Abstract
Background
The diagnosis of strongyloidiasis is challenging. Serological tests are acknowledged to have high sensitivity, but issues due to cross-reactions with other parasites, native parasite antigen supply and intrinsic test variability do occur. Assays based on recombinant antigens could represent an improvement. The aim of this study was to assess the sensitivity and specificity of two novel immunoglobulin (Ig)G and IgG4 enzyme-linked immunosorbent assays (ELISAs) based on the recombinant antigens NIE/SsIR for the diagnosis of strongyloidiasis.
Methods
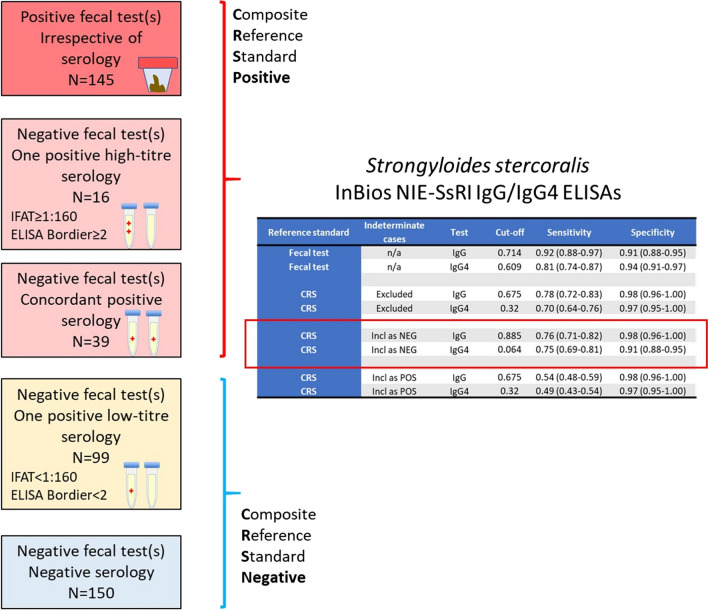
This was a retrospective diagnostic accuracy study. We included serum samples collected from immigrants from strongyloidiasis endemic areas for whom there was a matched result for Strongyloides stercoralis on agar plate culture and/or PCR assay, or a positive microscopy for S. stercoralis larvae. For the included samples, results were also available from an in-house indirect fluorescent antibody test (IFAT) and a commercial (Bordier ELISA; Bordier Affinity Products SA) ELISA. We excluded: (i) samples with insufficient serum volume; (ii) samples from patients treated with ivermectin in the previous 6 months; and (iii) sera from patients for whom only routine coproparasitology was performed after formol–ether concentration, if negative for S. stercoralis larvae. The performance of the novel assays was assessed against: (i) a primary reference standard, with samples classified as negative/positive on the basis of the results of fecal tests; (ii) a composite reference standard (CRS), which also considered patients to be positive who had concordant positive results for the IFAT and Bordier ELISA or with a single “high titer” positive result for the IFAT or Bordier ELISA. Samples with a single positive test, either for the IFAT or Bordier ELISA, at low titer, were considered to be “indeterminate,” and analyses were carried out with and without their inclusion.
Results
When assessed against the primary reference standard, the sensitivities of the IgG and IgG4 ELISAs were 92% (95% confidence interval [CI]: 88–97%) and 81% (95% CI: 74–87%), respectively, and the specificities were 91% (95% CI: 88–95%) and 94% (95% CI: 91–97%), respectively. When tested against the CRS, the IgG ELISA performed best, with 78% sensitivity (95% CI: 72–83%) and 98% specificity (95% CI: 96–100%), when a cut-off of 0.675 was applied and the indeterminate samples were excluded from the analysis.
Conclusion
The NIE-SsIR IgG ELISA demonstrated better accuracy than the IgG4 assay and was deemed promising particularly for serosurveys in endemic areas.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-021-04916-x.
Keywords: Strongyloides, Strongyloidiasis, ELISA, Recombinant antigen, NIE, SsIR, Accuracy, Diagnostic study
Background
Strongyloides stercoralis is a soil-transmitted helminth (STH) which affects about 600 million people in disadvantaged areas of the world [1], mainly in tropical/subtropical areas of South-East Asia, Africa, Western Pacific and Latin America. The infection is transmitted through contact with contaminated soil and perpetuates in the infected individuals due to the continuous re-exposure to the infective source in endemic areas and to a peculiar auto-infective cycle [2]. The latter is caused by larvae produced by the adult female worm in the human intestine that mature into the infective stage in the distal end of the bowel, subsequently penetrating the rectum wall or the perineal skin, leading to chronic infection. Strongyloidiasis often goes unnoticed due to intermittent, non-specific symptoms, and eosinophilia can sometimes be the only indicator of cryptic infection [2]. However, the infection is particularly significant in immunocompromised patients, in whom it can trigger severe disease, characterized by increased larval load (thus, worsening of symptoms) and dissemination of the parasite all over the body. This condition is almost invariably fatal [2].
The first-line treatment (a single dose of 200 µg/kg ivermectin) has an efficacy of around 86% for chronic uncomplicated infection [3]. Treatment of the disseminated disease is more challenging and relies on expert opinion [2].
Despite the potential for harm, the clinical burden and the prevalence of this parasitic infection have long been underestimated, mostly due to inadequacies of diagnostic tests [4]. Indeed, the “traditional” coproparasitological examination has an extremely low sensitivity for S. stercoralis. The sensitivity of other stool-based tests, such as agar plate culture (APC), Baermann method and PCR assay, is relatively better, but still unsatisfactory, particularly for low-burden infections [5].
Currently, serology testing is the most sensitive diagnostic method, with sensitivities of 83–92% for commercial enzyme-linked immunosorbent assay (ELISA) kits that use crude larval antigens, and 95% with an in-house immunofluorescence assay (IFAT) [6–8]. The latter detects immunoglobulin (Ig)G antibodies against intact S. stercoralis filariform larvae obtained from positive charcoal fecal culture [9]. A previous retrospective study demonstrated good accuracy, with 94.6% (95% conficence interval [CI]: 90.7–98.5%) sensitivity and 87.4% (95% CI: 83.4–91.3%) specificity [6]. An ELISA test used at the study site (Strongyloides ratti IgG ELISA) detects antibodies to Strongyloides spp. using somatic antigens from larvae of the closely related parasite S. ratti [7]. A previous retrospective study estimated its sensitivity and specificity at 90.8% (95% CI: 85.8–95.7%) and 94% (95% CI: 91.2–96.9%), respectively [6].
The need for a constant supply of parasites and the intrinsic variability of the antigenic source are two important limitations of seroassays based on either whole parasites (IFAT) or crude extracts (ELISA) [4].
In a recent World Health Organization meeting [10], seroassays based on recombinant antigens (which do not rely on supplies of larvae) have been identified as the most promising diagnostic method for large-scale use because of the better reproducibility of the results and the commercial availability.
An ELISA assay based on the recombinant S. stercoralis NIE antigen has to date showed a sensitivity in the range of 71–84% [6, 11, 12]. Another recombinant antigen, SsIR, has also been developed and evaluated in combination with a luciferase immunoprecipitation system (LIPS) [11, 13], showing excellent results in terms of sensitivity and specificity, both around 97%. Moreover, when the LIPS was used with the combination of NIE and SsIR antigens, there was a further improvement in diagnostic accuracy [13]. However, LIPS technology is not widely available and requires significant capital outlay.
Recently, two ELISA assays based on a combination of NIE and SsRI antigens were developed by InBios (InBios International Inc, Seattle, WA, USA), one for detection of IgG and the second for detection of IgG4 antibodies, for the diagnosis of S. stercoralis infection, with the goal of improving the performance of assays based on a single recombinant antigen. The two tests detect, respectively, specific IgG and IgG4 antibodies to Strongyloides recombinant antigens NIE and SsIR in serum using an enzymatically amplified sandwich-type immunoassay.
The aim of this study was to assess the sensitivity and specificity of the novel NIE/SsIR IgG and IgG4 ELISAs for the diagnosis of S. stercoralis infection using a panel of archived sera and using the results of specific fecal tests or a combination of conventional diagnostic tests (fecal and serology) for the diagnosis of S. stercoralis infection as reference.
Methods
Study design
This was a single-center, retrospective, diagnostic accuracy study. Sera were retrieved from the “Tropica Biobank” of the Department of Infectious Tropical diseases and Microbiology (DITM) of IRCCS Sacro Cuore Don Calabria hospital, Negrar, Verona, Italy. Results are reported following the STARD (Standards for Reporting Diagnostic Accuracy) guidelines [14].
Participants and inclusion/exclusion criteria
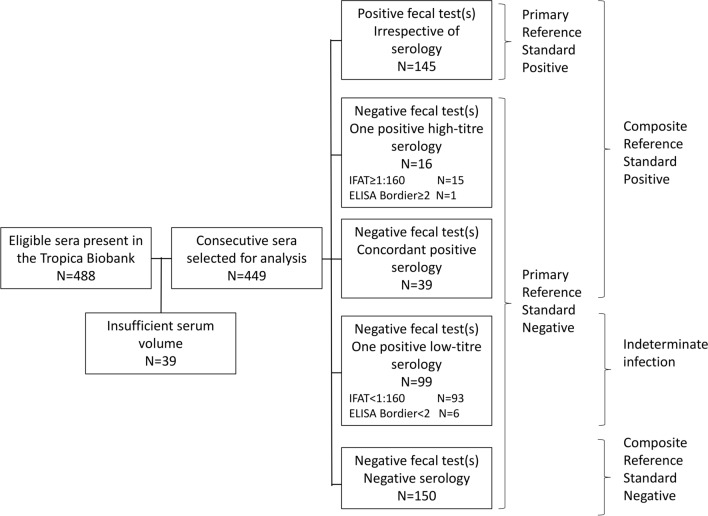
All available sera in the Tropica Biobank were considered eligible. The inclusion criteria were: (i) serum collected from immigrants from S. stercoralis endemic countries (i.e. individuals from Africa, Latin America, South-East Asia and Western Pacific regions); and (ii) serum with a matched result for APC and/or PCR assay for S. stercoralis (i.e. the latter test/s done on the same day or ± 30 days from the collection of the serum), or routine coproparasitology after formol–ether concentration if positive for S. stercoralis larvae. The exclusion criteria were: (i) unavailable/insufficient quantity of serum; (ii) sera from patients who received treatment with ivermectin in the previous 6 months; and (iii) sera from patients for whom only routine coproparasitology was performed after formol-ether concentration, if negative for S. stercoralis larvae.
A list of eligible samples was generated from the electronic database of the biobank, and all consecutive samples meeting the given criteria were included.
Test methods
Fecal tests
Both APC and the PCR assay for S. stercoralis are routinely used at the study site. The procedure for APC follows a modified Koga agar plate method. The cultures are incubated up to 6 days and visually inspected on day 2, 4 and 6. On day 6, cultures are flooded with saline and the sediment examined by microscopy. In a previous retrospective study, the sensitivity of the method was 45.4% (95% CI: 30.4–61.1%) [15].
The PCR is a real-time assay based on the method published by Verweij et al. [16]. Briefly, approximately 200 mg of unfixed feces was suspended in 200 µl of phosphate-buffered saline containing 2% polyvinylpolypyrolidone (Sigma-Aldrich Merck KGaA, Darmstadt, Germany) and frozen overnight at − 20 °C. After thawing, the samples were boiled for 10 min at 95 °C. The DNA was then extracted using Magnapure LC.2 extractor (F. Hoffmann-La Roche AG, Basel, Switzerland) following the protocol DNA_I_Blood_Cells_High performance_II, using the DNA isolation kit I (F. Hoffmann-La Roche AG), with a final elution volume of 100 µl. The real-time assay, targeting the small-subunit rRNA gene sequence of S. stercoralis, was performed as described. Appropriate positive and negative controls, as well as controls for PCR inhibitors and amplification quality using PhHV-1 control DNA in the same sample reaction, were included in each run. Thermocycling consisted of 40 cycles, and cycle threshold values < 40 were considered positive. The reactions, detection and data analysis were performed with the CFX96 detection system (Bio-Rad, Hercules, CA, USA). In a previous retrospective study the method demonstrated a sensitivity of 56.8% (95% CI: 41.0–71.6%) [15].
Serological tests
The following serological tests were used:
-
i.
The new Strongy Detect ™ IgG ELISA and IgG4 ELISA (InBios International, Inc., Seattle, WA, USA). Positive and negative control samples are provided in the kit. The tests were performed according to the manufacturer’s instructions. Interpretation of the results (positive/negative) was based on the analysis of the receiver operating characteristics (ROC) curve, as detailed below.
-
ii.
An in-house IFAT. This IFAT is routinely implemented at DITM for screening and individual diagnosis of strongyloidiasis. A positive result is defined as a titer ≥ 1:20.
-
iii.
The Strongyloides ratti IgG ELISA (Bordier ELISA; Bordier Affinity Products SA, Crissier, Switzerland). At DITM, the test is performed as per the manufacturer’s instructions. As the cut-off varies between runs, a normalized optical density (OD) ratio is used to compare the results obtained in different sessions. A ratio ≥ 1 defines positive results. The test is widely available and deployed for routine screening and diagnostic activities across Europe.
Test procedures
The new InBios ELISA assays were performed by staff of the DITM parasitology laboratory, which is a regional reference laboratory for parasitic infections. Sera were re-coded by staff not directly involved in the laboratory procedures. The laboratory staff were blinded to the results of any fecal and serological tests previously performed. The samples were tested shortly after thawing. For sera already tested using the Bordier ELISA and IFAT at the time of routine analysis, only the new InBios ELISA tests were performed. Conversely, in case results for either routine seroassay were missing, the Bordier ELISA and/or IFAT were carried out as needed.
Data on concomitant infections, assessed as per routine procedure in the parasitology laboratory, were retrieved from the electronic clinical records of DITM. These included routine microscopy after formol–ether concentration of feces for intestinal helminths and ELISA serology for filariasis and schistosomiasis (both assays from Bordier Affinity Products SA).
Data analysis
Reference standards and sera classification
The performances of the new InBios ELISA tests were first assessed against the results of fecal tests only (primary reference standard). For this endpoint, S. stercoralis infection was defined based on at least one positive fecal test among standard copromicroscopy of formol–ether concentrated feces, or on positive APC or PCR results.
As the included fecal tests are virtually 100% specific but lack sensitivity, a composite reference standard (CRS) based on the combination of the results of fecal and serological tests was also used to classify patients, as recommended for the evaluation of diagnostic tests in the absence of a gold standard [17]. Patients were classified as positive for S. stercoralis infection if they were: (i) positive on at least one fecal test among standard copromicroscopy of formol–ether-concentrated feces, APC or PCR; or (ii) negative on all fecal tests performed, including at least APC or PCR, but concordantly positive by Bordier ELISA and IFAT; or (iii) negative on all fecal tests performed, including at least APC or PCR, and positive only in one routine serology test but at a high titer (≥ 1:160 for IFAT, ≥ 2 for Bordier ELISA). These threshold values were chosen based on the results of a previous study [6]. Negativity for S. stercoralis infection was defined in the presence of negative fecal (all performed, including at least APC or PCR) and routine serology (Bordier ELISA and IFAT) tests. Finally, doubtful infection status (indeterminate cases) was defined when there was a negative fecal test (all performed, including at least APC or PCR) and positivity in only one of the two routine seroassays, at a low titer (< 1:160 for IFAT or < 2 for Bordier ELISA).
Sample size calculation
For sample size calculation, it was assumed that the new InBios ELISAs based on recombinant antigens NIE and SsIR would not have lower sensitivity and specificity than the existing NIE ELISA (75.4 and 89.5%, respectively [6]). For sensitivity, a sample size of 246 was estimated to be required based on a two-sided 95% CI of 0.10 width and test sensitivity of 0.80. For specificity, a sample size of 151 was calculated, assuming a specificity of 0.89 with a 95% CI with width of 0.10. Calculations were carried out using Power Analysis and Sample Size Software (PASS) version 2019 (NCSS Statistical Software, Kaysville, UT USA).
Statistical analysis
The optimal cut-off for the two index ELISA assays was obtained using the maximum sum of sensitivity and specificity (Youden’s J statistic), determined across all cut-off points of the ROC curve for each endpoint. Cut-off points obtained minimizing the distance to the upper-left corner of the ROC plot (D0-1) were also calculated. Individual samples analyzed with the InBios ELISA tests with OD exceeding the reading limit of the ELISA reader were assigned an OD = 5. Areas under the ROC curve (AUC) were compared by the DeLong test.
Tests results were displayed in contingency tables from which sensitivity and specificity were calculated against the two reference standards: fecal test (primary reference standard) and CRS. When applying the CRS, results obtained from sera of patients with doubtful infection status (i.e. indeterminate cases with negative fecal tests and positive serology at “low titer” in only one of the routine IFAT and Bordier ELISA tests) were first excluded from the analysis, then included while classifying them as either positive or negative.
The association between serology results of the index ELISA assays and presence of other intestinal helminth infections, schistosomiasis or filariasis was assessed using the Chi-square or Fisher Exact tests. Differences between sensitivities and specificities of the two tests were assessed using the McNemar test. Estimations were reported together with the 95% CI. Data analysis was performed using SAS software version 9.4 (SAS Institute, Cary, NC, USA).
Results
Tests were performed between the 4 and 31 March 2021. The study flow chart of sample selection is reported in Fig. 1.
Fig. 1.
Study flow chart of sample selection. ELISA Enzyme-linked immunosorbent assay, IFAT indirect fluorescent antibody test
Primary reference standard
Of the 449 (32.3%) cases examined, 145 were classified as positive based on at least one positive fecal test: 83 (57.2% of positive samples) had positive APC, 83 (57.2%) had positive stool microscopy and 77 (53.1%) had positive real-time PCR. Table 1 summarizes the results of the two index ELISA tests against this primary reference standard, considering cut-off points obtained by Youden’s J statistic; the results obtained using the cut-off calculated by the D0-1 method are reported in Additional file 1: Table S1.
Table 1.
Results of of the two NIE/SsIR antigen-based tests against the primary reference standard
| ELISA kit | Results of ELISA kit | Fecal tests (n) | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| InBios Strongy Detect™ IgG ELISA | Positive | 135 | 26 | 161 |
| Negative | 10 | 278 | 288 | |
| Total | 145 | 304 | 449 | |
| InBios Strongy Detect™ IgG4 ELISA | Positive | 118 | 18 | 136 |
| Negative | 27 | 286 | 313 | |
| Total | 145 | 304 | 449 | |
ELISA, Enzyme-linked immunosorbent assay, Ig, immunoglobulin
Of note, there were four cases classified as positive on the basis of fecal tests that had concordant negative IFAT and Bordier ELISA results. One of these also tested negative with the new NIE-SsIR; this sample originated from an immunocompetent 2-year-old child. The other three samples were positive for the IgG and IgG4 NIE-SsIR ELISA tests; these samples originated from a patient with tuberculosis, a patient with HTLV-1 infection and a patient with advanced metastatic cancer who developed disseminated strongyloidiasis, respectively.
The AUC of the ROC curves (Fig. 2) of the two index tests generated by the results reported in Table 1 was not statistically significantly different (P = 0.1210).
Fig. 2.
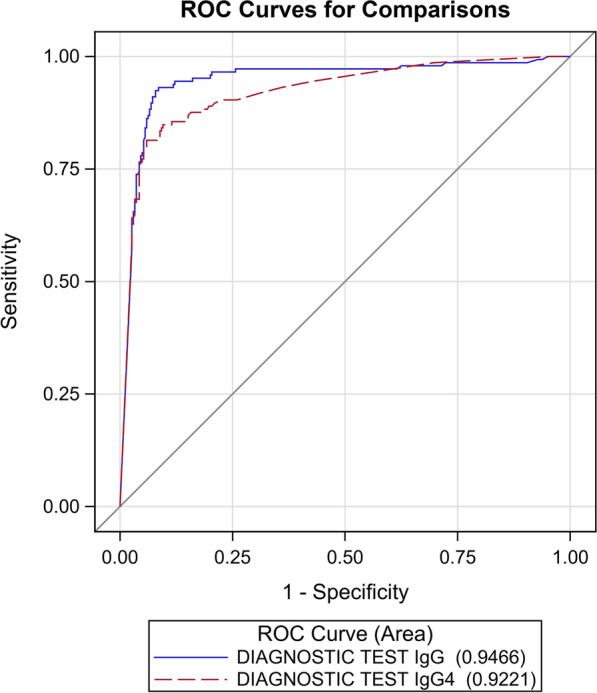
ROC curves for the IgG and IgG4 NIE-SsIR ELISA tests, calculated with the primary reference standard. Ig Immunoglobulin, ROC receiver operating characteristics
Composite reference standard
The entire panel of 449 sera was classified into 200 positive, 150 negative and 99 indeterminate cases, according to the CRS. First, the accuracy analysis was carried out, excluding the indeterminate cases. The results obtained when applying the cut-off calculated using Youden’s J statistic are reported in Table 2; the results obtained using the cut-off calculated by the D0-1 method are reported in Additional file 1: Table S1.
Table 2.
Results of the two NIE/SsIR antigen-based tests against the composite reference standard, excluding indeterminate cases
| ELISA kit | Results of ELISA kit | Composite reference standard—indeterminate cases excluded (n) | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| InBios Strongy Detect™ IgG ELISA | Positive | 155 | 3 | 158 |
| Negative | 45 | 147 | 192 | |
| Total | 200 | 150 | 350 | |
| InBios Strongy Detect™ IgG4 ELISA | Positive | 140 | 4 | 144 |
| Negative | 60 | 146 | 206 | |
| Total | 200 | 150 | 350 | |
Among cases classified as positives, some had OD values in the InBios ELISAs exceeding the reading limit of the ELISA plate reader: 68 cases for both IgG and IgG4, 23 cases for IgG only and 24 cases for IgG4 only. All of these samples had either positive fecal test results or a concordant “high titer” positive IFAT and Bordier ELISA. In addition, one case with an OD reading for IgG4 that exceeded the upper limit was observed in the strongyloidiasis-negative group; in this case, the IgG ELISA was also positive (OD 1.37). This sample originated from a patient who had been diagnosed with strongyloidiasis (positive APC and IFAT 1:20) and treated with ivermectin 7 years previously. At the time the sample included in this study was collected, the patient had come back for itching, had a normal eosinophil count and was negative for S. stercoralis by IFAT, APC and PCR
The AUC of the ROC curves (Fig. 3) generated by the results reported in Table 2 was statistically significantly different (P = 0.035).
Fig. 3.
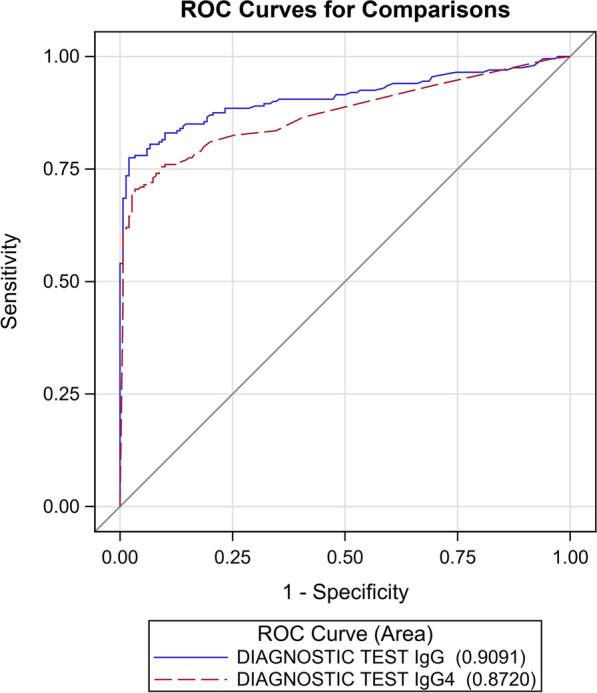
ROC curves for the IgG and IgG4 NIE-SsIR ELISA tests, calculated with the composite reference standard, excluding indeterminate cases
We then assessed the index ELISA cut-off and sensitivity and specificity against a CRS where indeterminate cases were classified as negative cases. The results obtained when applying the cut-off calculated using the Youden’s J statistics are reported in Table 3; the results obtained using the cut-off calculated by the D0-1 method are reported in Additional file 1: Table S1.
Table 3.
Results of the two NIE-SsIR antigen-based tests against the CRS, indeterminate cases were classified as negative cases
| ELISA kit | Results of ELISA kit | Composite reference standard—indeterminate cases included as negative cases (n) | ||
|---|---|---|---|---|
| Positive | Negative | Totals | ||
| InBios Strongy Detect™ IgG ELISA | Positive | 153 | 5 | 158 |
| Negative | 47 | 244 | 291 | |
| Total | 200 | 249 | 449 | |
| InBios Strongy Detect™ IgG4 ELISA | Positive | 150 | 22 | 172 |
| Negative | 50 | 227 | 277 | |
| Total | 200 | 249 | 449 | |
The AUC of the related ROC curves (Fig. 4) was significantly different (P = 0.0389).
Fig. 4.
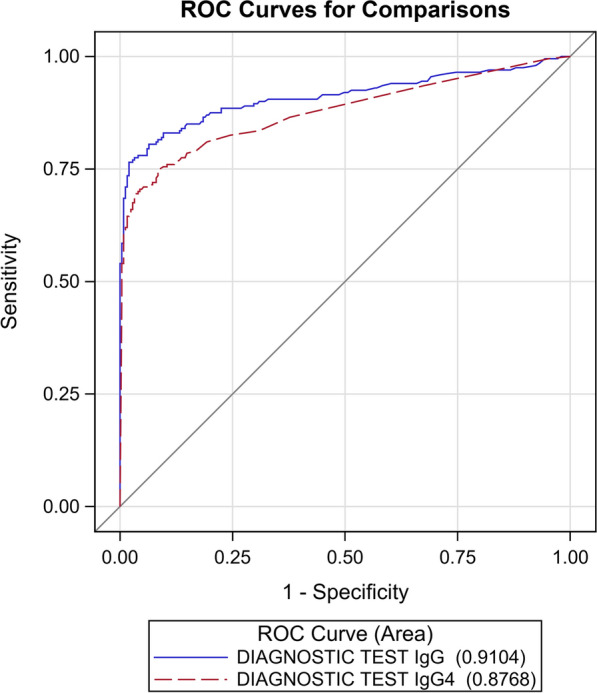
ROC curves for the IgG and IgG4 NIE-SsIR ELISA tests, calculated with the composite reference standard. Indeterminate cases were classified as negative cases
Finally, the sensitivity and specificity of the index ELISA cut-off were assessed against a CRS where indeterminate cases were classified as positive. The results obtained when applying the cut-off calculated using the Youden’s J statistics are reported in Table 4; the results obtained using the cut-off calculated by the D0-1 method are reported in Additional file 1: Table S1.
Table 4.
Results of the two NIE-SsIR antigen-based tests against the CRS, indeterminate cases were classified as positive cases
| ELISA kit | Results of ELISA kit | Composite reference standard—indeterminate cases included as positive cases (n) | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| InBios Strongy Detect™ IgG ELISA | Positive | 160 | 3 | 163 |
| Negative | 139 | 147 | 291 | |
| Total | 299 | 150 | 449 | |
| InBios Strongy Detect™ IgG4 ELISA | Positive | 146 | 4 | 150 |
| Negative | 153 | 146 | 277 | |
| Total | 299 | 150 | 449 | |
The AUC of the ROC curves (Fig. 5) of the two index tests generated by the results reported in Table 4 was not statistically significantly different (P = 0.1728).
Fig. 5.
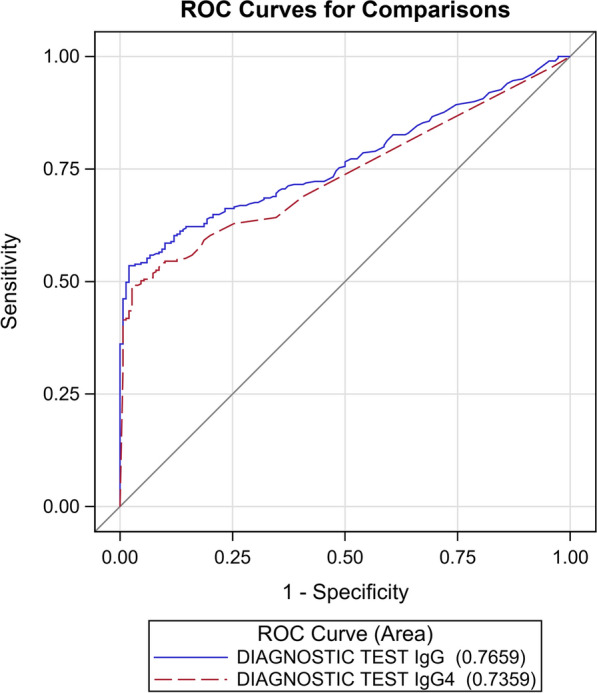
ROC curves for the IgG and IgG4 NIE-SsIR ELISA tests, calculated with the composite reference standard. Indeterminate cases were classified as positive cases
The cut-off values and the sensitivity and specificity calculated based on primary and composite reference standards and cut-off points generated using the Youdens’ J statistics, including when indeterminate cases were included as positive cases in the analysis, are summarized in Table 5; the cut-off and accuracy results calculated with the D0-1 method are reported in Additional File 1: Table S1.
Table 5.
Cut-off values, sensitivity and specificity of the index InBios ELISA tests using the primary (fecal test) and secondary (CRS) reference standards
| Reference standard | Indeterminate cases | Test | Cut-off value | Sensitivity (95% CI) | Pa | Specificity (95% CI) | Pa |
|---|---|---|---|---|---|---|---|
| Primary—fecal test | n/a | IgG | 0.714 | 0.93 (0.88–0.97) | < 0.0001 | 0.91 (0.88–0.95) | 0.0325 |
| Primary—fecal test | n/a | IgG4 | 0.609 | 0.81 (0.74–0.87) | 0.94 (0.91–0.97) | ||
| Secondary—CRS | Excluded | IgG | 0.675 | 0.78 (0.72–0.83) | 0.0011 | 0.98 (0.96–1.00) | 0.6547 |
| Secondary—CRS | Excluded | IgG4 | 0.32 | 0.70 (0.64–0.76) | 0.97 (0.95–1.00) | ||
| Secondary—CRS | Included as negative cases | IgG | 0.885 | 0.76 (0.71–0.82) | 0.5316 | 0.98 (0.96–1.00) | 0.0004 |
| Secondary—CRS | Included as negative cases | IgG4 | 0.064 | 0.75 (0.69–0.81) | 0.91 (0.88–0.95) | ||
| Secondary—CRS | Included as positive cases | IgG | 0.675 | 0.54 (0.48–0.59) | 0.0082 | 0.98 (0.96–1.00) | 0.6547 |
| Secondary—CRS | Included as positive cases | IgG4 | 0.32 | 0.49 (0.43–0.54) | 0.97 (0.95–1.00) |
CI, Confidence interval; CRS, composite reference standard, n/a, not available
aP-values comparing sensitivities and specificities were determined by McNemar’s test
Association between positive serology for S. stercoralis and concurrent helminth infections
In the whole cohort, 35 patients had one or more intestinal helminth infections, including hookworm (n = 22), Hymenolepis nana (n = 6), Trichuris trichiura (n = 7), Dicrocoelium dendriticum (n = 2), Strongyloides fuelleborni (n = 2), Trichostrongylus spp. (n = 2), Ascaris lumbricoides (n = 1) and Schistosoma spp. (n = 19 with eggs in stool). No significant association was found between the results of the IgG and IgG4 ELISA and concomitant infection with intestinal helminths, in neither the strongyloidiasis-negative or the strongyloidiasis-indeterminate group, or the two groups combined. Significant associations with Schistosoma infection and with serology for schistosomiasis (data available for 303 patients) were found only for the IgG4 ELISA in the strongyloidiasis-negative group (P = 0.0042 for eggs in stool and P = 0.0157 for anti-Schistosoma ELISA), and in the strongyloidiasis-negative + indeterminate groups combined (P = 0.0162 for eggs in stool and P = 0.0517 for anti-Schistosoma ELISA). Of note, all Schistosoma-positive samples originated from migrants from sub-Saharan Africa. A total of 234 patients had records of a routine ELISA serology exam for filariasis; no significant association was found between filarial serology and both IgG and IgG4 ELISA in the strongyloidiasis-negative or the strongyloidiasis-indeterminate groups, or the two groups combined.
Overall, similar results were found when the same analysis was applied to the routine serology tests for strongyloidiasis by IFAT and Bordier ELISA. No association was found between results of these tests and the presence of intestinal helminth infections in the strongyloidiasis-negative or the strongyloidiasis-indeterminate groups, or the two groups combined. The results of both routine seroassays were significantly associated with Schistosoma infection in the strongyloidiasis-indeterminate group (P = 0.0027 for both assays) and the Strongyloides-negative + indeterminate combined group (P = 0.0471 for IFAT and P = 0.0086 for Bordier ELISA). When schistosomiasis was assessed with the ELISA, significant associations with IFAT and Bordier ELISA results were found with the Strongyloides-indeteminate (P = 0.0153 both IFAT and Bordier ELISA) group and with the Strongyloides-negative + indeterminate group combined (P = 0.0248 Bordier ELISA).
No significant association was also found between IFAT and Bordier ELISA results and filariasis serology in the strongyloidiasis-negative and the strongyloidiasis-indeterminate groups.
Results of the association between positive serology for S. stercoralis and concurrent helminth infections are summarized in Additional file 2: Table S2.
Discussion
In this study, we evaluated two novel ELISAs based on the recombinant antigens NIE and SsIR using samples collected from a cohort of patients from S. stercoralis endemic areas, representative of people who would routinely be screened for strongyloidiasis and other parasitic infections in our setting. Due to the lack of a diagnostic gold standard for S. stercoralis infection [5], the two assays were evaluated not only against the results of fecal tests (primary reference standard), but also against a CRS using a panel of sera from patients with negative fecal tests results and variable results on routine serology assays performed at DITM. As expected, sensitivity was higher and specificity lower when using the primary reference standard than the CRS. For the latter analysis, we first excluded “indeterminate cases,” i.e. those with a single “low titer” positive routine serological test (IFAT or Bordier ELISA) result, with the aim of reducing the possible misclassification of positive and negative results. We then included the indeterminate cases so that the cohort could mirror real-life situations, where cases of uncertain classification can occur and cane be interpreted as positive or negative. Our results showed that when indeterminate cases were classified as negative, the accuracy of the tests was similar to that obtained when the indeterminate cases were not included in the CRS. This result indicates that classifying indeterminate results as negative was appropriate in terms of setting up the CRS; therefore, here we focus our discussion of the assays’ performance by referring to the results of this approach when addressing the CRS.
The AUC was similar for the two ELISAs when considering the primary reference standard, although sensitivity was significantly higher for IgG (93%; 95% CI: 88–97%) than for IgG4 (81%; 95% CI: 74–87%): P < 0.0001.
With the CRS, the AUC was instead statistically different for the two assays, favoring the IgG ELISA, with this assay showing excellent specificity (98%; 95% CI: 96–100%) and good sensitivity (76%, 95% CI: 71–82%) at a cut-off of 0.88 OD. Sensitivity of the IgG4 ELISA was similar to that of the IgG ELISA (P = 0.5316), but the specificity of the former was significantly lower (91%; 95% CI: 88–95%, P = 0.0004) and, in addition, this IgG4-detecting test showed a significant association with parasitological and serological diagnosis of schistosomiasis that was not shown by the IgG ELISA.
Overall, sensitivity of the two tests seems to be in line with that of the previous NIE-ELISA, while the specificity of the NIE-SsIR-IgG is improved compared with that of the NIE-ELISA [6, 12]. Other serological assays (namely our in-house IFAT, Bordier ELISA, IVD [in vitro diagnostic] ELISA) previously demonstrated higher sensitivity than these two assays [6, 7]; however, it must be noted that the two recombinant ELISAs detected three cases with positive fecal tests but with negative results to the other serological tests; these three cases were in patients with impaired immunity; hence it would be worthwhile to further explore this finding in a larger cohort of immunosuppressed patients, for whom other serological assays may have reduced sensitivity [18, 19].
The choice of tests with different sensitivity and specificity depends on the setting where the tests need to be applied and the diagnostic purpose. In the non-endemic setting, especially at the hospital level, test sensitivity would be prized over specificity, as it would be preferable to treat a falsely-positive patient with ivermectin, which is a well-tolerated and safe drug, than miss cases of this potentially life-threatening infection. In contrast, a high test specificity would be preferred in the context of control programs in endemic areas, where co-infections represent an issue when the prevalence and impact of control activities need to be assessed and the whole target population will receive the intervention(s) in any case, irrespective of the positive or negative result of the test [10, 20]. As mentioned, these programs entail testing large numbers of people; hence, in principle, ELISAs based on recombinant antigens, such as the one evaluated here, would be advantageous over those based on crude antigens on the basis of high reproducibility and supply capacity. Field studies should be carried out using the recombinant-based InBios ELISA assays to confirm their good performance in that setting. Another aspect that would encourage the deployment of these assays in the field is the possibility of performing the test on serum obtained from dried blood spots collected on filter paper, which eases sample collection and transport. Previous studies demonstrated that this is possible for the NIE-ELISA [21] and other serological assays [22], encouraging the evaluation of the NIE-SsIR ELISA tests with this material. Finally, other serological tests have also been demonstrated to be useful for post-treatment monitoring, both in the non-endemic (at individual level) [23, 24] and endemic (at population level) [25] settings. This aspect requires evaluation of these new tests as well in order to frame their usefulness both in the clinical and control program setting to reflect presence of ongoing infection.
The main limitation of this study is its retrospective design. While it was possible to review the clinical files of a few patients with unexpected results, we could not systematically collect clinical data from the entire cohort. In addition, as examinations for concurrent parasitic infections were not performed for all patients, the results of the association with serological test results for strongyloidiasis should be taken with caution and confirmed following a systematic approach. Moreover, the patients did not all undergo the same fecal tests, although we included only samples on which PCR and/or APC were performed, which have comparable accuracy, carried out at a matching time point. One of the major strengths of this study is that the cohort was representative of patients for whom these tests would be used in our non-endemic setting, making our results relevant for the evaluation of the performance of the index ELISA assays in clinical practice. Another strength is the use of a CRS, which permits a robust estimation of the accuracy of a test in the absence of a diagnostic gold standard [17]. However; consideration must be given to the fact that the CRS is still an imperfect tool and that it is possible that the values of sensitivity and specificity of the NIE-SsIR IgG and IgG4 ELISAs actually lay between those found with the primary reference standard and the CRS.
Conclusions
In conclusion, the NIE-SsIR IgG ELISA demonstrated better accuracy than the IgG4-detecting assay and the previous NIE-ELISA [6, 12]. The sensitivity of the IgG ELISA assay was lower than that demonstrated previously for other serological assays [6, 12], but its specificity was excellent. These characteristics, in addition to possible large-scale production and reproducibility, make the test promising for serosurveys in the field, also in the context of control programs. Further studies should evaluate its use in an endemic setting, preferably also on dried blood spots. In addition, performance for post-treatment monitoring should be assessed both in endemic and non-endemic settings.
Supplementary Information
Additional file 1: Table S1. Cut-off values calculated by the D0-1 method.
Additional file 2: Table S2. Results of the association between positive serology for S. stercoralis and concurrent helminth infections.
Acknowledgements
We wish to acknowledge Stefano Tais, Monica Degani and Eleonora Rizzi for technical support.
Abbreviations
- APC
Agar plate culture
- AUC
Area under curve
- CRS
Composite reference standard
- DITM
Department of Infectious Tropical Diseases and Microbiology
- ELISA
Enzyme-linked immunosorbent assay
- IFAT
Indirect fluorescent antibody test
- LIPS
Luciferase immunoprecipitation system
- OD
Optical density
- ROC
Receiver operating characteristics
- STH
Soil-transmitted helminthes
Authors' contributions
FT contributed to the study design, the data acquisition and interpretation and to the first draft of the manuscript. SL contributed to the data acquisition and interpretation and critically revised the manuscript. CM contributed to the data interpretation and analysis and critically revised the manuscript. SP contributed to the data acquisition and interpretation. AM contributed to the data interpretation and critically revised the manuscript. SM contributed to the data interpretation and critically revised the manuscript. ZB contributed to the data interpretation and critically revised the manuscript. DB contributed to the study design, the data interpretation and to the first draft of the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was partly funded by the Italian Ministry of Health “Fondi Ricerca Corrente-L2P4” to IRCCS Sacro Cuore Don Calabria Hospital, Negrar, Verona, Italy. The Strongy Detect™ IgG ELISA and IgG4 ELISA kits were donated by the Manufaturer, InBios International Inc, Seattle, WA, USA.
Availability of data and materials
The raw data will be published in Zenodo upon acceptance for publication of the present article.
Declarations
Ethics approval and consent to participate
The study protocol received clearance from the Ethics Committee of IRCCS Sacro Cuore Don Calabria hospital (Comitato Etico per la Sperimentazione Clinica delle Province di Verona e Rovigo) on 13 January 2021 (Prot. Negrar 2020-69). All samples used in the study were from patients who signed an informed consent form to donate their samples for research purpose at the time of blood sampling for routine diagnostic analyses; the results of this study did not influence the clinical management of patients whose sera were used. Before analysis, laboratory and demographic data were uncoupled and samples re-coded. Excel databases with demographic and laboratory data were stored separately, both protected by password.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Francesca Tamarozzi, Email: francesca.tamarozzi@sacrocuore.it.
Silvia Stefania Longoni, Email: silvia.longoni@sacrocuore.i.t.
Cristina Mazzi, Email: cristina.mazzi@sacrocuore.it.
Sofia Pettene, Email: sofiapettene@gmail.com.
Antonio Montresor, Email: montresora@who.int.
Siddhartha Mahanty, Email: siddhartha.mahanty@unimelb.edu.au.
Zeno Bisoffi, Email: zeno.bisoffi@sacrocuore.it.
Dora Buonfrate, Email: dora.buonfrate@sacrocuore.it.
References
- 1.Buonfrate D, Bisanzio D, Giorli G, Odermatt P, Fürst T, Greenaway C, et al. The global prevalence of Strongyloides stercoralis infection. Pathogens. 2020;9:468. doi: 10.3390/pathogens9060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nutman TB. Human infection with Strongyloides stercoralis and other related Strongyloides species. Parasitology. 2017;144:263–273. doi: 10.1017/S0031182016000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonfrate D, Salas-Coronas J, Muñoz J, Maruri BT, Rodari P, Castelli F, et al. Multiple-dose versus single-dose ivermectin for Strongyloides stercoralis infection (Strong Treat 1 to 4): a multicentre, open-label, phase 3, randomised controlled superiority trial. Lancet Infect Dis. 2019;19:1181–1190. doi: 10.1016/S1473-3099(19)30289-0. [DOI] [PubMed] [Google Scholar]
- 4.Krolewiecki AJ, Lammie P, Jacobson J, Gabrielli AF, Levecke B, Socias E, et al. A public health response against Strongyloides stercoralis: time to look at soil-transmitted helminthiasis in full. PLoS Negl Trop Dis. 2013;7:e2165. doi: 10.1371/journal.pntd.0002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buonfrate D, Formenti F, Perandin F, Bisoffi Z. Novel approaches to the diagnosis of Strongyloides stercoralis infection. Clin Microbiol Infect. 2015;21:543–552. doi: 10.1016/j.cmi.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Bisoffi Z, Buonfrate D, Sequi M, Mejia R, Cimino RO, Krolewiecki AJ, et al. Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLoS Negl Trop Dis. 2014;8:e2640. doi: 10.1371/journal.pntd.0002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doorn HR, Koelewijn R, Hofwegen H, Gilis H, Wetsteyn JC, Wismans PJ, et al. Use of enzyme-linked immunosorbent assay and dipstick assay for detection of Strongyloides stercoralis infection in humans. J Clin Microbiol. 2007;45:438–442. doi: 10.1128/JCM.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonfrate D, Marrone R, Silva R, Mirisola C, Ragusa A, Mistretta M, et al. Prevalence of strongyloidiasis in a cohort of migrants in Italy and accuracy of a novel ELISA assay for S. stercoralis infection, a cross-sectional study. Microorganisms. 2021;9:401. doi: 10.3390/microorganisms9020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boscolo M, Gobbo M, Mantovani W, Degani M, Anselmi M, Monteiro GB, et al. Evaluation of an indirect immunofluorescence assay for strongyloidiasis as a tool for diagnosis and follow-up. Clin Vaccine Immunol. 2007;14:129–133. doi: 10.1128/CVI.00278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Diagnostic methods for the control of strongyloidiasis. https://www.who.int/publications/i/item/9789240016538. Accessed 24 June 2021.
- 11.Krolewiecki AJ, Ramanathan R, Fink V, McAuliffe I, Cajal SP, Won K, et al. Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina. Clin Vaccine Immunol. 2010;17:1624–1630. doi: 10.1128/CVI.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fradejas I, Herrero-Martínez JM, Lizasoaín M, Rodríguez de Las Parras E, Pérez-Ayala A. Comparative study of two commercial tests for Strongyloides stercoralis serologic diagnosis. Trans R Soc Trop Med Hyg. 2018;112:561–567. doi: 10.1093/trstmh/try101. [DOI] [PubMed] [Google Scholar]
- 13.Ramanathan R, Burbelo PD, Groot S, Iadarola MJ, Neva FA, Nutman TB. A luciferase immunoprecipitation systems assay enhances the sensitivity and specificity of diagnosis of Strongyloides stercoralis infection. J Infect Dis. 2008;198:444–451. doi: 10.1086/589718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: An updated list ofessential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buonfrate D, Perandin F, Formenti F, Bisoffi Z. A retrospective study comparing agar plate culture, indirect immunofluorescence and real-time PCR for the diagnosis of Strongyloides stercoralis infection. Parasitology. 2017;144:812–816. doi: 10.1017/S0031182016002559. [DOI] [PubMed] [Google Scholar]
- 16.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EA, Polderman AM, et al. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans R Soc Trop Med Hyg. 2009;103:342–346. doi: 10.1016/j.trstmh.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Rutjes AW, Reitsma JB, Coomarasamy A, Khan KS, Bossuyt PM. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol Assess. 2007;11:iii, ix-51. doi: 10.3310/hta11500. [DOI] [PubMed] [Google Scholar]
- 18.Paula FM, Malta FM, Corral MA, Marques PD, Gottardi M, Meisel DM, et al. Diagnosis of Strongyloides stercoralis infection in immunocompromised patients by serological and molecular methods. Rev Inst Med Trop Sao Paulo. 2016;58:63. doi: 10.1590/S1678-9946201658063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascarello M, Gobbi F, Angheben A, Gobbo M, Gaiera G, Pegoraro M, et al. Prevalence of Strongyloides stercoralis infection among HIV-positive immigrants attending two Italian hospitals, from 2000 to 2009. Ann Trop Med Parasitol. 2011;105:617–623. doi: 10.1179/2047773211Y.0000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montresor A, Mupfasoni D, Mikhailov A, Mwinzi P, Lucianez A, Jamsheed M, et al. The global progress of soil-transmitted helminthiases control in 2020 and World Health Organization targets for 2030. PLoS Negl Trop Dis. 2020;14:e0008505. doi: 10.1371/journal.pntd.0008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mounsey K, Kearns T, Rampton M, Llewellyn S, King M, Holt D, et al. Use of dried blood spots to define antibody response to the Strongyloides stercoralis recombinant antigen NIE. Acta Trop. 2014;138:78–82. doi: 10.1016/j.actatropica.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Formenti F, Buonfrate D, Prandi R, Marquez M, Caicedo C, Rizzi E, et al. Comparison of S. stercoralis serology performed on dried blood spots and on conventional serum samples. Front Microbiol. 2016;7:1778. doi: 10.3389/fmicb.2016.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buonfrate D, Sequi M, Mejia R, Cimino RO, Krolewiecki AJ, Albonico M, et al. Accuracy of five serologic tests for the follow up of Strongyloides stercoralis infection. PLoS Negl Trop Dis. 2015;9:e0003491. doi: 10.1371/journal.pntd.0003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biggs BA, Caruana S, Mihrshahi S, Jolley D, Leydon J, Chea L, et al. Management of chronic strongyloidiasis in immigrants and refugees: is serologic testing useful? Am J Trop Med Hyg. 2009;80:788–791. doi: 10.4269/ajtmh.2009.80.788. [DOI] [PubMed] [Google Scholar]
- 25.Vargas P, Krolewiecki AJ, Echazú A, Juarez M, Cajal P, Gil JF, et al. Serologic monitoring of public health interventions against Strongyloides stercoralis. Am J Trop Med Hyg. 2017;97:166–172. doi: 10.4269/ajtmh.16-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Cut-off values calculated by the D0-1 method.
Additional file 2: Table S2. Results of the association between positive serology for S. stercoralis and concurrent helminth infections.
Data Availability Statement
The raw data will be published in Zenodo upon acceptance for publication of the present article.