Abstract
Although genome-wide association studies have identified more than eighty genetic variants associated with non-small cell lung cancer (NSCLC) risk, biological mechanisms of these variants remain largely unknown. By integrating a large-scale genotype data of 15,581 lung adenocarcinoma (AD) cases, 8,350 squamous cell carcinoma (SqCC) cases, and 27,355 controls, as well as multiple transcriptome and epigenomic databases, we conducted histology-specific meta-analyses and functional annotations of both reported and novel susceptibility variants. We identified 3,064 credible risk variants for NSCLC, which were overrepresented in enhancer-like and promoter-like histone modification peaks as well as DNase I hypersensitive sites. Transcription factor enrichment analysis revealed that USF1 was AD-specific while CREB1 was SqCC-specific. Functional annotation and gene-based analysis implicated 894 target genes, including 274 specifics for AD and 123 for SqCC, which were overrepresented in somatic driver genes (ER=1.95, P=0.005). Pathway enrichment analysis and Gene-Set Enrichment Analysis revealed that AD genes were primarily involved in immune-related pathways, while SqCC genes were homologous recombination deficiency related. Our results illustrate the molecular basis of both well-studied and new susceptibility loci of NSCLC, providing not only novel insights into the genetic heterogeneity between AD and SqCC but also a set of plausible gene targets for post-GWAS functional experiments.
Keywords: Lung cancer, genome-wide association study, function annotation, immune, homologous recombination repair deficiency, genetic heterogeneity
Introduction
Lung cancer is the leading cause of cancer morbidity and mortality worldwide[1]. Non-small cell lung cancer (NSCLC) is the main type of lung cancer, accounting for ~85% of all lung cancer cases[2]. Adenocarcinoma (AD) and squamous cell carcinoma (SqCC) represent the two major histological subtypes of NSCLC. Although tobacco smoking is generally considered as the major cause of lung cancer, genetic factors also play an important role in the development of lung cancer. Genome-wide association studies (GWASs) have previously identified 81 lung cancer susceptibility variants in 51 loci that robustly associated with lung cancer risk[3, 4]; however, only a relatively small proportion of lung cancer heritability (0.7%~2.4%) can be explained by the variants identified so far[5, 6]. Additionally, delineating the biological mechanism of susceptibility variants underlying the development of lung cancer has also lagged far behind[3].
The challenge of pinpointing predisposition genes in susceptibility loci lies in several aspects. First, most GWAS implicated variants are tag single-nucleotide polymorphisms (SNPs) which represent for all co-inherited SNPs in the same haplotype, and thus the direct inference of statistically associated SNPs rarely yields functional variants[7]. Second, as the vast majority (>80%) of the GWAS hits are located in the non-coding regions, distinguishing functional SNPs from non-functional ones can be a great challenge[8]. Third, as previous studies mainly focused on mixed NSCLC, the biological and genomic heterogeneity of lung AD and SqCC[9–11] also leads to a disparity of functional signals. Thus, further dissection of the genetic underpinnings of lung AD and SqCC is crucial for the understanding of lung cancer pathogenesis.
In recent years, the emergence of epigenomic datasets, such as the Encyclopedia of DNA Elements (ENCODE) project[12], the Functional Annotation of the Mammalian Genome (FANTOM) project[13], and the Roadmap Epigenomics project[14] provide a good opportunity to unveil the function of disease-associated signals from epigenomic level. By leveraging the wealth epigenomic data, many studies have attempted to illuminate the biological meanings of GWAS-implicated cancer risk loci. For example, a study from Michailidou et al.[15] examined the overlap of breast cancer variants with cis-regulatory elements (CREs) and observed a significant enrichment. Similar results were observed for the risk loci of colorectal, head and neck, ovary, and prostate cancers[16], and such CREs were active in disease-related cell lines[17]. Thus, incorporating functional information into association signals has the potential to improve our understanding of the biological consequences of human cancers.
In this study, we first conducted histology-specific genome-wide meta-analyses and then performed a comprehensive functional annotation of NSCLC susceptibility variants by integrating multiple in-house and publicly available databases. Our results first illustrate the molecular basis of all known susceptibility loci of lung cancer and provide novel insights into the genetic heterogeneity between lung AD and SqCC.
Materials and Methods
Study populations
We conducted a meta-analysis with 27,120 NSCLC cases (15,581 AD, 8,350 SqCC, and 3,189 other cases) and 27,355 controls. Of all subjects, 26,655 Chinese participants were from our previously published Nanjing Medical University (NJMU) lung cancer GWAS study[4] and 27,820 European participants were from the TRICL-ILCCO OncoArray project[18]. NJMU lung cancer GWAS study was consist of three datasets, including 19,546 participants (10,248 cases and 9,298 controls) from NJMU Global Screening Array (GSA) project[4], 5,203 participants (2,126 cases and 3,077 controls) from NJMU GWAS project[19], and 1,906 participants (953 cases and 953 controls) from NJMU OncoArray project[4]. Informed consent was obtained from all the participants included in this study, and each study was approved by the corresponding institutional review board. Detailed demographic characteristics of included participants have been described in our previous study[4], and were provided in Table 1.
Table 1.
Demographic characteristics of lung AD, SqCC and non-cancer controls included in this study
| NJMU project | TRICL-ILCCO OncoArray project | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lung AD | Lung SqCC | Control | Lung AD | Lung SqCC | Control | |||||||
| Number | % | Number | % | Number | % | Number | % | Number | % | Number | % | |
| Total | 8762 | 100.0 | 3860 | 100.0 | 13328 | 100.0 | 6819 | 100.0 | 4490 | 100.0 | 14027 | 100.0 |
| Age (Mean±S.D.) | 58.63±10.52 | 61.16±9.42 | 59.31±10.42 | 63.57±10.80 | 64.84±9.62 | 61.77±10.29 | ||||||
| Gender | ||||||||||||
| Male | 4650 | 53.1 | 3470 | 89.9 | 8605 | 64.6 | 3626 | 53.2 | 3489 | 77.7 | 8638 | 61.6 |
| Female | 4112 | 46.9 | 390 | 10.1 | 4723 | 35.4 | 3192 | 46.8 | 1001 | 22.3 | 5386 | 38.4 |
| Missing value | n/a | n/a | n/a | n/a | n/a | n/a | 1 | 0.0 | n/a | n/a | 3 | 0.0 |
| Smoking status | ||||||||||||
| Ever smoker | 3364 | 38.4 | 3172 | 82.2 | 5606 | 42.1 | 5771 | 84.6 | 4276 | 95.2 | 9339 | 66.6 |
| Life-long non-smoker | 5397 | 61.6 | 688 | 17.8 | 7720 | 57.9 | 974 | 14.3 | 156 | 3.5 | 4412 | 31.5 |
| Missing value | 1 | 0.0 | n/a | n/a | 2 | 0.0 | 74 | 1.1 | 58 | 1.3 | 276 | 1.9 |
Abbreviations: AD, adenocarcinoma; SqCC, squamous cell carcinoma.
n/a: No patients with missing information.
Quality control and imputation
Detailed imputation process has been described in our previous study[4]. Briefly, we first excluded samples with genotype completion rates <95%, gender discrepancies, familial relationships, extreme heterozygosity rates (6 S.D. from the mean), or population stratification (>6 S.D. from the mean on any one of the top ten principal components). Then, we excluded duplicate markers or SNPs with call rates <95%, minor allele frequencies (MAFs) <0.01 or Hardy-Weinberg equilibrium (HWE) P value <1×10−7 in controls or HWE P value <1×10−12 in cases. We phased the haplotypes with SHAPEIT v2[20] and imputed with IMPUTE2[21]. The 1000 Genomes Project (the Phase III integrated variant set release, across 2,504 samples) was set as the reference.
Identification of NSCLC risk loci
To perform functional annotation, we first conducted genome-wide meta-analyses for NSCLC, lung AD and SqCC respectively, and then derived a set of lung cancer risk associated index variants including both 81 previously reported SNPs (Table S1) and those with a genome-wide significant P value <1×10−6 in our overall NSCLC or histological meta-analyses. For 81 previously reported variants, index variants were defined as those met either of the following criteria: (1) MAF ≥0.01; and (2) SNPs in weak linkage disequilibrium (LD, r2 <0.6) with each other. For other independent variants, index variants were defined if met one or more of the following criteria in our meta-analyses: (1) MAF ≥0.01; (2) with a genome-wide significant P value <1×10−6 in NSCLC, lung AD or SqCC meta-analysis; and (3) SNPs in weak LD with each other and previously reported variants (r2 <0.01). If one SNP identified in histology-specific meta-analysis also showed association with the other histology of lung cancer (AD or SqCC) (P <0.05), it was considered as NSCLC related. Thus, 67 index variants derived from both 81 previously reported SNPs and those from our genome-wide meta-analyses were included in the following analysis. Then, we further mining SNPs in strong LD (r2 ≥0.6) with above defined index SNPs and physically within 500 kb upstream or downstream of the index SNP. All above defined index SNPs and associated SNPs in strong LD were considered as credible risk variants (CRVs) (Figure 1A).
Figure 1. Functional evaluation of 3,064 lung cancer related credible risk variants (CRVs) defined in this study.

A. Flowchart for the study design.
B. Genomic distribution of 3,064 lung cancer CRVs.
The x-axis indicates the number of CRVs included in the genomic region type.
C. Enrichment of defined lung cancer CRVs (1,020 for lung AD and 220 for SqCC) in histone modification peaks and DNase I hypersensitive sites.
The x-axis indicates different types of modification peaks in lung cancer cell line types. A549, lung AD cell line; NHLF, lung fibroblasts cell line; AG04450 and HPF, lung fibroblasts cell lines; Lung, normal lung tissue.
D. Enrichment of defined lung cancer CRVs (1,020 for lung AD and 220 for SqCC) in transcriptional factor binding sites.
The x-axis indicates binding sites of different transcriptional factors. IMR90, lung fibroblasts cell line.
Identification of target genes for NSCLC risk loci by functional annotation
To define candidate target genes for lung cancer risk loci, we performed functional annotation with an extended in silico prediction of GWAS targets (INQUISIT) strategy[15] and calculated a score for each gene-CRV pair representing for the coding impact or potential regulatory mechanisms (proximal or distal gene regulation) by integrating multiple lines of evidence (Table S2–3). Each target gene was scored based on distally regulation, proximally regulation, and coding sequence.
For the distally regulated gene, one score was given if: (1) the CRV was located in an enhancer element that predicted to physically interact with the promoter of the target gene by FANTOM5[13] or PreSTIGE [22]; (2) the enhancer element containing the CRV overlapped with the transcription factor binding sites (TFBS) of one transcription factor (TF) (two scores were given if overlapped with more than one TF); (3) the CRV is an expression quantitative trait loci (eQTL) for that gene in the Genotype-Tissue Expression (GTEx), The Cancer Genome Atlas (TCGA) or Nanjing Lung Cancer Cohort (NJLCC) [23] databases; or (4) the gene was listed as a lung cancer somatic driver gene (except for driver gene enrichment analysis). Additionally, two scores were added if the CRV was also located in an enhancer element that physically interact with the promoter of that gene based on Hi-C experiment. However, the score was down-weighted by multiplying by 0.05 if the target gene was separated from the CRV by topologically associating domain (TAD) boundaries, or down-weighted by multiplying by 0.1 if the gene was low expressed in the GTEx normal lung tissues (less than 1% samples with Transcripts Per Million [TPM] greater than 0.1), TCGA tumor/adjacent samples (less than 1% samples with Fragments Per Kilobase Million [FPKM] greater than 0.1) and NJLCC tumor/adjacent samples (less than 1% samples with TPM greater than 0.1).
Proximally regulated genes were defined as those with CRVs located 1 kb upstream and 100bp downstream surrounding the transcription start sites (TSSs). One score was given if: (1) the gene was overlapped with promoter histone modification peaks (H3K4me3 or H3K9ac); (2) the gene was listed as a lung cancer somatic driver gene (except for driver gene enrichment analysis); (3) the histone modification peak that the CRV resided was also intersected with the TFBS of TFs; or (4) the CRV is an eQTL for that gene in GTEx, TCGA or NJLCC databases. The down-weighted criteria were the same as that for distally regulated genes.
CRVs located in the exonic regions were evaluated for their impact on the protein function. Combined Annotation Dependent Depletion (CADD)[24], Functional Analysis through Hidden Markov Models (FATHMM)[25], LRT[26], MutationTaster[27], PolyPhen-2[28], and Sorting Tolerant from Intolerant (SIFT)[29] were used for the evaluation of missense variants. Other scoring strategy was the same as that for distally regulated genes.
Altogether, scores in the distal regulation category range from 0 to 7, in the promoter category from 0 to 4, and in the coding category from 0 to 3. We classified the candidate target genes into four levels based on the integrated scores: level 1 (distal score ≥4, promoter score ≥3, or coding score =3), level 2 (distal score 1–3, promoter score 1–2, or coding score 1–2), level 3 (any score greater than 0), and level 4 (score 0). Genes categorized into level 1–2 were considered as potential targets and were included in the following analysis.
Expression quantitative trait loci analysis based on NJLCC and TCGA data
In addition to the GTEx project (v7), we also performed eQTL analysis with data from NJLCC and TCGA projects. The NJLCC[23] project included 90 Chinese NSCLC samples with available clinical information, gene expression data, copy number variation profiles and matched genotyping data. Gene expression data was available for 98 tumor/adjacent pairs. NJLCC samples were genotyped with whole-genome sequencing and the expression data was quantified by RNA-seq (Illumina HiSeq 1500 platform)[23]. A systematic quality control (QC) procedure was performed to filter out samples with missing genotypes and duplicates. Principal component analysis (PCA) was also performed. Finally, all 90 NJLCC samples (55 lung ADs and 35 SqCCs) were included in the eQTL analysis.
Similarly, the TCGA project included 106 NSCLC samples with matched clinical information, gene expression data, copy number variation data, and genotyping data. TCGA samples were genotyped using Affymetrix Genome-Wide Human SNP Array 6.0 and the data was downloaded from the TCGA Firehose at the MIT Broad Institute. The RNA-seq (Illumina) based expression data of 106 TCGA matched adjacent normal samples was quantified by FPKM and obtained from the UCSC Xena website. For the genotyping data, we also performed a systematic QC procedure and PCA, and 3 TCGA samples with Asian ancestry were removed. As a result, 103 TCGA samples (55 lung ADs and 48 SqCCs) remained in the following analysis.
For eQTL analysis, we performed a linear regression using the R package Matrix eQTL[30] with default parameters. We set gene expression as the outcome, and SNP genotype as the covariate of interest with adjustment for age, gender, smoking status, the top ten principal components, and somatic copy number status.
Functional enrichment analysis of defined CRVs
To investigate the enrichment or depletion in chromatin modification peaks, we estimated the distribution of above CRVs in active promoter and enhancer regions identified in normal lung tissues, lung fibroblasts (NHLF), and lung cancer (A549) cell lines by using Variant Set Enrichment (VSE)[31]. The same analysis was performed in three lung fibroblasts (IMR90, HPF and AG04450) and A549 cell lines to evaluate the overrepresentation of CRVs in DNase I hypersensitive sites (DHS) regions or TFBS. All the histone modification peaks of promoter and enhancer marks (H3K4me3, H3K9ac, H3K4me1 and K3K27ac), DHS, and TFBS data were downloaded from the UCSC Genome Browser.
Gene-based analysis
We performed gene-based analysis with genome-wide gene-based association study (GWGAS) in MAGMA[32]. The P values from the GWAS meta-analyses for lung cancer, lung AD and SqCC were used as input, and all 19,427 protein-coding genes from the NCBI 37.3 gene definitions were used as the basis for GWGAS. We annotated all SNPs in our genome-wide meta-analyses to above genes, resulting in 18,233, 18,233 and 18,232 protein-coding genes that were represented by at least one SNP in the NSCLC, lung AD or SqCC meta-analyses, respectively. Genes with Benjamini-Hochberg (BH) adjusted P value <0.05 were considered as significant.
Driver gene enrichment analysis
A gene was considered as lung cancer somatic driver gene if met one of the following criteria: (1) the gene was included in the COSMIC Cancer Gene Census (v78) and showed evidence to be lung cancer related[33]; (2) the gene was categorized as lung cancer-related mutational drivers, somatic copy number alteration (SCNA) drivers, or fusion drivers in the IntOGen database[34]; or (3) the gene was identified as significantly mutated genes or SCNA-related genes in recent whole-genome or whole-exome sequencing studies[23, 35–39]. Finally, we listed a total 374 coding genes and 4 non-coding genes as lung cancer somatic driver genes. To evaluate the enrichment of these genes in our defined lung cancer target genes, we first re-scored genes implicated by INQUISIT, and then examined the overlap between this list of drivers and the target genes with different levels of evidence and performed fisher exact test to obtain the significance. To avoid the bias of non-coding genes, only protein-coding genes were included in this analysis.
Pathway enrichment analysis
We performed pathway enrichment analysis on the above defined candidate target genes as well as genes identified by gene-based analysis to evaluate their potential function in the development of lung cancer. The analysis was conducted with the Reactome Pathway Database[40] using R package clusterProfiler[41] and pathways with BH adjusted P value <0.05 were retained.
Gene-Set Enrichment Analysis (GSEA)
The immune infiltration proportions[42] and homologous recombination deficiency (HRD)[43] index of TCGA lung AD and lung SqCC samples were downloaded from previously published studies. We first calculated the correlation coefficients between the proportions of six types of immune cells (B cell, CD4 T cell, CD8 T cell, Neutrophils, Macrophages and Dendritic cells) or HRD index and the expression of all protein-coding genes. Then, a ranked list of correlation coefficient was analyzed by Gene-Set Enrichment Analysis (GSEA) with our predefined lung AD and SqCC genes. This analysis was performed with R package clusterProfiler[41].
Statistical analyses
Detailed description for the meta-analysis of GWAS data from Chinese and European populations was provided in our previous study[4]. Briefly, the association testing for each variant was performed using the SNPTEST software (v2.5.4) with adjustment for age, gender, and the principal components. Meta-analysis was performed with the fixed-effects inverse variance-weighting approach by using METAL [44]. Genetic variant with I2 ≥75% or P value for Cochran’s Q statistic ≤1.0×10−4 was considered with a high degree of heterogeneity, and was excluded from further analysis [45, 46]. The LD coefficients (R2 and D’) was calculated with PriorityPruner, and the genotyping data of the East Asian and European populations from the 1000 Genomes Project (the Phase III integrated variant set release) were set as the reference. General analyses were performed with R software (version 3.5.1). All statistical tests were two-sided.
Results
Definition of credible risk variants for non-small cell lung cancer
We performed overall and histological GWAS meta-analyses (Figure 1A, Table 1). In order to clarify potential functional signals, we first defined 67 index SNPs based on the following criteria: (1) 58 index SNPs represented for 81 previously reported SNPs (r2 ≥ 0.6) (Table S1); and (2) 9 independent index SNPs with a meta P value <1×10−6 in our overall NSCLC or histological meta-analyses (Table S4). Of 9 SNPs with a meta P value <1×10−6, two were located in previously reported loci but showed weak LD (r2 <0.01) with previously reported SNPs (6p22.1: rs1815741: OR=0.86, 95% CI=0.81–0.91, P=2.99×10−7; 11q23.3: rs4938515: OR=1.07, 95% CI=1.04–1.10, P=3.46×10−7), and the other seven SNPs included five for NSCLC (4p14: rs116205103: OR=0.83, 95% CI=0.77–0.89, P=1.82×10−7; 15q24.1: rs76354137: OR=0.92, 95% CI=0.88–0.95, P=5.74×10−7; 2q21.3: rs3217451: OR=0.90, 95% CI=0.87–0.94, P=7.35×10−7; 4q27: rs35661893: OR=0.93, 95% CI=0.91–0.96, P=3.56×10−6; 13q24: rs719739: OR=0.94, 95% CI=0.92–0.97, P=1.94×10−5), one for AD (9q31.3: rs12006500: OR=1.12, 95% CI=1.07–1.16, P=8.43×10−8), and one for SqCC (8p23.1: rs2945908: OR=0.89, 95% CI=0.85–0.93, P=7.92×10−7) (Table S4, Figure S1).
Then, we defined additional CRVs for further functional annotation (SNPs with r2 ≥ 0.6 with one of 67 independent index SNPs and within 500kb upstream or downstream of the corresponding index SNPs). Finally, we identified 3,064 CRVs in the following analysis, including 1,842 for NSCLC, 1,020 for AD and 220 for SqCC (Figure 1A).
Enrichment analysis of NSCLC CRVs
Most of the defined CRVs were in the intronic (intronic and ncRNA intronic: 1489; 48.60%) or intergenic regions (intergenic, downstream and upstream: 1444; 47.13%) (Figure 1B). We systematically evaluated the enrichment of these variants in histone modification peaks. Interestingly, we observed a significant enrichment of NSCLC related CRVs in promoter-like (H3K4me3 and H3K9ac) and enhancer-like (H3K4me1 and H3K27ac) histone modification peaks in normal lung tissues, lung fibroblasts (NHLF) and lung cancer (A549) cell lines (Figure 1C), and most (7/11) of the enrichment degrees were greater for lung AD related CRVs (Figure 1C). Additionally, the defined CRVs were also enriched in DHS regions in lung fibroblasts (AG04450) and lung cancer (A549) cell lines (Figure 1C).
Then, we conducted TF enrichment analysis and strong signals were observed for the binding sites of ATF3, POLR2A, TCF12, MAX, YY1, CTCF, and MAFK in lung fibroblasts (IMR90 and AG04450) and lung cancer (A549) cell lines at the significant level of PBH <0.05. Of these TFs, CTCF (HPF, AG04450 and IMR90), RAD21 (IMR90) and USF1 (A549) were special for lung AD while TCF12 (A549) and CREB1 (A549) were for lung SqCC (Figure 1D).
Additionally, 22 of the 39 exonic variants were non-synonymous, including two nonsense and 20 missense variants (Table S5, Figure 1B). The index SNP rs11571833 (K3326*, c.A9976T) in 13q13.1 was a nonsense variant in exon 27 of BRCA2. The T allele could significantly increase the risk of lung cancer (OR=1.50, 95% CI=1.26–1.78, P=4.63×10−6). Of the missense variants, rs17121881 (c.T281A, I94N) in AMICA1 was predicted with the highest CADD score (CADD score=23.5) and could lead to an isoleucine-to-asparagine change. Rs17121881 was in exon 3 of AMICA1 and showed strong LD with the index SNP rs55768116 (r2=0.78) in 11q23.3. The A allele of rs17121881 could significantly increase the risk of lung cancer (OR=1.08, 95% CI=1.05–1.11, P=2.10×10−9) as well as reduce the expression of AMICA1 (GTEx: β=−0.10, P=0.005; NJLCC: β=−0.18, P=0.02). Interestingly, AMICA1 also showed a decreased expression in both lung AD and SqCC samples when compared to adjacent normal samples in TCGA (AD: P=1.03×10−17; SqCC: P=7.75×10−23) and NJLCC (AD: P=1.76×10−18; SqCC: P=6.58×10−18) data (Figure S2), suggesting a tumor suppressor role during the development of lung cancer.
Systematic functional annotation of NSCLC CRVs
To link the candidate variants to genes, we then applied an extended INQUISIT functional annotation strategy and mapped CRVs to potential target genes by evaluating the impact on coding sequences, proximal promoter, and distal enhancer regulations (Figure 1A). Among all 3,064 CRVs, the coding impact evaluation strategy aligned CRVs to 25 genes, the proximal regulatory gene mapping strategy matched CRVs to 624 genes, and the distal regulatory gene mapping strategy annotated CRVs to 1,014 genes (Table S6). Above findings resulted in 1,047 unique mapped genes, among which 803 genes categorized as level 1 and 2 were considered as functional target genes and included in the following analysis (distal regulation strategy: 589 genes; proximal regulation strategy: 604 genes; coding impact strategy: 18 genes) (Figure 2A). Of these 803 genes, 395 were implicated by at least two mapping strategies, and 13 were implicated by all three (Figure 2B, Table S6). Additionally, 227 genes were implicated in lung AD samples while only 82 genes were in lung SqCC samples.
Figure 2. Implicated lung cancer target genes by functional annotation.
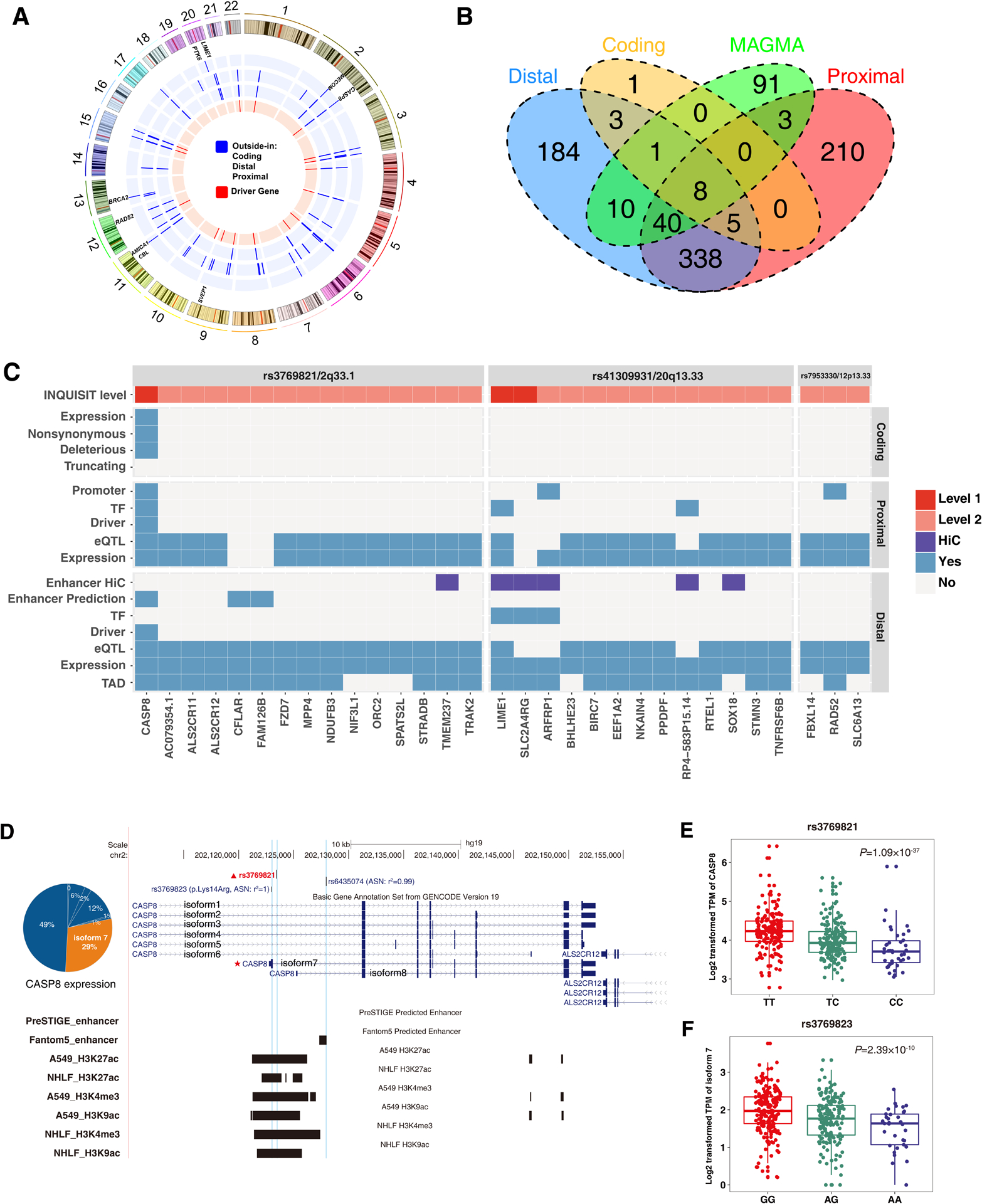
A. Circos plot showing 803 implicated genes by distal mapping, promoter mapping and coding mapping strategies.
Blue indicates the mapping strategy (from inside to outside: distal, promoter, and coding mapping) and red indicates if the implicated gene is a driver gene.
B. Venn diagram showing the number of overlapped genes implicated by distal mapping, promoter mapping, coding mapping strategies, and GWGAS.
C. Detailed functional annotation results for three risk loci of lung cancer.
The x-axis indicates the implicated genes, and y-axis indicates the annotation evidence types.
D. Genomic region of CASP8 in 2q33.1.
E-F. eQTL analysis of two CRVs (rs3769821 and rs3769823) and CASP8 expression in 383 GTEx lung tissues.
Of the newly identified genes, CASP8 was in a locus defined in our previous study[4] and was implicated by all three mapping strategies. The index SNP rs3769821 at 2q33.1, located in the histone modification marks targeting both promoters and enhancers in A549 and NHLF cell types (Figure 2C–D), was confirmed as a cis-eQTL variant for CASP8 in 383 GTEx lung tissues (P=1.09×10−37) (Figure 2E). Interestingly, we also identified a missense variant (rs3769823, c.41A>G, p.Lys14Arg), in strong LD (r2=1) with rs3769821, that was located in the first exon of CASP8 isoform 7 (Figure 2D). This isoform was highly expressed in normal lung tissues and was found to be regulated by rs3769823 (P=2.39×10−10) (Figure 2F). Additionally, we identified RAD52 as a lung SqCC related gene which was regulated by both distal and proximal elements (Figure S3). Rs11571376 (12p13.33) was located at the promoter region of RAD52 and the C allele could significantly increase the risk of lung cancer (OR=1.10, 95% CI=1.05–1.14, P=2.27×10−5). Interestingly, C allele of rs11571376 was also associated with the expression of RAD52 in normal lung tissues (beta=0.21, P=2.30×10−17), and the expression of RAD52 was significantly elevated in lung SqCC samples (TCGA: P=1.71×10−4; NJLCC: P=6.27×10−5) (Figure S3). For lung AD, we also identified a novel gene LIME1 with the highest INQUISIT score. Rs6122147, in strong LD with the index SNP rs41309931 (r2=0.98) in 20q13.3, was located in a distal enhancer element that physically interacted with the promoter of LIME1 in A549 cell line (Figure S4A). The T allele of rs6122147 could significantly increase the risk of lung cancer (OR=1.06, 95% CI=1.02–1.11, P=4.42×10−3) and decrease the expression of LIME1 in normal lung tissues (β=−0.14, P=4.20×10−4) (Figure S4B).
Gene-based analysis and driver gene enrichment analysis
To estimate the aggregated association of lung cancer, we performed GWGAS analysis using MAGMA and identified 154 lung cancer associated genes (Figure 3A, Table S7–9), of which 62 have been implicated in previous functional annotation analysis (Figure 2A). Of these 154 genes, 70 genes were specific for AD and 57 were specific for SqCC. When combined with 803 genes identified by INQUISIT, we implicated a total of 894 susceptibility genes for lung cancer (Figure 2A), of which 274 were specific for AD and 123 were for SqCC (Figure 3B).
Figure 3. Implicated lung cancer target genes by gene-based and pathway enrichment analyses.
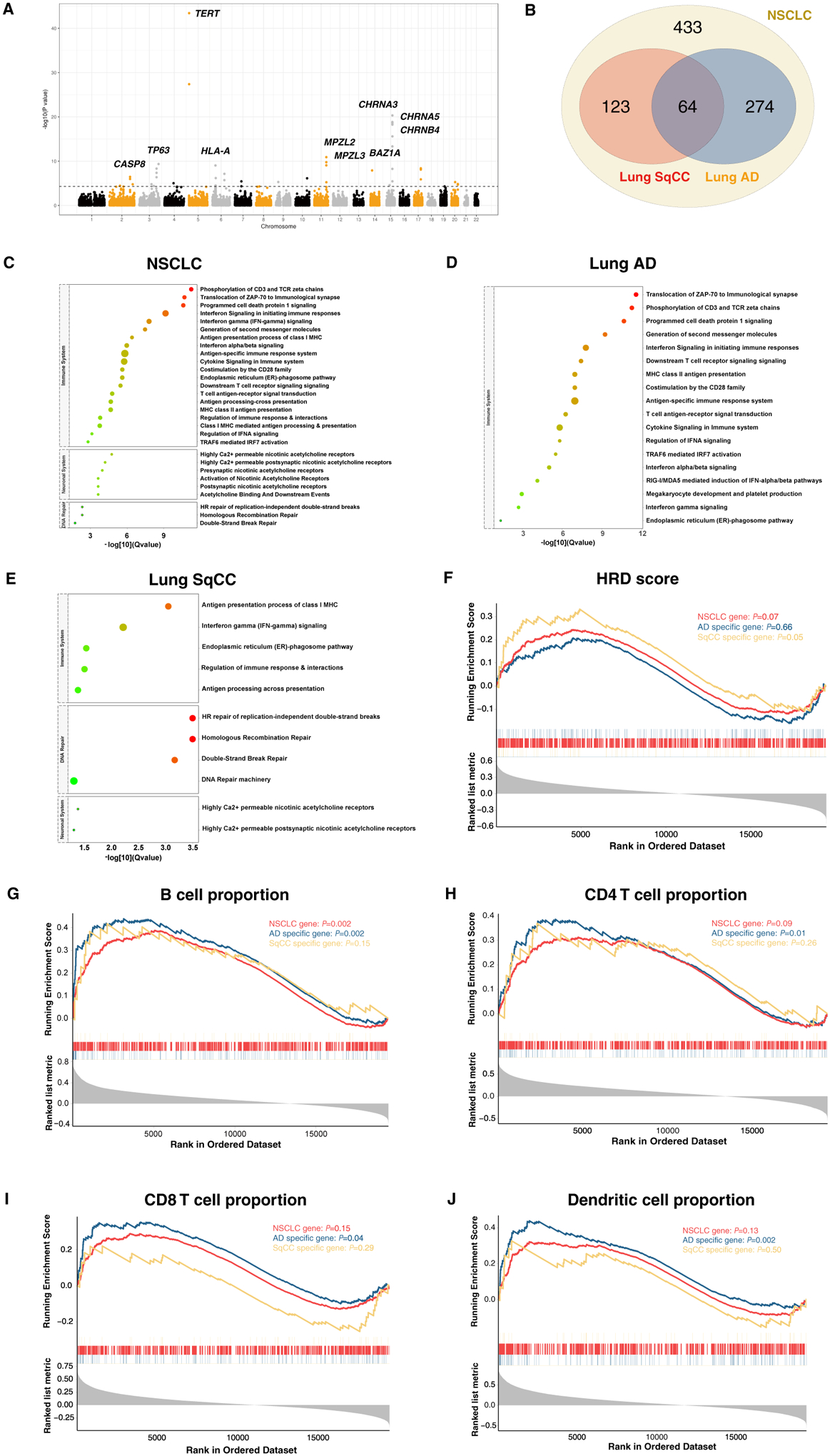
A. Manhattan plot of the GWGAS analysis for NSCLC meta-analysis.
The y axis shows the −log10 transformed two-tailed P value of each gene from a linear model, and chromosomal position is shown on the x axis.
B. Venn diagram showing the overlap of genes implicated by INQUISIT and MAGMA in NSCLC, lung AD and SqCC.
C. Pathway enrichment analysis of all genes implicated by INQUISIT and GMAMA for NSCLC.
D. Pathway enrichment analysis of lung AD genes.
E. Pathway enrichment analysis of lung SqCC genes.
F. GSEA analysis of NSCLC, lung AD and SqCC genes with homologous recombination deficiency.
G. GSEA analysis of NSCLC, lung AD and SqCC genes with B cell proportion.
H. GSEA analysis of NSCLC, lung AD and SqCC genes with CD4 T cell proportion.
I. GSEA analysis of NSCLC, lung AD and SqCC genes with CD8 T cell proportion.
J. GSEA analysis of NSCLC, lung AD and SqCC genes with dendritic cell proportion.
Then, we evaluated the association between our defined target genes and previously known somatic driver genes (Table S10). To avoid the bias of non-coding genes, only protein-coding genes were included in this analysis. Among the genes implicated by INQUISIT (level 1–4), we observed an enrichment of our defined target genes in 374 established lung cancer somatic-driver genes (23 out of 374 genes, ER=1.95, P=0.005), including EGFR, CDKN2A, CHEK2, and TP53BP1. The enrichment degree increased with the level of evidence (level 1: ER=14.05, P=6.90×10−5; level 2: ER=1.38, P=0.30; level 3: ER=2.22, P=0.06; level 4: ER=0.00, P=1.00; Ptrend=0.06). Similar results were found when genes implicated by distal regulation (ER=1.97, P=0.005; Ptrend=0.95), proximal regulation (ER=1.43, P=0.25; Ptrend=2.95×10−8), and coding impact (ER=4.64, P=0.08; Ptrend=0.13) were included. For genes derived from gene-based analysis, we also observed a significant enrichment in lung cancer somatic-driver genes (ER=4.48, P=3.87×10−5). Additionally, four somatic drivers (PTK6, CBL, MECOM, and SVEP1) were implicated specially for lung AD (ER=2.48, P=0.09), but no lung SqCC somatic drivers were detected.
Pathway enrichment analysis
To further explore biological pathways involved in the process of lung tumorigenesis, we performed pathway enrichment analysis of our defined target genes. To avoid the influence of non-coding genes, we only included 592 protein-coding genes in this analysis. The result revealed the involvement of 29 pathways (PBH<0.05) in the development of NSCLC, including 20 pathways related to immune function, such as PD-1 signaling (P=1.09×10−13) and interferon gamma signaling pathway (P=1.15×10−10), and six pathways in the neuronal system that related to nicotinic acetylcholine receptors, and three pathways in the DNA repair system (Figure 3C, Table S11). We also performed the same analysis for lung AD and SqCC genes, and identified that lung AD related genes were specifically enriched in immune related pathways (Figure 3D, Table S12), while lung SqCC genes were enriched in homologous recombination (HR)-related repair pathways (Figure 3E, Table S13), suggesting diverse mechanisms underlying the development of lung AD and SqCC.
Above findings indicated the importance of immune function and HR repair in the carcinogenesis of lung cancer. Thus, we evaluated the association of defined lung cancer genes with immune infiltration proportions and HRD index in TCGA lung AD and SqCC samples. Interestingly, we identified that lung SqCC genes were overrepresented in HRD-related genes (NES=1.33, P=0.05) (Figure 3F) while lung AD genes were significantly overrepresented in four types of immune cells-related genes (B cell: NES=1.39, P=0.002; CD4 T cell: NES=1.32, P=0.01; CD8 T cell: NES=1.31, P=0.04; Neutrophil: NES=1.19, P=0.10; Macrophage: NES=1.02, P=0.44; Dendritic: NES=1.46, P=0.002) while lung SqCC genes not (B cell: NES=1.22, P=0.15; CD4 T cell: NES=1.14, P=0.26; CD8 T cell: NES=−1.06, P=0.29; Neutrophil: NES=0.94, P=0.62; Macrophage: NES=−1.22, P=0.17; Dendritic: NES=1.00, P=0.50) (Figure 3G–J).
Discussion
In this study, we performed a combined strategy of large-scale genome-wide meta-analysis and functional annotation to identify biological significance of lung cancer susceptibility loci and implicated a total of 894 candidate target genes. These predisposition genes could modify the risk of lung cancer by both shared and histology-specific transcriptional (enhancers, promoters and TF) or translational (missense and nonsense variants) regulations. Furthermore, pathway enrichment and GSEA analysis indicated the importance of immune and HR-related DNA repair during the carcinogenesis process of lung AD and SqCC. These findings provided both a rich set of plausible gene targets for further functional studies and novel insights into understanding the biological underpinnings underlying the development of different histology types of lung cancer.
Consistent with previous studies, NSCLC CRVs were primarily mapped to the non-coding regions and showed a strong enrichment in CREs such as enhancer elements and histone modification peaks, suggesting that these variants contributed to the development of NSCLC mainly through transcriptional regulation. Interestingly, TFBS enrichment analysis implicated some pathology-specific TFs. CREB1 (cyclic AMP response element-binding protein) is a transcriptional coactivator which plays important roles in the differentiation of bronchial epithelial cells and is overexpressed in NSCLC samples[47]. Many studies have identified that CREB1 could be activated by nicotine exposure[48, 49], and the activated CREB1 recruits additional transcriptional machinery elements and leads to tumorigenesis[49]. Thus, CREB1 could be a possible target for the pathobiological process of smoking induced lung SqCC. As a member of the basic helix-loop-helix leucine zipper family, USF1 (upstream transcription factor 1) functions as a cellular transcription factor[50] and regulates the expression of SP-A[51]. SP-A is a lung-specific gene, especially for type II cells which is the origin of lung AD[51]. Thus, USF1 may regulate the risk of lung AD by modifying the expression of SP-A. Although exact roles of these TFs in the susceptibility to lung cancer have not been comprehensively studied, the overrepresented binding sites provides an improved understanding of the transcriptional regulation mechanism in NSCLC etiology.
In previous studies, the development of lung AD and SqCC were found to share many genetic factors[3, 4]. However, in recent years, accumulating evidence suggested that these two types of lung cancer also had large discrepancy in terms of both germline variations and somatic alterations[18]. In this study, we identified a set of predisposition genes both shared by lung AD and SqCC and specific to each histology. One of the most interesting result is the identification of CASP8 in 2q33.1, a NSCLC risk locus reported in our previous study[4]. CASP8 encodes caspase 8, which is a multivalent controller of innate immune signaling and inhibits inflammasome activation in dendritic cells, interferon-regulatory factor 3 activation and pro-inflammatory cell death[52]. CASP8 has been implicated as a susceptibility gene for multiple cancers except for lung cancer[53, 54]. In this study, we identified that the expression of CASP8 is regulated by both a cis-eQTL in the enhancer element and a missense variant in the major isoform, suggesting a potential joint modification mechanism by which the susceptibility variants affect NSCLC risk[55]. Additionally, we identified some pathologically specific genes, such as LIME1 for AD and RAD52 for SqCC. LIME1 encodes a membrane raft-associated adaptor protein which is an organizer of immunoreceptor signaling[56] and involves in CD4 and CD8 coreceptor signaling[57]. Thus, we speculate that LIME1 might regulate lung AD risk by modifying immune response. RAD52 is a key member in HR repair[58], which functions in DNA repair during S phase of the cell cycle[59] and act as a regulator of genomic stability[60]. In this study, we identified that RAD52 expression was elevated by cis-eQTLs in both proximal and distal regulatory elements in lung SqCC samples, suggesting a tumor-promoting role of RAD52 in the development of lung SqCC[61]. Above results indicate an important role of immune response during lung AD carcinogenesis as well as HR deficiency during lung SqCC carcinogenesis.
Interestingly, pathway enrichment and GSEA analysis also provided evidence for the importance of immune system and HR-related DNA repair during the development of lung AD and SqCC. Tumor-prompting inflammation is defined as a tumor-enabling hallmark of cancer[62] and has previously been implicated in lung cancer[63, 64]. Our recent work also provided evidence for the specific association of immune infiltration with lung AD, and identified that inflammatory microenvironments formed in the early stage of lung AD[23], suggesting that immune infiltration occurring in the initial stage is the major risk factor for lung AD. As an indication of genomic instability[65], elevated somatic HRD level has also been reported in lung SqCC[43]. The strong association between lung SqCC susceptibility genes and HRD observed in this study provided additional evidence that the somatic differences may have genetic ancestry origins and could regulate lung SqCC risk. As cigarette smoking is the major causal factor for lung SqCC[66], we propose that the continuous exposure to tobacco may directly lead to an increased level of HRD[66], and finally lead to lung SqCC. Thus, the use of PD1/PD-L1 and PARP inhibitors, which target immune infiltration and HRD[67, 68], implicate significant potential for the prevention of lung AD and SqCC.
However, the interpretation of our findings needs to be considered within the limitations of the study. First, the aggregation of susceptibility variants identified in both European and Chinese populations prevented us from capturing the genetic heterogeneity within different ethnic and individuals. Second, as the annotation data used in this study were obtained from different sources of tissues and cell types, further biological experiments are needed to elucidate the exact molecular mechanisms of these variants underlying the development of lung cancer.
In conclusion, by integrating GWAS information, in-house and publicly available biological data, we illustrate the molecular basis of both well-studied and newly identified lung cancer susceptibility loci and provide novel insights into the understanding of genetic heterogeneity between lung AD and SqCC, which may serve as guides for post-GWAS functional experiments and clinical drug target testing.
Supplementary Material
Acknowledgements
This study was supported by the Key international (regional) cooperative research project (81820108028), the National Natural Science Foundation of China (81521004, 81922061, 81973123 and 81803306), the Science Foundation for Distinguished Young Scholars of Jiangsu (BK20160046), and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions [Public Health and Preventive Medicine]. CARET is funded by the National Cancer Institute, National Institutes of Health of USA through grants U01-CA063673, UM1-CA167462, and U01-CA167462.
Abbreviations:
- NSCLC
non-small cell lung cancer
- AD
adenocarcinoma
- SqCC
squamous cell carcinoma
- GWAS
genome-wide association study
- SNP
single-nucleotide polymorphism
- ENCODE
Encyclopedia of DNA Elements
- FANTOM
Functional Annotation of the Mammalian Genome
- CRE
cis-regulatory element
- NJMU
Nanjing Medical University
- GSA
Global Screening Array
- MAF
minor allele frequency
- HWE
Hardy-Weinberg equilibrium
- LD
linkage disequilibrium
- CRV
credible risk variant
- TFBS
transcriptional factor binding sites
- TF
transcription factor
- eQTL
expression quantitative trait loci
- GTEx
Genotype-Tissue Expression
- TCGA
The Cancer Genome Atlas
- NJLCC
Nanjing Lung Cancer Cohort
- TAD
topologically associating domain
- TPM
Transcripts Per Million
- FPKM
Fragments Per Kilobase Million
- TSS
transcription start site
- CADD
Combined Annotation Dependent Depletion
- FATHMM
Functional Analysis through Hidden Markov Models
- SIFT
Sorting Tolerant from Intolerant
- QC
quality control
- PCA
principal component analysis
- NHLF
lung fibroblasts
- VSE
Variant Set Enrichment
- DHS
DNase I hypersensitive sites
- GWGAS
genome-wide gene-based association study
- BH
Benjamini-Hochberg
- SCNA
somatic copy number alteration
- HRD
homologous recombination deficiency
- GSEA
Gene-Set Enrichment Analysis
- HR
homologous recombination
Footnotes
Conflict of interest
Na Qin, Yuancheng Li, Cheng Wang, Meng Zhu, Juncheng Dai, Tongtong Hong, Demetrius Albanes, Stephen Lam, Adonina Tardon, Chu Chen, Gary Goodman, Stig E. Bojesen, Maria Teresa Landi, Mattias Johansson, Angela Risch, H-Erich Wichmann, Heike Bickeboller, Gadi Rennert, Susanne Arnold, Paul Brennan, John K. Field, Sanjay Shete, Loic Le Marchand, Olle Melander, Hans Brunnstrom, Geoffrey Liu, Rayjean J. Hung, Angeline Andrew, Lambertus A. Kiemeney, Shan Zienolddiny, Kjell Grankvist, Mikael Johansson, Neil Caporaso, Penella Woll, Philip Lazarus, Matthew B. Schabath, Melinda C. Aldrich, Victoria L. Stevens, Guangfu Jin, David C. Christiani, Zhibin Hu, Christopher I. Amos, Hongxia Ma, and Hongbing Shen declare that they have no conflict of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68(6): 394–424 [DOI] [PubMed] [Google Scholar]
- 2.Alberg AJ, Brock MV, Ford JG, Samet JM and Spivack SD: Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013; 143(5 Suppl): e1S–e29S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosse Y and Amos CI: A Decade of GWAS Results in Lung Cancer. Cancer Epidemiol Biomarkers Prev 2018; 27(4): 363–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, He YQ, Zhang R, Tan W, Fan J, Wang T, Zheng H, Sun Q, Wang L, Huang M, Ge Z, Yu C, Guo Y, Wang TM, Wang J, Xu L, Wu W, Chen L, Bian Z, Walters R, Millwood IY, Li XZ, Wang X, Hung RJ, Christiani DC, Chen H, Wang M, Wang C, Jiang Y, Chen K, Chen Z, Jin G, Wu T, Lin D, Hu Z, Amos CI, Wu C, Wei Q, Jia WH, Li L and Shen H: Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir Med 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai J, Shen W, Wen W, Chang J, Wang T, Chen H, Jin G, Ma H, Wu C, Li L, Song F, Zeng Y, Jiang Y, Chen J, Wang C, Zhu M, Zhou W, Du J, Xiang Y, Shu XO, Hu Z, Zhou W, Chen K, Xu J, Jia W, Lin D, Zheng W and Shen H: Estimation of heritability for nine common cancers using data from genome-wide association studies in Chinese population. Int J Cancer 2017; 140(2): 329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, Lan Q, Abnet CC, Amundadottir LT, Figueroa JD, Landi MT, Mirabello L, Savage SA, Taylor PR, De Vivo I, McGlynn KA, Purdue MP, Rajaraman P, Adami HO, Ahlbom A, Albanes D, Amary MF, An SJ, Andersson U, Andriole G Jr., Andrulis IL, Angelucci E, Ansell SM, Arici C, Armstrong BK, Arslan AA, Austin MA, Baris D, Barkauskas DA, Bassig BA, Becker N, Benavente Y, Benhamou S, Berg C, Van Den Berg D, Bernstein L, Bertrand KA, Birmann BM, Black A, Boeing H, Boffetta P, Boutron-Ruault MC, Bracci PM, Brinton L, Brooks-Wilson AR, Bueno-de-Mesquita HB, Burdett L, Buring J, Butler MA, Cai Q, Cancel-Tassin G, Canzian F, Carrato A, Carreon T, Carta A, Chan JK, Chang ET, Chang GC, Chang IS, Chang J, Chang-Claude J, Chen CJ, Chen CY, Chen C, Chen CH, Chen C, Chen H, Chen K, Chen KY, Chen KC, Chen Y, Chen YH, Chen YS, Chen YM, Chien LH, Chirlaque MD, Choi JE, Choi YY, Chow WH, Chung CC, Clavel J, Clavel-Chapelon F, Cocco P, Colt JS, Comperat E, Conde L, Connors JM, Conti D, Cortessis VK, Cotterchio M, Cozen W, Crouch S, Crous-Bou M, Cussenot O, Davis FG, Ding T, Diver WR, Dorronsoro M, Dossus L, Duell EJ, Ennas MG, Erickson RL, Feychting M, Flanagan AM, Foretova L, Fraumeni JF Jr., Freedman ND, Beane Freeman LE, Fuchs C, Gago-Dominguez M, Gallinger S, Gao YT, Gapstur SM, Garcia-Closas M, Garcia-Closas R, Gascoyne RD, Gastier-Foster J, Gaudet MM, Gaziano JM, Giffen C, Giles GG, Giovannucci E, Glimelius B, Goggins M, Gokgoz N, Goldstein AM, Gorlick R, Gross M, Grubb R 3rd, Gu J, Guan P, Gunter M, Guo H, Habermann TM, Haiman CA, Halai D, Hallmans G, Hassan M, Hattinger C, He Q, He X, Helzlsouer K, Henderson B, Henriksson R, Hjalgrim H, Hoffman-Bolton J, Hohensee C, Holford TR, Holly EA, Hong YC, Hoover RN, Horn-Ross PL, Hosain GM, Hosgood HD 3rd, Hsiao CF, Hu N, Hu W, Hu Z, Huang MS, Huerta JM, Hung JY, Hutchinson A, Inskip PD, Jackson RD, Jacobs EJ, Jenab M, Jeon HS, Ji BT, Jin G, Jin L, Johansen C, Johnson A, Jung YJ, Kaaks R, Kamineni A, Kane E, Kang CH, Karagas MR, Kelly RS, Khaw KT, Kim C, Kim HN, Kim JH, Kim JS, Kim YH, Kim YT, Kim YC, Kitahara CM, Klein AP, Klein RJ, Kogevinas M, Kohno T, Kolonel LN, Kooperberg C, Kricker A, Krogh V, Kunitoh H, Kurtz RC, Kweon SS, LaCroix A, Lawrence C, Lecanda F, Lee VH, Li D, Li H, Li J, Li YJ, Li Y, Liao LM, Liebow M, Lightfoot T, Lim WY, Lin CC, Lin D, Lindstrom S, Linet MS, Link BK, Liu C, Liu J, Liu L, Ljungberg B, Lloreta J, Di Lollo S, Lu D, Lund E, Malats N, Mannisto S, Le Marchand L, Marina N, Masala G, Mastrangelo G, Matsuo K, Maynadie M, McKay J, McKean-Cowdin R, Melbye M, Melin BS, Michaud DS, Mitsudomi T, Monnereau A, Montalvan R, Moore LE, Mortensen LM, Nieters A, North KE, Novak AJ, Oberg AL, Offit K, Oh IJ, Olson SH, Palli D, Pao W, Park IK, Park JY, Park KH, Patino-Garcia A, Pavanello S, Peeters PH, Perng RP, Peters U, Petersen GM, Picci P, Pike MC, Porru S, Prescott J, Prokunina-Olsson L, Qian B, Qiao YL, Rais M, Riboli E, Riby J, Risch HA, Rizzato C, Rodabough R, Roman E, Roupret M, Ruder AM, Sanjose S, Scelo G, Schned A, Schumacher F, Schwartz K, Schwenn M, Scotlandi K, Seow A, Serra C, Serra M, Sesso HD, Setiawan VW, Severi G, Severson RK, Shanafelt TD, Shen H, Shen W, Shin MH, Shiraishi K, Shu XO, Siddiq A, Sierrasesumaga L, Sihoe AD, Skibola CF, Smith A, Smith MT, Southey MC, Spinelli JJ, Staines A, Stampfer M, Stern MC, Stevens VL, Stolzenberg-Solomon RS, Su J, Su WC, Sund M, Sung JS, Sung SW, Tan W, Tang W, Tardon A, Thomas D, Thompson CA, Tinker LF, Tirabosco R, Tjonneland A, Travis RC, Trichopoulos D, Tsai FY, Tsai YH, Tucker M, Turner J, Vajdic CM, Vermeulen RC, Villano DJ, Vineis P, Virtamo J, Visvanathan K, Wactawski-Wende J, Wang C, Wang CL, Wang JC, Wang J, Wei F, Weiderpass E, Weiner GJ, Weinstein S, Wentzensen N, White E, Witzig TE, Wolpin BM, Wong MP, Wu C, Wu G, Wu J, Wu T, Wu W, Wu X, Wu YL, Wunder JS, Xiang YB, Xu J, Xu P, Yang PC, Yang TY, Ye Y, Yin Z, Yokota J, Yoon HI, Yu CJ, Yu H, Yu K, Yuan JM, Zelenetz A, Zeleniuch-Jacquotte A, Zhang XC, Zhang Y, Zhao X, Zhao Z, Zheng H, Zheng T, Zheng W, Zhou B, Zhu M, Zucca M, Boca SM, Cerhan JR, Ferri GM, Hartge P, Hsiung CA, Magnani C, Miligi L, Morton LM, Smedby KE, Teras LR, Vijai J, Wang SS, Brennan P, Caporaso NE, Hunter DJ, Kraft P, Rothman N, Silverman DT, Slager SL, Chanock SJ and Chatterjee N: Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. J Natl Cancer Inst 2015; 107(12): djv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D, James M, Liu P, Tichelaar JW, Vikis HG, You M and Mills IG: Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 2011; 43(6): 513–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009; 106(23): 9362–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garraway LA and Sellers WR: Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 2006; 6(8): 593–602 [DOI] [PubMed] [Google Scholar]
- 10.Sato M, Shames DS, Gazdar AF and Minna JD: A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007; 2(4): 327–43 [DOI] [PubMed] [Google Scholar]
- 11.Shen H, Zhu M and Wang C: Precision oncology of lung cancer: genetic and genomic differences in Chinese population. NPJ Precis Oncol 2019; 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.E. P. Consortium: The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306(5696): 636–40 [DOI] [PubMed] [Google Scholar]
- 13.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest ARR, Carninci P, Rehli M and Sandelin A: An atlas of active enhancers across human cell types and tissues. Nature 2014; 507(7493): 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. Roadmap Epigenomics, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T and Kellis M: Integrative analysis of 111 reference human epigenomes. Nature 2015; 518(7539): 317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A, Bolla MK, Wang Q, Tyrer J, Dicks E, Lee A, Wang Z, Allen J, Keeman R, Eilber U, French JD, Qing Chen X, Fachal L, McCue K, McCart Reed AE, Ghoussaini M, Carroll JS, Jiang X, Finucane H, Adams M, Adank MA, Ahsan H, Aittomaki K, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Arun B, Auer PL, Bacot F, Barrdahl M, Baynes C, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Borresen-Dale AL, Brand JS, Brauch H, Brennan P, Brenner H, Brinton L, Broberg P, Brock IW, Broeks A, Brooks-Wilson A, Brucker SY, Bruning T, Burwinkel B, Butterbach K, Cai Q, Cai H, Caldes T, Canzian F, Carracedo A, Carter BD, Castelao JE, Chan TL, David Cheng TY, Seng Chia K, Choi JY, Christiansen H, Clarke CL, N. Collaborators, Collee M, Conroy DM, Cordina-Duverger E, Cornelissen S, Cox DG, Cox A, Cross SS, Cunningham JM, Czene K, Daly MB, Devilee P, Doheny KF, Dork T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Elvira M, Engel C, Eriksson M, Fasching PA, Figueroa J, Flesch-Janys D, Fletcher O, Flyger H, Fritschi L, Gaborieau V, Gabrielson M, Gago-Dominguez M, Gao YT, Gapstur SM, Garcia-Saenz JA, Gaudet MM, Georgoulias V, Giles GG, Glendon G, Goldberg MS, Goldgar DE, Gonzalez-Neira A, Grenaker Alnaes GI, Grip M, Gronwald J, Grundy A, Guenel P, Haeberle L, Hahnen E, Haiman CA, Hakansson N, Hamann U, Hamel N, Hankinson S, Harrington P, Hart SN, Hartikainen JM, Hartman M, Hein A, Heyworth J, Hicks B, Hillemanns P, Ho DN, Hollestelle A, Hooning MJ, Hoover RN, Hopper JL, Hou MF, Hsiung CN, Huang G, Humphreys K, Ishiguro J, Ito H, Iwasaki M, Iwata H, Jakubowska A, Janni W, John EM, Johnson N, Jones K, Jones M, Jukkola-Vuorinen A, Kaaks R, Kabisch M, Kaczmarek K, Kang D, Kasuga Y, Kerin MJ, Khan S, Khusnutdinova E, Kiiski JI, Kim SW, Knight JA, Kosma VM, Kristensen VN, Kruger U, Kwong A, Lambrechts D, Le Marchand L, Lee E, Lee MH, Lee JW, Neng Lee C, Lejbkowicz F, Li J, Lilyquist J, Lindblom A, Lissowska J, Lo WY, Loibl S, Long J, Lophatananon A, Lubinski J, Luccarini C, Lux MP, Ma ESK, MacInnis RJ, Maishman T, Makalic E, Malone KE, Kostovska IM, Mannermaa A, Manoukian S, Manson JE, Margolin S, Mariapun S, Martinez ME, Matsuo K, Mavroudis D, McKay J, McLean C, Meijers-Heijboer H, Meindl A, Menendez P, Menon U, Meyer J, Miao H, Miller N, Taib NAM, Muir K, Mulligan AM, Mulot C, Neuhausen SL, Nevanlinna H, Neven P, Nielsen SF, Noh DY, Nordestgaard BG, Norman A, Olopade OI, Olson JE, Olsson H, Olswold C, Orr N, Pankratz VS, Park SK, Park-Simon TW, Lloyd R, Perez JIA, Peterlongo P, Peto J, Phillips KA, Pinchev M, Plaseska-Karanfilska D, Prentice R, Presneau N, Prokofyeva D, Pugh E, Pylkas K, Rack B, Radice P, Rahman N, Rennert G, Rennert HS, Rhenius V, Romero A, Romm J, Ruddy KJ, Rudiger T, Rudolph A, Ruebner M, Rutgers EJT, Saloustros E, Sandler DP, Sangrajrang S, Sawyer EJ, Schmidt DF, Schmutzler RK, Schneeweiss A, Schoemaker MJ, Schumacher F, Schurmann P, Scott RJ, Scott C, Seal S, Seynaeve C, Shah M, Sharma P, Shen CY, Sheng G, Sherman ME, Shrubsole MJ, Shu XO, Smeets A, Sohn C, Southey MC, Spinelli JJ, Stegmaier C, Stewart-Brown S, Stone J, Stram DO, Surowy H, Swerdlow A, Tamimi R, Taylor JA, Tengstrom M, Teo SH, Beth Terry M, Tessier DC, Thanasitthichai S, Thone K, Tollenaar R, Tomlinson I, Tong L, Torres D, Truong T, Tseng CC, Tsugane S, Ulmer HU, Ursin G, Untch M, Vachon C, van Asperen CJ, Van Den Berg D, van den Ouweland AMW, van der Kolk L, van der Luijt RB, Vincent D, Vollenweider J, Waisfisz Q, Wang-Gohrke S, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett W, Winqvist R, Wolk A, Wu AH, Xia L, Yamaji T, Yang XR, Har Yip C, Yoo KY, Yu JC, Zheng W, Zheng Y, Zhu B, Ziogas A, Ziv E, A. Investigators, A. I. ConFab, Lakhani SR, Antoniou AC, Droit A, Andrulis IL, Amos CI, Couch FJ, Pharoah PDP, Chang-Claude J, Hall P, Hunter DJ, Milne RL, Garcia-Closas M, Schmidt MK, Chanock SJ, Dunning AM, Edwards SL, Bader GD, Chenevix-Trench G, Simard J, Kraft P and Easton DF: Association analysis identifies 65 new breast cancer risk loci. Nature 2017; 551(7678): 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang X, Finucane HK, Schumacher FR, Schmit SL, Tyrer JP, Han Y, Michailidou K, Lesseur C, Kuchenbaecker KB, Dennis J, Conti DV, Casey G, Gaudet MM, Huyghe JR, Albanes D, Aldrich MC, Andrew AS, Andrulis IL, Anton-Culver H, Antoniou AC, Antonenkova NN, Arnold SM, Aronson KJ, Arun BK, Bandera EV, Barkardottir RB, Barnes DR, Batra J, Beckmann MW, Benitez J, Benlloch S, Berchuck A, Berndt SI, Bickeboller H, Bien SA, Blomqvist C, Boccia S, Bogdanova NV, Bojesen SE, Bolla MK, Brauch H, Brenner H, Brenton JD, Brook MN, Brunet J, Brunnstrom H, Buchanan DD, Burwinkel B, Butzow R, Cadoni G, Caldes T, Caligo MA, Campbell I, Campbell PT, Cancel-Tassin G, Cannon-Albright L, Campa D, Caporaso N, Carvalho AL, Chan AT, Chang-Claude J, Chanock SJ, Chen C, Christiani DC, Claes KBM, Claessens F, Clements J, Collee JM, Correa MC, Couch FJ, Cox A, Cunningham JM, Cybulski C, Czene K, Daly MB, deFazio A, Devilee P, Diez O, Gago-Dominguez M, Donovan JL, Dork T, Duell EJ, Dunning AM, Dwek M, Eccles DM, Edlund CK, Edwards DRV, Ellberg C, Evans DG, Fasching PA, Ferris RL, Liloglou T, Figueiredo JC, Fletcher O, Fortner RT, Fostira F, Franceschi S, Friedman E, Gallinger SJ, Ganz PA, Garber J, Garcia-Saenz JA, Gayther SA, Giles GG, Godwin AK, Goldberg MS, Goldgar DE, Goode EL, Goodman MT, Goodman G, Grankvist K, Greene MH, Gronberg H, Gronwald J, Guenel P, Hakansson N, Hall P, Hamann U, Hamdy FC, Hamilton RJ, Hampe J, Haugen A, Heitz F, Herrero R, Hillemanns P, Hoffmeister M, Hogdall E, Hong YC, Hopper JL, Houlston R, Hulick PJ, Hunter DJ, Huntsman DG, Idos G, Imyanitov EN, Ingles SA, Isaacs C, Jakubowska A, James P, Jenkins MA, Johansson M, Johansson M, John EM, Joshi AD, Kaneva R, Karlan BY, Kelemen LE, Kuhl T, Khaw KT, Khusnutdinova E, Kibel AS, Kiemeney LA, Kim J, Kjaer SK, Knight JA, Kogevinas M, Kote-Jarai Z, Koutros S, Kristensen VN, Kupryjanczyk J, Lacko M, Lam S, Lambrechts D, Landi MT, Lazarus P, Le ND, Lee E, Lejbkowicz F, Lenz HJ, Leslie G, Lessel D, Lester J, Levine DA, Li L, Li CI, Lindblom A, Lindor NM, Liu G, Loupakis F, Lubinski J, Maehle L, Maier C, Mannermaa A, Marchand LL, Margolin S, May T, McGuffog L, Meindl A, Middha P, Miller A, Milne RL, MacInnis RJ, Modugno F, Montagna M, Moreno V, Moysich KB, Mucci L, Muir K, Mulligan AM, Nathanson KL, Neal DE, Ness AR, Neuhausen SL, Nevanlinna H, Newcomb PA, Newcomb LF, Nielsen FC, Nikitina-Zake L, Nordestgaard BG, Nussbaum RL, Offit K, Olah E, Olama AAA, Olopade OI, Olshan AF, Olsson H, Osorio A, Pandha H, Park JY, Pashayan N, Parsons MT, Pejovic T, Penney KL, Peters WHM, Phelan CM, Phipps AI, Plaseska-Karanfilska D, Pring M, Prokofyeva D, Radice P, Stefansson K, Ramus SJ, Raskin L, Rennert G, Rennert HS, van Rensburg EJ, Riggan MJ, Risch HA, Risch A, Roobol MJ, Rosenstein BS, Rossing MA, De Ruyck K, Saloustros E, Sandler DP, Sawyer EJ, Schabath MB, Schleutker J, Schmidt MK, Setiawan VW, Shen H, Siegel EM, Sieh W, Singer CF, Slattery ML, Sorensen KD, Southey MC, Spurdle AB, Stanford JL, Stevens VL, Stintzing S, Stone J, Sundfeldt K, Sutphen R, Swerdlow AJ, Tajara EH, Tangen CM, Tardon A, Taylor JA, Teare MD, Teixeira MR, Terry MB, Terry KL, Thibodeau SN, Thomassen M, Bjorge L, Tischkowitz M, Toland AE, Torres D, Townsend PA, Travis RC, Tung N, Tworoger SS, Ulrich CM, Usmani N, Vachon CM, Van Nieuwenhuysen E, Vega A, Aguado-Barrera ME, Wang Q, Webb PM, Weinberg CR, Weinstein S, Weissler MC, Weitzel JN, West CML, White E, Whittemore AS, Wichmann HE, Wiklund F, Winqvist R, Wolk A, Woll P, Woods M, Wu AH, Wu X, Yannoukakos D, Zheng W, Zienolddiny S, Ziogas A, Zorn KK, Lane JM, Saxena R, Thomas D, Hung RJ, Diergaarde B, McKay J, Peters U, Hsu L, Garcia-Closas M, Eeles RA, Chenevix-Trench G, Brennan PJ, Haiman CA, Simard J, Easton DF, Gruber SB, Pharoah PDP, Price AL, Pasaniuc B, Amos CI, Kraft P and Lindstrom S: Shared heritability and functional enrichment across six solid cancers. Nat Commun 2019; 10(1): 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA and Bernstein BE: Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015; 518(7539): 337–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, Caporaso NE, Johansson M, Xiao X, Li Y, Byun J, Dunning A, Pooley KA, Qian DC, Ji X, Liu G, Timofeeva MN, Bojesen SE, Wu X, Le Marchand L, Albanes D, Bickeboller H, Aldrich MC, Bush WS, Tardon A, Rennert G, Teare MD, Field JK, Kiemeney LA, Lazarus P, Haugen A, Lam S, Schabath MB, Andrew AS, Shen H, Hong YC, Yuan JM, Bertazzi PA, Pesatori AC, Ye Y, Diao N, Su L, Zhang R, Brhane Y, Leighl N, Johansen JS, Mellemgaard A, Saliba W, Haiman CA, Wilkens LR, Fernandez-Somoano A, Fernandez-Tardon G, van der Heijden HFM, Kim JH, Dai J, Hu Z, Davies MPA, Marcus MW, Brunnstrom H, Manjer J, Melander O, Muller DC, Overvad K, Trichopoulou A, Tumino R, Doherty JA, Barnett MP, Chen C, Goodman GE, Cox A, Taylor F, Woll P, Bruske I, Wichmann HE, Manz J, Muley TR, Risch A, Rosenberger A, Grankvist K, Johansson M, Shepherd FA, Tsao MS, Arnold SM, Haura EB, Bolca C, Holcatova I, Janout V, Kontic M, Lissowska J, Mukeria A, Ognjanovic S, Orlowski TM, Scelo G, Swiatkowska B, Zaridze D, Bakke P, Skaug V, Zienolddiny S, Duell EJ, Butler LM, Koh WP, Gao YT, Houlston RS, McLaughlin J, Stevens VL, Joubert P, Lamontagne M, Nickle DC, Obeidat M, Timens W, Zhu B, Song L, Kachuri L, Artigas MS, Tobin MD, Wain LV, SpiroMeta C, Rafnar T, Thorgeirsson TE, Reginsson GW, Stefansson K, Hancock DB, Bierut LJ, Spitz MR, Gaddis NC, Lutz SM, Gu F, Johnson EO, Kamal A, Pikielny C, Zhu D, Lindstroem S, Jiang X, Tyndale RF, Chenevix-Trench G, Beesley J, Bosse Y, Chanock S, Brennan P, Landi MT and Amos CI: Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet 2017; 49(7): 1126–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D and Shen H: A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011; 43(8): 792–6 [DOI] [PubMed] [Google Scholar]
- 20.Delaneau O, Marchini J and Zagury JF: A linear complexity phasing method for thousands of genomes. Nat Methods 2011; 9(2): 179–81 [DOI] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P and Marchini J: A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5(6): e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal lari R, Lupien M, Markowitz S and Scacheri PC: Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 2014; 24(1): 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Yin R, Dai J, Gu Y, Cui S, Ma H, Zhang Z, Huang J, Qin N, Jiang T, Geng L, Zhu M, Pu Z, Du F, Wang Y, Yang J, Chen L, Wang Q, Jiang Y, Dong L, Yao Y, Jin G, Hu Z, Jiang L, Xu L and Shen H: Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat Commun 2018; 9(1): 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM and Shendure J: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46(3): 310–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN and Gaunt TR: Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 2013; 34(1): 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun S and Fay JC: Identification of deleterious mutations within three human genomes. Genome Res 2009; 19(9): 1553–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz JM, Rodelsperger C, Schuelke M and Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7(8): 575–6 [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7(4): 248–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P, Henikoff S and Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4(7): 1073–81 [DOI] [PubMed] [Google Scholar]
- 30.Shabalin AA: Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012; 28(10): 1353–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH and Lupien M: Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012; 44(11): 1191–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Leeuw CA, Mooij JM, Heskes T and Posthuma D: MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11(4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N and Stratton MR: A census of human cancer genes. Nat Rev Cancer 2004; 4(3): 177–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A and Lopez-Bigas N: IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods 2013; 10(11): 1081–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer N Genome Atlas Research: Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489(7417): 519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer N Genome Atlas Research: Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511(7511): 543–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, Imielinski M, Hu X, Ling S, Akbani R, Rosenberg M, Cibulskis C, Ramachandran A, Collisson EA, Kwiatkowski DJ, Lawrence MS, Weinstein JN, Verhaak RG, Wu CJ, Hammerman PS, Cherniack AD, Getz G, N. Cancer Genome Atlas Research, Artyomov MN, Schreiber R, Govindan R and Meyerson M: Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48(6): 607–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, Moy C, Fan Y, Albertyn Z, Huang X, Zhang J, Qiu Y, Platero S, Lorenzi MV, Zudaire E, Yang J, Cheng Y, Xu L and Wu YL: Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 2019; 10(1): 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G and Meyerson M: Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150(6): 1107–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P and Stein L: Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 2011; 39(Database issue): D691–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu G, Wang LG, Han Y and He QY: clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16(5): 284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, N. Cancer Genome Atlas Research, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG and Shmulevich I: The Immune Landscape of Cancer. Immunity 2018; 48(4): 812–830 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, Liu Y, Akbani R, Feng B, Donehower LA, Miller C, Shen Y, Karimi M, Chen H, Kim P, Jia P, Shinbrot E, Zhang S, Liu J, Hu H, Bailey MH, Yau C, Wolf D, Zhao Z, Weinstein JN, Li L, Ding L, Mills GB, Laird PW, Wheeler DA, Shmulevich I, N. Cancer Genome Atlas Research, Monnat RJ Jr., Xiao Y and Wang C: Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep 2018; 23(1): 239–254 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer CJ, Li Y and Abecasis GR: METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26(17): 2190–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK, C. International Multiple Sclerosis Genetics, I. B. D. G. C. International, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA and Weersma RK: Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47(9): 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, Staley JR, Tragante V, Tukiainen T, Yaghootkar H, Masca N, Freitag DF, Ferreira T, Giannakopoulou O, Tinker A, Harakalova M, Mihailov E, Liu C, Kraja AT, Fallgaard Nielsen S, Rasheed A, Samuel M, Zhao W, Bonnycastle LL, Jackson AU, Narisu N, Swift AJ, Southam L, Marten J, Huyghe JR, Stancakova A, Fava C, Ohlsson T, Matchan A, Stirrups KE, Bork-Jensen J, Gjesing AP, Kontto J, Perola M, Shaw-Hawkins S, Havulinna AS, Zhang H, Donnelly LA, Groves CJ, Rayner NW, Neville MJ, Robertson NR, Yiorkas AM, Herzig KH, Kajantie E, Zhang W, Willems SM, Lannfelt L, Malerba G, Soranzo N, Trabetti E, Verweij N, Evangelou E, Moayyeri A, Vergnaud AC, Nelson CP, Poveda A, Varga TV, Caslake M, de Craen AJ, Trompet S, Luan J, Scott RA, Harris SE, Liewald DC, Marioni R, Menni C, Farmaki AE, Hallmans G, Renstrom F, Huffman JE, Hassinen M, Burgess S, Vasan RS, Felix JF, C. H.-H. F. Consortium, Uria-Nickelsen M, Malarstig A, Reily DF, Hoek M, Vogt T, Lin H, Lieb W, EchoGen C, Traylor M, Markus HF, M. Consortium, Highland HM, Justice AE, Marouli E, G. Consortium, Lindstrom J, Uusitupa M, Komulainen P, Lakka TA, Rauramaa R, Polasek O, Rudan I, Rolandsson O, Franks PW, Dedoussis G, Spector TD, E. P.-I. Consortium, Jousilahti P, Mannisto S, Deary IJ, Starr JM, Langenberg C, Wareham NJ, Brown MJ, Dominiczak AF, Connell JM, Jukema JW, Sattar N, Ford I, Packard CJ, Esko T, Magi R, Metspalu A, de Boer RA, van der Meer P, van der Harst P, Lifelines Cohort S, Gambaro G, Ingelsson E, Lind L, de Bakker PI, Numans ME, Brandslund I, Christensen C, Petersen ER, Korpi-Hyovalti E, Oksa H, Chambers JC, Kooner JS, Blakemore AI, Franks S, Jarvelin MR, Husemoen LL, Linneberg A, Skaaby T, Thuesen B, Karpe F, Tuomilehto J, Doney AS, Morris AD, Palmer CN, Holmen OL, Hveem K, Willer CJ, Tuomi T, Groop L, Karajamaki A, Palotie A, Ripatti S, Salomaa V, Alam DS, Shafi Majumder AA, Di Angelantonio E, Chowdhury R, McCarthy MI, Poulter N, Stanton AV, Sever P, Amouyel P, Arveiler D, Blankenberg S, Ferrieres J, Kee F, Kuulasmaa K, Muller-Nurasyid M, Veronesi G, Virtamo J, Deloukas P, C. Wellcome Trust Case Control, Elliott P, G. Understanding Society Scientific, Zeggini E, Kathiresan S, Melander O, Kuusisto J, Laakso M, Padmanabhan S, Porteous D, Hayward C, Scotland G, Collins FS, Mohlke KL, Hansen T, Pedersen O, Boehnke M, Stringham HM, E.-C. Consortium, Frossard P, Newton-Cheh C, C. E. C. B. P. Consortium, Tobin MD, Nordestgaard BG, T. D. G. Consortium, T. D. C. Go, B. P. C. Exome, C. H. D. E. Consortium, Caulfield MJ, Mahajan A, Morris AP, Tomaszewski M, Samani NJ, Saleheen D, Asselbergs FW, Lindgren CM, Danesh J, Wain LV, Butterworth AS, Howson JM and Munroe PB: Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet 2016; 48(10): 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, Wistuba II and Koo JS: Cyclic AMP response element-binding protein overexpression: a feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res 2008; 68(15): 6065–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters CL, Cleck JN, Kuo YC and Blendy JA: Mu-opioid receptor and CREB activation are required for nicotine reward. Neuron 2005; 46(6): 933–43 [DOI] [PubMed] [Google Scholar]
- 49.Srinivasan S, Totiger T, Shi C, Castellanos J, Lamichhane P, Dosch AR, Messaggio F, Kashikar N, Honnenahally K, Ban Y, Merchant NB, VanSaun M and Nagathihalli NS: Tobacco Carcinogen-Induced Production of GM-CSF Activates CREB to Promote Pancreatic Cancer. Cancer Res 2018; 78(21): 6146–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung CC, Kuo CW, Wang WH, Chang TH, Chang PJ, Chang LK and Liu ST: Transcriptional activation of Epstein-Barr virus BRLF1 by USF1 and Rta. J Gen Virol 2015; 96(9): 2855–66 [DOI] [PubMed] [Google Scholar]
- 51.Gao E, Wang Y, Alcorn JL and Mendelson CR: The basic helix-loop-helix-zipper transcription factor USF1 regulates expression of the surfactant protein-A gene. J Biol Chem 1997; 272(37): 23398–406 [DOI] [PubMed] [Google Scholar]
- 52.Ho PK and Hawkins CJ: Mammalian initiator apoptotic caspases. FEBS J 2005; 272(21): 5436–53 [DOI] [PubMed] [Google Scholar]
- 53.MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP and Cox A: Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst 2004; 96(24): 1866–9 [DOI] [PubMed] [Google Scholar]
- 54.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, Guo Y, Yang M, Zhang X, Zhang Q, Zeng C and Lin D: A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet 2007; 39(5): 605–13 [DOI] [PubMed] [Google Scholar]
- 55.Castel SE, Cervera A, Mohammadi P, Aguet F, Reverter F, Wolman A, Guigo R, Iossifov I, Vasileva A and Lappalainen T: Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat Genet 2018; 50(9): 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horejsi V, Zhang W and Schraven B: Transmembrane adaptor proteins: organizers of immunoreceptor signalling. Nat Rev Immunol 2004; 4(8): 603–16 [DOI] [PubMed] [Google Scholar]
- 57.Brdickova N, Brdicka T, Angelisova P, Horvath O, Spicka J, Hilgert I, Paces J, Simeoni L, Kliche S, Merten C, Schraven B and Horejsi V: LIME: a new membrane Raft-associated adaptor protein involved in CD4 and CD8 coreceptor signaling. J Exp Med 2003; 198(10): 1453–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lok BH and Powell SN: Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res 2012; 18(23): 6400–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lisby M, Rothstein R and Mortensen UH: Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A 2001; 98(15): 8276–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galanos P, Pappas G, Polyzos A, Kotsinas A, Svolaki I, Giakoumakis NN, Glytsou C, Pateras IS, Swain U, Souliotis VL, Georgakilas AG, Geacintov N, Scorrano L, Lukas C, Lukas J, Livneh Z, Lygerou Z, Chowdhury D, Sorensen CS, Bartek J and Gorgoulis VG: Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol 2018; 19(1): 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieberman R, Xiong D, James M, Han Y, Amos CI, Wang L and You M: Functional characterization of RAD52 as a lung cancer susceptibility gene in the 12p13.33 locus. Mol Carcinog 2016; 55(5): 953–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanahan D and Weinberg RA: Hallmarks of cancer: the next generation. Cell 2011; 144(5): 646–74 [DOI] [PubMed] [Google Scholar]
- 63.Chen DS and Mellman I: Elements of cancer immunity and the cancer-immune set point. Nature 2017; 541(7637): 321–330 [DOI] [PubMed] [Google Scholar]
- 64.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, Meinhof K, Chow A, Kim-Shulze S, Wolf A, Medaglia C, Li H, Rytlewski JA, Emerson RO, Solovyov A, Greenbaum BD, Sanders C, Vignali M, Beasley MB, Flores R, Gnjatic S, Pe’er D, Rahman A, Amit I and Merad M: Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017; 169(4): 750–765 e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiwari S, Tripathy BC, Jajoo A, Das AB, Murata N, Sane PV and Govindjee: Prasanna K. Mohanty (1934–2013): a great photosynthetiker and a wonderful human being who touched the hearts of many. Photosynth Res 2014; 122(3): 235–60 [DOI] [PubMed] [Google Scholar]
- 66.Hecht SS: Lung carcinogenesis by tobacco smoke. Int J Cancer 2012; 131(12): 2724–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G Jr., Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M and Garon EB: Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016; 387(10027): 1540–1550 [DOI] [PubMed] [Google Scholar]
- 68.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR and K.−. Investigators: Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016; 375(19): 1823–1833 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


