Abstract
Purpose
Age-related macular degeneration (AMD) is associated with altered gene and protein expression in the retina. We characterize the aqueous humor (AH) proteome in AMD to gain insight into the pathogenesis of the disease and identify potential biomarkers.
Methods
AH was collected from age and gender matched neovascular AMD (nvAMD; n = 10) patients and controls (n = 10). AH was pooled to create two samples (nvAMD and control), followed by intensity-based label-free quantification (MS1). Functional and bioinformatic analysis were then performed. A validation set (20 controls, 15 atrophic AMD and 15 nvAMD) was tested via multiplex ELISA for nine differentially expressed proteins according to the MS1 findings.
Results
MS1 identified 674 proteins in the AH. 239 proteins were upregulated in nvAMD (nvAMD/control > 2, peptide tags (PT) > 2), and 86 proteins were downregulated (nvAMD/control < 0.5, PT > 2). Functional analysis of proteins upregulated in AMD demonstrated enrichment for platelet degranulation (enrichment score (ES):28.1), negative regulation of endopeptidase activity (ES:18.8), cellular protein metabolic process (ES:11.8), epidermal growth factor-like domain (ES:10.3), sushi/SCR/CCP (ES:10.1), and complement/coagulation cascades (ES:9.2). AMD protein clusters were upregulated for 3/6 (χ2 < 0.05 compared to randomization). Validation via ELISA confirmed MS1 in 2/9 proteins (Clusterin and Serpin A4, P < 0.05), while 3/9 showed differential expression between aAMD and nvAMD (Clusterin, Serpin A4, and TF P < 0.05). Receiver operating characteristic curve calculation identified the area under the curve of 0.82 for clusterin as a biomarker for distinction of AMD.
Conclusions
AH proteomics in AMD patients identified several proteins and functional clusters with altered expression. Further research should confirm if these proteins may serve as biomarkers or therapeutic target for the disease.
Keywords: age- related macular degeneration, aqueous humor, retinal degeneration
Immune response and oxidative injury are implicated in the pathogenesis of age-related macular degeneration (AMD), yet, comprehensive understanding of the process is lacking, and biomarkers to diagnose and grade the disease are still needed.
The aqueous humor (AH) contains nutrients and metabolic wastes from the eye. Changes in the AH protein and cytokine profile may play a role and reflect pathological processes of the eye.1,2 Research has shown that there are differences in the levels of several AH proteins between AMD patients and controls.2–4 These studies examined differences of specific proteins. However, a comprehensive comparison of AH proteomic in AMD patients and controls could provide additional insights into the pathogenesis of the disease and identify potential biomarkers for AMD.
In recent years, novel proteomic technologies became available, which facilitate unbiased characterization of tissue proteomics. A number of studies have already employed proteome analysis of AH for the study of a variety of ocular disorders, including two which focused on AMD.5–8 Very few studies have also examined vitreous humor in AMD,9,10 either via a full proteome approach or via specific protein analyses.
We aim to confirm and extend previous observations on AH proteomics in neovascular AMD (nvAMD) and atrophic AMD (aAMD) and to identify candidate biomarkers for the disease using an unbiased proteome analysis.
Methods
Patients
AMD patients and controls (n = 70) were recruited from the retina clinic of the Department of Ophthalmology at the Hadassah-Hebrew University Medical Center. Criteria for inclusion of nvAMD patients included age over 55 years, diagnosis of AMD according to the AREDS criteria,11 and diagnosis of choroidal neovascularization (CNV) according to fluorescein angiogram (FA) and optical coherence tomography (OCT). Eyes with neovascular lesions comprised of less than 50% active CNV, subretinal hemorrhage greater than 25% of the lesion size, or presence of other retinal diseases were excluded from the study. Specifically, eyes with any other potential cause for CNV, such as myopia over (>6 diopter), trauma, or uveitis were excluded. Also excluded were patients with a major systemic illness, such as cancer, autoimmune disease, congestive heart failure, or uncontrolled diabetes. The research followed the tenets of the Declaration of Helsinki, all patients signed an informed consent form, and the study was approved by the institutional ethics committee (Hadassah Medical Center).
Collection of Human Aqueous Humor
AH samples were collected from patients with AMD and age-matched controls during cataract surgery. The samples were collected using a 1cc syringe and a cannula. After collection, samples of 50 to 150 µl were transferred into eppendorf tubes, and frozen at –80°C until further analysis.
Intensity-Based Label-Free Quantification (MS1)
AH samples from nvAMD patients (and age-matched controls) were collected. AH was taken per sample (10 µl) and pooled together to create two main samples indicated as pooled-nvAMD (P-nvAMD) and pooled-controls (P-controls). Intensity-based label-free quantification (MS1) was then performed at the Weizmann Institute of Science (Rechovot, Israel). MS1 was performed in two ways: 40 µl AH from each pooled sample were subjected to depletion of the 14 most abundant proteins found in human serum (MARS14, Agilent Technologies, Inc, Wilmington, MA, USA): albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin. The 60 µl of AH that remained in the two samples were tested without depletion of proteins.
MS1 was performed on both pooled samples with and without depletion.12 The AH samples were read via a quadrupole orbitrap mass spectrometer (Q Exactive Plus machine, Thermo Scientific, Rockford, IL, USA), and data was acquired via the XCalibur v3.0 program. Raw data was interpreted, filtered, smoothed, and aligned via the Expressionist program (Genedata, Basel, Switzerland). Protein intensity was obtained using the Hi-3 method.13 Unbiased protein results were obtained via a datafile indicating UniProt name (via the UniProtKB, human version 2015_07, http://www.uniprot.org/),14 gene name, peptide value, coverage, peptide sequence, and protein intensity for both the P-nvAMD and P-control samples.
Proteome Data Filtering
To obtain informative results, we included only proteins that had a peptide value greater than 2. To remove background bias, we only included proteins that had a ratio between the P-nvAMD and the P-control greater than 2 or smaller than 0.5. Then, we were able to analyze the proteins that were differentially expressed in the P-nvAMD set as opposed to the P-control. For data mining, we only used the main isoforms of proteins that were directly related to the official gene symbol via UniProt.
Proteome Functional Analysis
Functional analysis was performed via DAVID 6.8 (https://david.ncifcrf.gov/).15 Functional categories included UniProt Keywords, Sequence features, GO Terms, Bicarta, BBID and Kegg Pathways, InterPro Protein interactions, and others. Significance was set via a P-Benjamini-Hochberg (BH) < 0.0516 and an enrichment score (ES) > 5. To validate the association of functional clusters identified by DAVID, several randomized datasets, each encompassing 10 permutations of randomization algorithms, were used to validate the null hypothesis as we have described previously.17 Significant clusters were determined via χ2. In addition, we also compared the identified proteins to previous studies performed on human AMD AH samples and human AMD vitreous samples.
Multiplex ELISA Array
A validation set was evaluated using Custom Quantibody Arrays, (RayBiotech, Inc. Norcross, GA, USA) for nine proteins and performed according to the manufacturer's instructions. Briefly, 50 µl of individual AH was mixed with 50 µl of sample diluent buffer and added to each well of a 16-well that was fitted onto a glass slide. After an initial 30-minute incubation and 30-minutes of blocking buffer, the sample was added to the well and left on a rotator overnight (ON) at 4°C. After ON incubation, the samples were decanted from the wells and rinsed with wash buffer seven times for five minutes on a rotator. Detection antibody cocktail in the amount of 8 µl was added to each well for two hours. The same amount of Cy3-equivalent dye-conjugated streptavidin was added after washes as described and incubated for one hour at room temperature in the dark. Analyses were performed in accordance with the manufacturer's instructions. The validation set included 50 additional samples: 20 controls (mean age 74.8 ± 7.1, female/male = 10/10), 15 samples from patients with atrophic AMD and 15 with nvAMD.
Statistical Analysis
Data was processed using the biostatistical package InStat (GraphPad, San Diego, CA, USA). P < 0.05 was considered to indicate statistical significance. Values over two standard deviations from the average were excluded from statistical analysis. Appropriate statistical tests, as detailed in the results section, were applied according to the results of a normalcy test for continuous and categorical parameters as we have described previously.18
Area Under the Curve
Area under the curve (AUC) calculation using receiver operating characteristic curves (ROC) analysis was performed on data obtained from the validation set samples to assess diagnostic accuracy. A ROC curve was calculated for each of the proteins which showed differential expression levels according to the multiplex ELISA assay results. Calculations were performed with Web-based software (http://www.biosoft.hacettepe.edu.tr/easyROC/). For input data purposes, patients with AMD were defined as “0” and controls as “1.” To calculate the AUC value for differentiation between aAMD and nvAMD, patients with AMD were defined as “0” and nvAMD as “1” as we have previously described.19
Results
Analysis of Aqueous Humor Proteome
A total of 674 proteins were identified in AH following application of filters and processing in the depleted and nondepleted AH sets combined. Upregulation was defined as the ratio between the P-nvAMD (n = 10, mean age 81.8 ± 5.69, female /male = 5/5) and the P-control (n = 10, mean age 78.3 ± 6.22, female/male = 5/5) greater than 2. According to this definition, 239 protein-coding genes were upregulated in P-nvAMD (depleted and nondepleted) and 86 protein-coding genes were upregulated in P-control (depleted and nondepleted). Of these proteins, 20 from the nondepleted sample and 219 proteins from the depleted sample were upregulated in P-nvAMD. Forty-four proteins from the nondepleted sample were upregulated in P-control, and an additional 42 proteins from the depleted sample (Supplementary Table S1). Five proteins were expressed only in P-nvAMD: APOL1 (O14791), RELN (P78509), IL1RAP (Q9NPH3), IGFBP2 (P18065), and NAGLU (P54802).
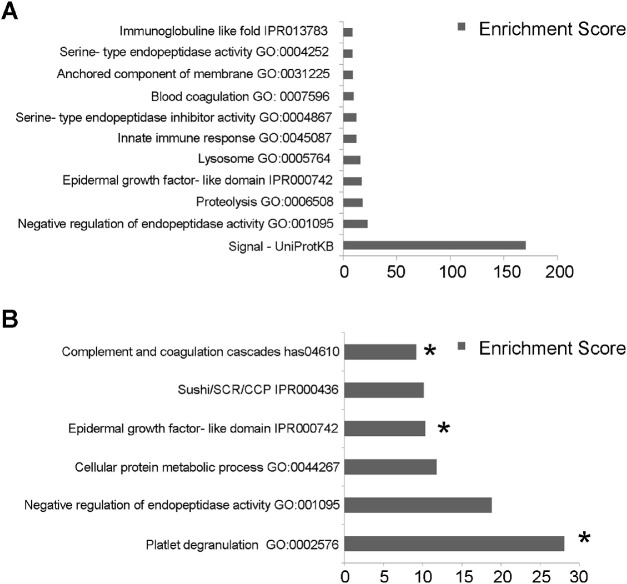
Functional analysis via DAVID of all 674 proteins identified in AH demonstrated enrichment for 11 functional clusters (BH-P < 0.05, enrichment score (ES) > 5) (Fig. 1A). Functional analysis for proteins upregulated in P-nvAMD (Fig. 1B) identified enrichment for six clusters: platelet degranulation (enrichment score (ES): 28.1, GO:0002576), negative regulation of endopeptidase activity (ES:18.82 GO:0010951), cellular protein metabolic process (ES: 11.78, GO:0044267), epidermal growth factor-like domain (ES: 10.34, IPR000742), Sushi/SCR/CCP (ES:10.14, IPR000436), and complement and coagulation cascades (ES:9.16, hsa04610) (Fig. 1B).
Figure 1.
Functional analysis. DAVID algorithm was applied to identify functional protein clusters in the AH. Functional annotations were detected from multiple databases including UniProt, Gene Ontology (GO) (see Method section). The functional analysis was performed on the total proteins discovered in AH according to MS1 (11 clusters) (A), and on proteins which were upregulated in nvAMD (six clusters) (B). Association of nvAMD and the functional clusters was confirmed by χ2 test as compared to randomized protein clusters. Three clusters showed differential expression in this analysis (B). * indicates P < 0.05.
To validate the association between the P-nvAMD significant clusters and the disease, χ2 was calculated on the P-nvAMD clusters as compared to randomized proteins clusters. We randomized by proteins name, peptide value, and ratio between P-nvAMD and P-control as describe above. Three clusters of proteins were found to correlate with P-nvAMD in this analysis: platelet degranulation (GO:0002576), cellular protein metabolic process (GO:0044267), and complement and coagulation cascades (GO:0004857) (P = 0.0122, P = 0.006 and P = 0.024, respectively).
Multiplex ELISA Assay
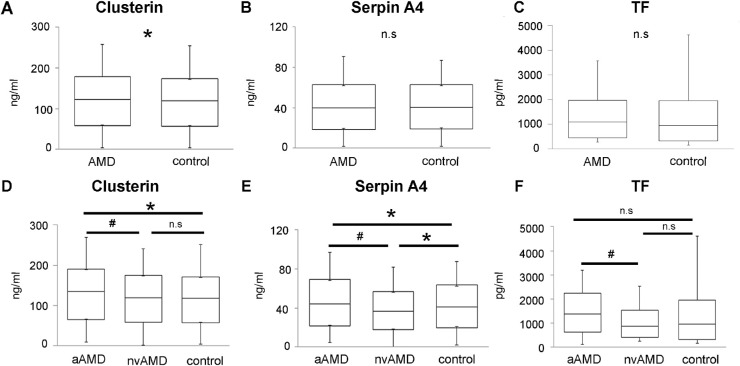
Nine proteins that were upregulated in the pooled samples were tested in the multiplex ELISA assay on an additional set of nvAMD (n = 15, mean age 78.6 ± 7.4, female/male = 9/6), aAMD (n = 15, mean age 77.3 ± 10.5, female/male = 5/10), and control (n = 20, mean age 74.8 ± 7.1, female/male = 10/10) samples to validate the MS1 results. These proteins included: TF, clusterin, Serpin A4, IGFBP-2, IL1 R3, RBP4, CFD, CRP, and FETUB. Of these nine, eight were upregulated in P-nvAMD and one (TF) was upregulated in P-control according to MS1 (Table 2). ELISA results demonstrated increased levels of clusterin in AMD as compared to controls (1.06-fold, P = 0.032) (Fig. 2A). Clusterin levels was also found to be higher in aAMD subtype as compared to controls (1.09-fold, P = 0.022) (Figs. 2B, D). Serpin A4 protein level was elevated in controls as compared to nvAMD (1.11-fold, P = 0.007) (Fig. 2E). Some of the tested proteins showed differential levels between aAMD and nvAMD. Clusterin, Serpin A4, and TF were increased in aAMD as compared to nvAMD (1.12-fold, P = 0.0016; 1.17-fold, P = 0.002; and 1.66-fold, P = 0.016, respectively) (Figs. 2D, E, and F).
Table 2.
List of the Proteins That Validated via Multiplex ELISA Assay. Multiplex ELISA was Performed for Nine Proteins to Validate the MS1 Results. Nine Proteins Shown in the Table. Fetuin B Protein was Found not Detectable via the Multiplex ELISA and not Present
| Protein name | AMD vs. Control | nvAMD vs. Control | aAMD vs. Control | aAMD vs. nvAMD | |
|---|---|---|---|---|---|
| IGFBP-2 | mean (SD) | 0.975492808 | 1.008362027 | 1.008362027 | 1.072410923 |
| P value* | 0.5401 | 0.7508 | 0.9629 | 0.6414 | |
| IL-1 R3 | mean (SD) | 0.535656929 | 0.498382857 | 0.695160229 | 1.394831742 |
| P value | 0.0418 | 0.1073 | 0.346 | 0.25 | |
| Clusterin | mean (SD) | 1.062251272 | 0.995116819 | 1.112383554 | 1.11784218 |
| P value | 0.0321 | 0.87 | 0.0045 | 0.0016 | |
| Serpin A4 | mean (SD) | 0.980970015 | 0.928417489 | 1.09016161 | 1.174214858 |
| P value | 0.6221 | 0.0491 | 0.0414 | 0.002 | |
| CRP | mean (SD) | 1.084106393 | 1.342750368 | 0.742959842 | 0.553311963 |
| P value | 0.7869 | 0.46 | 0.38 | 0.1154 | |
| RBP-4 | mean (SD) | 0.798257282 | 0.970020658 | 1.24505538 | 0.82800002 |
| P value | 0.123 | 0.8399 | 0.3744 | 0.2759 | |
| Adipsin | mean (SD) | 1.013083426 | 1.064145886 | 1.046463066 | 0.983383087 |
| P value* | 0.8514 | 0.39 | 0.622 | 0.844 | |
| TF | mean (SD) | 0.651255947 | 0.478581978 | 0.797364689 | 0.016 |
| P value* | 0.1437 | 0.1293 | 0.5217 | 1.666098446 | |
Figure 2.
Validation of differential protein expression. Multiplex ELISA was utilized to measure expression levels of nine proteins identified as differentially expressed in nvAMD AH according to MS1. The box plots show confirmed differential expression (the full data is presented in Supplementary Table S1). The comparison between the control group (n = 20), and AMD samples (aAMD, n = 15, and nvAMD, n = 15) shows in panels A, B and C. Panels D, E, and F shows protein expression in AMD subtypes as compared to the control group. * indicates P < 0.05. # indicates P < 0.05 between aAMD and nvAMD.
ROC Analysis
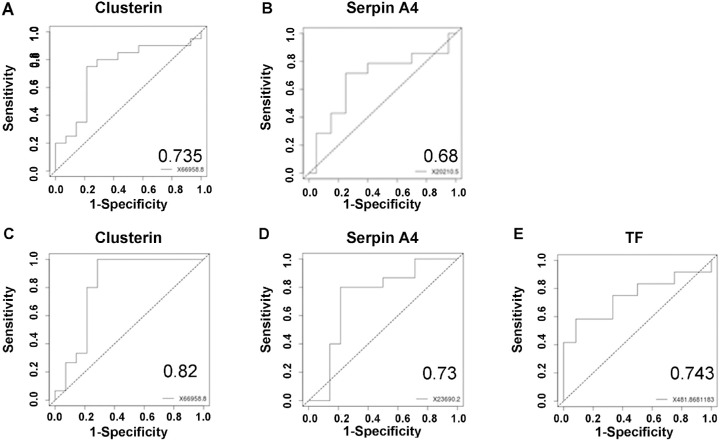
To assess the feasibility of using measurements of the protein levels as biomarkers for AMD, we fitted the multiplex ELISA results for each of the significant proteins on a ROC curve. AUC was calculated via comparing aAMD, nvAMD, and controls. We also combined aAMD and nvAMD (indicated as AMD) and controls for the proteins that were differentially expressed between them according to the multiplex ELISA. Comparing AMD and control clusterin had low AUC values (0.4). When we tested the differences in protein levels between controls and aAMD, the proteins clusterin and Serpin A4 had AUC values of 0.73 and 0.61, respectively. When we compared controls with nvAMD for Serpin A4, the AUC value was 0.68. AUC calculation was also performed between nvAMD and aAMD. Clusterin, Serpin A4, and TF had AUC values of 0.82, 0.73, and 0.74, respectively, for that comparison (Fig. 3).
Figure 3.
ROC analysis. To validate the clinical significance differential protein expression in AH, we performed ROC analysis (area under the curve, AUC) of all nine proteins tested via ELISA based on 20 controls, 15 aAMD, and 15 nvAMD. Presented here are the top ranking AUCs (the complete list of values is presented in Supplementary Table S2). Panel A shows the AUC of clusterin for distinction of aAMD patients from controls. Panel B shows the AUC of Serpin A4 for distinction of controls from nvAMD patients, and Panels C, D, and E show the AUCs of clusterin, Serpin A4, and TF, respectively, for distinction of aAMD patients from nvAMD patients. Each AUC graph represents sensitivity versus 1-specificity.
Comparing Results With Previous Studies
We have compared results of proteomics in our study to previous reports of unbiased proteomics in AH and vitreous from AMD eyes.9,10,20 Kim et al. reported of 10 novel proteins in nvAMD AH, which were not previously reported, and we confirmed the presence of six of them in the AH of AMD patients and controls. The comparison between the different research shows the number of common proteins that discovered in most of the studies. In the AH analysis, C3 protein overlapped with our AMD increasing proteins. Kim et al. also reported about SERPINA5 as a novel protein in nvAMD AH, and we found SERPINA4, from the same family as a potential biomarker. Furthermore, comparison between vitreous analysis results and our results, both revealed increasing levels of complement factors and apolipoprotein proteins in nvAMD versus control group (Table 12 and Supplementary Table S3).
Table 1.
Comparison of Previous Unbiased Proteomic Studies of AH and Vitreous in AMD (The Full List of the Proteins Presented in Supplementary Table S3)
| Study | Rinsky et al. (2021) | Kim et al. (2012) | Koss et al. (2016) | Siwy et al. (2014) |
|---|---|---|---|---|
| Source of Sample | Aqueous | Aqueous | Vitreous | Vitreous |
| Mass spectrometry analysis | ||||
| AMD patients | 10 | 3 | 72 | 73 |
| Controls | 10 | 3 | 16 | 15 |
| Total proteins identified | 674 | 154 | 101 | 97 |
| Proteins upregulated in AMD | 239 | 52 | 4 (up or down) | 19 |
| Proteins upregulated in control | 86 | 22 | ||
| Overlap with Rinsky et al. | N/A | |||
| Validation set and potential biomarkers * | ||||
| AMD patients | 30** | 6 | 36 | 4 |
| Controls | 20 | 5 | 8 | 4 |
| Number of proteins tested | 9 | 7 | 4 | 5 |
| AMD Proteins Overlaping | 6 discovered in this study | 10 new proteins compared to previous studies | 40 | 8 |
aAMD and nvAMD.
Discussion
We performed an unbiased whole proteome analysis of AH samples collected from AMD patients and controls. A total of 674 proteins were detected in the AH; of these, 239 were upregulated in nvAMD, including five proteins that were only found in nvAMD, and 86 proteins were upregulated in controls. Validation via multiplex ELISA for nine of these proteins on another 50 samples confirmed that two proteins were significantly increased in AMD compared to controls, and that three proteins showed differential expression across the stages of the disease. In addition, AUC calculation suggested that measurement of the level of clusterin may facilitate distinction between eyes at the atrophic and neovascular stages of AMD. Finally, the analysis provided insights into functional clusters of proteins which show altered expression in AH from AMD patients.
Previous studies have examined the proteome of human AH,21,22 and also the alterations in AH protein levels in diseases such as glaucoma,6 type 2 diabetes,5 and diabetic retinopathy.23 Several studies have also been performed on AH in AMD. The majority of these previous studies focused on specific proteins. For example, Kramer et al. found an increase in CCL2 levels in nvAMD as compared to controls.3 Few studies have performed an unbiased proteomic analysis of AH in patients with AMD.7,8 Our study confirmed several findings from previous studies, but we have also identified new proteins which show increased expression in AH from AMD. We also compared proteins found in aAMD and nvAMD, indicating differential expression of proteins between the different stages of the disease.
Our functional analysis showed a connection between upregulated proteins in AMD and pathways involving the complement and coagulation cascades (GO:0004857). The complement system was demonstrated to have an important role in the pathogenesis of AMD.24–29 Schick and colleagues have previously investigated complement activation in the AH of AMD patients via measurements of the levels of six complement proteins in the AH by ELISA. They reported an increase in several complement factors in the AH in AMD compared to controls. No differences were shown in levels of these proteins in serum samples of the same individuals.30 Taken together, our findings are consistent with previous studies and expand current knowledge by providing information of additional complement proteins which are differentially expressed in AMD AH.
There were several specific proteins of interest which were differentially expressed in AH from AMD patients. For example, clusterin level was increased in AMD samples compared to controls in both assays used. Clusterin is an extracellular chaperone encoded by a single nine-exon gene on chromosome 8 and consists of two subunits. From an evolutionary point of view, clusterin has possibly a high biological significance, as this gene is highly conserved among species and is expressed in nearly all human tissues. Interestingly, clusterin was found in drusen in retinas affected by AMD.31 In addition, suppressing expression of clusterin using antisense oligonucleotides led to a reduction of the angiogenesis level in vitro.32 Kim et al. performed an unbiased proteomic analysis on AH and reported a high level of clusterin in nvAMD samples compared to controls.8 Additionally, recent studies indicated an increased level of clusterin in vitreous samples from nvAMD as compared to controls, with an AUC value of 0.747.9 Our research demonstrated increased clusterin levels in aAMD (and an AUC value of 0.73), but not in nvAMD. These results further implicate clusterin in AMD, however, determination of the role of this protein as a potential biomarker for the disease requires additional research.
Serpin A4 (kallistatin) is a protein from the serine protease inhibitor family which was increased in aAMD as compared to nvAMD in our study and decreased in nvAMD as compared to controls.33 This protein family inhibits coagulation and inflammatory processes. The serine protease inhibitor family has other members, including PEDF (SERPINF1), which is known to have antiangiogenic activity.34 PEDF level in vitreous samples from AMD patients were also reported to be decreased as compared to controls.2 A further member of this family of proteins is Serpin G1. A SNP variant in this protein was reported to have an association with AMD.35 Serpin A4 was reported previously in connection to AMD. A previous study reported that Serpin A4 was decreased in AMD patients’ plasma as compared to controls.33 Another study reported that Serpin A4 is high in expression in dry AMD patients’ AH to compared to control patients.36 Furthermore, a recent study revealed the connection between Serpin A4 and macrophages in context of atherosclerotic inflammation.37 The involvement of monocytes and macrophages in the pathogenesis of AMD is suggested by several lines of evidence from humans, and from experimental models in rodents.38–40 These results may suggest an important role of Serpin A4 in the pathogenesis of AMD and further studies should be done to understand the Serpin A4 role.
Our research has several limitations. The unbiased screening was performed on pooled samples. Thus, the variation in protein expression levels could not be evaluated in the initial screen. We performed multiplex ELISA for only nine proteins. Only two of these were confirmed to be increased in the validation set of samples. These discrepancies may be explained by the fact that we have tested a pooled sample by the MS1 screen. However, the results of the study are supported by its consistence with previous works in the field and with current understanding of the pathogenesis of AMD.
In conclusion, the study suggests a specific proinflammatory proteomic profile in the aqueous humor of AMD patients as compared to controls. These changes in AH are also associated with the stages of AMD. Some of the protein level alterations may serve as biomarkers for AMD and its stages.
Supplementary Material
Acknowledgments
Supported in part by a grant from the Israel Science Foundation (ISF; Grant 1006/13).
Disclosure: B. Rinsky, None; G. Beykin, None; M. Grunin, None; R. Amer, None; S. Khateb, None; L. Tiosano, None; D. Almeida, None; S. Hagbi-Levi, None; S. Elbaz-Hayoun, None; I. Chowers, None
References
- 1.Zhu X, Zhang K, He W, et al.. Proinflammatory status in the aqueous humor of high myopic cataract eyes. Exp Eye Res. 2014; 142: 13–18, 10.1016/j.exer.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 2.Holekamp NM, Bouck N, Volpert O.. Pigment epithelium-derived factor is deficient in the vitreous of patients with choroidal neovascularization due to age-related macular degeneration. Am J Ophthalmol. 2002; 134(2): 220–227, 10.1016/S0002-9394(02)01549-0. [DOI] [PubMed] [Google Scholar]
- 3.Kramer M, Hasanreisoglu M, Feldman A, et al.. Monocyte chemoattractant protein-1 in the aqueous humour of patients with age-related macular degeneration. Clin Exp Ophthalmol. 2012; 40(6): 617–625, 10.1111/j.1442-9071.2011.02747.x. [DOI] [PubMed] [Google Scholar]
- 4.Fauser S, Viebahn U, Muether PS.. Intraocular and systemic inflammation-related cytokines during one year of ranibizumab treatment for neovascular age-related macular degeneration. Acta Ophthalmol. 2015; 93(8): 734–738, 10.1111/aos.12770. [DOI] [PubMed] [Google Scholar]
- 5.Kulaksızoglu S, Karalezli A.. Aqueous humour and serum levels of nitric oxide, malondialdehyde and total antioxidant status in patients with type 2 diabetes with proliferative diabetic retinopathy and nondiabetic senile cataracts. Can J Diabetes. 2016; 40(2): 115–119, 10.1016/j.jcjd.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Kaeslin MA, Killer HE, Fuhrer CA, Zeleny N, Huber AR, Neutzner A.. Changes to the aqueous humor proteome during glaucoma. PLoS One. 2016; 11(10): 1–15, 10.1371/journal.pone.0165314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao J, Liu X, Yang Q, et al.. Proteomic analysis of the aqueous humor in patients with wet age-related macular degeneration. Proteomics Clin Appl. 2013; 7(7-8): 550–560, 10.1002/prca.201200012. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, Kang JW, Ahn J, et al.. Proteomic analysis of the aqueous humor in age-related macular degeneration (AMD) patients. J Proteome Res. 2012; 11(8): 4034–4043, 10.1021/pr300080s. [DOI] [PubMed] [Google Scholar]
- 9.Nobl M, Reich M, Dacheva I, et al.. Proteomics of vitreous in neovascular age-related macular degeneration. Exp Eye Res. 2016; 146: 107–117, 10.1016/j.exer.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Koss MJ, Hoffmann J, Nguyen N, et al.. Proteomics of vitreous humor of patients with exudative age-related macular degeneration. PLoS One. 2014; 9(5): e96895, 10.1371/journal.pone.0096895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Age-Related Eye Disease Study Research Group . The Age-Related Eye Disease Study (AREDS): design implications. AREDS report no. 1. Control Clin Trials. 1999; 20(6): 573–600, http://www.ncbi.nlm.nih.gov/pubmed/10588299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalit T, Elinger D, Savidor A, Gabashvili A, Levin Y.. MS1-based label-free proteomics using a quadrupole orbitrap mass spectrometer. J Proteome Res. 2015; 14(4): 1979–1986, 10.1021/pr501045t. [DOI] [PubMed] [Google Scholar]
- 13.Silva JC.Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2005; 5(1): 144–156, 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Wasmuth E V, Lima CD.. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2016; 45(November 2016): 1–12, 10.1093/nar/gkw1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4(1): 44–57, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19131956. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y, Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995; 57(1): 289–300, https://www.stat.purdue.edu/∼doerge/BIOINFORM.D/FALL06/Benjamini%20and%20Y%20FDR.pdf. [Google Scholar]
- 17.Grunin M, Tiosano L, Jaouni T, Averbukh E, Sharon D, Chowers I.. Evaluation of the association of single nucleotide polymorphisms in the PRPH2 gene with adult-onset foveomacular vitelliform dystrophy. Ophthalmic Genet. 2016; 37(3): 285–289, 10.3109/13816810.2015.1059456. [DOI] [PubMed] [Google Scholar]
- 18.Hagbi-Levi S, Grunin M, Jaouni T, et al.. Pro-angiogenic characteristics of activated macrophages from patients with age-related macular degeneration. Neurobiol Aging. 2016; 51: 71–82, 10.1016/j.neurobiolaging.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Lederman M, Weiss A, Chowers I.. Association of neovascular age-related macular degeneration with specific gene expression patterns in peripheral white blood cells. Invest Ophthalmol Vis Sci. 2010; 51(1): 53–58, 10.1167/iovs.08-3019. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Ko Y, Kwon K, et al.. Analysis of monocyte subsets and toll-like receptor 4 expression in peripheral blood monocytes of women in preterm labor. J Reprod Immunol. 2012; 94(2): 190–195, 10.1016/j.jri.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury UR, Madden BJ, Charlesworth MC, Fautsch MP.. Proteome analysis of human aqueous humor. Invest Ophthalmol Vis Sci. 2010; 51(10): 4921–4931, 10.1167/iovs.10-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson MR, Price MO, Price FW, et al.. Proteomic analysis of human aqueous humor using multidimensional protein identification technology. Mol Vis. 2009; 15(November): 2740–2750, https://pubmed.ncbi.nlm.nih.gov/20019884/. [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Zhang X, Liao N, Wen F.. Increased levels of IL-6, sIL-6R, and sgp130 in the aqueous humor and serum of patients with diabetic retinopathy. Mol Vis. 2016; 22: 1005–1014. [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson DH, Mullins RF, Hageman GS, Johnson L V.. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002; 134(3): 411–431, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12208254. [DOI] [PubMed] [Google Scholar]
- 25.Edwards AO, Malek G.. Molecular genetics of AMD and current animal models. Angiogenesis. 2007; 10(2): 119–132, 10.1007/s10456-007-9064-2. [DOI] [PubMed] [Google Scholar]
- 26.Haines JL, Hauser MA, Schmidt S, et al.. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308(5720): 419–421, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15761120. [DOI] [PubMed] [Google Scholar]
- 27.Jia Y, Bailey ST, Wilson DJ, et al.. Quantitative optical coherence tomography angiography of choroidal neovascularization in age-related macular degeneration. Ophthalmology. 2014; 121(7): 1435–1444, 10.1016/j.ophtha.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81(3): 559–575, 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollok-Kopp B, Borncke F, Scholl HP, et al.. Systemic complement activation in age-related macular degeneration (AMD). Wien Klin Wochenschr. 2008; 120: 58, https://pubmed.ncbi.nlm.nih.gov/18596911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schick T, Steinhauer M, Aslanidis A, et al.. Local complement activation in aqueous humor in patients with age-related macular degeneration. Eye. 2017; 49(December 2016): 1–4, https://pubmed.ncbi.nlm.nih.gov/28128795/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crabb JW, Miyagi M, Gu X, et al.. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA. 2002; 99(23): 14682–14687, https://doi.org/10.1073/pnas.222551899222551899 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson JK, Gleave ME, Gleave J, Burt HM.. The inhibition of angiogenesis by antisense oligonucleotides to clusterin. Angiogenesis. 2005; 8(3): 229–238, 10.1007/s10456-005-9018-5. [DOI] [PubMed] [Google Scholar]
- 33.Tuo J, Wang Y, Cheng R, et al.. Wnt signaling in age-related macular degeneration: human macular tissue and mouse model. J Transl Med. 2015; 13(1): 330, 10.1186/s12967-015-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson DW, Volpert O V, Gillis P, et al.. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999; 285(July): 245–248, 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 35.Ennis S, Jomary C, Mullins R, et al.. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008; 372(9652): 1828–1834, 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baek J, Lim D, Park KH, Chae J, Jang H, Lee J. Quantitative proteomic analysis of aqueous humor from patients with drusen and reticular pseudodrusen in age-related macular degeneration. BMC Ophthalmol. 2018; 18: 1–13, https://pubmed.ncbi.nlm.nih.gov/30404605/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Sheng Z, Liu C, et al.. Kallistatin inhibits atherosclerotic inflammation by regulating macrophage polarization. Hum Gene Ther. 2019; 30(87): 339–351, 10.1089/hum.2018.084. [DOI] [PubMed] [Google Scholar]
- 38.Sarks SH, Van Driel D, Maxwell L, Killingsworth M.. Softening of drusen and subretinal neovascularization. Trans Ophthalmol Soc UK. 1980; 100(3): 414–422, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=6171074. [PubMed] [Google Scholar]
- 39.Grunin M, Burstyn-Cohen T, Hagbi-Levi S, Peled A, Chowers I.. Chemokine receptor expression in peripheral blood monocytes from patients with neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012; 53(9): 5292–5300, 10.1167/iovs.11-9165. [DOI] [PubMed] [Google Scholar]
- 40.Grunin M, Hagbi-Levi S-, Rinsky B, et al.. Transcriptome analysis on monocytes from patients with neovascular age-related macular degeneration. Sci Rep. 2016; 6: 29046, 10.1038/srep29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.