5 (10.0, 1.0 - 22.0) | 12.7 (13.0, 2.0 - 22.0) | 13.6 (14.0, 2.0 - 24.0) | 15.3 (16.0, 6.0 - 24.0) | <0.001 | 12.1 (12.0, 1.0 - 24.0) |
| Thickness (mm)[mean (median, range)] | 4.4 (3.5, 1.0 - 14.0) | 6.2 (5.2, 1.3 - 15.0) | 6.7 (6.0, 0.7 - 16.0) | 7.6 (7.1, 2.1 - 20.4) | <0.001 | 5.6 (4.7, 0.7 - 20.4) |
| Tumor epicenter | ||||||
| Choroid | 457 (94) | 126 (89) | 227 (87) | 103 (90) | 0.008 | 913 (91) |
| Ciliary body | 20 (4) | 11 (8) | 26 (10) | 11 (10) | 68 (7) | |
| Iris | 9 (2) | 4 (3) | 7 (3) | 0 (0) | 20 (2) | |
| Anterior margin | ||||||
| Macula | 32 (7) | 9 (6) | 6 (2) | 4 (4) | <0.001 | 51 (5) |
| Macula to equator | 245 (50) | 40 (28) | 64 (25) | 22 (19) | 371 (37) | |
| Equator to ora serrata | 131 (27) | 44 (31) | 81 (31) | 38 (33) | 294 (29) | |
| Ciliary body | 62 (13) | 40 (28) | 82 (32) | 47 (41) | 231 (23) | |
| Iris | 16 (3) | 8 (6) | 27 (10) | 3 (3) | 54 (5) | |
| Posterior margin | ||||||
| Macula | 305 (63) | 79 (56) | 124 (48) | 63 (55) | <0.001 | 571 (57) |
| Macula to equator | 156 (32) | 54 (38) | 115 (44) | 48 (42) | 373 (37) | |
| Equator to ora serrata | 10 (2) | 2 (1) | 12 (5) | 3 (3) | 27 (3) | |
| Ciliary body | 7 (1) | 2 (1) | 5 (2) | 0 (0) | 14 (1) | |
| Iris | 8 (2) | 4 (3) | 4 (2) | 0 (0) | 16 (2) | |
Bold values indicate significant P. TCGA=The Cancer Genome Atlas
Outcomes for melanoma-related metastasis and death are listed in Table 3. Overall mean follow-up duration was 41.0 months (median 30.6, range < 0.1–184.9), and tumors with greater TCGA classification had a shorter follow-up duration (46.4 vs. 45.0 vs. 32.4 vs. 32.7, P < 0.001).
Table 3.
Ten-Year Outcomes of Uveal Melanoma Based on The Cancer Genome Atlas (TCGA) Classification in 1001 Cases. Outcomes.
| Outcomes | TCGA Class | Total Population | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| A (n=486) [n (%)] | B (n=141) [n (%)] | C (n=260) [n (%)] | D (n=114) [n (%)] | Odds Ratio (95% Confidence Interval) | P | (n=1001) [n (%)] | |
| Follow-up (months) (median, range) | 46.4 (40.1, <0.1 - 161.0) | 45.0 (33.9, 0.1 - 184.9) | 32.4 (23.9, 0.1 - 173.1) | 32.7 (28.0, 0.0 - 117.4) | --- | <0.001 | 41.0 (30.6, <0.1 - 184.9) |
| Metastasis | 14 (3) | 12 (9) | 53 (20) | 52 (46) |
A vs. B: 2.48 (1.11-5.56)
A vs. C: 6.21 (3.32 - 11.62) A vs. D: 22.25 (11.54 - 42.90) |
0.026
0.027 <0.001 |
131 (13) |
| Mean time to metastasis (months) (median, range) | 37.4 (40.2, 0.5 - 81.0) | 38.7 (38.3, 1.1 - 77.3) | 27.7 (19.1, 0.2 - 85.6) | 21.5 (18.0, 0.0 - 107.2) | --- | 0.009 | 27.3 (19.9, 0.0 - 107.2) |
| Liver metastasis | 10 (2) | 12 (9) | 51 (20) | 52 (46) |
A vs. B: 3.20 (1.32 - 7.74)
A vs. C: 7.32 (3.55 - 15.07) A vs. D: 25.04 (11.81 - 53.11) |
0.026
0.009 <0.001 |
125 (12) |
| Mean time to metastasis (months) (median, range) | 30.2 (22.2, 0.5 - 65.7) | 38.9 (38.3, 1.1 - 77.3) | 26.7 (18.7, 0.2 - 85.6) | 21.9 (18.0, 0.0 - 107.2) | --- | 0.032 | 26.2 (19.5, 0.0 - 107.2) |
| Lung metastasis | 1 (<1) | 2 (1) | 10 (4) | 11 (10) | A vs. B: 4.91 (0.44 - 55.23) A vs. C: 11.50 (1.43 - 92.32) A vs. D: 34.75 (4.40 - 274.59) |
0.615 0.047 <0.001 |
24 (2) |
| Mean time to metastasis (months) (median, range) | 28.4 (28.4, 28.4 - 28.4) | 47.4 (47.4, 17.5 - 77.3) | 31.5 (25.1, 1.3 - 65.7) | 36.8 (37.5, 5.6 - 72.3) | --- | 0.896 | 34.8 (32.9, 1.3 - 77.3) |
| Other metastasis* | 5 (1) | 5 (4) | 13 (5) | 16 (14) | A vs. B: 1.98 (0.53 - 7.37) A vs. C: 2.10 (0.67 - 6.57) A vs. D: 7.30 (2.39 - 22.25) |
0.659 0.709 <0.001 |
39 (4) |
| Mean time to metastasis (months) (median, range) | 48.2 (53.9, 0.5 - 81.0) | 42.8 (50.7, 6.9 - 65.9) | 41.7 (41.8, 13.6 - 73.7) | 35.3 (35.2, 3.6 - 72.3) | --- | 0.263 | 40.2 (43.4, 0.5 - 81.0) |
| Death from UM metastasis | 2 (<1) | 0 (0) | 4 (2) | 8 (7) | A vs. B: na A vs. C: 2.26 (0.36 - 14.07) A vs. D: 9.65 (1.67 - 55.73) |
-- 0.634 0.003 |
14 (1) |
| Mean time to death (months) (median, range) | 48.0 (48.0, 27.2 - 68.8) | 0.0 (0.0, 0.0 - 0.0) | 47.4 (46.0, 38.2 - 59.4) | 26.5 (24.3, 3.5 - 50.5) | --- | 0.097 | 35.5 (38.3, 3.5 - 68.8) |
Bold values indicate significant P. UM = Uveal melanoma, NA= not available. *Sites of other metastasis include bone, brain, breast, intestine, distant lymph nodes, mesentery, muscle, skin
By comparison, increasing category (A vs. B vs. C vs. D) was associated with the increased rate of any melanoma metastasis (3% vs. 9% vs. 20% vs. 46%, P < 0.001), shorter mean time to any metastasis (37.4 vs. 38.7 vs. 27.7 vs. 21.5, P = 0.009), and specifically the increased rate of liver metastasis (2% vs. 9% vs. 20% vs. 46%, P < 0.001), lung metastasis (<1% vs. 1% vs. 4% vs. 10%, P < 0.001), metastasis to other systemic locations (bone, brain, breast, intestine, distant lymph nodes, mesentery, muscle, skin) (1% vs. 4% vs. 5% vs. 14%, P < 0.001), and melanoma-related death (<1% vs. 0% vs. 2% vs. 7%, P = 0.003). By comparison, increasing specific category was associated with the increased odds ratio (OR) for any metastasis (A vs. B, OR 2.48 (P = 0.026); A vs. C, OR 6.21 (P = 0.027); A vs. D, OR 22.25 (P < 0.001)) and for liver metastasis (A vs. B, OR 3.20 (P = 0.026); A vs. C, OR 7.32 (P = 0.009); A vs. D, OR 25.04 (P < 0.001)). A similar increasing risk (A vs. D) was observed for lung metastasis (OR 34.75, P < 0.001) and metastasis to other locations (OR 7.30, P < 0.001).
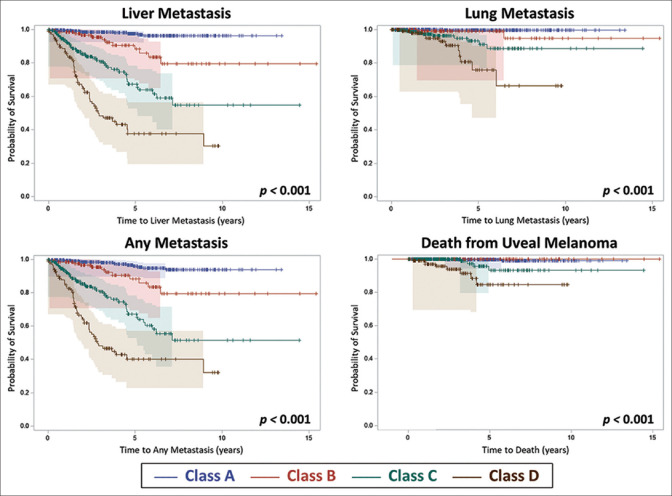
Kaplan–Meier analysis of outcomes of metastasis and death is listed in Table 4 [Fig. 1]. By comparison, increasing category (A vs. B vs. C vs. D) was associated with greater risk for any melanoma-related metastasis (P < 0.001) at 1-year (1% vs. 2% vs. 7% vs. 15%), at 2-years (2% vs. 3% vs. 15% vs. 36%), at 5-years (4% vs. 12% vs. 33% vs. 60%), and at 10-years (6% vs. 20% vs. 49% vs. not available). A similar increasing risk for liver (P < 0.001), lung (P < 0.001), and other metastases as well as melanoma-related death (P < 0.001) over time were documented [Table 4 and Fig. 1].
Table 4.
Ten-Year Outcomes for Uveal Melanoma Based on The Cancer Genome Atlas (TCGA) Classification in 1001 Cases. Event-Free Survival Analysis of Metastasis and Death
| Event-Free Survival | TCGA Class | Total Population | |||
|---|---|---|---|---|---|
|
|
|
||||
| A (n=486) [n (%)] | B (n=141) [n (%)] | C (n=260) [n (%)] | D (n=114) [n (%)] | (n=1001) [n (%)] | |
| Overall Metastasis | |||||
| 1 Year | 394 (99) | 123 (98) | 185 (93) | 83 (85) | 785 (96) |
| 2 Years | 315 (98) | 92 (97) | 118 (85) | 52 (64) | 577 (91) |
| 5 Years | 160 (96) | 40 (88) | 41 (67) | 12 (40) | 254 (82) |
| 10 Years | 16 (94) | 6 (80) | 5 (51) | NA | 27 (75) |
| Liver Metastasis | |||||
| 1 Year | 394 (99) | 123 (98) | 185 (93) | 83 (86) | 785 (96) |
| 2 Years | 315 (98) | 92 (97) | 118 (85) | 52 (65) | 577 (91) |
| 5 Years | 160 (98) | 40 (88) | 41 (67) | 12 (42) | 254 (82) |
| 10 Years | 17 (96) | 6 (80) | 5 (55) | NA | 28 (77) |
| Lung Metastasis | |||||
| 1 Year | 394 (100) | 123 (100) | 195 (99) | 91 (99) | 801 (>99) |
| 2 Years | 316 (100) | 91 (>99) | 130 (97) | 64 (95) | 601 (99) |
| 5 Years | 161 (99) | 41 (99) | 42 (93) | 13 (76) | 257 (96) |
| 10 Years | 17 (99) | 6 (95) | 5 (89) | NA | 28 (94) |
| Other Metastasis* | |||||
| 1 Year | 394 (>99) | 121 (99) | 196 (100) | 92 (99) | 802 (>99) |
| 2 Years | 316 (>99) | 91 (99) | 131 (99) | 64 (93) | 604 (99) |
| 5 Years | 160 (98) | 40 (95) | 42 (88) | 14 (71) | 256 (93) |
| 10 Years | 16 (97) | 6 (90) | 5 (83) | NA | 27 (90) |
| Death from Uveal Melanoma | |||||
| 1 Year | 394 (100) | 123 (100) | 196 (100) | 92 (99) | 802 (>99) |
| 2 Years | 316 (100) | 92 (100) | 131 (100) | 66 (96) | 604 (99) |
| 5 Years | 162 (>99) | 41 (100) | 42 (93) | 14 (85) | 259 (97) |
| 10 Years | 17 (99) | 6 (100) | 5 (93) | NA | 28 (97) |
TCGA=The Cancer Genome Atlas, NA=not available. *Sites of other metastasis include bone, brain, breast, intestine, distant lymph nodes, mesentery, muscle, skin
Figure 1.
Kaplan–Meier estimates of metastasis and survival according to The Cancer Genome Atlas (TCGA) classification into Group A, B, C, or D. With increasing group, there was increasing risk for liver metastasis (P < 0.001), lung metastasis (P < 0.001), any metastasis (P < 0.001), and death (P < 0.001)
Discussion
The Cancer Genome Atlas (TCGA) has provided a comprehensive “cancer atlas” through wide-ranging multi-platform analyses of over 30 human cancers, including uveal melanoma.[2] This project involved a multicenter organization including tissue source sites for the collection of blood and tissue samples; biospecimen core resources for coordination of sample delivery and cataloging; genome sequencing centers for high-throughput sequencing and identification of DNA alterations; cancer genome characterization centers for description of alterations in miRNA, gene expression, single nucleotide polymorphisms, and others; proteome characterization centers for the identification of cancer-specific proteins; data coordinating centers for the collection and transfer of data to public databases; cancer genomics hub for storage, cataloging, and access to information; and genome data analysis centers for the development of informatics tools for processing data across the entire genome.[2] This unprecedented effort was then made available for public access, providing researchers the opportunity to evaluate this data.
In 2017, Robertson et al. were the first to publish results of TCGA regarding uveal melanoma in 80 cases.[3] They identified four molecularly-distinct groups of uveal melanoma, with two groups demonstrating favorable prognosis related to disomy 3 (D3) and two groups showing poor prognosis related to monosomy 3 (M3). Within each of the four groups there were unique gene alterations, DNA methylation, mRNA expression levels, and other findings that accounted for increasing the risk for melanoma metastasis. Jager et al. later clarified this distinct molecular grouping of uveal melanoma as A, B, C, and D, whereby the criteria for Group A included D3 and disomy of chromosome 8, Group B showed D3 and 8q gain, Group C showed M3 and often with 8q gain, and Group D showed M3 with multiple 8q gains manifesting as an isochromosome for 8q.[4] The simplicity of this 4-category DNA-based classification scheme provides a straightforward approach for stratifying uveal melanoma prognosis, as previous investigations have included complex multiple factors[9,10,11,12,13,14] such as over 50 combinations of DNA alterations in chromosomes 3, 6, and 8 giving a stepwise, graded prognosis,[15,16] whereas others have blended in the AJCC classification[17,18,19] with DNA alterations[20,21] further refining prognosis, and still others have combined DNA alterations of chromosomes 3 and 8 plus mitotic activity, closed loops, epithelioid cells, basal tumor diameter, extraocular spread, and optic disc and ciliary body involvement in the equation for an all-inclusive prognosis.[22,23]
In 2019, Vichitvejpaisal et al. validated the 4-category TCGA classification in an analysis of 658 cases, demonstrating that the 5-year Kaplan–Meier cumulative percentage of distant metastasis based on DNA results for Group A was 4%, B was 20%, C was 33%, and D was 63%.[5] In that cohort, longer-term, 10-year data was not available. In this current analysis, we explore a larger cohort of 1001 cases of uveal melanoma with more robust 10-year Kaplan–Meier outcomes. Based on TCGA classification using DNA results, we found that the 5-year Kaplan–Meier rate of any distant metastasis for Group A was 4%, B was 12%, C was 33%, and D was 60%, whereas the 10-year Kaplan–Meier rate of any distant metastasis for Group A was 6%, B was 20%, C was 49%, and D was not available due to the small cohort number. The odds ratios for any distant metastasis (vs. Group A) were 2.48 for Group B, 6.21 for Group C, and 22.25 for Group D. Furthermore, the 10-year Kaplan–Meier rates of specific liver metastasis revealed Group A at 4%, B at 20%, C at 45%, and D not available with odds ratios for liver metastasis (vs. Group A) at 3.20 for Group B, 7.32 for Group C, and 25.04 for Group D.
Understanding the uveal melanoma prognosis is an important driver for adjuvant therapies to prevent metastasis. Before TCGA data were released, a previous study of adjuvant sunitinib[24] for patients at a high-risk of metastases, defined as M3 and 8q gain or M3 and large tumor size[15] or gene expression profiling of Class 2,[25,26] revealed a better overall survival in the treatment arm, particularly if the patient was younger than 60 years of age. Currently, we employ TCGA classification for high-risk adjuvant trials, and those that meet the criteria for Group C or Group D are considered for adjuvant therapy.
There are limitations to this study including its retrospective nature and the rarity of uveal melanoma. However, in our practice of ocular oncology, we specialize in the management of uveal melanoma and have offered FNAB for DNA prognostication to all patients undergoing therapy, with subsequent monitoring of patient outcomes for decades. When we began our FNAB program for genetic testing in the mid-2000s, analysis was performed only on chromosome 3; thus the data from these patients could not be incorporated into TCGA as information on chromosome 8 is required. Nowadays, we sample for multiple chromosomes and our large cohort of 1001 eyes with uveal melanoma includes information on chromosomes 3 and 8 and provides a robust sample for validation of outcomes. Another limitation of this dataset is that outcomes for metastasis and death were per report by the patient, family, or physician. We realize that there can be gaps with this method of information collection, but this represents “real world” data analysis. Some metastatic and death events could be under-reported.
Conclusion
In conclusion, we have updated our database regarding prognostic classification of uveal melanoma by the 4-category TCGA and now provide 10-year outcomes for any metastasis and specifically for metastasis to the liver, lung, and other sites. We have shown that increasing TCGA grouping leads to a significantly (P < 0.001) increased risk for metastatic events and death over time. This classification system is practical and highly predictive of uveal melanoma metastatic risk.
Financial support and sponsorship
Support was provided in part by the Eye Tumor Research Foundation, Philadelphia, PA (CLS). The funders had no role in the design and conduct of the study, in the collection, analysis and interpretation of data, and in the preparation, review or approval of the manuscript. Carol L. Shields, M.D. has had full access to all the data in the study and takes responsibility for the integrity of the data.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA):An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–20. e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jager MJ, Brouwer NJ, Esmaeli B. The Cancer Genome Atlas Project:An integrated molecular view of uveal melanoma. Ophthalmology. 2018;125:1139–42. doi: 10.1016/j.ophtha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Vichitvejpaisal P, Dalvin LA, Mazloumi M, Ewens KG, Ganguly A, Shields CL. Genetic analysis of uveal melanoma in 658 patients using the cancer genome atlas classification of uveal melanoma as A, B, C, and D. Ophthalmology. 2019;126:1445–53. doi: 10.1016/j.ophtha.2019.04.027. [DOI] [PubMed] [Google Scholar]
- 6.Shields CL, Dalvin LA, Vichitvejpaisal P, Mazloumi M, Ganguly A, Shields JA. Prognostication of uveal melanoma is simple and highly predictive using The Cancer Genome Atlas (TCGA) classification:A review. Indian J Ophthalmol. 2019;67:1959–63. doi: 10.4103/ijo.IJO_1589_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazloumi M, Vichitvejpaisal P, Dalvin LA, Yaghy A, Ewens KG, Ganguly A, et al. Accuracy of The Cancer Genome Atlas (TCGA) classification versus American Joint Committee on Cancer (AJCC) classification for prediction of metastasis in patients with uveal melanoma. JAMA Ophthalmol. 2020;138:260–7. doi: 10.1001/jamaophthalmol.2019.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jager MJ, Shields CL, Cebulla CM, Abdel-Rahman MH, Grossniklaus HE, Stern MH, et al. Uveal melanoma [Primer] Nat Rev Dis Primers. 2020;6:24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 9.Damato B, Coupland SE. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol. 2009;127:423–9. doi: 10.1001/archophthalmol.2009.40. [DOI] [PubMed] [Google Scholar]
- 10.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–92. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 11.Ewens KG, Kanetsky PA, Richards-Yutz J, Al-Dahmash S, De Luca MC, Bianciotto CG, et al. Genomic profile of 320 uveal melanoma cases:Chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci. 2013;54:5721–9. doi: 10.1167/iovs.13-12195. [DOI] [PubMed] [Google Scholar]
- 12.Dogrusoz M, Jager MJ, Damato B. Uveal melanoma treatment and prognostication. Asia Pac J Ophthalmol (Phila) 2017;6:186–96. doi: 10.22608/APO.201734. [DOI] [PubMed] [Google Scholar]
- 13.Vaquero-Garcia J, Lalonde E, Ewens KG, Ebrahimzadeh J, Richard-Yutz J, Shields CL, et al. PRiMeUM:A model for predicting risk of metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2017;58:4096–105. doi: 10.1167/iovs.17-22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogrusoz M, Jager MJ. Genetic prognostication in uveal melanoma. Acta Ophthalmol. 2018;96:331–47. doi: 10.1111/aos.13580. [DOI] [PubMed] [Google Scholar]
- 15.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Personalized prognosis of uveal melanoma based on cytogenetic profile in 1059 patients over an 8-year period:The 2017 Harry S. Gradle Lecture. Ophthalmology. 2017;124:1523–31. doi: 10.1016/j.ophtha.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Cytogenetic abnormalities in uveal melanoma based on tumor features and size in 1059 patients:The 2016 W. Richard Green Lecture. Ophthalmology. 2017;124:609–18. doi: 10.1016/j.ophtha.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Shields CL, Kaliki S, Furuta M, Fulco E, Alarcon C, Shields JA. American Joint Committee on Cancer classification of uveal melanoma (tumor size category) predicts prognosis in 7731 patients. Ophthalmology. 2013;120:2066–71. doi: 10.1016/j.ophtha.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Shields CL, Kaliki S, Furuta M, Fulco E, Alarcon C, Shields JA. American Joint Committee on Cancer classification of uveal melanoma (anatomic stage) predicts prognosis in 7731 patients. The 2013 Zimmerman Lecture. Ophthalmology. 2015;122:1180–6. doi: 10.1016/j.ophtha.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 19.AJCC Ophthalmic Oncology Task Force. International validation of the American Joint Committee on Cancer's 7th ed.ition classification of uveal melanoma. JAMA Ophthalmol. 2015;133:376–83. doi: 10.1001/jamaophthalmol.2014.5395. [DOI] [PubMed] [Google Scholar]
- 20.Bagger M, Andersen MT, Andersen KK, Heegaard S, Andersen MK, Kiilgaard JF. The prognostic effect of American Joint Committee on Cancer staging and genetic status in patients with choroidal and ciliary body melanoma. Invest Ophthalmol Vis Sci. 2014;56:438–44. doi: 10.1167/iovs.14-15571. [DOI] [PubMed] [Google Scholar]
- 21.Dogrusoz M, Bagger M, van Duinen SG, Kroes WG, Ruivenkamp CA, Böhringer S, et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Invest Ophthalmol Vis Sci. 2017;58:833–42. doi: 10.1167/iovs.16-20212. [DOI] [PubMed] [Google Scholar]
- 22.Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, et al. Cytogenetics of uveal melanoma:A 7-year clinical experience. Ophthalmology. 2007;114:1925–31. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Berus T, Halon A, Markiewicz A, Orlowska-Heitzman J, Romanowska-Dixon B, Donizy P. Clinical, histopathological and cytogenetic prognosticators in uveal melanoma–A comprehensive review. Anticancer Res. 2017;37:6541–9. doi: 10.21873/anticanres.12110. [DOI] [PubMed] [Google Scholar]
- 24.Valsecchi ME, Orloff M, Sato R, Chervoneva I, Shields CL, Shields JA, et al. Adjuvant sunitinib in high-risk patients with uveal melanoma:Comparison with institutional controls. Ophthalmology. 2018;125:210–7. doi: 10.1016/j.ophtha.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, et al. Collaborative Ocular Oncology Group report number 1:Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry D, Seider M, Stinnett S, Mruthyunjaya P, Schefler AC Ocular Oncology Study Consortium. Relationship of clinical features and baseline tumor size with gene expression profile status in uveal melanoma:A multi-institutional study. Retina. 2019;39:1154–64. doi: 10.1097/IAE.0000000000002113. [DOI] [PubMed] [Google Scholar]