Abstract

Endophilin, which is a member of the Bin-amphiphysin-Rvs (BAR) domain protein superfamily, contains a homodimeric N-BAR domain of a characteristic crescent shape. The N-BAR domain comprises a six-helix bundle and is known to sense and generate membrane curvature. Here, we characterize aspects of the unfolding mechanism of the endophilin A1 N-BAR domain during thermal denaturation and examine factors that influence the thermal stability of this domain. Far-UV circular dichroism (CD) spectroscopy was applied to monitor changes in the secondary structure above room temperature. The protein’s conformational changes were further characterized through Foerster resonance energy transfer and cross-linking experiments at varying temperatures. Our results indicate that thermal unfolding of the endophilin N-BAR is (minimally) a two-step process, with a dimeric intermediate that displays partial helicity loss. Furthermore, a thermal shift assay and temperature-dependent CD were applied to compare the unfolding processes of several truncated versions of endophilin. The melting temperature of the N-BAR domain decreased when we deleted either the N-terminal H0 helix or the unstructured linker of endophilin. This result suggests that these intrinsically disordered domains may play a role in structurally stabilizing the functional N-BAR domain in vivo. Finally, we show that single-site mutations can also compromise endophilin’s thermal stability.
Introduction
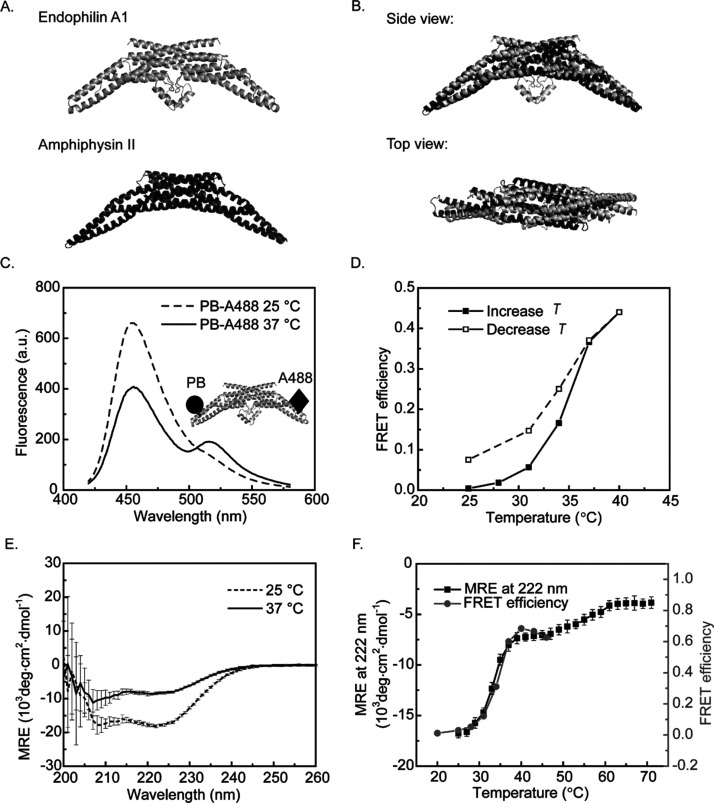
As a member of the Bin-amphiphysin-Rvs (BAR) domain protein superfamily, endophilin is known to bind cellular membranes and to facilitate endocytosis.1−7 An endophilin monomer consists of an N-terminal BAR domain, an intrinsically disordered middle region and a C-terminal SRC homology 3 (SH3) domain.8,9 Crystallization studies have shown that endophilin N-BAR domains (endo_N-BAR) dimerize to form a crescent structure (Figure 1A), which is able to induce and stabilize membrane curvature and to “sense” (i.e., bind selectively to) curved membranes.9−13
Figure 1.
Endo_N-BAR thermal denaturation induces tip distance change. (A) Crystal structure of rat endophilin A1 (PDB: 2C08,9 H0 of 1–32 is not included since it only forms a helix structure upon membrane binding) and human amphiphysin II (PDB: 2FIC(43)). (B) Structure alignment of endophilin A1 and amphiphysin II. The representation and alignments were prepared using PyMol. (C) Comparison of fluorescence spectra of PB and A488 dual-labeled endo_N-BAR 187C (C108A) at 25 and 37 °C. The amino acid L187C is on the tip of endo_N-BAR. No significant FRET peak at 520 nm was observed at T = 25 °C since the fluorophore distance expected from the endo_N-BAR crystal structure is 109 Å, which is larger than the Förster distance R0 = 51.0 Å. At 37 °C, the donor emission peak at 457 nm decreased and the acceptor emission peak at 520 nm increased, implying energy transfer between the donor–acceptor pair. (D) FRET efficiency shows sigmoidal temperature dependence and high reversibility comparing heating followed by cooling. (E) CD spectra of endo_N-BAR 187C (C108A) at 25 and 37 °C indicates that the protein helicity decreased from 47 to 22% when the temperature increased to 37 °C (determined by the online tool Bestsel), which suggests that the protein unfolded under thermal stress. (F) FRET efficiency and CD signal [mean residue ellipticity (MRE) at 222 nm] were measured for endo_N-BAR 187C (C108A) with increasing temperature. Both signals showed sigmoidal change between 30 and 40 °C. The MRE signal shows a second sigmoidal change at a higher temperature, suggesting a two-step unfolding process. The error bar of CD (mean residue ellipticity) measurements was the standard deviation from three measurements on the same sample. All experiments shown in this figure were carried out with endo_N-BAR C108A L187C.
Since the dimerization of endo_N-BAR is important for its membrane remodeling function, a variety of experimental thermodynamic and kinetic studies, as well as molecular dynamics simulations,14 have focused on its dimerization affinity.9,15,16 At room temperature (RT), a sub-nanomolar affinity has been determined.14,15 With temperature (T) varying between 4 and 37 °C, the endo_N-BAR dissociation constant varies from 6.8 × 10–13 to 5.1 × 10–9 M.16 Although the dimeric structure is dominant under these conditions, the question whether the rigid crescent shape of the N-BAR domain can be maintained in this temperature range has been unresolved. It is also unclear how the dimerization step is involved in the folding process of endo_N-BAR. Moreover, no reported studies have focused on the detection of this dimeric structure at more severe conditions for endophilin (such as at higher temperature or under chemical denaturation).
Generally speaking, dimerization has been suggested to be a key step for the proper folding of dimeric proteins.17−19 The structural changes of various dimeric proteins have been studied under thermal denaturation and chemical denaturation.20−24 Small proteins tend to adopt one-step unfolding mechanisms, where dimerization and folding occur simultaneously.18,25,26 Larger proteins show a multiple-step folding/unfolding process, with the formation of either a monomeric intermediate or a dimeric intermediate.17,27−29 Proteins with a large dimerization interface area tend to show high dimerization affinity and to form a dimeric intermediate at the beginning of denaturation.19,30 Such dimeric intermediates can also be detected in the corresponding refolding studies.17,29 Similar folding/unfolding studies have been carried out on one BAR protein, amphiphysin II (structure shown in Figure 1A,B).31 It has been shown to maintain the dimeric structure at the initial stage of chemical denaturation. Through kinetic studies, Gruber and Balbach proposed that the folding of amphiphysin is a multistep process and that dimerization happened in the early stages.31 However, their approach did not reveal the structural details of the unfolding and refolding intermediates. Moreover, it remains to be investigated if their findings are universal for all BAR proteins.
Studies of endophilin’s dimerization mechanism can reveal the intramolecular interactions between different domains.16 Dissociation rates were observed to increase when either the SH3 domain or the N-terminal H0 (residue 1–32 of endophilin A1) were deleted, which suggested an intramolecular interaction between the H0 and the SH3 domain.32−34 To further investigate if there are potential interactions between other domains, we compared the melting temperature of similar endophilin truncations with deletion of the SH3 domain, the H0 helix, or the flexible linker (residue 248–292 of endophilin A1).
In this study, we first aim to reveal the structural changes of endo_N-BAR during thermal unfolding. Temperature-dependent circular dichroism (CD) spectra for this protein are consistent with a two-step unfolding mechanism. The dimeric structure is maintained in the first step while the distance decreases between the two tips of the crescent-shaped endo_N-BAR, suggesting the tips unfold earlier than the dimerization interface during thermal denaturation. Then, we characterize how the thermal stability of endophilin is influenced by single-site mutations and the presence of additional domains and motifs. Even though the H0 motif and the flexible linker are both unstructured in the aqueous environment, deleting either of them results in a decreased melting temperature of endophilin.
Results
FRET Allows the Monitoring of Structural Transitions of the Endophilin N-BAR Domain (endo_N-BAR) during Denaturation
We used Foerster resonance energy transfer (FRET) as a probe to monitor structural changes of endophilin during thermal denaturation. In order to focus on folding transitions of the endophilin BAR domain, we used a truncated version of endophilin, referred to as endo_N-BAR, which contains the N-terminal BAR domain without the SH3 domain and the flexible linker in between the two domains. To enable FRET, two different fluorescent dyes that form an acceptor–donor pair were attached to the tips of the endo_N-BAR (Figure 1C). To label the endo_N-BAR protein, its natural leucine (L) at position 187 was mutated to cysteine (C). We also mutated the wild-type cysteine at position 108 to alanine (A) to avoid fluorescence conjugation in that position. We note that this C108A mutation can disturb the thermal stability of endo_N-BAR, which will be discussed in more detail further below.
Next, endo_N-BAR was labeled using a mixture of Pacific Blue (PB) and Alexa Fluor 488 (A488) dyes by thiol conjugation to the cysteine. Samples labeled with two dyes are referred to as dual-labeled endo_N-BAR throughout this text. PB served as the donor and A488 served as the acceptor. PB and A488 in the same BAR protein dimer can show intradimer FRET only if the two tips of the crescent-shaped dimer reduce their relative distance (such as during an unfolding transition) compared to that which they assume in the protein’s crystal structure. Only a minor fraction of BAR protein dimers is capable in principle of intradimer FRET as the majority of dimers is labeled either with two identical fluorophores, with just one fluorophore, or unlabeled. However, dual- and single-labeled dimers may cause FRET in the absence of any structural transitions if endo_N-BAR dimers closely approach other dimers in processes such as protein oligomerization. Below, FRET efficiency was calculated without distinguishing intradimer or interdimer situations, as described in the Materials and Methods section.
Most of the in vitro studies on the function of endophilin have been carried out at RT.12,15,35,36 However, the physiologically relevant temperature in mammals is 37 °C. Therefore, we measured the emission spectra of the dual-labeled endo_N-BAR at both, 25 and 37 °C. Before each measurement, endo_N-BAR (which was stored at 4 °C) was first incubated at 25 and 37 °C for 20 min to ensure that potential structural changes reached equilibrium.15 At 25 °C, no change in the FRET efficiency was observed in the emission spectra after 20 min incubation. PB dye was excited at 405 nm. It shows an emission peak around 455 nm, while the A488 dye is barely excited at 405 nm. The Foerster distance of the PB-A488 FRET pair is 51.0 Å.37 According to the crystal structure of endo_N-BAR,9 the distance between the two labeled positions (L187C) in the dimer is around 109 Å. The FRET efficiency at this distance is estimated to be around 1%. At 25 °C, a peak at 455 nm and a smaller hump at around 520 nm were observed in the fluorescence spectra (Figure 1C). These correspond to the main emission peak of the donor (PB) and the acceptor (A488), respectively. However, when the temperature was increased to 37 °C, the donor emission peak dropped while the acceptor peak increased. At this temperature, we determined a FRET efficiency of 36%. This observation suggests that the distance between the two fluorophores decreased when the temperature was increased to 37 °C.
Next, we performed these FRET experiments over a wider temperature range to determine if there is a systematic temperature dependence of the FRET signal (Figure 1D). The FRET efficiency of the dual-labeled endo_N-BAR increased when the temperature was increased from 25 to 40 °C. Before taking the measurement at each temperature, we incubated the sample for 5 min at that temperature. Interestingly, the FRET efficiency was almost fully reversible when the temperature was decreased from 40 to 25 °C.
We hypothesized that the increase of FRET efficiency of the dual-labeled endo_N-BAR can be explained by conformational changes. To understand how these potential conformational changes are related to the temperature increase, we monitored the secondary structure of endo_N-BAR via CD spectroscopy as a function of temperature.
CD spectra of endo_N-BAR L187C (C108A) show that the protein helicity decreases from 47 to 22% when the temperature is increased from 25 to 37 °C (Figure 1E, the helicities were determined by the online tool Bestsel39,40), which is consistent with the notion of protein unfolding as a consequence of thermal stress at 37 °C. This finding is surprising since presumably endo_N-BAR is supposed to maintain its structural integrity at the body temperature of mammals to function properly in cellular transportation phenomena.1,4,10 This observation could have resulted from truncations of endophilin introduced to yield the endo_N-BAR mutant and the single-site mutation C108A. The effect of truncations and mutations on the thermal stability of endophilin will be discussed further below.
We systematically monitored protein helicity changes during the thermal denaturation process. Temperature-dependent CD signals at 222 nm were obtained for a larger range, as shown in Figure 1F. Interestingly, the CD spectra suggest a two-step denaturation process—one between 30 and 40 °C and the other between 50 and 60 °C. There is a significant loss of helicity in both steps. Temperature-dependent FRET was obtained at temperatures varying from 20 to 46 °C. As shown in Figure 1F, the FRET efficiency follows the CD signal obtained at 222 nm in the first unfolding step. This suggests that FRET efficiency changes are correlated with structural changes induced by thermal stress. We note that the FRET efficiencies at the same temperature were different in Figure 1D,E, which is most likely due to the difference in labeling efficiencies of fluorescent dyes in the two protein samples. The agreement between the temperature-dependent observations with complementary techniques suggests that the conformational change contributing to the FRET signal was induced by protein unfolding.
All the experiments presented so far were carried out with an endo_N-BAR L187C (C108A) mutant. We next asked if the observations were related specifically to the L187C mutation. The molecular weight of endo_N-BAR L187C (C108A) was determined to be 58.0 ± 2.9 kDa by analytical ultracentrifugation (AUC), while the molecular weight of endo_N-BAR monomer is 28.2 kDa. This result implies that this mutant forms a dimer similar to the wild-type protein (Figure S1A,B).15 We evaluated the possibility of the L187C mutant affecting protein folding stability through denaturation studies based on CD spectroscopy. We found essentially the same temperature-dependent ellipticity for the endo_N-BAR +/– L187C species, suggesting that the protein unfolding process was not disturbed by this mutation (Figure S1C). An alternative tip mutant, endo_N-BAR L183C (C108A), was also labeled with the PB-A488 FRET pair. Comparison of the emission spectra (EX: 405 nm) of this L183C mutant at 25 and 37 °C showed FRET efficiency increase at higher temperature, as observed for the L187C mutant, further suggesting that the temperature-dependent FRET change was not induced by mutagenesis in the protein’s tip region (Figure S2).
To summarize, we confirmed that the endo_N-BAR denaturation-induced FRET signal change is not an artifact that is caused by specific artificial mutations in the tip regions but rather is explained by a natural conformational change of the BAR protein in response to thermal stress. We then asked exactly what conformational change was induced to cause the FRET change. The FRET signal can either be caused by interdimer or intradimer interactions between donors and acceptors. In the first situation, the proteins might oligomerize, leading to energy transfer from the donor on one dimer to the acceptor on another dimer. In an alternative hypothesis, the endo_N-BAR dimer may start to unfold from the tip region (“tip unfolding”). The disordered tips may be more flexible compared to the folded state, and thus, the two tips might have higher tendency to remain in closer proximity to each other. A schematic summary of these two hypotheses is shown in Figure 2A.
Figure 2.
Endo_N-BAR tip unfolding through thermal denaturation does not result in significant oligmerization. (A) Two hypotheses of endo_N-BAR conformational changes could explain the temperature-dependent FRET during endo_N-BAR thermal denaturation. Hypothesis 1 is that endo_N-BAR may oligomerize during protein denaturation to induce interdimer FRET. Hypothesis 2 is that two tips of the dimer unfold first to increase intradimer FRET. (B) SDS PAGE gels of endo_N-BAR 187C (C108A) BS3-cross-linked at RT and at 37 °C show a similar amount of oligomerization. The dominant species in both cases were dimers or monomers. For [BS3] = 0.5 and 5 mM with different incubation temperatures, the dimer was the main species. Detection of monomers was due to <100% cross-linking efficiency. The arrows point to bubbles that were generated when the gel image was taken. (C) AUC VS experiments show the dimer of endo_N-BAR 187C (C108A) to be the dominant species at both 20 and 37 °C. Only one peak around 56 kDa was observed for both 20 and 37 °C (endo_N-BAR monomer MW = 28 kDa). (D) Cross-linking comparison of endo_N-BAR 187C, endo_N-BAR 183C, and endo_N-BAR 241C (all with C108A mutation) confirmed that the distance of two tips of endo_N-BAR was reduced at high temperature. Full gel images with ladders can be found in Figure S3. The arm spacer of the linker Bis-MAL-dPEG has a length of 48.7 Å. Endo_N-BAR 241C served as control with the distance between two cysteines being less than 49 Å. Also, 48 and 51% detected dimer percentages at RT and 37 °C suggest that the cross-linking efficiency is around 50%. For both endo_N-BAR 187C and 183C, around 20% of close cysteine pairs were detected, while no such cross-linked cysteine pairs were observed for RT incubation. This result suggested that the dimer structure was intact at RT. However, during thermal denaturation, the two arms of endo_N-BAR became flexible and reduced their tip distance.
During Thermal Denaturation of endo_N-BAR, Unfolding of the Tip Region Precedes the Unfolding of the Dimerization Interface
To explain the denaturation-induced FRET signal change of dual labeled endo_N-BAR, we further tested both the oligomerization and the “tip unfolding” hypotheses.
We first carried out experiments to show that endo_N-BAR does not oligomerize significantly at the temperature where thermal denaturation is observed (T = 30–40 °C). We observed that cross-linking with BS3 shows negligible oligomerization induced by temperature increase. To arrive at that conclusion, different concentrations of BS3 linkers were mixed with endo_N-BAR L187C. After incubation at RT or 37 °C, the mixture was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). As shown in Figure 2B, there is no significant difference in the extent of oligomer formation when comparing the samples incubated at different temperatures. To further confirm this, we also used AUC to study N-BAR oligomerization.44 Velocity sedimentation (VS) experiments indicate that the endo_N-BAR 187C (C108A) dimer is the dominant species at both 20 and 37 °C (Figure 2C).
Cross-linking experiments with the Bis-MAL-dPEG linker confirmed that the average distance decreases between the two tips of the N-BAR dimer during thermal denaturation. The spacer of the crosslinker Bis-MAL-dPEG is 48.7 Å, which is similar to the Foerster distance R0 = 51.0 Å of the PB-A488 FRET pair. Three endo_N-BAR samples with cysteine at different positions (187C, 183C, and 241C) were treated with the crosslinker for comparison. Incubations at both RT and 37 °C were carried out to study the temperature effect. Endo_N-BAR 241C served as a control with the distance between the two cysteines less than 49 Å. The previously discussed AUC experiments (Figure 2C) confirm that the dimeric structure is maintained at both temperatures. The cross-linking efficiency is around 50% for the 241C mutant at both RT and 37 °C. For endo_N-BAR 187C and 183C, the cross-linking position was at the tip of the N-BAR domain. Around 20% of close cysteine pairs were detected at 37 °C, while no such cross-linked cysteine pairs were observed for RT incubation (Figure 2D). These observations agree with our “tip unfolding” hypothesis. In this hypothesis, we propose that the two arms of endo_N-BAR reduce their tip-to-tip distance at the temperature above which thermal unfolding occurs. Alternatively, one might argue that the observed tip-to-tip distance reduction may be induced by formation of dimers with a tail-to-tail interaction at higher temperatures. The notion that this scenario is unlikely is supported by both simulation and in vitro experimental studies for BAR protein dimerization in the aqueous environment. MD simulations compared the free energy of different dimeric structures of a BAR protein and confirmed that the dimer corresponding to the crystal structure (see Figure 1A) is by far the most stable state.14 Dimerization studies on endophilin have used FRET-pair-labeling near the dimerization interface—the experimental findings are also consistent with the conclusion that the structure shown in Figure 1A is preferred in aqueous solutions.15,16
All the evidence presented so far supports our hypothesis that tip unfolding is the first step of endo_N-BAR denaturation, followed by the unfolding of the dimerization interface. This hypothesis is also consistent with the reported endo_N-BAR dissociation study by Chen et al.(16) By varying temperature from 27 to 40 °C, they obtained a constant equilibrium enthalpy and entropy change of endo_N-BAR dimer dissociation. According to our T-dependent CD measurements, the protein started to unfold at 30 °C. If the unfolding process started near the interface of the dimer, we would expect the dissociation enthalpy to be varied at a temperature above 30 °C. The two studies together suggest that the integrity of the dimerization interface was not affected during thermal denaturation with T < 40 °C. We therefore conclude that early helicity loss with increasing temperature occurs primarily in the tip region at the beginning of the protein unfolding process.
Gruber and Balbach have carried out chemical denaturation studies with the N-BAR domain of amphiphysin (Figure 1A,B), another member in the BAR protein family.31 They have suggested a denaturation model in which an unspecified intermediate unfolded state was proposed, followed by dimer dissociation. We propose that the “tip unfolding” structure may correspond to the first-stage unfolding product.
In summary, we used a FRET technique to monitor endophilin N-BAR unfolding under thermal stress. Our observations are consistent with the hypothesis that the tips of the endo_N-BAR unfold at the initial stage of thermal denaturation. However, the study above only focused on the endophilin N-BAR domain instead of the full-length protein. Other domains could also affect the folding stability of N-BAR. This will be addressed in the following sections.
Unstructured Domains of Endophilin Influence Its Thermal Stability
We have shown that the truncated version of rat endophilin, endo_N-BAR, partially unfolds at physiological temperature (37 °C). We therefore asked if full-length endophilin (endo_FL) also unfolds at body temperature.
Similar to how we examined endo_N-BAR in the FRET study discussed above, we implemented an L187C mutation in endo_FL and labeled the protein with the PB-A488 FRET pair. We mutated the cysteine at position 108 to serine (S) instead of alanine (A), as had been done for the endo_N-BAR construct discussed in the previous section. Emission spectra were recorded with EX: 410 nm at different temperatures. In contrast to endo_N-BAR, the dual-labeled endo_FL L187C (C108S) showed no detectable FRET peak (Figure 3A) at either 25 or 37 °C. Expanding the study to higher temperatures showed that the FRET efficiency started to increase at 40 °C (Figure 3A,B), which is significantly higher than the temperature of 30 °C at the onset of unfolding for endo_N-BAR. Temperature-dependent CD (Figure 3B) showed a similar trend at around T = 40 °C. This finding confirms that endo_FL is thermodynamically more stable than endo_N-BAR. Moreover, the consistency of temperature-dependent CD and FRET observations suggested that the N-BAR tips in the same dimer reduce their distance during denaturation for endo_FL, as was observed before for endo_N-BAR.
Figure 3.
Endophilin truncations influence its thermal stability. (A) Fluorescence emission spectra of dual-labeled endo_FL 187C (C108S) with excitation at 401 nm at different temperatures. Contrary to endo_N-BAR (Figure 1), no FRET was observed at T = 37 °C. The fluorescence spectra (EX: 401 nm) for 25 and 37 °C were almost identical, but FRET was observed when the temperature was further increased to 42 °C. (B) T-dependent FRET suggests tip unfolding to initiate around T = 40 °C, which explains why no FRET is observed at 37 °C in panel A. T-dependent CD showed that full-length endophilin started to unfold at 40 °C, which was consistent with the FRET data. (C) In the TSA, the SYPRO orange signal was tracked during temperature ramping for full-length endophilin and N-BAR (C108S E241C). Full-length endophilin displayed increased thermal stability compared to N-BAR with Tm being around 5 °C higher. (D) CD signals at 222 nm were measured for full-length endophilin C108S E241C and N-BAR C108S E241C when the temperature was increased from 25 to 55 °C. The full-length protein started to unfold when temperature was increased to 40 °C, while the N-BAR started to unfold at T = 35 °C. The CD data were consistent with the TSA data shown in (C). (E) Tm of various endophilin truncations obtained from TSA. Deletion of the disordered linker and the disordered H0 in aqueous solution showed decreased endophilin melting temperature. To note, all of the samples [in (E)] contained the mutations C108S and E241C except for #6 endo_N-BAR dH0, which remained C108 and E241. The Tm of endo_N-BAR C108 E241 is 40.3 ± 0.2 °C (not shown in the table), which is also higher than the Tm = 38.7 °C of endo_N-BAR dH0. According to TSA results, the C108S and E241C mutations did not impact the protein thermal stability significantly (Table S1).
We used a thermal shift assay (TSA) to directly compare the thermal stabilities of the N-BAR domain and the full-length protein. SYPRO orange fluorescence intensity was monitored during temperature ramping after mixing with two proteins. To avoid the influence of different single-site mutations, we used endo_N-BAR and endo_FL that contained the same mutation: C108S and E241C. The fluorescence signal of SYPRO orange reflected the denaturation of the protein. Consistent with the FRET data as well as CD data, endo_FL displayed increased thermal stability compared to endo_N-BAR, with its melting temperature (Tm) being around 5 °C higher (Figure 3C). The temperature-dependent CD spectra further confirmed this conclusion (Figure 3D). CD signals at 222 nm were measured for endo_FL and N-BAR when the temperature was increased from 25 to 55 °C. The full-length protein started to unfold when the temperature was increased to 40 °C, while the N-BAR domain started to unfold at T = 35 °C.
Why do the endo_FL and endo_N-BAR show significantly different thermal stabilities? To address this question, we systematically compared endophilin mutants with/without H0 deletion or linker deletion and examined their thermal stability. We found that both domains contributed to the thermal stability of endophilin, as per results determined via TSA and thermal denaturation experiments characterized by CD spectroscopy. We compared endo_FL and the H0 deletion mutant (endo_dH0) to show that Tm decreased with H0 deletion, suggesting that the presence of the H0 domain stabilized the protein (Figure 3E). The difference of Tm values of endo_FL and endo_dH0 was also confirmed by CD spectroscopy (Figure S4). A similar comparison was performed between endophilin-ΔSH3 (endo_dSH3, containing the N-BAR domain and the linker) and endophilin ΔSH3 ΔH0 (endo_dH0 dSH3) to focus on the denaturation of the BAR domain (the only structured domain remaining after SH3 deletion). Consistently, the results also showed that the deletion of H0 reduced the melting temperature. The same result was obtained by comparison of endophilin N-BAR and endophilin N-BAR H0 deletion with further deletion of the linker. Comparison of endophilin N-BAR and endophilin N-BAR plus linker showed that the unstructured linker had a similarly stabilizing effect as the H0 domain.
Differential scanning calorimetry (DSC) also indicated that the deletion of the unstructured linker decreased Tm of endophilin. To show this, we compared endo_FL, endo_dSH3, and endo_N-BAR. The deletion of the disordered linker decreased the Tm by around 5 °C compared to the full-length protein. Moreover, the denaturation enthalpy change was also different for endo_dSH3 (N-BAR + linker) and endo_N-BAR. The enthalpy change decreased by more than 20% with the linker deletion, consistent with the proposed effect of the flexible linker in stabilizing the protein N-BAR structure (Figure S5).
Variable influences of intrinsically disordered domains (IDRs) on the protein thermal stability have been observed in other protein systems. It is commonly observed that the protein stability decreases in the presence of unstructured domains.45,46 However, in some cases, IDRs enhance protein stability through facilitating protein oligomerization or protein interactions with a binding partner.47,48 Association among IDRs, or an interaction between IDRs and the structured regions, can also increase the protein thermal stability.49,50 Since we have observed that both the H0 region and the unstructured linker enhance the thermal stability of endo_N-BAR, our next step was to investigate the mechanism of such an effect. In the discussion above, we have excluded oligomerization during endo_N-BAR denaturation. We hypothesize that the flexible linker and the H0 domain may interact with other domains in the endophilin protein. The interaction between the two flexible linkers on the endophilin dimer was of low probability according to the study of the dimerization kinetics of several endophilin truncations by Chen and his co-workers.16 They have shown that the deletion of the linkers has negligible influence on endo_N-BAR dimerization. Interactions between two H0 domains on the same dimer are not possible due to the large distance between the two domains in the crystal structure. An interaction between H0 and the linker can also be excluded because the linker stabilized the BAR domain with H0 deletion in the TSA study. Thus, we hypothesize that H0 and the linker can both bind to the structured portion of the N-BAR domain via weak interactions.
To further confirm that the linker does influence the N-BAR thermal stability and investigate the potential mechanism, we created several mutations in the linker region. The length of the linker is 45 amino acids. We purified mutants of N-BAR plus a truncated linker with 24, 15, or 8 amino acids (truncations involved residues 248–271, 248–262, and 248–255, respectively, of the endophilin sequence). Only the last one (with residues 248–255 of the linker remaining) showed a >2 °C decrease in the melting temperature measured by TSAs, which suggests that cutting residues 256–262 of the linker, but not the residues after them, destabilizes endo_N-BAR. To confirm that residues 256–262 of the linker play a key role in influencing the N-BAR domain stability, single-site mutations (P257A, P259A, and R260E) were also created in this region to result in the endo_ N-BAR + linker 257PKPR → AKAE mutant. This mutant showed a 2 °C decrease of the melting temperature compared to endo_N-BAR + linker (or ΔSH3) (Figure S6). Amino acids arginine (R) and proline (P) have frequently been reported to function in key positions to recruit protein-binding partners.51 Proline is a hydrogen-bond acceptor, and proline-rich motifs (PRMs) are often involved in protein interactions.52 While arginine is positively charged, the surface of the N-BAR arms are covered with negatively charged residues. Electrostatic interactions between the linker and the N-BAR domain may therefore stabilize the folded core of the N-BAR domain under thermal stress. Even though the precise roles of prolines and the arginine would require further investigation, together with the linker truncation comparison, this 257PKPR → AKAE mutation result suggests that a region close to the N-terminus of the linker (256–262) plays the key role in enhancing the thermal stability of endo_N-BAR, and P and R residues may be involved in this stabilizing effect.
Single-Site Mutation in the BAR Domain Core Influences Endophilin’s Thermal Stability
As discussed above, truncating endophilin influences its thermal stability. We further looked at the role of an amino acid mutation that has been used for protein labeling and functional studies of endophilin,15,53 as well as used in our studies presented above.
In the previously discussed thrusts, we had created fluorophore conjugating positions through cysteine mutations and mutated the cysteine at 108 position to an alanine or a serine. We then asked if these different exchanges at the 108-position influence protein stability to varied extents. The C108 position is within a short loop between helices (Figure 4A). The C108S mutation did not change the Tm significantly, while the mutation to alanine decreased the Tm by 5 °C (Figure 4C). Hydrogen bonds are predicted to form between N109 and D208 as well as between F110 and D208, as per the online tool PDBePISA.54 The increased hydrophobicity of alanine at the 108 position compared to cysteine and serine may interfere with hydrogen bonding interactions that stabilize the protein structure. The CD signal determined during endo_N-BAR thermal denaturation also suggests that the C108A mutation decreases endo_N-BAR thermal stability while E241C does not.
Figure 4.
Cysteine replacement at 108 position influences endo_N-BAR thermal stability. (A) Position of C108 and E241 in the N-BAR crystal structure. (B) Structure formula of cysteine, alanine, and serine. The side chain of alanine is more hydrophobic compared to cysteine and serine. (C) Tm comparison of 108 cysteine replacement in endo_N-BAR. The mutation C108S did not change Tm significantly, while mutation to C108A decreased the Tm by 5 °C. Errors: standard deviations for repeated trials for the same protein preparation (n = 3–9). (D) CD signals at 222 nm were measured for endo_N-BAR C108S, endo_N-BAR C108A, endo_N-BAR C108S + E241C, and endo_N-BAR C108A + E241C when the temperature was increased from 20 to 50 °C. The E241C mutation did not influence the protein thermal stability, while the C108A mutation induced >5 °C difference in the melting temperature compared to the C108S mutation, which agrees with the TSA results in Figure 4C.
Discussion
In the present study, we investigated thermal denaturation of the endophilin N-BAR domain (endo_N-BAR). Based on a variety of complementary experimental approaches, we proposed a two-step unfolding mechanism with a dimeric intermediate. Furthermore, we showed evidence that the deletion of the H0 domain and the flexible linker decreases the thermal stability of endo_N-BAR.
Endo_N-BAR is a crescent-shaped dimeric protein. Dimerization is involved in the final formation of the functional structure. This complicates the mechanism of endophilin folding and unfolding compared to monomeric proteins.19 We were able to study the conformational changes and the changes of protein secondary structure during thermal denaturation of endo_N-BAR. First, the temperature-dependent helicity change detected by CD spectroscopy indicates a two-step denaturation process: one between 30 and 40 °C and the other between 50 and 60 °C. This suggests that at least one intermediate exists, which suffers partial loss of secondary structure compared to the natural protein.19 Furthermore, we found that the dimeric structure is maintained during the first unfolding step. Finally, via FRET, we observed that the distance between two tips within the same BAR domain dimer decreases during thermal denaturation. Based on the findings presented above, we propose that during endo_N-BAR thermal denaturation, a dimeric intermediate exists and is characterized by the presence of unfolded tips of the elongated protein structure.
The folding and unfolding of BAR proteins during chemical denaturation has been studied by Gruber and Balbach.31 They monitored the fluorescence signal of tryptophans in amphiphysin II to carry out kinetic studies. They proposed the following model for a folding mechanism of the BAR domain: in the unfolded protein, there is an equilibrium between prolyl cis- and trans-conformations, resulting in two parallel pathways.55 In each pathway, there is a rapid formation of a monomeric intermediate, followed by a relatively slower dimerization to form a dimeric intermediate, before finally forming the natural structure. Conversely, in the unfolding process, the first step is the formation of the dimeric intermediate, followed by its dissociation into monomeric intermediates. Our finding of the dimeric intermediate formation in the first denaturation step appears to be consistent with their proposed folding/unfolding model. We further characterized the structural nature of the dimeric intermediate: the two tips unfold, while the dimeric interface appears to remain intact.
The folding/unfolding pathway of dimeric proteins can be classified into three major groups: a two-state mechanism, a three-state mechanism with a monomeric intermediate and a three-state mechanism with a dimeric intermediate.19 The size of the protein as well as the ratio of the interface area to total surface area (% buried area) seem to be the key factors determining the unfolding mechanism.19,56,57
Two-state unfolding is common for small dimeric proteins whose monomers contain less than ∼100 residues. Monomeric intermediates tend to be formed by proteins that have relatively larger subunits (chain lengths between ∼100 and ∼250 amino acids) while dimeric intermediates can be detected for even larger proteins (chain lengths >250 amino acids).17,18,26 The N-BAR domains of endophilin A1 and amphiphysin II consist of 247 and 249 residues, respectively. Dimeric intermediates are detected for both proteins, roughly consistent with the above-mentioned estimates.
The absolute interface area and fraction buried area can contribute to the unfolding mechanism.19 For the dimers forming dimeric intermediates, the absolute interface areas tend to be larger. However, the fraction of buried area is comparatively small due to the large protein size, which seems to suggest the formation of a stable monomeric intermediate instead of the dimeric one. These controversial observations can be explained by the independent unfolding of different domains of a large dimeric protein.20,24 Protein dimerization can contribute to the stability of the domain containing the dimer interface,24,58,59 and thus, this domain unfolds at a later stage of protein denaturation. In the case of endo_N-BAR, the arm region and the interface-forming region are connected via kinks, which may allow for an independent unfolding of the two regions separated by the kinks.11 Specific molecular properties of the interface may also affect the protein unfolding mechanism and the formation of intermediates.21,60,61
There are also some disagreements between our results on endophilin and those reported by Gruber and Balbach observed for amphiphysin.31 First, they showed no difference of the stability comparing wild-type protein and a variant without the H0 domain, suggesting that the H0 domain has no influence on the thermodynamic stability of the BAR domain. Second, in their study, no significant CD signal change was detected between the natural protein and the dimeric intermediate, while we observed significant helicity loss for the intermediate of endo_N-BAR. There can be two major reasons for these differences: first, the difference in studied proteins and second, the difference in denaturation methods.
Endophilin and amphiphysin belong to the N-BAR subfamily, with a similar curvature of the BAR domain.9,43 The core of a BAR domain is a six-helix bundle formed by three helices of each monomer (helix 1–3). Two arms protrude from the central helix bundle.62 It is possible that endophilin and amphiphysin have similar dimeric intermediates during denaturation. However, the two proteins share little sequence similarity. Moreover, endophilin contains an extra H1i helix in the middle of the helix 1 compared to amphiphysin. These differences may introduce variations to their unfolding behaviors. As for the helicity loss from natural protein to intermediates, the dimeric intermediates of different proteins do show varied extents in the loss of secondary structure, with some of the changes not detectable by CD spectroscopy.22,23
The different denaturation methods applied can also influence the state of the unfolding intermediates. Both chemical denaturation and thermal denaturation weaken hydrophobic cores and disrupt protein secondary structures.63,64 The free energy changes upon protein unfolding have been claimed to be similar by these two methods.65 Urea, a common chemical denaturant, was used in Gruber’s study on amphiphysin. Urea is proposed to interact directly with polar residues and the peptide backbone, thereby stabilizing nonnative conformations. It has been confirmed by MD simulations that urea can also stabilize the unfolded structure indirectly by altering the solvent environment.66,67 Thermal denaturation of proteins is caused by a strong increase of the entropy change of protein unfolding at high temperatures.68,69 The main contributions to protein entropy arise from conformational fluctuations of the protein backbones and the side chains, as well as the ordering of water molecules around the side chains. With increased temperature, a stronger difference in structural entropy between the folded and the unfolded state have been observed. A comparison of the far-UV CD signal at 222 nm for a group of alpha-helical proteins between the highest temperature (in the absence of denaturant) and highest denaturant concentration (at a fixed temperature) reveals a consistent difference between the two denaturation conditions, with a significant mean absolute difference of ∼3100 deg·cm2·dmol–1.70 The final status of the unfolded protein may also be different when comparing chemical denaturation and thermal denaturation, especially for proteins that tend to aggregate upon unfolding.71 These differences indicate that the two denaturation methods contribute to different final conformational subensembles within the denatured state. Therefore, it is unsurprising that the conformation of the intermediates may also vary under two conditions. It may explain why we observe helicity loss of the dimeric intermediate during thermal denaturation, while Gruber et al. did not detect changes in the secondary structure for the dimeric intermediate in their chemical denaturation studies.
As mentioned above, we show that the H0 contributes to the thermal stability of endo_N-BAR, while Gruber and Balbach suggest no influence of the H0 in the amphiphysin BAR domain.31 Our finding is supported by the shift of the melting temperature with H0 deletion in both TSA experiments and temperature-dependent CD spectrum measurements. We have compared endo_dH0 to endo_FL, endo_dH0 dSH3 to endo_dSH3, as well as endo_N-BAR dH0 and endo_N-BAR. All of the comparisons show that the melting temperatures decrease by 1–5 °C with H0 deleted, suggesting that the H0 helix can stabilize the BAR domain of endophilin. Given Gruber et al.’s observations, however, this stabilizing effect does not appear to be generalizable for all BAR proteins.
Similar to the H0 domain, we find that the flexible linker between the BAR domain and the SH3 domain of endophilin also has a stabilizing effect. The deletion of the linker induces a 6–8 °C decrease of the melting temperatures when comparing endo_N-BAR to endo_dSH3 (N-BAR + linker) and endo_N-BAR dH0 to endo_dSH3 dH0. We hypothesize that the protein stabilizing effect results from potential molecular interactions between the linker and the N-BAR arm. This hypothesis is supported by the loss of such a stabilizing effect when the PKPR sequence in the linker region is removed by linker truncation or single-site mutations. The surface of the N-BAR arm is covered with negatively charged side chains.9 The positively charged arginine in PKPR is located around 10 residues (∼3.5 nm) away from the BAR domain along the amino acid sequence, contributing to a potential electrostatic interaction between the linker and the BAR arm. The arginine and the prolines located near it were mutated to create an AKAE sequence from the original PKPR. Proline is known to be a hydrogen-bond acceptor.52 Indeed, PRMs often function as binding modules functionally linking proteins in signaling events.72 The prolines may also contribute to the potential interactions between the linker and the BAR domain. Chen and co-workers have indicated that the truncation of the linker has negligible influence on the dimerization of endo_N-BAR, suggesting that there should not be any interactions between the linker in one monomer and the BAR domain of the other monomer of an N-BAR dimer.16 Thus, we hypothesize that the proposed interaction between the linker and the N-BAR arm may be within the same monomer or it may become significant only when protein denaturation is initiated.
Besides different domains in the endophilin structure, single-site mutations can also influence the protein thermal stability, even if they are in the unstructured loop region.73−76 Such an effect may arise from varying motilities of the loops and nearby secondary structures. In our study, the C108A or C108S mutation is in the loop connecting the helix 1 and the helix 2 of endo_N-BAR, and E241C mutation is located at the C-terminus of helix 3. The C108A mutation decreases the melting temperature of endo_N-BAR by 6 °C, while C108S and E241C show no significant effect. Several online tools have been developed to predict protein stability upon point mutations.77,78 However, they can only claim an accuracy below 90%, predicting the stability of a protein mutant.77 These prediction tools can offer guidance when working on mutant design, but they may fail to reveal the full molecular consequences of the effect of the point mutation.
We have focused on the thermal denaturation and thermal stability of the BAR protein, endophilin. An open question is how these thermodynamic properties relate to endophilin’s membrane binding and remodeling function in vivo. The crescent BAR domain is proposed to bend the membrane via a scaffolding mechanism.8,10,79 The concave face of endo_N-BAR binds to the negatively charged lipid membrane by electrostatic interactions.12,80,81 The structure of this BAR domain may in turn be protected by the stabilizing effect of membrane binding.82,83 Our studies have indicated that the flexible linker and H0 can also contribute to the stability of the BAR domain, which may protect the crescent domain shape before interacting with membranes. However, how the structural stability of BAR proteins influences the protein functions remains an interesting question for future investigation.
Conclusions
Our study has indicated that a dimeric intermediate is generated during thermal denaturation of endophilin. We propose that the tip region of endo_N-BAR unfolds before dimer dissociation occurs to a significant extent. The rigid dimer structure can be maintained even under thermal stress, which may be explained by the presence of multiple hydrogen bonds and salt bridges on the dimer interface. In vivo, a fast and stable formation of the dimeric or oligomeric intermediate might avoid an unwarranted association of monomers with other cellular proteins and guarantee proper folding.84 Moreover, the stable dimerization enables membrane reshaping by endophilin through the scaffolding effect.
A stable tip region is also important to maintain the rigid crescent shape of the BAR domain, which contributes to endophilin’s membrane curvature generation function through the scaffolding mechanism.12 Interestingly, our study suggests a potential role of H0 and the flexible linker in protecting the endo_N-BAR structure. It remains an open question if these IDRs indirectly contribute to endophilin’s membrane-related functions through such stabilizing effects.
Materials and Methods
Materials
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was obtained from Sigma-Aldrich; sodium chloride (NaCl), Tris(2-carboxyethyl)phosphine (TCEP), Coomassie Plus (Bradford) protein assay reagent, bovine serum albumin (BSA) standards, bis(sulfosuccinimidyl)suberate (BS3), dithiothreitol (DTT), and β-casein were obtained from Fisher scientific; Bis-MAL-dPEG11 was obtained from Quanta BioDesign; the dyes Pacific Blue (PB) C5-maleimide, Alexa Fluor 488 (A488) C5-maleimide, and SYPRO Orange Protein Gel Stain were obtained from Invitrogen/Life Technologies. All commercial reagents were used without further purification.
Plasmids and Protein Purification
A plasmid encoding rat endophilin A1, UniProtKB O35179 (SH3G2_RAT), kindly provided by P. De Camilli, served as the template to generate all the endophilin mutants used in this work. Endo_N-BAR (residues 1–247) was used to study the unfolding mechanism of the N-BAR domain. Mutations of C108A + L187C and C108A + L183C were created to study the tip unfolding with cysteines in the tip regions. Mutations of C108A + E241C served as a comparison for cross-linking experiments. The full-length protein, endo_FL (1–352), containing C108S + L187C + C294S + C295S mutations was created to compare with the endo_N-BAR (C108A + L187C) for the unfolding study. To study influences of different protein domains on the thermal stability of endophilin, endo_FL (1–352), endo_dH0 (33–352), endo_dSH3 (1–292), endo_dH0 dSH3 (33–352), endo_N-BAR (1–247), and endo_N-BAR dH0 (33–247) were created with C108S + E241C mutations, except for endo_N-BAR dH0, which contained no single-site mutations (C108 + E241). To further investigate which parts of the flexible linker influence the melting temperature of endophilin, linker truncations were created to generate endo_N-BAR+1/2 linker (1–271), endo_N-BAR+1/3 linker (1–262), and endo_N-BAR+ 1/4 linker (1–255). Single-site mutations P257A, P259A, and R260E on the linker were created to generate endo_dSH3 257PKPR → AKAE (1–292). These proteins with linker truncations also contained C108S and E241C mutations to compare with other endophilin truncations. Endo_N-BAR (1) of wild type (C108) or with mutations of (2) C108A, (3) C108S, (4) C108A + E241C, and (5) C108S + E241C were used to study the impact of different single-site mutations on the protein thermal stability. All sequences were verified by DNA sequencing.
Glutathione-S-transferase (GST) fusion proteins were purified from bacterial lysates [BL21(DE3), RIL CodonPlus, Stratagene] using glutathione affinity chromatography. PreScission protease was added to the fusion proteins with a 1:50 molar ratio for the cleavage of the GST moiety. The mixture was then shaken at 4 °C for 8 h to achieve complete cleavage. The proteins were further purified by ion exchange with a linear NaCl gradient and size exclusion chromatography (SEC) [Superdex200 (GE Healthcare)]. Protein identity and purity was assessed by SDS-PAGE after each purification step. The products were concentrated and flash-frozen in liquid nitrogen and stored at −80 °C. For measurements with thawed samples, we removed potential aggregates via ultracentrifugation. Concentrations were determined by the Bradford assay using BSA solution as a standard. Concentrations indicated refer to total endophilin/N-BAR in terms of monomeric units. Labeling was conducted at introduced Cys residues for N-BAR_187C, N-BAR_183C and FL_187C. Fivefold excess of maleimide dye reagent was used via a dimethyl sulfoxide solution for reaction at 4 °C, in 20 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 7.4 solution (HN150T buffer). Reactions were quenched with excess DTT, and the excess dye reagent was removed via three 5 mL HiTrap desalting columns (GE Healthcare) connected in series. To create dual-labeled proteins, endo_N-BAR or endo_FL were simultaneously reacted with PB and A488 by mixing protein samples with 10-fold concentration of each dye, shaken at 4 °C for 8 h. The mixture was then applied to a set of three desalting columns (GE Histrap) via fast protein liquid chromatography to remove unreacted dyes. The protein concentration was determined via Bradford assay. The concentrations of PB and A488 were determined by measuring absorption at 402 and 493 nm. Labeling efficiencies obtained were in the range of 40–80%.
Fluorescence Spectroscopy
Measurements employed a Cary Eclipse fluorometer with a Peltier-controlled temperature block. Donor (PB) and acceptor (A488) conjugates (D and A, respectively) were examined for FRET tests at different temperatures in HN150T buffer. To avoid protein adsorption to the cuvette walls, quartz cuvettes were treated with casein solution for 1 h and washed once with the sample buffer. The casein solution comprised the soluble portion of a 2.5 mg/mL suspension in the sample buffer. The FRET efficiency was determined by the loss of PB fluorescence compared to the original fluorescence when no FRET was observed. To collect spectra at a given temperature, the sample was incubated at the temperature of interest for 20 min. Within uncertainties, it took around 5 min for FRET changes to reach equilibrium at 37 °C after heating from 25 °C. For that heating interval, it took around 1 min for the sample cell to reach the temperature of 37 °C. For studies of FRET reversibility for temperature-dependent folding and unfolding experiments, the incubation time at each temperature was reduced to 5 min.
Protein Cross-Linking
Cross-linking with amine-reactive BS3 was carried out to test protein oligomerization. Site-specific cross-linking with Bis-MAL-dPEG11 (Quanta BioDesign, Powell, OH; linker length, 48.74 Å) via cysteine coupling was conducted to estimate the intradimer cysteine–cysteine distance. Protein samples of 5 μM concentration were incubated at RT or at 37 °C in HN150T buffer for 30 min before adding BS3 comparing concentrations of [BS3] = 0, 0.5, 5 mM. Then, all the mixtures were incubated at 37 °C for 10 min, after which the reactions were terminated by adding twofold excess of DTT. For intramolecular cross-linking with Bis-MAL-dPEG11, 5 μM endophilin samples were incubated with 100 μM of the cross-linker at RT or 37 °C for 30 min, and the reactions were terminated with twofold excess of DTT. Cross-linked samples were examined via SDS-PAGE to quantify non-cross-linked monomers, cross-linked dimers, and cross-linked oligomers.
Far-UV CD Measurements
Far-UV CD spectra were recorded with temperature control on an AVIV model 425 CD spectrometer (Biomedical, Inc., Lakewood, NJ, USA) using a 1 mm path-length cell in HN150T buffer. A full spectrum was collected every 1 nm at 3 nm/min from 200 to 260 nm with a bandwidth of 1 nm, and the results are averages of 10 scans. The contribution of the buffer to the measured ellipticity was subtracted as blank. Ellipticity at 222 nm during temperature increase was also measured to monitor thermal denaturation, taking steps of 2 °C and using an equilibration time of 1 min at each temperature. Molar ellipticity values (ϕ) were calculated using the expression ϕ = ϵ/10cnl, where ϵ is the ellipticity (millidegrees), c is the protein concentration (mol/liter), l is the cuvette path length, and n is the number of amino acid residues in the protein. The amount of secondary structural elements was estimated with the online tool BestSel.85
Analytical Ultracentrifugation
Sedimentation equilibrium of the N-BAR_187C labeled with A488 was carried out in HN150T buffer at concentrations of 3.2, 6.5, 13 μM at 4 °C in a Beckman Optima XL-I analytical ultracentrifuge (Beckman Coulter, Palo Alto, CA). Absorbance at 280 nm was taken using quartz windows in six-chamber charcoal centerpiece in an AN-60 Ti four-hole rotor (Beckman Coulter) with 6 h scans with a radial step size of 0.001 cm. The speeds used were 9000, 12,000, and 16,000 rpm. Molecular masses were calculated by fitting to the equation
Here T is the temperature, ω is the spin rate, r is the radial length, r0 is the radius of the reference position, c(r) denotes the protein distribution, c0 is protein concentration at reference position, and Mb is the molecular weight of the protein species.
Sedimentation velocity experiments were carried out in HN150T buffer at a concentration of 5 μM at both 20 and 37 °C. The experiments were run at 42,000 rpm with a radial step size of 0.001 cm and a 5 min delay between scans. The data were analyzed by SEDFIT with a continuous c(M) distribution model with mass conservation and a confidence level (F ratio) of 0.95, where the frictional ratio (f/f0) was allowed to float.86
Thermal Stability Assay
Individual 20 μL reactions were set up with 4 μM endophilin and a 1:1000 dilution of 5000× concentrated SYPRO orange solution in HN150T buffer. For each protein mutant, three parallel samples were prepared each time and added to separate wells in a 96-well assay plate specific for real PCR instrument. Thermal melting curves were obtained by heating the protein from 20 to 95 °C with a ramping rate of 2 min/°C and monitoring fluorescence at 580 nm using a 7900HT Fast Real Time PCR System (Applied Biosystems). The fluorescence was normalized by the peak value. Tm was defined as the temperature at which the slope of the SYPRO fluorescence change reached a maximum.
Differential Scanning Calorimetry
Around 1 mg/mL protein samples in HN150T buffer were used for DSC measurements. The samples were degassed while stirring in an evacuation chamber for 5 min at 10 °C before loading 400 μL into the cell. All the samples were analyzed by MicroCal VP-Capillary DSC (Malvern Panalytical), with HN150T buffer in the reference cell. The samples were scanned from 10.0 to 90.0 °C with a 10 min pre-equilibration at 10.0 °C and a 30 or 60 °C h–1 ramp rate. The data were analyzed with the ORIGIN 7.0 VP-DSC package. The data were baseline-corrected through a linear connect model, followed by manual adjustment. The Tm and ΔH were calculated through a non-two-state Levenberg–Marquardt fitting model and evaluated by reduced χ2 analysis.
Acknowledgments
The authors thank Dr. Pietro De Camilli for generously providing the template of rat endophilin A1 and Jaclyn Robustelli for the help in designing mutations for the endophilin template. They are grateful to Jeffery G. Saven lab for the use of the fluorimeter. The authors thank Tyler Reagle, Karthik B Narayan, and Sankalp Shukla for critical reviews during the preparation of the manuscript and helpful comments and suggestions from Zachary Zimmerman and Samsuzzoha Mondal.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01905.
More TSA, CD, static fluorescence and AUC data; noncropped gel images for cross-linking experiments; and DSC results (PDF)
Accession Codes
UniProtKB: O35179.
Author Present Address
∥ Bristol Myers Squibb in Lawrenceville, NJ, USA
Author Contributions
R.J., R.M., and T.B. designed the research, and R.J. and T.B. wrote the paper. R.J. developed methods, performed experiments, and analyzed data. M.G. and M.Z. performed TSA and DSC experiments and analyzed data.
This work was financially supported by the National Institutes of Health, grant no. GM 097552.
The authors declare no competing financial interest.
Supplementary Material
References
- Boucrot E.; Ferreira A. P. A.; Almeida-Souza L.; Debard S.; Vallis Y.; Howard G.; Bertot L.; Sauvonnet N.; McMahon H. T. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 2015, 517, 460–465. 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- Etxebarria A.; Terrones O.; Yamaguchi H.; Landajuela A.; Landeta O.; Antonsson B.; Wang H.-G.; Basañez G. Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large scale membrane morphological rearrangements. J. Biol. Chem. 2009, 284, 4200–4212. 10.1074/jbc.m808050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hak L. C. W.; Khan S.; Di Meglio I.; Law A.-L.; Lucken-Ardjomande Häsler S.; Quintaneiro L. M.; Ferreira A. P. A.; Krause M.; McMahon H. T.; Boucrot E. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat. Cell Biol. 2018, 20, 1023. 10.1038/s41556-018-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundborger A.; Soderblom C.; Vorontsova O.; Evergren E.; Hinshaw J. E.; Shupliakov O. An endophilin-dynamin complex promotes budding of clathrin-coated vesicles during synaptic vesicle recycling. J. Cell Sci. 2011, 124, 133–143. 10.1242/jcs.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K.; Ringstad N.; Takei K.; Floyd S. R.; Rose K.; De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 2001, 155, 193–200. 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N.; Gad H.; Löw P.; Di Paolo G.; Brodin L.; Shupliakov O.; De Camilli P. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron 1999, 24, 143–154. 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Llobet A.; Gallop J. L.; Burden J. J. E.; Camdere G.; Chandra P.; Vallis Y.; Hopkins C. R.; Lagnado L.; McMahon H. T. Endophilin drives the fast mode of vesicle retrieval in a ribbon synapse. J. Neurosci. 2011, 31, 8512–8519. 10.1523/jneurosci.6223-09.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff O.; Brodin L.; Jung A. The structure and function of endophilin proteins. Cell Biochem. Biophys. 2011, 60, 137–154. 10.1007/s12013-010-9137-5. [DOI] [PubMed] [Google Scholar]
- Gallop J. L.; Jao C. C.; Kent H. M.; Butler P. J. G.; Evans P. R.; Langen R.; McMahon H. T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C.; Cui H.; Gawronski-Salerno J. A.; Frost A.; Lyman E.; Voth G. A.; Unger V. M. Structural Basis of Membrane Bending by the N-BAR Protein Endophilin. Cell 2012, 149, 137–145. 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J. Mol. Biol. 2005, 351, 653–661. 10.1016/j.jmb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Masuda M.; Takeda S.; Sone M.; Ohki T.; Mori H.; Kamioka Y.; Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006, 25, 2889–2897. 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P.; Jin R.; Baumgart T.; Deserno M.; Dimova R.; Frolov V. A.; Bashkirov P. V.; Grubmüller H.; Jahn R.; Risselada H. J.; Johannes L.; Kozlov M. M.; Lipowsky R.; Pucadyil T. J.; Zeno W. F.; Stachowiak J. C.; Stamou D.; Breuer A.; Lauritsen L.; Simon C.; Sykes C.; Voth G. A.; Weikl T. R. The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D: Appl. Phys. 2018, 51, 343001. 10.1088/1361-6463/aacb98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri A.; Maisuria D.; Varga M.; Mohammadyani D.; Johnson M. E. Thermodynamics and Free Energy Landscape of BAR-Domain Dimerization from Molecular Simulations. J. Phys. Chem. B 2021, 125, 3739–3751. 10.1021/acs.jpcb.0c10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capraro B. R.; Shi Z.; Wu T.; Chen Z.; Dunn J. M.; Rhoades E.; Baumgart T. Kinetics of endophilin N-BAR domain dimerization and membrane interactions. J. Biol. Chem. 2013, 288, 12533–12543. 10.1074/jbc.m112.435511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Chang K.; Capraro B. R.; Zhu C.; Hsu C.-J.; Baumgart T. Intradimer/Intermolecular interactions suggest autoinhibition mechanism in endophilin A1. J. Am. Chem. Soc. 2014, 136, 4557–4564. 10.1021/ja411607b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping T. B.; Hoch D. A.; Gloss L. M. Folding mechanism of FIS, the intertwined, dimeric factor for inversion stimulation. J. Mol. Biol. 2004, 335, 1065–1081. 10.1016/j.jmb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Zeeb M.; Lipps G.; Lilie H.; Balbach J. Folding and association of an extremely stable dimeric protein from Sulfolobus islandicus. J. Mol. Biol. 2004, 336, 227–240. 10.1016/j.jmb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Rumfeldt J. A. O.; Galvagnion C.; Vassall K. A.; Meiering E. M. Conformational stability and folding mechanisms of dimeric proteins. Prog. Biophys. Mol. Biol. 2008, 98, 61–84. 10.1016/j.pbiomolbio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Ramstein J.; Hervouet N.; Coste F.; Zelwer C.; Oberto J.; Castaing B. Evidence of a thermal unfolding dimeric intermediate for the Escherichia coli histone-like HU proteins: thermodynamics and structure. J. Mol. Biol. 2003, 331, 101–121. 10.1016/s0022-2836(03)00725-3. [DOI] [PubMed] [Google Scholar]
- Cervelli M.; Leonetti A.; Cervoni L.; Ohkubo S.; Xhani M.; Stano P.; Federico R.; Polticelli F.; Mariottini P.; Agostinelli E. Stability of spermine oxidase to thermal and chemical denaturation: comparison with bovine serum amine oxidase. Amino Acids 2016, 48, 2283–2291. 10.1007/s00726-016-2273-5. [DOI] [PubMed] [Google Scholar]
- Kishore D.; Kundu S.; Kayastha A. M. Thermal, chemical and pH induced denaturation of a multimeric β-galactosidase reveals multiple unfolding pathways. PLoS One 2012, 7, e50380 10.1371/journal.pone.0050380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss L. M.; Matthews C. R. Urea and thermal equilibrium denaturation studies on the dimerization domain of Escherichia coli Trp repressor. Biochemistry 1997, 36, 5612–5623. 10.1021/bi970056e. [DOI] [PubMed] [Google Scholar]
- Fessas D.; Iametti S.; Schiraldi A.; Bonomi F. Thermal unfolding of monomeric and dimeric β-lactoglobulins. Eur. J. Biochem. 2001, 268, 5439–5448. 10.1046/j.0014-2956.2001.02484.x. [DOI] [PubMed] [Google Scholar]
- Milla M. E.; Sauer R. T. P22 Arc repressor: folding kinetics of a single-domain, dimeric protein. Biochemistry 1994, 33, 1125–1133. 10.1021/bi00171a011. [DOI] [PubMed] [Google Scholar]
- Topping T. B.; Gloss L. M. Stability and folding mechanism of mesophilic, thermophilic and hyperthermophilic archael histones: the importance of folding intermediates. J. Mol. Biol. 2004, 342, 247–260. 10.1016/j.jmb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Mok Y.-K.; Bycroft M.; Prat-Gay G. d. The dimeric DNA binding domain of the human papillomavirus E2 protein folds through a monomeric intermediate which cannot be native-like. Nat. Struct. Biol. 1996, 3, 711–717. 10.1038/nsb0896-711. [DOI] [PubMed] [Google Scholar]
- Nájera H.; Costas M.; Fernández-velasco D. A. Thermodynamic characterization of yeast triosephosphate isomerase refolding: insights into the interplay between function and stability as reasons for the oligomeric nature of the enzyme. Biochem. J. 2003, 370, 785–792. 10.1042/bj20021439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek B. J.; Gloss L. M. Three-state kinetic folding mechanism of the H2A/H2B histone heterodimer: the N-terminal tails affect the transition state between a dimeric intermediate and the native dimer. J. Mol. Biol. 2005, 345, 827–836. 10.1016/j.jmb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Nooren I. M. A.; Thornton J. M. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 2003, 325, 991–1018. 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- Gruber T.; Balbach J. Protein Folding Mechanism of the Dimeric AmphiphysinII/Bin1 N-BAR Domain. PLoS One 2015, 10, e0136922 10.1371/journal.pone.0136922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez F. X.; Unger V. M.; Voth G. A. Autoinhibition of endophilin in solution via interdomain interactions. Biophys. J. 2013, 104, 396–403. 10.1016/j.bpj.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Kaan H. Y. K.; Hooda R. N.; Goh S. L.; Sondermann H. Structure and plasticity of Endophilin and Sorting Nexin 9. Structure 2008, 16, 1574–1587. 10.1016/j.str.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Meinecke M.; Boucrot E.; Camdere G.; Hon W.-C.; Mittal R.; McMahon H. T. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J. Biol. Chem. 2013, 288, 6651–6661. 10.1074/jbc.m112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Zhu C.; Kuo C. J.; Robustelli J.; Baumgart T. The N-terminal amphipathic helix of endophilin does not contribute to its molecular curvature generation capacity. J. Am. Chem. Soc. 2016, 138, 14616–14622. 10.1021/jacs.6b06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.; Das S. L.; Baumgart T. Nonlinear sorting, curvature generation, and crowding of endophilin N-BAR on tubular membranes. Biophys. J. 2012, 102, 1837–1845. 10.1016/j.bpj.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M. Förster resonance energy transfer—FRET what is it, why do it, and how it’s done. Lab. Tech. Biochem. Mol. Biol. 2009, 33, 1–57. 10.1016/s0075-7535(08)00001-6. [DOI] [Google Scholar]
- Louis-Jeune C.; Andrade-Navarro M. A.; Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 2012, 80, 374–381. 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- Greenfield N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal E.; Federici L.; Zhang W.; Fernandez-Recio J.; Priego E.-M.; Miguel R. N.; DuHadaway J. B.; Prendergast G. C.; Luisi B. F.; Laue E. D. The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition. Biochemistry 2006, 45, 12917–12928. 10.1021/bi060717k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. L.; Lary J. W.; Moody T.; Laue T. M. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008, 84, 143–179. 10.1016/s0091-679x(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chana M. S.; Tripet B. P.; Mant C. T.; Hodges R. Stability and specificity of heterodimer formation for the coiled-coil neck regions of the motor proteins Kif3A and Kif3B: the role of unstructured oppositely charged regions. J. Pept. Res. 2005, 65, 209–220. 10.1111/j.1399-3011.2005.00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon J. J.; McManus E. J.; Yao S.; DeSouza D. P.; Mielke L. A.; Sprigg N. S.; Willson T. A.; Hilton D. J.; Nicola N. A.; Baca M.; Nicholson S. E.; Norton R. S. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol. Cell 2006, 22, 205–216. 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Chana M.; Tripet B. P.; Mant C. T.; Hodges R. S. The role of unstructured highly charged regions on the stability and specificity of dimerization of two-stranded α-helical coiled-coils: Analysis of the neck-hinge region of the kinesin-like motor protein Kif3A. J. Struct. Biol. 2002, 137, 206–219. 10.1006/jsbi.2002.4446. [DOI] [PubMed] [Google Scholar]
- Nagi A. D.; Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Folding Des. 1997, 2, 67–75. 10.1016/s1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- Kundu M.; Sen P. C.; Das K. P. Structure, stability, and chaperone function of αA-crystallin: Role of N-terminal region. Biopolymers 2007, 86, 177–192. 10.1002/bip.20716. [DOI] [PubMed] [Google Scholar]
- Constantinescu D. C.; Makbul C.; Koturenkiene A.; Lüdemann M.-B.; Herrmann C. Dimerization-induced folding of MST1 SARAH and the influence of the intrinsically unstructured inhibitory domain: low thermodynamic stability of monomer. Biochemistry 2011, 50, 10990–11000. 10.1021/bi201110h. [DOI] [PubMed] [Google Scholar]
- Cestra G.; Castagnoli L.; Dente L.; Minenkova O.; Petrelli A.; Migone N.; Hoffmüller U.; Schneider-Mergener J.; Cesareni G. The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J. Biol. Chem. 1999, 274, 32001–32007. 10.1074/jbc.274.45.32001. [DOI] [PubMed] [Google Scholar]
- Ball L. J.; Kühne R.; Schneider-Mergener J.; Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem., Int. Ed. 2005, 44, 2852–2869. 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- Yoon Y.; Zhang X.; Cho W. Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains. J. Biol. Chem. 2012, 287, 34078–34090. 10.1074/jbc.m112.372789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute. J. Mol. Biol. 2007, 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Martin A.; Schmid F. X. A proline switch controls folding and domain interactions in the gene-3-protein of the filamentous phage fd. J. Mol. Biol. 2003, 331, 1131–1140. 10.1016/s0022-2836(03)00864-7. [DOI] [PubMed] [Google Scholar]
- Jackson S. E. How do small single-domain proteins fold?. Folding Des. 1998, 3, R81–R91. 10.1016/s1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- Li L.; Gunasekaran K.; Gan J. G.-K.; Zhanhua C.; Shapshak P.; Sakharkar M. K.; Kangueane P. Structural features differentiate the mechanisms between 2S (2 state) and 3S (3 state) folding homodimers. Bioinformation 2005, 1, 42. 10.6026/97320630001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huus K.; Havelund S.; Olsen H. B.; van de Weert M.; Frokjaer S. Thermal dissociation and unfolding of insulin. Biochemistry 2005, 44, 11171–11177. 10.1021/bi0507940. [DOI] [PubMed] [Google Scholar]
- Pollegioni L.; Iametti S.; Fessas D.; Caldinelli L.; Piubelli L.; Barbiroli A.; Pilone M. S.; Bonomi F. Contribution of the dimeric state to the thermal stability of the flavoprotein D-amino acid oxidase. Protein Sci. 2003, 12, 1018–1029. 10.1110/ps.0234603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden E. M.; Owen B. A. L.; Peterson F. C.; Volkman B. F.; Ramirez-Alvarado M.; Thompson J. R. Altered dimer interface decreases stability in an amyloidogenic protein. J. Biol. Chem. 2008, 283, 15853–15860. 10.1074/jbc.m705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci E.; Vitagliano L.; Barone R.; Sorrentino S.; D’Alessio G.; Graziano G. On the thermal stability of the two dimeric forms of ribonuclease A. Biophys. Chem. 2005, 116, 89–95. 10.1016/j.bpc.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Qualmann B.; Koch D.; Kessels M. M. Let’s go bananas: revisiting the endocytic BAR code. EMBO J. 2011, 30, 3501–3515. 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.; Daggett V. Identification and characterization of the unfolding transition state of chymotrypsin inhibitor 2 by molecular dynamics simulations. J. Mol. Biol. 1996, 257, 412–429. 10.1006/jmbi.1996.0172. [DOI] [PubMed] [Google Scholar]
- Naganathan A. N.; Doshi U.; Muñoz V. Protein folding kinetics: barrier effects in chemical and thermal denaturation experiments. J. Am. Chem. Soc. 2007, 129, 5673–5682. 10.1021/ja0689740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramprakash J.; Doseeva V.; Galkin A.; Krajewski W.; Muthukumar L.; Pullalarevu S.; Demirkan E.; Herzberg O.; Moult J.; Schwarz F. P. Comparison of the chemical and thermal denaturation of proteins by a two-state transition model. Anal. Biochem. 2008, 374, 221–230. 10.1016/j.ab.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Solvent Denaturation. Biopolymers 1978, 17, 1305–1322. 10.1002/bip.1978.360170515. [DOI] [Google Scholar]
- Bennion B. J.; Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 5142–5147. 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter J. A measure of conformational entropy change during thermal protein unfolding using neutron spectroscopy. Biophys. J. 2003, 84, 3924–3930. 10.1016/s0006-3495(03)75120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada A. G.; Díaz-Salazar A. J.; Cabrera N.; Pérez-Montfort R.; Piñeiro Á.; Costas M. Interplay between protein thermal flexibility and kinetic stability. Structure 2017, 25, 167–179. 10.1016/j.str.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Narayan A.; Bhattacharjee K.; Naganathan A. N. Thermally versus Chemically Denatured Protein States. Biochemistry 2019, 58, 2519–2523. 10.1021/acs.biochem.9b00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzwolak W.; Ravindra R.; Lendermann J.; Winter R. Aggregation of bovine insulin probed by DSC/PPC calorimetry and FTIR spectroscopy. Biochemistry 2003, 42, 11347–11355. 10.1021/bi034879h. [DOI] [PubMed] [Google Scholar]
- Kay B. K.; Williamson M. P.; Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. 10.1096/fasebj.14.2.231. [DOI] [PubMed] [Google Scholar]
- Hait S.; Mallik S.; Basu S.; Kundu S. Finding the generalized molecular principles of protein thermal stability. Proteins 2020, 88, 788–808. 10.1002/prot.25866. [DOI] [PubMed] [Google Scholar]
- Berezovsky I. N.; Zeldovich K. B.; Shakhnovich E. I. Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput. Biol. 2007, 3, e52 10.1371/journal.pcbi.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartalesi I.; Bertini I.; Di Rocco G.; Ranieri A.; Rosato A.; Vanarotti M.; Vasos P. R.; Viezzoli M. S. Protein stability and mutations in the axial methionine loop of a minimal cytochrome c. J. Biol. Inorg Chem. 2004, 9, 600–608. 10.1007/s00775-004-0558-8. [DOI] [PubMed] [Google Scholar]
- Ricagno S.; Colombo M.; Rosa M. d.; Sangiovanni E.; Giorgetti S.; Raimondi S.; Bellotti V.; Bolognesi M. DE loop mutations affect β2-microglobulin stability and amyloid aggregation. Biochem. Biophys. Res. Commun. 2008, 377, 146–150. 10.1016/j.bbrc.2008.09.108. [DOI] [PubMed] [Google Scholar]
- Parthiban V.; Gromiha M. M.; Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006, 34, W239–W242. 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-W.; Lin M.-H.; Liao C.-C.; Chang H.-P.; Chu Y.-W. iStable 2.0: Predicting protein thermal stability changes by integrating various characteristic modules. Comput. Struct. Biotechnol. J. 2020, 18, 622–630. 10.1016/j.csbj.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T.; Capraro B. R.; Zhu C.; Das S. L. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu. Rev. Phys. Chem. 2011, 62, 483–506. 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B. J.; Kent H. M.; Mills I. G.; Vallis Y.; Butler P. J. G.; Evans P. R.; McMahon H. T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Blood P. D.; Voth G. A. Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 15068–15072. 10.1073/pnas.0603917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsdottir H.; Hunte C. Lipids in membrane protein structures. Biochim. Biophys. Acta 2004, 1666, 2–18. 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Gupta K.; Donlan J. A. C.; Hopper J. T. S.; Uzdavinys P.; Landreh M.; Struwe W. B.; Drew D.; Baldwin A. J.; Stansfeld P. J.; Robinson C. V. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 2017, 541, 421–424. 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J.; Wyman J.; Changeux J.-P. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965, 12, 88–118. 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Micsonai A.; Wien F.; Kernya L.; Lee Y.-H.; Goto Y.; Réfrégiers M.; Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, E3095–E3103. 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P.; Zhao H.; Brautigam C. A.; Ghirlando R.. Basic Principles of Analytical Ultracentrifugation; CRC Press, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.