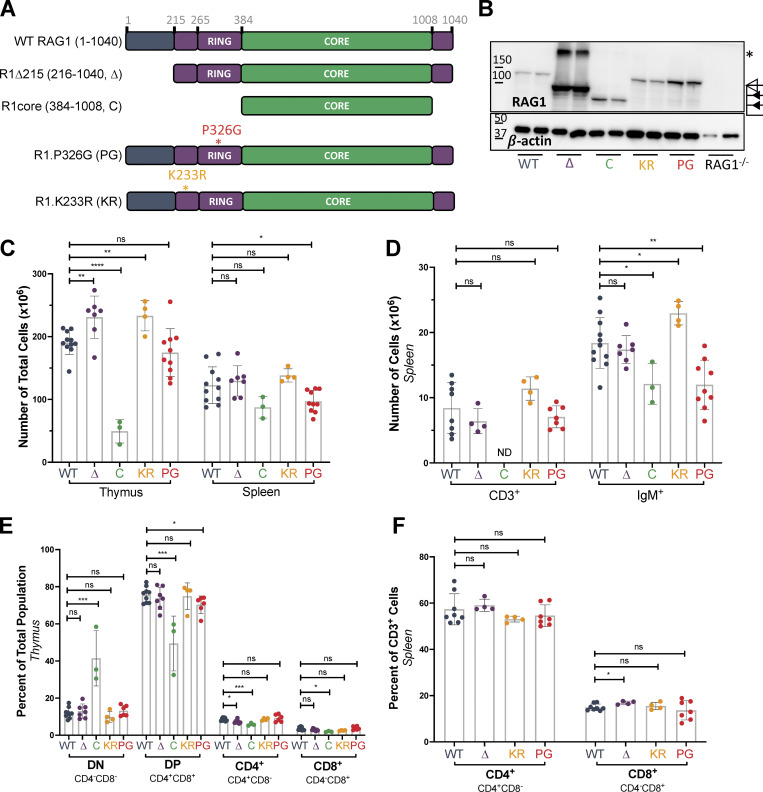
Studies of novel RAG1 N-terminal region mutants reveal a role for the first 215 amino acids in establishing the balance between short- and long-range V(D)J recombination and a distinct role for RAG1 ubiquitin ligase activity in promoting efficient recombination and lymphocyte development.
Abstract
Immunoglobulin and T cell receptor gene assembly depends on V(D)J recombination initiated by the RAG1-RAG2 recombinase. The RAG1 N-terminal region (NTR; aa 1–383) has been implicated in regulatory functions whose influence on V(D)J recombination and lymphocyte development in vivo is poorly understood. We generated mice in which RAG1 lacks ubiquitin ligase activity (P326G), the major site of autoubiquitination (K233R), or its first 215 residues (Δ215). While few abnormalities were detected in R1.K233R mice, R1.P326G mice exhibit multiple features indicative of reduced recombination efficiency, including an increased Igκ+:Igλ+ B cell ratio and decreased recombination of Igh, Igκ, Igλ, and Tcrb loci. Previous studies indicate that synapsis of recombining partners during Igh recombination occurs through two pathways: long-range scanning and short-range collision. We find that R1Δ215 mice exhibit reduced short-range Igh and Tcrb D-to-J recombination. Our findings indicate that the RAG1 NTR regulates V(D)J recombination and lymphocyte development by multiple pathways, including control of the balance between short- and long-range recombination.
Introduction
The diversity of jawed vertebrate adaptive immune responses depends on programmed assembly and hypermutation of antigen receptor (AgR) genes (Cooper and Alder, 2006). The first AgR gene diversification process to occur in developing lymphocytes is V(D)J recombination, which assembles Ig and TCR genes from arrays of V, in some cases D, and J gene segments (Schatz and Swanson, 2011). V(D)J recombination is initiated by the endonuclease RAG, a heterotetramer made up of RAG1 and RAG2. RAG binds to and cleaves DNA at recombination signal sequences (RSSs) that flank rearranging gene segments. RAG-mediated DNA cleavage requires the synapsis of two RSSs and leads to simultaneous generation of DNA double-strand breaks (DSBs) at the two sites (Schatz and Swanson, 2011). The DNA ends are processed and ligated by nonhomologous end joining repair factors (Rooney et al., 2004).
RAG1 is the primary DNA-binding and cleaving component of RAG, while RAG2 is an essential accessory factor. The minimal portion of RAG1 required for RSS binding and cleavage is called the “core” domain (R1core), consisting of aa 384–1008 of the 1040-aa protein (numbering according to mouse RAG; Fig. 1 A; Sadofsky et al., 1993; Silver et al., 1993). RAG1 also contains two noncore regions, one at the N-terminus (aa 1–383; hereafter RAG1 N-terminal region [NTR]) and a short C-terminal tail (aa 1009–1040) that modulates RAG cleavage activity in vitro (Grundy et al., 2010; Kim et al., 2015) and is not a focus of this study. Mice expressing R1core exhibit a partial block in early B and T cell development, reduced numbers of mature lymphocytes, an altered Vβ gene segment repertoire in Tcrb recombination events, and an increase in aberrant V(D)J recombination products, demonstrating the importance of RAG1 noncore regions (Dudley et al., 2003; Horowitz and Bassing, 2014; Talukder et al., 2004). The functional significance of the RAG1 NTR is highlighted by the wide array of atypical SCID-like phenotypes found in human patients with mutations in this region (Notarangelo et al., 2016). While multiple regulatory functions have been associated with the RAG1 NTR, largely through in vitro and cell line studies, little is known about its roles in the context of lymphocyte development and AgR repertoire formation in vivo (Jones and Simkus, 2009).
Figure 1.
RAG1 NTR regulates RAG1 protein levels and has minimal effect on T cell development. (A) Schematic of RAG1 mutants. Figure not to scale. (B) Western blot of RAG1 in whole thymic lysates from mice 4–6 wk of age. Open triangle, full-length protein; closed triangles, truncated proteins. Asterisk, band approximately twice the molecular weight of R1Δ215 that likely represents a RAG1 dimer resistant to dissociation, as observed previously (Leu and Schatz, 1995). (C) Total numbers of nucleated cells in thymus and spleen. (D) Numbers of live CD3+ T cells or IgM+ B cells in the spleen. (E) Percentages of DN (CD4−CD8−), DP (CD4+CD8+), CD4+CD8−, and CD4−CD8+ thymocytes in whole thymuses. (F) Percentages of CD4+CD8− and CD4−CD8+ T cells among live CD3+ cells in the spleen. Data are presented as mean with error bars indicating SEM. Statistical significance determined by two-tailed Welch’s t test (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Distinct functions have been attributed to different portions of the RAG1 NTR. RAG1 aa 287–351 constitute a zinc–RING finger domain that binds four zinc atoms and possesses ubiquitin ligase activity (Jones and Gellert, 2003; Yurchenko et al., 2003). Mutation of the zinc-coordinating residue C325 (C325Y) abrogates ubiquitin ligase activity, dramatically reduces recombination activity, disrupts protein tertiary structure, and almost completely blocks lymphocyte development in mice (Deng et al., 2015; Simkus et al., 2007). The corresponding mutation in human RAG1 (C328Y) also results in severe B and T cell lymphopenia (Villa et al., 2001). However, because this mutation disrupts RAG1 folding and structure (Simkus et al., 2007), it is difficult to determine the contribution of ubiquitin ligase activity per se to the phenotypes observed (Deng et al., 2015). Mutation of a proline residue within the RING domain to glycine (P326G) disrupts ubiquitin ligase activity with only minor perturbation to the structural integrity of the domain and modestly reduces recombination of extrachromosomal plasmid substrates (Simkus et al., 2007). Numerous targets of RAG1-mediated ubiquitination have been identified in vitro, including the nuclear import factor KPNA1, histone H3, and histone variant H3.3, with histone ubiquitination implicated in recruitment of post-cleavage repair proteins (Deng et al., 2015; Grazini et al., 2010; Jones et al., 2011; Kassmeier et al., 2012; Simkus et al., 2009). In addition, RAG1 autoubiquitination, primarily at residue K233, stimulates RAG cleavage activity in vitro and has been implicated in increased recombination of extrachromosomal substrates in transfected cells (Singh and Gellert, 2015). A region mapping to the first ∼200 aa of RAG1 is involved in the down-regulation of RAG1 protein levels through interactions with VprBP, a subunit of the cullin E3 ubiquitin ligase complex (Kassmeier et al., 2012; Schabla et al., 2018). The RAG1 NTR also regulates RAG1 nucleolar sequestration, with aa 243–249 required for nucleolar import and aa 1–215 required for release from the nucleolus and efficient V(D)J recombination (Brecht et al., 2020).
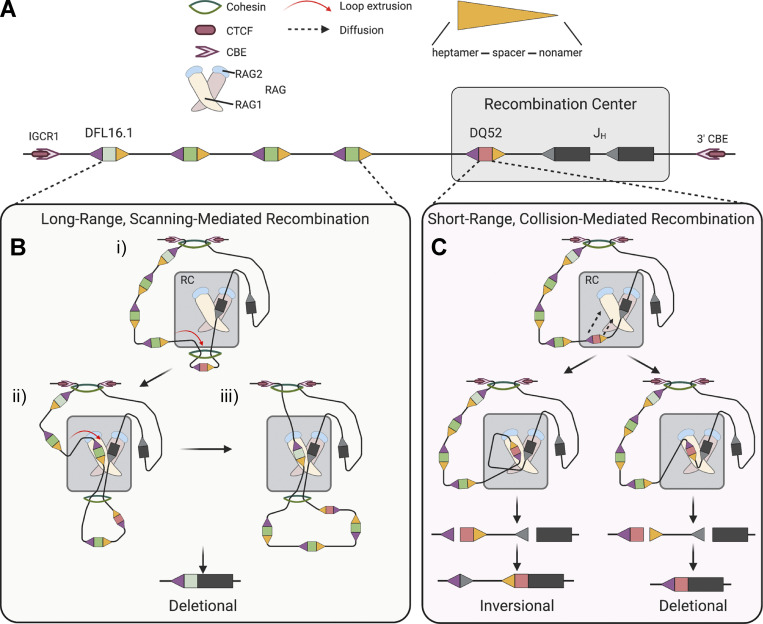
At AgR loci, RAG accumulates in recombination centers (RCs) encompassing J and J-proximal D gene segments (Schatz and Ji, 2011). RAG is thought to first bind to an RSS within the RC, after which a partner RSS is brought into the RC for synapsis and cleavage, a process long assumed to involve chromatin looping and random collision. Recently however, a model of RAG chromatin scanning has emerged to explain partner RSS capture, based initially on the observation of a strong orientation bias for recombination to cryptic RSSs located >5–10 kb away from a reference RSS (Hu et al., 2015). Subsequent analyses of Ig heavy chain gene (Igh) recombination provided strong evidence that V(D)J recombination can occur through a mechanism involving RAG chromatin scanning driven by cohesin-dependent loop extrusion (Ba et al., 2020; Dai et al., 2021; Hill et al., 2020; Jain et al., 2018; Zhang et al., 2019). Short-range recombination events between gene segments within the Igh RC were shown to be cohesin independent and governed largely by RSS sequence and not RSS orientation (Ba et al., 2020; Zhang et al., 2019). In contrast, longer-range recombination events were found to require the cohesin component Rad21 and a convergent orientation of the recombining RSSs (Ba et al., 2020; Dai et al., 2021; Jain et al., 2018; Zhang et al., 2019) and to be regulated by the chromatin looping factor CCCTC-binding factor (CTCF; Ba et al., 2020), CTCF-binding elements (CBEs; Ba et al., 2020; Guo et al., 2011; Jain et al., 2018; Lin et al., 2015), and the cohesin loader WAPL (Dai et al., 2021; Hill et al., 2020). Together, these data strongly support the existence of two distinct modes of recombination that differ in the mechanism leading to RSS synapsis—short-range recombination within or close to an RC that involves random RSS collision and long-range scanning driven by cohesin-mediated loop extrusion. It is unknown whether the RAG proteins exert control over the choice between these two modes of recombination.
To study the in vivo roles of the RAG1 NTR in V(D)J recombination and lymphocyte development, we generated and characterized mice in which RAG1 lacks ubiquitin ligase activity (P326G), the major site of autoubiquitylation (K233R), or its first 215 aa (Δ215). We found that RAG1 ubiquitin ligase activity, the K233 autoubiquitination site, and the first 215 aa help restrict RAG1 protein levels. We show that the P326G, but not the K233R, mutation results in impaired B cell development and decreased Igh, Igκ, Igλ, and Tcrb recombination, with no evidence that RAG1 or its ubiquitin ligase activity regulates AgR locus accessibility, arguing that RAG1 ubiquitin ligase activity stimulates recombination efficiency. We also demonstrate that the RAG1 NTR regulates the balance between short- and long-range recombination, with the first 215 aa facilitating short-range D-to-J recombination. Overall, our findings identify multiple, distinct roles played by the RAG1 NTR in regulating V(D)J recombination and lymphocyte development.
Results
Post-transcriptional regulation of RAG1 protein levels by the RAG1 NTR
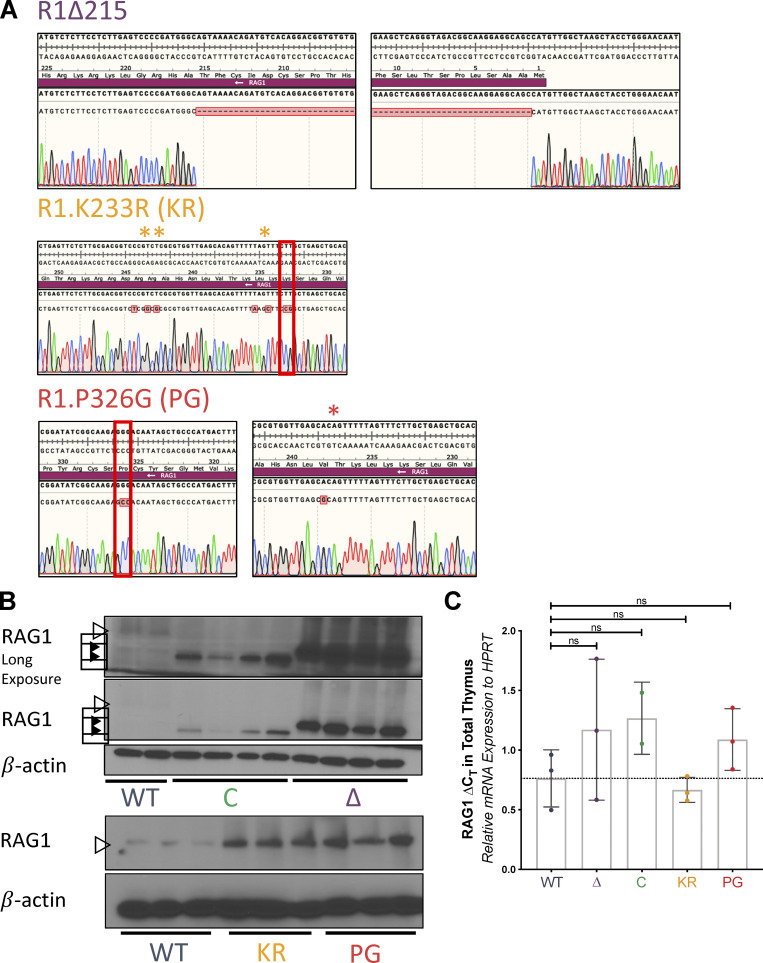
To assess the function of the RAG1 NTR in vivo, we generated and analyzed three mouse lines on the C57BL/6 background with alterations in previously identified functional elements of the NTR. The RAG1 NTR is composed of two subregions, with the first 215 aa contributing to protein stability and intranuclear trafficking and the remainder, aa 216–383, consisting of a basic region harboring the major autoubiquitination site and a zinc–RING finger domain with ubiquitin ligase activity (Fig. 1 A; Brecht et al., 2020; Jones and Gellert, 2003; Jones and Simkus, 2009; Kassmeier et al., 2012; Yurchenko et al., 2003). To distinguish the contributions of these two subregions, a mouse line lacking the first 215 aa was generated (R1Δ215; Fig. 1 A and Fig. S1 A). The role of RAG1-mediated ubiquitination was assessed in a mouse line in which RAG1 lacks ubiquitin ligase activity (R1.P326G, hereafter R1.PG), while the function of autoubiquitination was evaluated using a mouse line in which the major site of RAG1 autoubiquitination was mutated (R1.K233R, hereafter R1.KR; Fig. 1 A and Fig. S1 A; Jones and Gellert, 2003; Simkus et al., 2007).
Figure S1.
RAG1 protein and mRNA expression in WT and RAG1 mutant mice. (A) Sequencing tracks of PCR products amplified from the genomes of RAG1 NTR mutant mice. Red boxes indicate site of indicated mutation. Single asterisk indicates K234 and L235, where silent mutations were introduced to disrupt PAM site in R1.KR mice. Double asterisks indicate R243 and R244, where silent mutations were introduced to add a site for HindIII for genotyping purposes. The red asterisk indicates a passenger V238A mutation in R1.PG mice. This mutation is conservative, and V238 is not highly conserved and is replaced by alanine or isoleucine in several jawed vertebrates (Fig. S4 B from Brecht et al., 2020). It is not predicted to have an impact on RAG activity. (B) Western blot of RAG1 in whole thymic lysates from additional independent WT and RAG1 mutant mice. Open triangles, full-length protein; closed triangles, truncated proteins. (C) RAG1 mRNA expression in whole thymic lysates from mice of indicated genotypes, calculated relative to that of HPRT. Each dot indicates data from an independent mouse, with data presented as mean ± SEM. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; * P, ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Western blots of whole thymic lysates revealed elevated RAG1 protein levels in all RAG1 mutant strains compared with WT, with levels dramatically elevated in R1Δ215 (Fig. 1 B and Fig. S1 B). The increase in protein levels was not due to changes in RAG1 mRNA levels (Fig. S1 C). Elevated levels of RAG1 protein in R1.KR and R1.PG thymocytes are consistent with the hypothesis that autoubiquitination serves to down-regulate RAG1 protein levels (Singh and Gellert, 2015), while increased levels of the R1Δ215 protein are consistent with loss of VprBP-cullin–mediated protein degradation (Kassmeier et al., 2012; Schabla et al., 2018). R1Δ215, R1.PG, and R1.KR mice could be maintained to at least 18 mo of age without overt evidence of pathologies (minimum of five mice per genotype), indicating that their elevated RAG1 protein levels do not severely compromise life span or physiology.
RAG1 ubiquitin ligase activity is required for efficient Igκ and Igλ recombination and production of normal numbers of Igλ+ B cells
To assess the role of the RAG1 NTR in T and B cell development, lymphocyte subpopulations were analyzed by flow cytometry in thymus, bone marrow, and spleen in WT and RAG1 mutant mouse strains. Gross αβ T cell development was largely unperturbed in R1Δ215, R1.PG, and R1.KR mice: thymocyte and splenic T cell numbers and percentages were either normal or slightly elevated (Fig. 1, C, D, and F; and Fig. S2, A and B), and percentages of CD4−/CD8− double-negative (DN) and CD4+/CD8+ double-positive (DP) thymocytes were not different from those in WT mice, with the exception of a small (∼5%) decrease in DP thymocytes in R1.PG mice (Fig. 1 E; Burn et al., 2021 Preprint). As expected, R1core mice displayed reduced thymocyte numbers and a relative accumulation of DN thymocytes and reduction in DP thymocytes (Fig. 1, C and E; Dudley et al., 2003).
Figure S2.
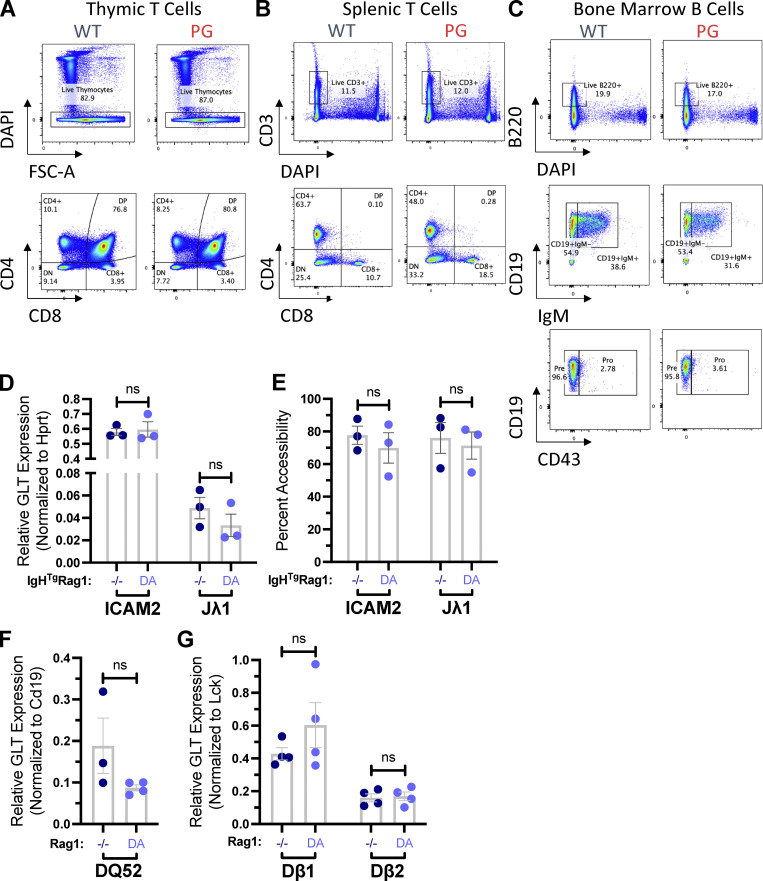
Flow cytometry analyses of B and T cell development in WT and R1.PG mice and characterization of germline transcripts in developing lymphocytes in WT and R1.D708A mice. (A–C) Representative flow cytometry data of WT and R1.PG mice showing the populations from thymic T cells (A), splenic T cells (B), and bone marrow B cells (C). (D–G) Quantification of RNA transcripts (D, F, and G) or DNA accessibility (E) of ICAM2 or Jλ1 (D and E), DQ52 (F), or Dβ1 and Dβ2 (G) conducted on samples from three or four independent samples of sorted preB cells (D and E), CD19+ cells from total bone marrow (F), or total thymocytes (G) from RAG1−/− and R1.D708A (DA) mice with (D and E) or without (F) an IgH transgene. Signals from each assay were normalized to values from assay of the HPRT (D), CD19 (F), or Lck (G) gene. Shown are mean values ± SEM. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). FSC-A, forward scatter-A.
B cell development was largely unperturbed in R1Δ215 and R1.KR mice, with normal numbers of pro-, pre-, and immature B cells in the bone marrow (although the percentages of preB and immature B cells were slightly reduced in R1.KR mice) and normal or somewhat elevated numbers of IgM+ cells in the spleen (Fig. 1 D; and Fig. 2, A and B). In contrast, numbers of bone marrow preB and immature B cells and of mature splenic B cells were reduced in R1.PG mice (Fig. 1 D and Fig. 2 A), with the reduction most evident in the immature B cell population, which also exhibited a relative decline (Fig. 2, A and B; and Fig. S2 C). Furthermore, the Igκ/Igλ ratio on IgM+ bone marrow and splenic B cells was significantly elevated in R1.PG and RAG1core, but not in R1Δ215 or R1.KR, mice relative to WT mice (Fig. 2 D). This elevated Igκ/Igλ ratio is due to decreased genesis of Igλ B cells in the bone marrow, which is also reflected in the spleen (Fig. 2, C–F). Given the normal parameters of B cell development in R1.KR mice, the RAG1 ubiquitin ligase domain appears to perturb B cell development by a mechanism that is independent of RAG1 autoubiquitination.
Figure 2. .
RAG1 ubiquitin ligase promotes Igλ+ B cell development. (A) Numbers of pro-, pre-, and immature B cells in bone marrow from mice 4–6 wk of age. (B) Percentages of pro-, pre-, and immature B cells among live B220+ cells in the bone marrow. (C) Representative flow cytometry data showing the populations of B220+IgM+ B cells of the Igκ or Igλ lineage in bone marrow or spleen of WT and R1.PG mice. (D) Igκ:Igλ ratio in bone marrow and spleen. Calculated by taking exact number of IgM+ Igκ+ divided by exact number of IgM+ Igλ+ cells. All analyses were done from organs isolated from mice aged 4–6 wk. Each dot in B and D indicates data from an independent mouse, with data presented as mean ± SEM. (E and F) Graphed flow cytometry data depicting the frequency or number of B220+IgM+ bone marrow (E) or spleen (F) cells that express Igλ on their surface. Shown are individual data points collected from three mice from each genotype, with the average ± SEM. Statistical significance determined by two-tailed Welch’s t test (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Igλ B cells develop from preB cells that fail to generate a nonautoreactive B cell receptor from Igκ recombination and proceed to Igλ recombination (Gorman and Alt, 1998). In preB cells, RAG-mediated DSBs up-regulate expression of the Pim2 kinase (Bednarski et al., 2012), whose activity enhances preB cell survival, thereby facilitating Igλ recombination and development of Igλ B cells (Derudder et al., 2009). The decreased production of Igλ B cells and reduced numbers of developing and mature B cells in R1.PG mice are consistent with decreased efficiency of V(D)J recombination, a specific impairment in Igλ rearrangement, and/or diminished survival of preB cells. To investigate V(D)J recombination, we conducted Taqman PCR to quantify Igκ and Igλ rearrangements in sorted preB cells of WT and R1.PG mice. Recombination of both loci is reduced in R1.PG mice relative to WT (Fig. 3 A). The decrease in Igλ recombination is somewhat more pronounced than at Igκ, which might reflect the ordered nature of Igκ and Igλ rearrangements rather than a specific defect in Igλ recombination.
Figure 3.
RAG1 ubiquitin ligase stimulates Igκ and Igλ gene recombination. (A) Taqman qPCR quantification of Vκ-Jκ1 or Vλ1-Jλ1 recombination conducted on genomic DNA from sorted preB cells. Signals from each sample were normalized to values from an assay for the invariant CD19 gene. Shown are the average values ± SEM from three independent isolations of preB cells. Statistical significance determined by two-tailed unpaired t test. (B–E) Characterization of the cellular response to RAG cleavage of Igκ alleles in primary preB cell cultures 48 h after IL-7 withdrawal. (B) Taqman qPCR quantification of intact germline Jκ1 segment calculated by dividing germline Jκ1 signals after IL-7 withdrawal by signals before IL-7 withdrawal. CD19 was used to normalize the input DNA. (C) Quantitative real-time–PCR quantification of the induction of Pim2 transcripts calculated by dividing mRNA signals after IL-7 withdrawal by signals before IL-7 withdrawal. CD19 was used to normalize input cDNA. (D) Western quantification of the induction of Pim2 protein calculated by dividing combined signals of all three Pim2 isoforms after IL-7 withdrawal by signals before IL-7 withdrawal. (E) Representative Western blot with the locations of each Pim2 isoform and a nonspecific band (asterisk) indicated. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05).
We considered that RAG1 ubiquitin ligase activity might stimulate recombination by driving transcription-associated chromatin accessibility. To investigate this possibility, we analyzed Igh, Igλ, and Tcrb locus transcriptional accessibility in developing lymphocytes lacking RAG1 protein or expressing a DNA cleavage-incompetent R1.D708A mutant (Ji et al., 2010). We observed equivalent levels of Jλ1 germline transcripts and genome accessibility (Fig. S2, D and E) in cells of each genotype. Similarly, we observed equivalent levels of germline transcripts in RAG1−/− and R1.D708A mice in the Igh RC in proB cells and in the two Tcrb RCs in DN thymocytes (Fig. S2, F and G). These data demonstrate that parameters of accessibility are not altered by the absence of RAG1 and argue that R1.PG and the other RAG1 mutants under study are unlikely to affect recombination by altering locus accessibility.
To determine whether R1.PG preB cells exhibit decreased induction of Pim2 mRNA and protein, we used an approach that isolates preB cell responses to RAG cleavage of Igκ loci (Bednarski et al., 2012). The approach involves culturing preB cells from mice lacking the Artemis DSB repair factor and containing the anti-apoptotic EμBCL2 transgene and an IgH transgene that drives development of preB cells. Withdrawal of the IL-7 cytokine initiates Igκ recombination, which leads to accumulation of RAG DSBs at Igκ loci and signals from these lesions. We generated EμBCL2:IgH:Artemis−/− mice on both the WT RAG1 (BIA) and R1.PG (BIAPG) backgrounds and analyzed preB cells cultured from each genotype. Consistent with our recombination TaqMan PCR data, we detected a modest increase in germline Jκ1, indicating a reduction of RAG Igκ cleavage, in BIAPG cells (Fig. 3 B). Despite this decreased RAG cleavage, BIAPG cells exhibited no difference from BIA cells in up-regulation of Pim2 transcripts or protein (Fig. 3, C–E). Together, our data indicate that RAG1 ubiquitin ligase activity is required for normal efficiency of Igκ and Igλ recombination and that loss of ubiquitin ligase activity impairs the progression from Igκ to Igλ recombination in preB cells and the development of B cells through subsequent stages.
Alterations in Igh repertoire in R1Δ215 and R1.PG but not R1.KR mice
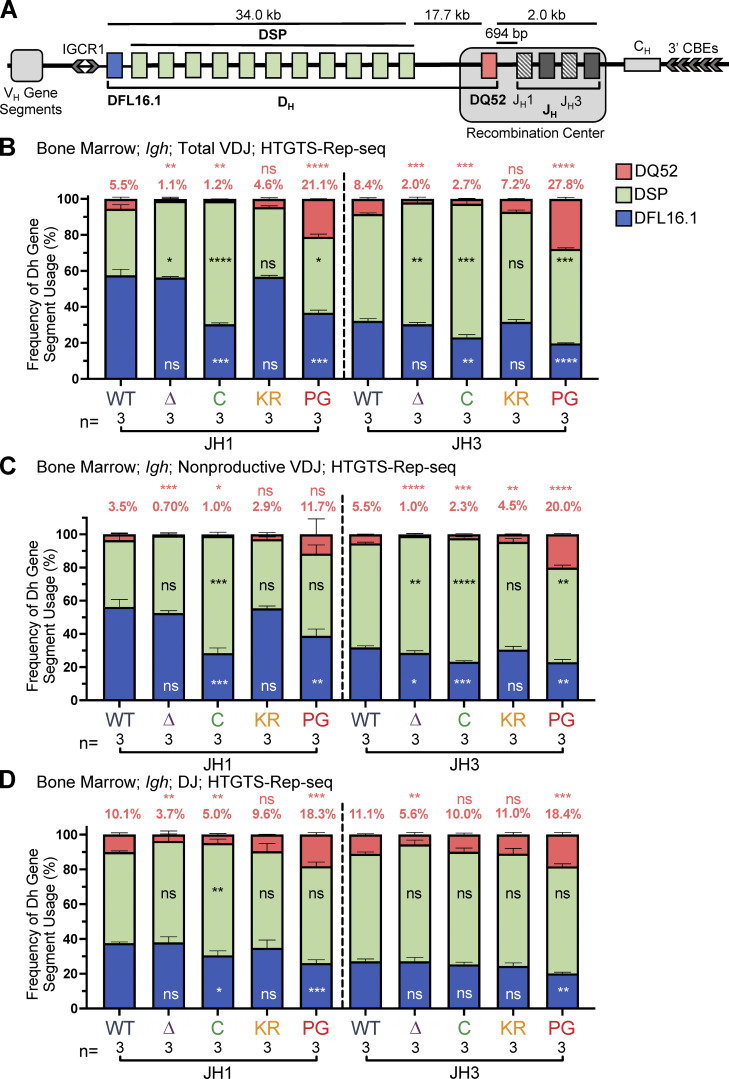
The murine Igh locus spans 2.7 Mb, with more than 100 VH gene segments located ∼100 kb upstream of 13 DH and 4 JH gene segments (Fig. 4 A). Igh recombination is initiated in proB cells by recruitment of RAG to the Igh RC, which encompasses the JH segments and the most downstream DH gene segment, DQ52 (Fig. 4 A; Ji et al., 2010; Teng et al., 2015). DQ52-to-JH recombination occurs through a topologically unconstrained collision-based mechanism (Zhang et al., 2019), whereas recombination of other DH gene segments to JH involves a distinct mechanism in which a JH-bound RAG complex scans chromatin, driven by cohesin-mediated loop extrusion (Ba et al., 2020; Zhang et al., 2019). JH-bound RAG chromatin scanning is arrested by the CBE-containing element IGCR1, which lies immediately upstream of DFL16.1 (Fig. 4 A), explaining why DFL16.1 is the most frequently used DH gene segment (Ba et al., 2020; Jain et al., 2018; Zhang et al., 2019).
Figure 4.
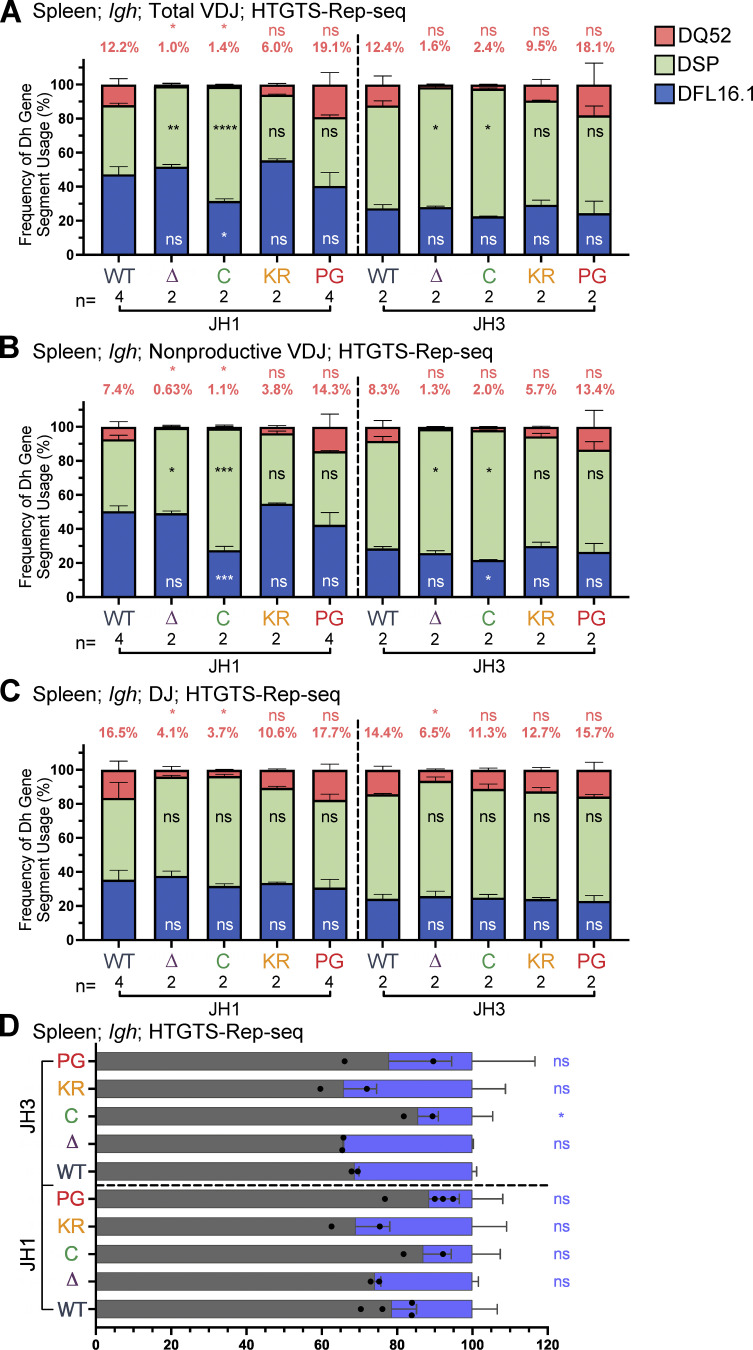
DH gene segment usage in Igh rearrangements in bone marrow assessed by HTGTS–Rep-seq. (A) Schematic of murine Igh with the RC containing the JH-proximal DQ52 gene segment and four JH segments highlighted. Figure not to scale. Chevrons indicate the orientation of CBEs. CBEs in VH portion of locus not depicted. (B–D) Igh HTGTS–Rep-seq data from mice 5–7 wk of age were analyzed for frequency of DFL16.1 (blue), DQ52 (salmon), and intervening 11 DH gene segments (green) in total (B), nonproductive VDJH (C), or DJ recombination events (D) with JH1 or JH3 in bone marrow. Percentage of DQ52 usage is indicated above each bar. n, number of mice analyzed per genotype. Number of recombination events analyzed can be found in Table S4, Table S5, and Table S6. Data are presented as mean with error bars indicating SEM. Statistical significance relative to WT determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). DSP, intervening DH gene segments considered as a group.
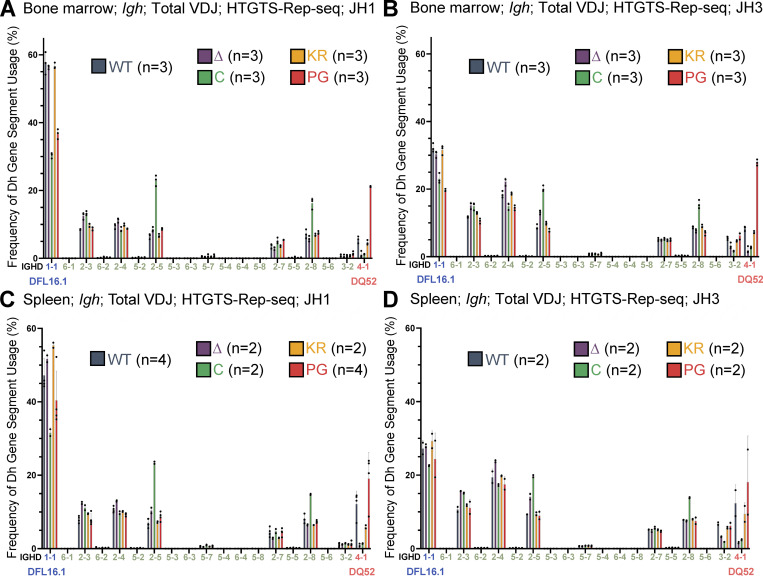
To investigate the possibility that the RAG1 NTR mutations alter gene segment usage, we used high-throughput genome-wide translocation sequencing–adapted repertoire sequencing (HTGTS–Rep-seq) to characterize the Igh repertoire in B220+IgM− bone marrow B-lineage cells (predominantly preB cells) and B220+ spleen cells (predominantly naive, mature B cells) using JH1 and JH3 as baits (Lin et al., 2016). This approach reveals the repertoire of VH and DH gene segments that become joined to these JH gene segments. We did not observe major alterations in VH gene segment usage in the RAG1 NTR mutant mice generated for this study. Thus, we focus here on the DH gene segment repertoire, with usage frequency presented for DQ52, DFL16.1, and the intervening DH gene segments considered as a group (DSP; Fig. 1 A). The full dataset of VH and DH gene segment usage is provided in Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6.
R1Δ215 mice exhibited a striking decrease of up to ∼10-fold in the frequency of usage of DQ52 in rearrangements involving JH1 and JH3 and in both the bone marrow and spleen relative to WT mice (data for bone marrow and spleen are shown in Fig. 4, B–D; and Fig. S3, A–C, respectively). This was true in total VDJ rearrangements, nonproductive (VDJ−) rearrangements, and DJ rearrangements. Data for productive (VDJ+) rearrangements closely resemble those for total VDJ rearrangements and are not shown. In contrast, usage of DFL16.1 was not substantially or consistently altered in R1Δ215 relative to WT mice (Fig. 4, B–D; and Fig. S3, A–C). DFL16.1 was the most frequently used DH gene segment in all genotypes analyzed, with usage higher in JH1 than JH3 rearrangements, as reported previously (Lin et al., 2016; Fig. S4). We conclude that deletion of the first 215 aa of RAG1 strongly reduces DQ52 usage and that this phenotype arises at the step of DH-to-JH rearrangement and is not due to selection for productive rearrangements.
Figure S3.
DH gene segment usage and analysis of productive and nonproductive rearrangements in VDJH rearrangements in spleen assessed by HTGTS–Rep-seq. (A–C) Igh HTGTS–Rep-seq data from mice 5–7 wk of age were analyzed for frequency of DFL16.1 (blue), DQ52 (salmon), and intervening 11 DH gene segments (green) in total (A) or nonproductive (B) VDJH or DJ recombination events (C) with JH1 or JH3 in spleen. Percentage of DQ52 usage is indicated above each bar. n, number of mice analyzed per genotype. (D) Frequency of productive (VDJ+) and nonproductive (VDJ−) recombination events at Igh in spleen with JH1 or JH3 using HTGTS–Rep-seq data. Number of recombination events analyzed can be found in Table S1, Table S2, and Table S3. Data are presented as mean with error bars indicating SEM. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Figure S4.
DH gene segment usage in total VDJH recombination events assessed by HTGTS–Rep-seq. (A–D) Frequency of DH gene segment usage in total VDJ recombination events in mice 5–7 wk of age with JH1 (A and C) or JH3 (B and D) in bone marrow (A and B) and spleen (C and D) using HTGTS–Rep-seq. n represents number of mice analyzed per genotype. Each dot indicates data from an independent mouse, with data presented as mean ± SEM. Number of recombination events analyzed can be found in Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6.
R1.PG mice also exhibited perturbations in Igh gene segment repertoire. DQ52 usage was increased and DFL16.1 usage was decreased relative to WT in total VDJ, VDJ-, and DJ rearrangements in bone marrow (Fig. 4, B–D). A similar trend was observed in spleen rearrangements, although the magnitudes of the changes were smaller and were not statistically significant (Fig. S3, A–C). In contrast, DH segment usage in R1.KR mice closely resembled that of WT in DJH and VDJH joints in bone marrow and spleen (Figs. 4, B–D; and Fig. S3, A–C). We conclude that loss of RAG1 ubiquitin ligase activity leads to increased representation of DQ52 and decreased representation of DFL16.1 in the Igh repertoire, most strikingly in preB cells, and that this phenotype is not due to loss of RAG1 autoubiquitination at K233. AgR repertoire alterations in R1core mice are considered together at the end of the Results.
Reduced secondary D-to-JH recombination contributes to the R1.PG Igh repertoire phenotype
An attractive explanation for increased DQ52 and decreased DFL16.1 usage in R1.PG mice, suggested by our finding of reduced Igκ and Igλ recombination in these mice, is decreased recombinase activity leading to a decrease in secondary D-to-JH recombination, which should preserve DQ52-JH alleles at the expense of DFL16.1. If this explanation is correct, then R1.PG mice should exhibit increased usage of 5′ and decreased usage of 3′ JH gene segments relative to WT. To test this prediction, we assessed Igh repertoires using ImmunoSEQ (Adaptive Biotechnologies), an approach that provides the repertoire of VH, DH, and JH segments in VDJH rearrangements. The analysis was performed on bone marrow B220+IgM− cells from WT, R1Δ215, R1.PG, and R1core mice (two mice of each genotype).
The ImmunoSEQ results (Fig. 5) recapitulate the HTGTS–Rep-seq findings regarding DH segment usage in R1Δ215 and R1.PG mice: in R1Δ215, DQ52 usage was reduced ∼10 fold compared with WT, with little change observed for DFL16.1, while in R1.PG, DQ52 usage was increased and DFL16.1 usage was decreased compared with WT (Fig. 5 A). These differences were seen in total and nonproductive VDJH rearrangements (Fig. 5 A) and were recapitulated at each of the four JH segments (Fig. 5 B). JH repertoire analysis revealed increased JH1 and JH2 and decreased JH3 and JH4 usage in R1.PG mice relative to WT in total VDJ and VDJ− rearrangements, as predicted by the model that R1.PG mice perform secondary D-to-JH recombination inefficiently (Fig. 5 C). The JH repertoire of R1Δ215 mice exhibited only minor differences from that of WT, with small increases in JH3 and JH4, decreased JH2, and slightly decreased JH1 usage, consistent with a normal or slightly elevated efficiency of secondary D-to-JH recombination (Fig. 5 B). Together, our data argue strongly that the altered DH segment repertoire in R1.PG mice is due in part or entirely to reduced secondary D-to-JH recombination and that altered secondary D-to-JH recombination makes at most a minor contribution to the R1Δ215 phenotype.
Figure 5.
DH and JH gene segment usage in VDJH rearrangements in bone marrow assessed by ImmunoSEQ. (A–C) ImmunoSEQ data from mice 5–7 wk of age were analyzed for frequency of DFL16.1 (blue), DQ52 (salmon), and intervening 11 DH gene segments (green) in total or nonproductive VDJH rearrangements (A) or VDJH rearrangements to individual JH gene segments (B) and for JH gene segments in total and nonproductive VDJH recombination events (C). Percentage of DQ52 usage is indicated above each bar in A and B. n, number of mice analyzed per genotype. Number of recombination events analyzed can be found in Table S7. Data are presented as mean with error bars indicating SEM. Statistical significance relative to WT determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Reduced nonproductive VDJH alleles further support inefficient recombination in R1.PG mice
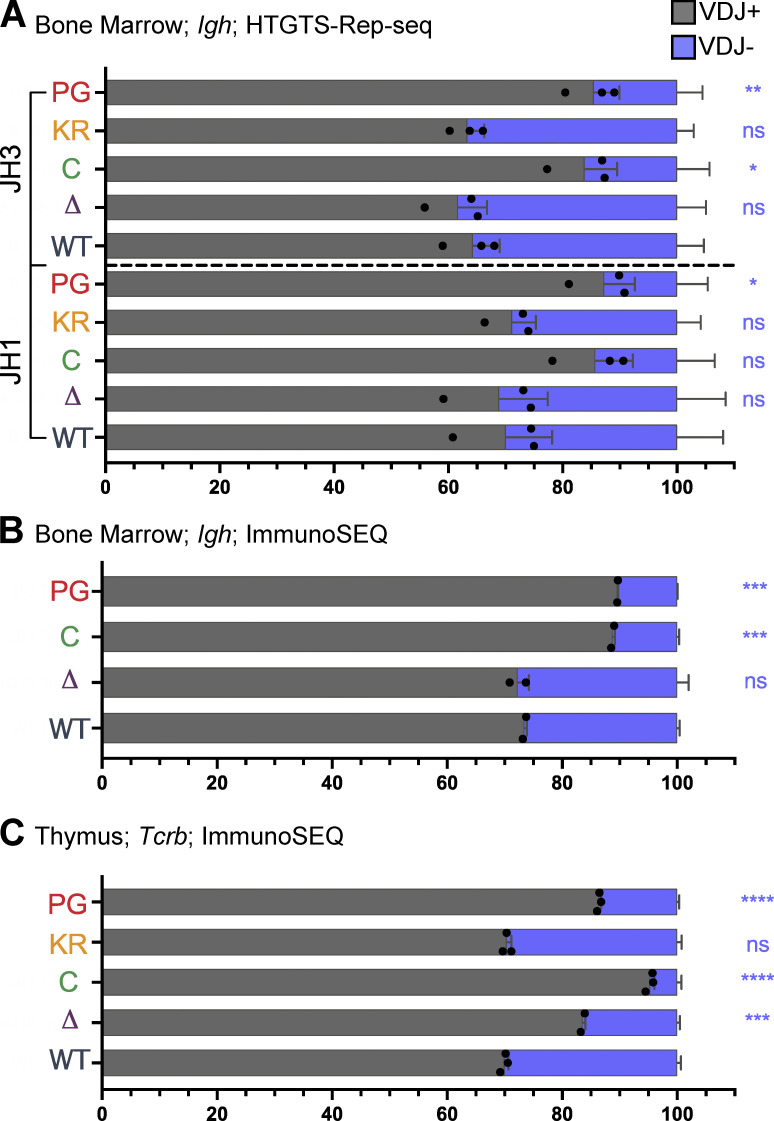
Reduced recombinase activity during Igh assembly in proB cells might reduce the frequency of VDJH− alleles because such alleles can only be carried forward to subsequent stages of B cell development in cells able to perform V-to-DJ recombination on both alleles. In support of reduced recombinase activity in proB cells due to the P326G mutation, the frequency of VDJH− alleles in R1.PG mice was consistently lower than in WT mice in both the HTGTS–Rep-seq and ImmunoSEQ analyses (Fig. 6, A and B; and Fig. S3 D). Indeed, the frequency of VDJH− alleles was as low or lower than in R1core mice, which are known to perform Igh gene assembly inefficiently (Dudley et al., 2003). In both R1.KR and R1Δ215 mice, the frequency of VDJH− alleles was similar to that in WT (Fig. 6, A and B; and Fig. S3 D), suggesting a normal efficiency of Igh gene assembly and consistent with the normal parameters of B cell development observed in these mice.
Figure 6.
Frequency of productive and nonproductive VDJH and VDJβ rearrangements. (A and B) Frequency of productive (VDJ+) and nonproductive (VDJ−) recombination events at Igh in bone marrow with JH1 or JH3 using HTGTS–Rep-seq data (A) and using ImmunoSEQ data (B). Number of recombination events analyzed can be found in Table S4, Table S5, Table S6, and Table S7. The frequency of VDJ+ alleles in mature B cells is predicted to be ∼71% and was measured to be 73% in splenic B cells (Lin et al., 2016). (C) Frequency of VDJ+ and VDJ− recombination events in thymus using ImmunoSEQ data. Data are presented as mean with error bars indicating SEM. Number of recombination events analyzed can be found in Table S8. Statistical significance relative to WT determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
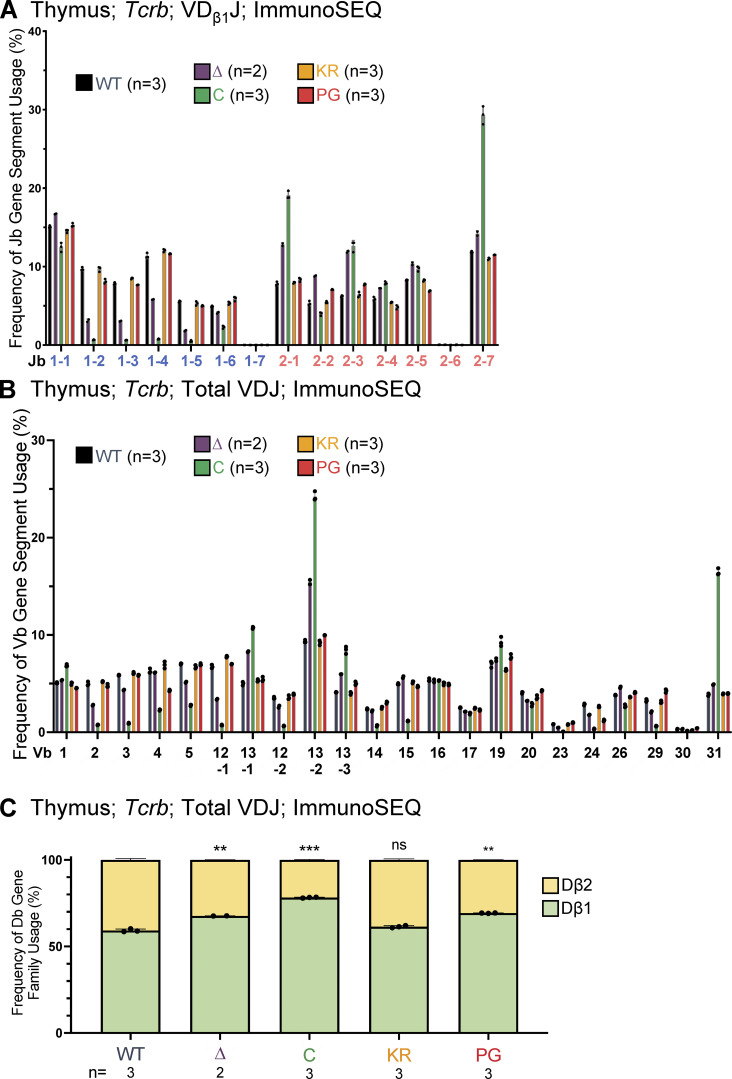
Analysis of Tcrb repertoires reveals reduced Dβ1-to-Jβ1 recombination in R1Δ215 mice
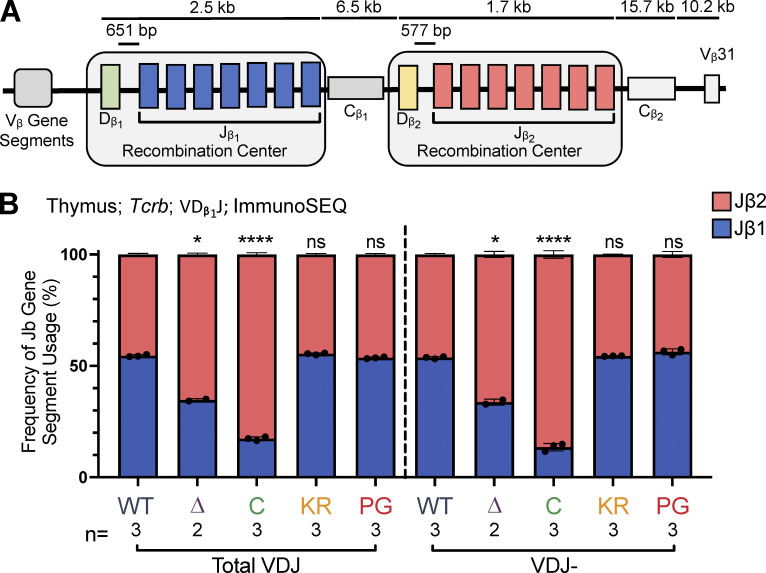
Tcrb is made up of 21 functional Vβ gene segments upstream and 1 Vβ gene segment downstream of two Dβ-Jβ-Cβ clusters (Fig. 7 A). Recombination occurs within two RCs, each containing a single Dβ gene segment followed by seven Jβ gene segments. The relative contributions of the RAG chromatin scanning versus collisional modes of recombination have not yet been established at Tcrb. If Tcrb follows the precedent set by Igh, then intra-RC Dβ1-to-Jβ1 and Dβ2-to-Jβ2 recombination events would be predicted to occur through a short-range collisional mechanism, whereas inter-RC Dβ1-to-Jβ2 recombination events, which span at least 9 kb, might occur by RAG chromatin scanning. These assumptions, together with our finding that the R1Δ215 protein is selectively defective in short-range recombination, lead to the prediction that recombination events involving Dβ1 should be biased toward Jβ2 gene segments over Jβ1 gene segments in R1Δ215 mice relative to WT mice.
Figure 7.
Jβ gene segment usage in VDJβ rearrangements. (A) Schematic of murine Tcrb with the two RCs containing Dβ1 and Jβ1 or Dβ2 and Jβ2 gene segments highlighted. Figure not to scale. (B) Tcrb ImmunoSEQ data from mice 5–7 wk of age in CD4+CD8+CD3εlo thymic T cells were analyzed for frequency of Jβ1 and Jβ2 gene cluster usage in VDJ recombination events using the Dβ1 gene segment. Number of recombination events analyzed can be found in Table S8. Data are presented as mean with error bars indicating SEM. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; *, P ≤ 0.05; ****, P ≤ 0.0001).
To test this prediction, we characterized Tcrb repertoires in sorted CD3εlo DP thymocytes using ImmunoSEQ. The data revealed that in R1Δ215 mice, VDJβ rearrangements involving Dβ1 were indeed biased away from Jβ1 and toward Jβ2 gene segments, with usage of Jβ1 dropping from 55% in WT to 35% in R1Δ215 mice (Fig. 7 B). This bias in J cluster rearrangements involving Dβ1 was seen in nonproductive rearrangements (Fig. 7 B) and at all individual Jβ1 and Jβ2 gene segments with the curious exception of Jβ1.1 (Fig. S5 A; see Discussion). No such bias was observed in R1.KR and R1.PG mice, indicating that disrupting RAG1 ubiquitin ligase activity or autoubiquitination does not perturb Dβ1’s preferences for the Jβ1 versus Jβ2 cluster. These findings argue that deletion of the first 215 aa of RAG1 disfavors intra-RC Dβ1-to-Jβ1 recombination.
Figure S5.
Jβ, Vβ, and Dβ gene segment usage in VDJβ recombination events in thymocytes assessed using ImmunoSEQ. (A) Frequency of Jβ gene segment usage in VDJ recombination events containing Dβ1 in CD4+CD8+CD3εlo thymic T cells. (B) Frequency of Vβ gene segment usage in total VDJ recombination events in CD4+CD8+CD3εlo thymic T cells. Repertoires were analyzed in mice of 5–7 wk of age. n represents number of mice analyzed per genotype. (C) Frequency of Dβ1 and Dβ2 gene segment usage in total VDJβ recombination events. Each dot indicates data from an independent mouse, with data presented as mean ± SEM. Number of recombination events analyzed can be found in Table S8. Statistical significance determined by two-tailed unpaired t tests (ns, P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Analysis of the relative use of the Dβ1 and Dβ2 gene segments revealed that R1Δ215, R1.PG, and R1core mice exhibited significant increases in usage of Dβ1 over Dβ2 relative to WT in VDJβ recombination events (Fig. S5 C). Notably, these increases parallel the reductions in the proportions of nonproductive VDJβ rearrangement alleles observed in these three genotypes relative to WT (Fig. 6 C). R1.KR mice displayed a normal proportion of nonproductive VDJβ rearrangements (Fig. 6 C). These data argue that RAG1 aa 1–215 and ubiquitin ligase activity, but not autoubiquitination, promote efficient recombination of Tcrb, and they raise the possibility that inefficient recombination contributes to preferential usage of Dβ1 over Dβ2. They also indicate that inefficient Tcrb recombination is unlikely to explain the bias toward Jβ2 over Jβ1 recombination observed in R1Δ215 mice since recombination is at least as inefficient in R1.PG as in R1Δ215 (Fig. 6 C), but no Jβ2 > Jβ1 bias is seen in R1.PG (Fig. 7 B).
Igh and Tcrb repertoire changes in R1core mice
Igh and Tcrb repertoire alterations in R1core mice exhibited features resembling those in R1Δ215 and R1.PG mice, as expected given the complete absence of the RAG1 NTR. DFL16.1 usage was reduced in R1core relative to WT in a pattern that resembled that in R1.PG, whereas DQ52 usage in R1core was reduced relative to WT in a pattern that resembled that in R1Δ215 (Fig. 4, B–D; Fig. 5 A; and Fig. S3, A–C). As in R1Δ215 mice, R1core Tcrb rearrangements involving Dβ1 were biased away from Jβ1 and toward Jβ2 gene segments (Fig. 7 B).
We observed a substantially altered repertoire of Vβ usage in R1core thymocytes (Fig. S5 B), consistent with a prior study (Horowitz and Bassing, 2014). A similar pattern of changes in Vβ repertoire was observed in R1Δ215 thymocytes, although they are more modest in magnitude than in R1core (Fig. S5 B). The Vβ repertoires in R1.KR and R1.PG thymocytes closely resemble that of WT thymocytes (Fig. S5 B; Burn et al., 2021 Preprint).
Discussion
RAG1 ubiquitin ligase activity contributes to efficient recombination and lymphocyte development
Our data indicate that the RAG1 P326G mutation decreases V(D)J recombinase activity during both B and T cell development. This reduced activity manifests in vivo as reduced secondary D-to-JH recombination, reductions in nonproductive VDJH and VDJβ alleles, decreased Igκ and Igλ recombination, and diminished development of Igλ+ B cells. Consistent with our observations, Burn et al. reported lower than normal levels of Vβ and Vα rearrangements in DN and DP thymocytes, respectively, of R1.PG mice (Burn et al., 2021 Preprint). The decreased cleavage of Igκ loci that we detected in R1.PG preB cells argues that the RAG1 P326G mutation reduces RAG endonuclease activity. Impaired repair of RAG DSBs in the absence of RAG1-mediated ubiquitylation of H3 histones might also contribute to reduced levels of completed V(D)J rearrangements (Grazini et al., 2010). Reduced endonuclease activity could result from altered folding of the RAG1 N-terminus, abrogation of RAG1 ubiquitin ligase activity, or both. A change in structure of the RING domain could negatively impact the RAG1 endonuclease catalytic site or destabilize RAG1 dimerization and thereby stability of the heterotetrameric RAG enzyme. The normal lymphocyte development and V(D)J rearrangements observed in R1.KR mice argue that the RAG1 ubiquitin ligase does not stimulate RAG endonuclease activity through RAG1 autoubiquitination, at least at K233. We cannot rule out that elevated R1.KR protein levels mask the reduction in recombination activity that would have been predicted from biochemical and cell line studies (Singh and Gellert, 2015). Notably, our finding of substantial levels of lymphocyte development in R1.PG mice argues strongly that the severe lymphocyte developmental defects reported previously in R1.C325Y mice and in a human bearing a RAG1 C328Y mutation were a consequence of perturbations of RAG1 function that extend beyond abrogation of ubiquitin ligase activity (Deng et al., 2015; Villa et al., 2001). Finally, considering that Burn et al. observed decreased expression of signaling molecules in R1.PG thymocytes, including the Syk protein that mediates B cell receptor signaling (Burn et al., 2021 Preprint), altered proB and preB cell proteomes could contribute to impaired development of Igκ+ and Igλ+ B cells in R1.PG mice. In this regard, while DSBs up-regulate Pim2 expression normally in R1.PG preB cells, we cannot rule out a contribution of decreased cell survival to the R1.PG phenotype.
The most striking alterations in AgR gene segment repertoire observed in R1.PG mice are increased usage of DQ52 and decreased usage of DFL16.1, accompanied by increased usage of JH1 and JH2 and decreased usage of JH4. These findings are fully consistent with, and plausibly explained by, reduced secondary D-to-JH recombination in R1.PG mice relative to WT, although other mechanisms might also contribute.
Modes of recombination and the RAG1 NTR
Deletion of the first 215 aa of RAG1 results in strongly decreased usage of DQ52 in Igh rearrangements and reduced usage of Jβ1 gene segments in Tcrb rearrangements involving Dβ1 without grossly perturbing B or αβ T cell development. Our data argue strongly that reduced usage of DQ52 is not due to increased secondary D-to-JH recombination. In particular, R1Δ215 mice exhibit levels of DFL16.1 usage that are, if anything, lower than in WT and levels of JH1 usage that are only slightly reduced relative to WT (Fig. 4 and Fig. 5), neither of which is consistent with a large increase in secondary D-to-JH recombination. The possibility that reduced DQ52 usage in R1Δ215 mice is due to a small increase in secondary D-to-JH recombination that selectively targets DQ52-JH alleles is ruled out by the finding that DQ52 usage is strongly reduced in rearrangements involving all four JH segments, including JH4 (Fig. 5 B); D-JH4 alleles cannot be targets of secondary recombination. We conclude that decreased usage of DQ52 in R1Δ215 mice is due primarily to mechanism(s) distinct from altered secondary D-to-JH recombination.
The high levels of expression of the R1Δ215 protein might have been predicted to lead to a hyper-recombination phenotype, but we see little evidence for this. Indeed, the decreased frequency of nonproductive Tcrb alleles observed in R1Δ215 mice suggests reduced recombination activity in thymocytes, despite the high levels of expression of the R1Δ215 protein. We propose that potential hyper-recombination effects of elevated R1Δ215 protein levels are counterbalanced by the inability of this protein to efficiently escape the repressive environment of the nucleolus. In our previous study, R1Δ215 was ∼10-fold less active than WT RAG1 for recombination of a chromosomal substrate even though expressed at higher levels (Brecht et al., 2020). In this context, we emphasize that decreased recombination per se does not result in reduced usage of the Jβ1 gene segment cluster, since Jβ1 gene segment usage is normal in P326G thymocytes. While we cannot rule out the possibility that deletion of the first 215 aa perturbs folding of other portions of the RAG1 protein, aa 215 corresponds closely to an NTR domain boundary (Arbuckle et al., 2011), and robust RAG1 protein accumulation and normal lymphocyte development in R1Δ215 mice argue against major structural perturbations.
There are multiple possible explanations for the defect in short-range D-to-J recombination observed for the R1Δ215 protein. The first 215 aa of RAG1 have been implicated in regulation of RAG1 protein stability (Schabla et al., 2018), interaction with the splicing factor SF3A2 (Maitra and Sadofsky, 2009), zinc binding involving three conserved pairs of cysteine residues (Arbuckle et al., 2011), and release of RAG1 from the nucleolus (Brecht et al., 2020). It is unknown if these functions contribute to the phenotypes we observe in R1Δ215 mice. RAG1 noncore regions influence the targeting of RAG1 in the genome through chromatin interactions (Maman et al., 2016; Teng et al., 2015), and the NTR has been implicated in histone binding (Deng et al., 2015; Grazini et al., 2010; Jones et al., 2011). It will be important to determine whether aa 1–215 contribute to RAG1–chromatin interactions.
We propose that the first 215 aa of RAG1 are required to maintain the proper balance between short-range, collisional recombination and long-range recombination mediated by RAG chromatin scanning. How this balance is established is not known, and to our knowledge, no prior evidence has implicated RAG in its control. Collisional and chromatin scanning–mediated recombinations differ in the constraints placed on partner RSS capture by a RAG–RSS complex in the RC (Fig. 8). While collisional recombination is largely unconstrained topologically (Fig. 8 C), in RAG chromatin scanning the cohesin ring is envisioned to bring potential partner RSSs into close proximity of RSS-bound RAG in the RC in a topologically constrained manner (Fig. 8 B; Zhang et al., 2019). For RSSs separated by a short distance from one another in the RC (2–3 kb for the Igh and Tcrb RCs), loop extrusion (measured to occur at 0.5–2 kb/s; Davidson et al., 2019; Kim et al., 2019) would be predicted to move a potential partner RSS to the opposite side of the cohesin ring from the RAG-bound RSS very rapidly, likely sequestering it from the RC (as depicted for DQ52 in Fig. 8 B, panel i). Substantial collision-mediated recombination of gene segments that have been extruded through the cohesin ring adjacent to the RAG–RSS complex would be inconsistent with the powerful orientation bias observed for recombination of VH, most DH, and cryptic RSS sequences (Ba et al., 2020; Dai et al., 2021; Hill et al., 2020; Hu et al., 2015; Jain et al., 2018; Zhang et al., 2019). Cohesin-mediated loop extrusion might thereby compete with and inhibit short-range collisional recombination. RAG1 residues 1–215 might act to facilitate collisional recombination, perhaps by stabilizing the 12RSS–23RSS synaptic complex. We propose that such stabilization would be less important for scanning-mediated recombination due to the presence of the cohesin ring. Alternatively, or in addition, residues 1–215 might inhibit loop extrusion, allowing more time for collisional recombination to occur. We note that in the current model of RAG chromatin scanning (Zhang et al., 2019), the cohesin ring encounters the RAG-bound RSS at the nonamer end of the RSS, where the nonamer binding domain and the RAG1 NTR would be located. Our data argue that ubiquitin ligase activity is not required for the ability of the RAG1 NTR to facilitate short-range recombination. Further mechanistic insights might require reconstitution of RAG chromatin scanning in vitro.
Figure 8.
Schematic diagrams depicting models for short-range collision– and long-range scanning–mediated D-to-J recombination. (A) Schematic of murine Igh depicting some DH and JH gene segments. Gray box represents the RC, encompassing JH gene segments and DQ52. Gene segments are depicted as rectangles, 23RSSs as gray triangles, and 3′ and 5′ DH 12RSSs as orange and purple triangles, respectively. (B) Long-range recombination is thought to occur through RAG chromatin scanning in which the cohesin ring brings RSSs into the RC in close proximity of RSS-bound RAG in a topologically constrained manner. Panel i: DQ52, which resides very close to the JH gene segments, has already been extruded through the cohesin ring and now is separated from JH by cohesin. Long-range recombination (panels ii and iii) occurs predominantly by deletion due to the topological constraint imposed by cohesin. (C) Short-range recombination occurs through collision-mediated recombination, largely topologically unconstrained. Short-range recombination can result in either inversion or deletion; however, DQ52-to-JH recombination occurs almost exclusively by deletion due to the different sequences of its 5′ and 3′ RSSs (Zhang et al., 2019). Models in B and C adapted from Zhang et al. (2019). This figure was created with BioRender.com.
We note that, because all VH recombination involves RAG chromatin scanning (Dai et al., 2021; Hill et al., 2020), an altered balance between collisional and scanning modes of recombination might not be expected to perturb VH repertoire. Consistent with this, we did not observe substantial alterations in VH repertoire in R1Δ215 mice.
Support for our model regarding the role of RAG1 aa 1–215 in establishing the balance between short- and long-range recombination derives primarily from our findings with the Igh locus, where collisional and scanning modes of recombination have been firmly established and mapped to particular classes of rearrangements. Because a similar mechanistic understanding is not yet available for Tcrb recombination, findings from this locus should be interpreted cautiously. Our observation that R1Δ215 thymocytes exhibit a skewing of Dβ1 recombination toward the Jβ2 cluster and away from the Jβ1 cluster should be considered only preliminary corroboration of our findings at Igh. Jβ1.1 is the only Jβ segment that does not show this skewing, suggesting that its recombination is influenced by additional mechanisms. One speculative possibility is that Fos-driven RAG loading onto the Dβ1 3′RSS (Wang et al., 2008), combined with the close proximity of Jβ1.1 to Dβ1, compensates for the R1Δ215 short-range recombination defect.
Insights into the phenotype of R1core mice
While the B cell developmental defects observed in R1core mice are mirrored in R1.PG mice (albeit in a milder form), the substantial defects in T cell development were not observed in any of the NTR mutant mice analyzed here and likely reflect the combined loss of multiple noncore domains/activities. Igh repertoire alterations in R1core B cells appear to represent a combination of loss of the first 215 aa, abrogation of ubiquitin ligase activity, and likely other deficits as well. Tcrb repertoire perturbations in R1core mice, such as reduced usage of Jβ1 gene segments and Vβ repertoire changes, are mirrored to a substantial extent in R1Δ215 mice, and loss of ubiquitin ligase activity appears to make less of a contribution. The severe reduction in nonfunctional VDJβ rearrangements observed in R1core thymocytes appears to be due to a combination of the defects observed in R1Δ215 and R1.PG thymocytes.
The generally more severe phenotypes observed in R1core mice compared with the RAG1 NTR mutants created for this study could be due to one or more additional domains/activities missing in the R1core protein, including a positively charged region between residues 216 and 238 that enhances recombination activity (McMahan et al., 1997), a signal for nucleolar localization between aa 243 and 249 (Brecht et al., 2020), the RAG1 C-terminal tail, loss of the strong dimerization activity provided by the zinc–RING finger, and the ability to interact with Ku (Raval et al., 2008) and chromatin (Maman et al., 2016), which have yet to be mapped within the RAG1 noncore regions.
Conclusions
Our findings implicate different portions/activities of the RAG1 NTR in regulating distinct functions: (i) maintenance of proper RAG1 protein levels (residues 1–215, ubiquitin ligase activity, major site of autoubiquitination), (ii) efficient V(D)J recombination and lymphocyte development (ubiquitin ligase activity), and (iii) efficient short-range recombination within RCs (residues 1–215). The RAG1 NTR is proposed to be derived from a transposable element known as N-RAG-TP (Panchin and Moroz, 2008) and to have become incorporated into a RAG1-like protein early in the evolution of RAG-like transposons (transposons that encode RAG1-like and RAG2-like proteins; Carmona and Schatz, 2017). The NTR of extant RAG1-like proteins shows considerable variation in the domains that have been retained (Fugmann et al., 2006; Huang et al., 2016; Martin et al., 2020), suggesting that the NTR has been a flexible substrate for the evolution of new functions. Given that known RAG-like transposons are <10 kb in size (Martin et al., 2020), an activity to ensure efficient short-range recombination might have arisen in a RAG-like transposon to facilitate synapsis and cleavage of the transposon terminal inverted repeats. In this scenario, the domain would have subsequently been repurposed for short-range recombination in jawed vertebrates. Alternatively, such an activity might have evolved only after RAG had begun to function as a recombinase in jawed vertebrates, to help address the complexities of assembling large AgR loci containing many gene segments. It will now be important to determine which residues in the first 215 aa of RAG1 contribute to this activity and whether this activity can be detected in RAG-like transposases.
Materials and methods
Mouse strains
WT C57BL/6 mice were purchased from The Jackson Laboratory. R1core mice have been described previously (Dudley et al., 2003). R1Δ215 and R1.KR mice were generated by the Yale Genome Editing Center, as previously described, with CRISPR-Cas9 technology into C57BL/6N embryos (Wang et al., 2013). To generate R1Δ215 mice, two guide RNAs (Table S9) targeting the far N-terminus–encoding region of Rag1 were coinjected into fertilized WT C57BL/6N eggs with a single-strand DNA Ultramer (IDT) donor template (Table S9). The knock-in cassette replaces the sequence encoding the initial 215 aa of RAG1, such that upon homology-directed repair-mediated genomic integration, the protein will begin at aa 216. To generate R1.KR mice, a guide RNA (Table S9) and single-strand DNA Ultramer (IDT; Table S9) were coinjected into fertilized WT C57BL/6N eggs. This sequence introduces the K233R point mutation into Rag1 and introduces a HindIII site that does not affect the amino acid sequence of the protein to facilitate genotyping. For both lines, single heterozygous males were generated and backcrossed to C57BL/6 females for eight generations. Heterozygote-by-heterozygote breeding was then conducted to establish homozygote lines of mice. R1Δ215 and R1.KR lines were confirmed to have only the designated mutations by sequencing of the Rag1 locus. R1.PG mice were generated as described (Burn et al., 2021 Preprint). Sequencing revealed that R1.PG mice harbor an additional V238A mutation in Rag1 (Fig. S1 A). This mutation is conservative, and V238 is not well conserved in vertebrates. These mice are deposited in the Mutant Mouse Resource & Research Centers Repository (strain RAG1-tm1; #37105) and are cryo-archived.
All animal studies were conducted in accordance with national guidelines and were approved by the Institutional Animal Care and Use Committee of the Children's Hospital of Philadelphia or by the Yale Institutional Animal Care and Use Committee. Female and male mice were used for all experiments.
Western blotting
For Western blots for Fig. 1 B and Fig. S1 B, whole thymuses were lysed in RIPA buffer containing 1× cOmplete protease inhibitor cocktail (Sigma-Aldrich; 4693159001) and Pierce Universal Nuclease for Cell Lysis (Thermo Fisher Scientific; 88700) and incubated on ice for 10 min. 25 µg of lysates was resolved on 8% SDS-PAGE gels. Immunoblotting was performed according to standard procedures with mAbs for RAG1 (mAb 23; Coster et al., 2012) and β-actin (Sigma-Aldrich; A5441).
For Western blots for Fig. 3 E, 6–10 million preB cells were harvested at indicated time points, washed with PBS, and resuspended in ice-cold lysis RIPA buffer with protease and phosphatase inhibitor cocktail (Cell Signaling) and benzonase (Sigma-Aldrich). Laemmli buffer was added to the lysate; then samples were boiled for 5 min. 20 μg of protein was loaded and run on ExpressPlus PAGE 4–12% gels (Genscript). Electrophoresed proteins were transferred to Immobilon-FL polyvinylidene fluoride membrane (EMD Millipore). Membranes were blocked with a blocking buffer (Tri base 1X, Tween 0.1%, and BSA 5%) and incubated overnight with a primary antibody. Primary antibodies were monoclonal mouse anti-Pim2 antibody (Santa Cruz Biotechnology; clone 1D12) and monoclonal mouse anti-βactin antibody (Sigma-Aldrich; clone AC-15). Blots were washed and incubated with appropriate HRP-conjugated secondary antibodies. After washing, proteins were revealed using WesternBright ECL HRP Substrate Kits (Advansta). All blots were acquired with the G:box Chemi-XRQ. Quantification was performed using ImageJ software (Wayne Rasband, National Institutes of Health, version 1.52q).
RAG1 mRNA analysis
RNA was isolated from whole thymuses with TRIzol reagent (Invitrogen) and was converted to cDNA with SuperScript III and random hexamers according to the manufacturer’s instructions (Life Technologies). Transcripts were quantified by iTaq Universal SYBR Green (Bio-Rad) with primers RAG1 Forward and RAG1 Reverse. Analysis was normalized using Hprt expression (primers Hprt Forward 1 and Hprt Reverse 1). Primer sequences are listed in Table S9.
Flow cytometry and cell sorting
Lymphocyte development was analyzed in age-matched mice aged 4–6 wk. Both males and females were analyzed, with no differences noted. The analyses for Fig. 1, C–F; Fig. 2, A, B, and D; and Fig. S2, A–C were performed as follows. Total thymocytes were harvested from whole thymuses, total splenocytes were harvested from whole spleens, and total bone marrow was harvested from whole bone marrow. Total cells were calculated based on number of nucleated cells after erythrocytes were lysed using ZAP-OGLOBIN II Lytic Reagent (Beckman Coulter; 7546138). Lymphocyte development analyses were conducted using flow cytometry. Thymic and splenic T cells were analyzed using PE anti-mouse CD3 (BioLegend clone 17A2), APC anti-mouse CD4 (BioLegend; clone GK1.5), and FITC anti-mouse CD8a (BioLegend; clone 53–6.7) antibodies and DAPI (BioLegend; 422801). B cell development in the bone marrow was analyzed using FITC anti-mouse CD43 (BD BioSciences; clone S7), PE anti-mouse CD19 (BioLegend; clone 6D5), PerCP anti-mouse B220 (BioLegend; clone RA3-6B2), and APC anti-mouse IgM (BioLegend; clone RMM-1) antibodies and DAPI. Igκ and Igλ staining was conducted in the bone marrow and spleen using PerCP anti-mouse B220, APC anti-mouse IgM, PE anti-mouse Igκ (BD Biosciences; 559940), and FITC anti-mouse Igλ (BD BioSciences; clone R26-46) antibodies and DAPI. Samples were run on BD LSRII or CytoFlex LX machines, and data were analyzed using FlowJo software (Tree Star).
The analyses for Fig. 2, C, E, and F were performed as follows. Cells isolated from bone marrow and spleens were depleted of red blood cells with NH4Cl lysis buffer, and FC receptors were blocked using anti-CD16/CD32 (BD Biosciences; 2.4G2). Single-cell suspensions from bone marrow cells or splenocytes were stained in PBS containing 2% BSA, 0.5 mM EDTA with BUV395-conjugated anti-B220 (clone RA3-6B2), APC- or PE-conjugated anti-IgM (clone 11/41), BV421- or APC-conjugated anti-CD43 (clone S7), PE-conjugated anti-Igκ (clone 187.1), and FITC-conjugated anti-Igλ (clone R26-46) antibodies. Dead cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen). Data were acquired on an LSRFortessa (BD Biosciences) using Diva software (BD Biosciences) and analyzed using FlowJo software. Cell sorting was conducted using a MoFlo Astrios (Beckman Coulter, Inc.) using PE-Cy7 anti-mouse CD19 and FITC anti-mouse CD43 to identify preB cells.
Taqman PCR quantification of Ig light chain recombination
Genomic DNA was isolated from sorted preB cells using the QiaAMP DNA mini kit (Qiagen). Taqman quantitative PCR (qPCR) assays were performed using conditions previously described (Gopalakrishnan et al., 2013) and the following reagents: Vk forward primer, Jk1 reverse primer, and Jk1 probe or Vλ1 forward primer, Jλ1 reverse primer, and Jλ1 probe (Integrated DNA Technologies; Table S9). Vk-Jk1 and Vλ1-Jλ1 rearrangements were normalized to an unrearranged region of the genome (CD19; using primers CD19 Forward, CD19 Reverse, and CD19 probe).
Quantification of germline Jλl transcripts
Total RNA was isolated from sorted preB cells using the Trizol Reagent (Ambion). RNA was incubated with DNase (Promega) to destroy contaminating DNA. cDNA was synthesized using ProtoScript First Strand cDNA synthesis Kit (New England BioLabs) as described (Brady et al., 2010). Primers used to detect Jλ1 germline transcripts were the Jλ1 Forward 1 and Jλ1 Reverse 1 primers (Table S9). Levels of Jλ1 transcripts were calculated using ΔΔCt analysis with values normalized to levels of HPRT.
Quantification of germline Jλl accessibility
Nuclei were isolated from the sorted preB cells and resuspended into 1× restriction digestion buffer (Mandal et al., 2015). Samples were divided into two equal parts and either treated as undigested control or digested with 50 U of BseYI restriction enzyme (New England BioLabs) in 100 µl volume at 37°C for 1 h. Genomic DNA was extracted using QiaAMP DNA mini kit (Qiagen). The isolated DNA was subjected to quantitative analysis by SYBR Green PCR using the primer pairs flanking the BseYI restriction site. CD19 was used to normalize the input DNA. ICAM2 served as a control reference. Primers used were CD19 Forward, CD19 Reverse, ICAM2 Forward 2, ICAM2 Reverse 2, Jλ1 Forward 2, and Jλ1 Reverse 2 (Table S9).
Primary preB cell culture
Primary bone marrow cells were harvested by flushing bone marrow from all leg bones of at least three mice of the appropriate genotype for each experiment. These bone marrow cells were cultured for 4 d in RPMI 1640 supplemented with 10% FBS, 10 mM Hepes, 13 nonessential aa, 1 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin-streptomycin, 50 mM 2-ME, and 5 ng/ml IL-7. Cells were plated at a density of 5 million cells per milliliter of media. Each 2 d, cells were harvested and put back into culture in fresh media at a density of 5 million cells per milliliter. After 4 d of culture, B cells were sorted by depletion using EasySep mouse B cell isolation kit according to the manufacturer’s instructions (StemCell Technologies, Inc.). PreB cell purity was assessed by flow cytometry and was above 90% (live B220+CD43−IgM− cells). To induce G1 arrest and activate transcription of Rag1 and Rag2 by IL-7 withdrawal, we pelleted cells by centrifugation, resuspended them in the same media lacking IL-7 at a density of 2 million cells per milliliter. Cells were harvested after 48 h.
Real-time PCR quantification of Pim2 mRNA and DQ52, Dβ1, and Dβ2 transcripts
For Pim2 (Fig. 3 B), 3 million cultured preB cells were harvested at indicated time points and immediately lysed in TRIzol (Life Technologies). For DQ52 transcripts (Fig. S2 F), CD19+ cells were purified from either RAG1−/− or R1.D708A total bone marrow using the EasySep Mouse CD19 Positive Selection Kit II (cat no. 18954) according to the manufacturer’s directions and immediately lysed in TRIzol. For Dβ1 and Dβ2 transcripts (Fig. S2 G), total thymocytes from either RAG1−/− or R1.D708A mice were isolated and lysed in TRIzol. Total RNA was isolated using RNeasy mini kit (Qiagen), treated with DNase (RNase-Free DNase Set; Qiagen), and reverse transcribed to generate cDNA with High-Capacity RNA-to-cDNA Kit (Thermo-Fisher Scientific) according to the manufacturer’s directions. The cDNAs were then used as a template for real-time PCRs performed with SYBR Green Mastermix (Applied Biosystems) and run on a Quant Studio Flex 7 machine using the following corresponding primers: Pim2 Forward, Pim2 Reverse, DQ52 Forward, DQ52 Reverse, Dβ1 Forward, Dβ1 Reverse, Dβ2 Forward, and Dβ2 Reverse primers (Table S9). Pim2 and DQ52 values were normalized to CD19 using CD19 forward and CD19 reverse primers. Dβ1 and Dβ2 values were normalized to Lck using Lck Forward and Lck Reverse primers. Fold change was determined by ΔΔCt analysis.
HTGTS–Rep-seq B cell repertoire library construction
B cells were isolated from mice aged 5–7 wk. Splenic B cells were isolated using B220 magnetic beads (Miltenyi Biotec) or by FACS sorting. B220+IgM− cells were purified via FACS sorting from bone marrow. Genomic DNA samples were obtained using phenol-chloroform extraction of whole-cell lysates. HTGTS–Rep-seq libraries were prepared as previously described (Hu et al., 2016; Lin et al., 2016). Briefly, genomic DNA was sonicated and subjected to linear amplification–mediated PCR using biotinylated JH1 or JH3 bait primers (Lin et al., 2016). Linear amplification–mediated PCR products were purified using Dynabeads MyONE C1 streptavidin beads (Life Technologies; 65002) and ligated to bridge adaptors. Adaptor-ligated products were amplified by nested PCR with indexed JH1/JH3 primers (8-bp indexes were unique to each sample; Table S9) and primer annealed to the adaptor. PCR products were further tagged with Illumina sequencing adaptor sequences and size-selected via gel extraction. Libraries were sequenced by paired-end 300-bp sequencing on an Illumina MiSeq by the Yale Center for Genome Analysis. Primers were previously described and are listed in Table S9 (Hu et al., 2016; Lin et al., 2016).
HTGTS–Rep-seq B cell repertoire analysis
To quantify Igh repertoires, an HTGTS pipeline was implemented that provides the all-inclusive steps for the analysis of HTGTS–Rep-seq data (Lin et al., 2016). In brief, the pipeline involves fastq-multx tool–based (https://expressionanalysis.github.io/ea-utils/) demultiplexing followed by the implementation of cutadapt (https://cutadapt.readthedocs.io/en/stable/) to trim the adaptors in the metadata file for each sample. The high-quality (Phred score >20) adaptor-trimmed reads were then joined using fastq-join tool if, and only if, the reads from both sides had at least a 10-bp overlap with mismatch rate ≤8%. Both joined and unjoined reads were mapped against the mouse (mm10) IGH V(D)J gene database (IMGT) using default parameters of the IgBLAST. The aligned reads (now assigned with V, D, and J genes) were further filtered to have high IgBLAST score >150 with total alignment length >100, comprising overall mismatch ratio <0.1. The usage of V and D genes was computed based on the processed IgBLAST results as provided by the pipeline. To quantitate DJH rearrangements, we implemented the transloc_pipeline (https://github.com/robinmeyers/transloc_pipeline) that involves DH-to-JH junction detection based on the Optimal Query Coverage algorithm, as previously described (Hu et al., 2016). The identified junctions were then intersected with expanded RSS locations (±40 bp) to annotate the region with their respective D and J gene segments. After gene segments were called, all quantitative analysis was conducted in R.
ImmunoSEQ B and T cell repertoire library building and repertoire analysis
DNA was extracted from FACS-sorted bone marrow B220+IgM− cells and thymic CD4+CD8+CD3εlo cells using the Qiagen DNeasy Blood & Tissue Kit (Qiagen; 69506). High-throughput DNA sequencing of rearranged Igh and Tcrb genes was performed using ImmunoSEQ and was performed by Adaptive Biotechnologies. Gene segment usage was analyzed by ImmunoSEQ Analyzer software (Adaptive Biotechnologies) and R.
Online supplemental material
Fig. S1 shows RAG1 mutant mouse sequencing tracks and protein and mRNA expression. Fig. S2 shows FACS gating strategies and germline transcription and genome accessibility in D708A mice. Fig. S3 presents HTGTS–Rep-seq data from splenic B cells. Fig. S4 shows individual DH gene segment usage from HTGTS–Rep-seq experiments. Fig. S5 shows individual Jβ and Vβ gene segment usage from thymic ImmunoSEQ experiments. Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6 detail data for Igh HTGTS–Rep-seq experiments from spleen and bone marrow. Table S7 and Table S8 show detailed data for Igh and Tcrb ImmunoSEQ experiments from bone marrow and thymus, respectively. Table S9 lists oligonucleotides used in these studies and their sequences.
Supplementary Material
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 1).
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 2).
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 3).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 1).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 2).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 3).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (ImmunoSEQ).
shows utilization of Tcrb gene segments in VDJ rearrangements in primary CD8+CD4+CD3εlo thymocytes (ImmunoSEQ).
lists oligos used and their sequences.
Acknowledgments
The authors would like to thank J. Horowitz for initial discovery of the altered Igκ/Igλ B cell ratio in R1core mice; R. Jiang, S. Kleinstein, and other members of the Kleinstein laboratory for help and advice in repertoire analysis; and G. Teng for initial characterization of the R1.PG mouse.
This work was funded by the Yale University Interdisciplinary Immunology Training Grant T32 AI-007019 (H.A. Beilinson), University of Pennsylvania Cell and Molecular Biology Training Grant T32 GM-07229 (R.A. Glynn), Fondation ARC pour la Recherche sur le Cancer (C. Miot), La Ligue Contre le Cancer (C. Miot), Fondation Pasteur Mutualite (C. Miot), and National Institutes of Health grants R01 AI032524 (D.G. Schatz), R21 AI135435 (C.H. Bassing), and RO1 AI112621 (C.H. Bassing).
Author contributions: H.A. Beilinson, C.H. Bassing, and D.G. Schatz designed experiments, which were performed by H.A. Beilinson, R.A. Glynn, J. Xiao, E. Corbett, H. Saribasak, R. Arya, and C. Miot. H.A. Beilinson, A.D. Yadavalli, and D.G. Schatz designed computational analyses, which were performed by H.A. Beilinson and A.D. Yadavalli. J.M. Jones generously provided the R1.PG mouse generated by A. Bhattacharyya. J.M.R. Pongubala provided mentorship to A.D. Yadavalli. H.A. Beilinson, R.A. Glynn, C.H. Bassing, and D.G. Schatz assembled figures and wrote the paper with input from other authors.
Data availability
The accession no. for the HTGTS–Rep-seq datasets reported in this paper is Gene Expression Omnibus no. GSE180734. The ImmunoSEQ datasets reported can be found at https://doi.org/10.21417/HAB2021JEM and https://clients.adaptivebiotech.com/pub/beilinson-2021-jem.
References
- Arbuckle, J.L., Rahman N.S., Zhao S., Rodgers W., and Rodgers K.K.. 2011. Elucidating the domain architecture and functions of non-core RAG1: the capacity of a non-core zinc-binding domain to function in nuclear import and nucleic acid binding. BMC Biochem. 12:23. 10.1186/1471-2091-12-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba, Z., Lou J., Ye A.Y., Dai H.Q., Dring E.W., Lin S.G., Jain S., Kyritsis N., Kieffer-Kwon K.R., Casellas R., and Alt F.W.. 2020. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature. 586:305–310. 10.1038/s41586-020-2578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarski, J.J., Nickless A., Bhattacharya D., Amin R.H., Schlissel M.S., and Sleckman B.P.. 2012. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre-B cell survival and limit proliferation. J. Exp. Med. 209:11–17. 10.1084/jem.20112078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, B.L., Steinel N.C., and Bassing C.H.. 2010. Antigen receptor allelic exclusion: an update and reappraisal. J. Immunol. 185:3801–3808. 10.4049/jimmunol.1001158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht, R.M., Liu C.C., Beilinson H.A., Khitun A., Slavoff S.A., and Schatz D.G.. 2020. Nucleolar localization of RAG1 modulates V(D)J recombination activity. Proc. Natl. Acad. Sci. USA. 117:4300–4309. 10.1073/pnas.1920021117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, T.N., Miot C., Kreiger P., Hayer K.E., Bhattacharyya A., Jones J.M., Bassing C.H., and Behrens E.M.. 2021. The RAG1 Ubiquitin Ligase Domain Enhances T Cell Receptor Gene Assembly and Thymic Selection. bioRxiv. (Preprint posted June 29, 2021) 10.1101/2021.01.04.425211 [DOI]
- Carmona, L.M., and Schatz D.G.. 2017. New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J. 284:1590–1605. 10.1111/febs.13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, M.D., and Alder M.N.. 2006. The evolution of adaptive immune systems. Cell. 124:815–822. 10.1016/j.cell.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Coster, G., Gold A., Chen D., Schatz D.G., and Goldberg M.. 2012. A dual interaction between the DNA damage response protein MDC1 and the RAG1 subunit of the V(D)J recombinase. J. Biol. Chem. 287:36488–36498. 10.1074/jbc.M112.402487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, H.Q., Hu H., Lou J., Ye A.Y., Ba Z., Zhang X., Zhang Y., Zhao L., Yoon H.S., Chapdelaine-Williams A.M., et al. 2021. Loop extrusion mediates physiological Igh locus contraction for RAG scanning. Nature. 590:338–343. 10.1038/s41586-020-03121-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, I.F., Bauer B., Goetz D., Tang W., Wutz G., and Peters J.M.. 2019. DNA loop extrusion by human cohesin. Science. 366:1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- Deng, Z., Liu H., and Liu X.. 2015. RAG1-mediated ubiquitylation of histone H3 is required for chromosomal V(D)J recombination. Cell Res. 25:181–192. 10.1038/cr.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derudder, E., Cadera E.J., Vahl J.C., Wang J., Fox C.J., Zha S., van Loo G., Pasparakis M., Schlissel M.S., Schmidt-Supprian M., and Rajewsky K.. 2009. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat. Immunol. 10:647–654. 10.1038/ni.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, D.D., Sekiguchi J., Zhu C., Sadofsky M.J., Whitlow S., DeVido J., Monroe R.J., Bassing C.H., and Alt F.W.. 2003. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med. 198:1439–1450. 10.1084/jem.20030627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann, S.D., Messier C., Novack L.A., Cameron R.A., and Rast J.P.. 2006. An ancient evolutionary origin of the Rag1/2 gene locus. Proc. Natl. Acad. Sci. USA. 103:3728–3733. 10.1073/pnas.0509720103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan, S., Majumder K., Predeus A., Huang Y., Koues O.I., Verma-Gaur J., Loguercio S., Su A.I., Feeney A.J., Artyomov M.N., and Oltz E.M.. 2013. Unifying model for molecular determinants of the preselection Vβ repertoire. Proc. Natl. Acad. Sci. USA. 110:E3206–E3215. 10.1073/pnas.1304048110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, J.R., and Alt F.W.. 1998. Regulation of immunoglobulin light chain isotype expression. Adv. Immunol. 69:113–181. 10.1016/S0065-2776(08)60607-0 [DOI] [PubMed] [Google Scholar]
- Grazini, U., Zanardi F., Citterio E., Casola S., Goding C.R., and McBlane F.. 2010. The RING domain of RAG1 ubiquitylates histone H3: a novel activity in chromatin-mediated regulation of V(D)J joining. Mol. Cell. 37:282–293. 10.1016/j.molcel.2009.12.035 [DOI] [PubMed] [Google Scholar]
- Grundy, G.J., Yang W., and Gellert M.. 2010. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc. Natl. Acad. Sci. USA. 107:22487–22492. 10.1073/pnas.1014958107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, C., Yoon H.S., Franklin A., Jain S., Ebert A., Cheng H.L., Hansen E., Despo O., Bossen C., Vettermann C., et al. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature. 477:424–430. 10.1038/nature10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, L., Ebert A., Jaritz M., Wutz G., Nagasaka K., Tagoh H., Kostanova-Poliakova D., Schindler K., Sun Q., Bönelt P., et al. 2020. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature. 584:142–147. 10.1038/s41586-020-2454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, J.E., and Bassing C.H.. 2014. Noncore RAG1 regions promote Vβ rearrangements and αβ T cell development by overcoming inherent inefficiency of Vβ recombination signal sequences. J. Immunol. 192:1609–1619. 10.4049/jimmunol.1301599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Zhang Y., Zhao L., Frock R.L., Du Z., Meyers R.M., Meng F.L., Schatz D.G., and Alt F.W.. 2015. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 163:947–959. 10.1016/j.cell.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Meyers R.M., Dong J., Panchakshari R.A., Alt F.W., and Frock R.L.. 2016. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 11:853–871. 10.1038/nprot.2016.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Tao X., Yuan S., Zhang Y., Li P., Beilinson H.A., Zhang Y., Yu W., Pontarotti P., Escriva H., et al. 2016. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 166:102–114. 10.1016/j.cell.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S., Ba Z., Zhang Y., Dai H.Q., and Alt F.W.. 2018. CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell. 174:102–116.e14. 10.1016/j.cell.2018.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., Resch W., Corbett E., Yamane A., Casellas R., and Schatz D.G.. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 141:419–431. 10.1016/j.cell.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.M., and Gellert M.. 2003. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc. Natl. Acad. Sci. USA. 100:15446–15451. 10.1073/pnas.2637012100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.M., and Simkus C.. 2009. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch. Immunol. Ther. Exp. (Warsz.). 57:105–116. 10.1007/s00005-009-0011-3 [DOI] [PubMed] [Google Scholar]
- Jones, J.M., Bhattacharyya A., Simkus C., Vallieres B., Veenstra T.D., and Zhou M.. 2011. The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol. Lett. 136:156–162. 10.1016/j.imlet.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Kassmeier, M.D., Mondal K., Palmer V.L., Raval P., Kumar S., Perry G.A., Anderson D.K., Ciborowski P., Jackson S., Xiong Y., and Swanson P.C.. 2012. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 31:945–958. 10.1038/emboj.2011.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.S., Lapkouski M., Yang W., and Gellert M.. 2015. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 518:507–511. 10.1038/nature14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Shi Z., Zhang H., Finkelstein I.J., and Yu H.. 2019. Human cohesin compacts DNA by loop extrusion. Science. 366:1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, T.M., and Schatz D.G.. 1995. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol. Cell. Biol. 15:5657–5670. 10.1128/MCB.15.10.5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.G., Guo C., Su A., Zhang Y., and Alt F.W.. 2015. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc. Natl. Acad. Sci. USA. 112:1815–1820. 10.1073/pnas.1424936112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.G., Ba Z., Du Z., Zhang Y., Hu J., and Alt F.W.. 2016. Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc. Natl. Acad. Sci. USA. 113:7846–7851. 10.1073/pnas.1608649113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra, R., and Sadofsky M.J.. 2009. A WW-like module in the RAG1 N-terminal domain contributes to previously unidentified protein-protein interactions. Nucleic Acids Res. 37:3301–3309. 10.1093/nar/gkp192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maman, Y., Teng G., Seth R., Kleinstein S.H., and Schatz D.G.. 2016. RAG1 targeting in the genome is dominated by chromatin interactions mediated by the non-core regions of RAG1 and RAG2. Nucleic Acids Res. 44:9624–9637. 10.1093/nar/gkw633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M., Hamel K.M., Maienschein-Cline M., Tanaka A., Teng G., Tuteja J.H., Bunker J.J., Bahroos N., Eppig J.J., Schatz D.G., and Clark M.R.. 2015. Histone reader BRWD1 targets and restricts recombination to the Igk locus. Nat. Immunol. 16:1094–1103. 10.1038/ni.3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E.C., Vicari C., Tsakou-Ngouafo L., Pontarotti P., Petrescu A.J., and Schatz D.G.. 2020. Identification of RAG-like transposons in protostomes suggests their ancient bilaterian origin. Mob. DNA. 11:17. 10.1186/s13100-020-00214-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan, C.J., Difilippantonio M.J., Rao N., Spanopoulou E., and Schatz D.G.. 1997. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol. Cell. Biol. 17:4544–4552. 10.1128/MCB.17.8.4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo, L.D., Kim M.S., Walter J.E., and Lee Y.N.. 2016. Human RAG mutations: biochemistry and clinical implications. Nat. Rev. Immunol. 16:234–246. 10.1038/nri.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchin, Y., and Moroz L.L.. 2008. Molluscan mobile elements similar to the vertebrate Recombination-Activating Genes. Biochem. Biophys. Res. Commun. 369:818–823. 10.1016/j.bbrc.2008.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval, P., Kriatchko A.N., Kumar S., and Swanson P.C.. 2008. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic Acids Res. 36:2060–2072. 10.1093/nar/gkn049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, S., Chaudhuri J., and Alt F.W.. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131. 10.1111/j.0105-2896.2004.00165.x [DOI] [PubMed] [Google Scholar]
- Sadofsky, M.J., Hesse J.E., McBlane J.F., and Gellert M.. 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21:5644–5650. 10.1093/nar/21.24.5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabla, N.M., Perry G.A., Palmer V.L., and Swanson P.C.. 2018. VprBP (DCAF1) Regulates RAG1 Expression Independently of Dicer by Mediating RAG1 Degradation. J. Immunol. 201:930–939. 10.4049/jimmunol.1800054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz, D.G., and Ji Y.. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263. 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- Schatz, D.G., and Swanson P.C.. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202. 10.1146/annurev-genet-110410-132552 [DOI] [PubMed] [Google Scholar]
- Silver, D.P., Spanopoulou E., Mulligan R.C., and Baltimore D.. 1993. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl. Acad. Sci. USA. 90:6100–6104. 10.1073/pnas.90.13.6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkus, C., Anand P., Bhattacharyya A., and Jones J.M.. 2007. Biochemical and folding defects in a RAG1 variant associated with Omenn syndrome. J. Immunol. 179:8332–8340. 10.4049/jimmunol.179.12.8332 [DOI] [PubMed] [Google Scholar]
- Simkus, C., Makiya M., and Jones J.M.. 2009. Karyopherin alpha 1 is a putative substrate of the RAG1 ubiquitin ligase. Mol. Immunol. 46:1319–1325. 10.1016/j.molimm.2008.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., and Gellert M.. 2015. Role of RAG1 autoubiquitination in V(D)J recombination. Proc. Natl. Acad. Sci. USA. 112:8579–8583. 10.1073/pnas.1510464112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder, S.R., Dudley D.D., Alt F.W., Takahama Y., and Akamatsu Y.. 2004. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 32:4539–4549. 10.1093/nar/gkh778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, G., Maman Y., Resch W., Kim M., Yamane A., Qian J., Kieffer-Kwon K.R., Mandal M., Ji Y., Meffre E., et al. 2015. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 162:751–765. 10.1016/j.cell.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, A., Sobacchi C., Notarangelo L.D., Bozzi F., Abinun M., Abrahamsen T.G., Arkwright P.D., Baniyash M., Brooks E.G., Conley M.E., et al. 2001. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 97:81–88. 10.1182/blood.V97.1.81 [DOI] [PubMed] [Google Scholar]
- Wang, X., Xiao G., Zhang Y., Wen X., Gao X., Okada S., and Liu X.. 2008. Regulation of Tcrb recombination ordering by c-Fos-dependent RAG deposition. Nat. Immunol. 9:794–801. 10.1038/ni.1614 [DOI] [PubMed] [Google Scholar]
- Wang, H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., and Jaenisch R.. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 153:910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko, V., Xue Z., and Sadofsky M.. 2003. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 17:581–585. 10.1101/gad.1058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang X., Ba Z., Liang Z., Dring E.W., Hu H., Lou J., Kyritsis N., Zurita J., Shamim M.S., et al. 2019. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature. 573:600–604. 10.1038/s41586-019-1547-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 1).
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 2).
shows utilization of Igh gene segments in primary B220+ splenocytes (HTGTS–Rep-seq, experiment 3).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 1).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 2).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (HTGTS–Rep-seq, experiment 3).
shows utilization of Igh gene segments in primary B220+IgM− bone marrow cells (ImmunoSEQ).
shows utilization of Tcrb gene segments in VDJ rearrangements in primary CD8+CD4+CD3εlo thymocytes (ImmunoSEQ).
lists oligos used and their sequences.
Data Availability Statement
The accession no. for the HTGTS–Rep-seq datasets reported in this paper is Gene Expression Omnibus no. GSE180734. The ImmunoSEQ datasets reported can be found at https://doi.org/10.21417/HAB2021JEM and https://clients.adaptivebiotech.com/pub/beilinson-2021-jem.