“Intelligence is based on how efficient a species became at doing the things they need to survive.” - Charles Darwin
Uveal melanoma is the most common primary malignant intraocular tumor. Radioactive plaque brachytherapy and proton beam radiation for most medium-sized tumors and enucleation for large-sized tumors provide impressive local tumor control. However, about half of the treated patients ultimately succumb to systemic metastasis, irrespective of the primary treatment modality.[1] Although therapeutic options for metastatic uveal melanoma are limited, patients and the treating medical teams may still desire reliable prognostic information to stratify the risk and personalize the management and surveillance plan.
Prognostic Factors in Uveal Melanoma
Patient demographic, clinical, and histopathological features have been traditionally used to determine the risk for metastasis of uveal melanoma. Older age, male gender, an association of oculocutaneous melanocytosis, ciliary body involvement, ring melanoma, larger tumor basal diameter and thickness, diffuse growth pattern, optic nerve involvement, invasion of the sclera, extraocular extension, closed periodic acid–Schiff-positive loops, degree of pigmentation, epithelioid cell type, high mitotic rate, inflammation, lymphocyte infiltration, macrophage infiltration, mean diameter of 10 largest nucleoli, higher expression of insulin-like growth factor-1 receptors, tumor necrosis, and vascular invasion are some of the factors associated with a relatively higher incidence of metastasis and tumor-related mortality.[2]
American Joint Committee on Cancer (AJCC) Tumor-Node-Metastasis (TNM) Staging of Uveal Melanoma
The AJCC TNM staging is conventionally used to prognosticate uveal melanoma.[3] In the eighth edition of AJCC, T is classified based on the largest basal diameter and thickness into four categories and then into 17 sub-categories based on ciliary body involvement and extraocular extension.[3] N is defined by regional lymph node metastasis, and M is dictated by distant metastasis and the size of the metastatic tumor.[3] The AJCC system further organizes possible combinations of T, N, and M into four prognostic stages, with increased risk for metastasis and mortality.[3] Histopathological staging includes spindle cell, mixed cell, and epithelioid cell types, but is not a part of the prognostic staging.[3] Uveal melanoma may not follow the stepwise anatomical progression on which the AJCC system is based. Early and nonlinear micrometastasis is unique to uveal melanoma, and thus, it can defy accurate and personalized prognostication by the AJCC system.
Prognostication of Uveal Melanoma Based on Tumor Genetics
The ability to perform reliable multiparametric genetic analysis using the fine-needle aspiration biopsy samples has been a paradigm change in the prognostication of uveal melanoma.[4,5,6,7] Onken et al.[8] found that tumors cluster into two groups: Class 1, having a good prognosis, and Class 2, with a higher rate of metastasis and disease-related mortality. In 2010, Damato et al.[9] advocated the use of chromosomes 1, 3, 6, and 8 abnormalities to determine the prognosis. There is evidence that monosomy 3 is associated with an increased risk of metastasis,[6] loss-of-function mutations in BAP1 located on 3p21 is associated with monosomy 3,[10] and consequently decreased BAP1 mRNA and protein expression predict metastasis.[11] Currently, monosomy 3 or a 15-gene microarray-based panel is used to determine the prognosis.[12] There have been attempts to enhance the prognostic value of AJCC staging by co-opting genetic parameters.[13,14] Personalized risk estimation has been attempted by the online tools – Liverpool Uveal Melanoma Prognosticator Online (LUMPO)[13] and Predicting Risk of Metastasis in Uveal Melanoma (PriMeUM).[15]
The National Cancer Institute Center for Cancer Genomics and the National Human Genome Research Institute-initiated The Cancer Genome Atlas (TCGA) project data is based on integrated multidimensional molecular–computational characterization of 80 primary uveal melanoma. This could help categorize four molecularly distinct, clinically relevant subtypes based on the presence or absence of monosomy 3 and the presence and degree of 8q gain. The best prognostic class (class A) demonstrated disomy 3 and 8, and subsequent classes showed disomy 3 and 8q gain (class B), monosomy 3 and 8q gain (class C), and monosomy 3 and multiple 8q gains (class D) correlating with increased risk of metastasis [Fig. 1].[15] Jager et al.[16] provided clarity to the four main prognostic categories of uveal melanoma as identified by TCGA [Table 1]. The superiority of the simplified TGCA classification over AJCC in predicting the risk of metastasis at 5 years has been established.[17] Published in this issue of Indian Journal of Ophthalmology, Shields et al.[18] have provided evidence that a simplified four-category tumor genetics-based classification of uveal melanoma using TCGA is highly predictive of the risk of metastasis at 10 years.
Figure 1.
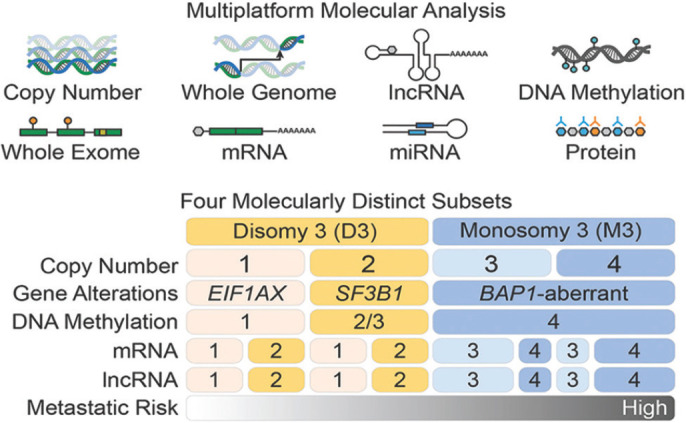
Comprehensive multi-platform analysis of uveal melanoma unravels four molecularly distinct, clinically relevant subtypes: two associated with poor-prognosis monosomy 3 and two with better-prognosis disomy 3. Two subsets of disomy 3 are based on EIF1AX or SF3B1 mutations with consequent somatic copy number alterations and DNA methylation profiles. BAP1 aberration and global DNA methylation are seen to be associated with monosomy 3. Two subsets of monosomy 3 are based on genomic aberrations and transcriptional features. There is a linear increase in metastatic risk from subtype 1 through 4. Reproduced with permission from Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204-220
Table 1.
| Group | Genetic/molecular Profile | Prognosis |
|---|---|---|
| Group A | mRNA class 1, infrequent chromosomal aberrations, disomy 3, extra 6p, normal 8q, no inflammation | Favorable |
| Group B | mRNA class 1, infrequent chromosomal aberrations, disomy 3, extra 6p, partial extra 8q, no inflammation | Less favorable |
| Group C | mRNA class 2, frequent chromosomal aberrations, monosomy 3, extra 8q, some inflammation | Unfavorable |
| Group D | mRNA class 2, frequent chromosomal aberrations, monosomy 3, >>extra 8q, severe inflammation | Unfavorable |
Adapted from Jager MJ, Brouwer NJ, Esmaeli B. The Cancer Genome Atlas Project: an integrated molecular view of uveal melanoma. Ophthalmology. 2018;125:1139-1142
Tumor genetics-based simplified classification of uveal melanoma[16] appears to dovetail well with prognosis for metastasis and may help in selecting categories of patients for tight surveillance to enable early detection of metastasis, as well as in designing and customizing adjuvant therapies. The future AJCC staging systems may propose a simplified approach incorporating the important genetic parameters into clinicopathological criteria. Meanwhile, we owe it to our patients to build tumor genetics inseparably into the management algorithm of uveal melanoma.
“Prediction is very difficult” said Niels Bohr, and we must agree, but armed with the vital information that tumor genetics provides, we may get closer to reality.
References
- 1.Damato B. Progress in the management of patients with uveal melanoma. The 2012 Ashton Lecture. Eye. 2012;26:1157–72. doi: 10.1038/eye.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill HS, Char DH. Uveal melanoma prognostication:From lesion size and cell type to molecular class. Can J Ophthalmol. 2012;47:246–53. doi: 10.1016/j.jcjo.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 3.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual:Continuing to build a bridge from a population-based to a more “personalized“approach to cancer staging. CA Cancer J Clin. 2017;67:93–9. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 4.Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, et al. Cytogenetics of uveal melanoma:A 7-year clinical experience. Ophthalmology. 2007;114:1925–31. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Shields JA, Shields CL, Materin M, Sato T, Ganguly A. Role of cytogenetics in management of uveal melanoma. Arch Ophthalmol. 2008;126:416–9. doi: 10.1001/archopht.126.3.416. [DOI] [PubMed] [Google Scholar]
- 6.Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2010;118:396–401. doi: 10.1016/j.ophtha.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 7.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Cytogenetic abnormalities in uveal melanoma based on tumor features and size in 1059 patients:The 2016 W. Richard Green lecture. Ophthalmology. 2017;124:609–18. doi: 10.1016/j.ophtha.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–92. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 10.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111:1373–80. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014;1102:427–40. doi: 10.1007/978-1-62703-727-3_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eleuteri A, Damato B, Coupland SE, Taktak AF. Enhancing survival prognostication in patients with choroidal melanoma by integrating pathologic, clinical and genetic predictors of metastasis. Int J Biomed Eng Technol. 2012;8:18–35. [Google Scholar]
- 14.Bagger M, Andersen MT, Andersen KK, Heegaard S, Andersen MK, Kiilgaard JF. The prognostic effect of American Joint Committee on Cancer staging and genetic status in patients with choroidal and ciliary body melanoma. Invest Ophthalmol Vis Sci. 2014;56:438–44. doi: 10.1167/iovs.14-15571. [DOI] [PubMed] [Google Scholar]
- 15.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–20. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jager MJ, Brouwer NJ, Esmaeli B. The Cancer Genome Atlas project:An integrated molecular view of uveal melanoma. Ophthalmology. 2018;125:1139–42. doi: 10.1016/j.ophtha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Mazloumi M, Vichitvejpaisal P, Dalvin LA, Yaghy A, Ewens KG, Ganguly A, et al. Accuracy of The Cancer Genome Atlas classification vs American Joint Committee on Cancer classification for prediction of metastasis in patients with uveal melanoma. JAMA Ophthalmol. 2020;138:260–7. doi: 10.1001/jamaophthalmol.2019.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields CL, Mayro EL, Bas Z, Dockery PW, Yaghy A, Lally SE, et al. Ten-year outcomes of uveal melanoma based on The Cancer Genome Atlas (TCGA) classification in 1001 cases. Indian J Ophthalmol. 2021;69:1839–45. doi: 10.4103/ijo.IJO_313_21. [DOI] [PMC free article] [PubMed] [Google Scholar]


