Abstract
Background
The C2H2-type zinc finger proteins (C2H2-ZFPs) are one of major classes of transcription factors that play important roles in plant growth, development and stress responses. Limit information about the C2H2-ZF genes hinders the molecular breeding in bread wheat (Triticum aestivum).
Results
In this study, 457 C2H2-ZFP proteins (including 253 splice variants), which contain four types of conserved domain (named Q, M, Z, and D), could be further classified into ten subsets. They were identified to be distributed in 21 chromosomes in T. aestivum. Subset-specific motifs, like NPL-, SFP1-, DL- (EAR-like-motif), R-, PL-, L- and EK-, might make C2H2-ZFP diverse multifunction. Interestingly, NPL- and SFP1-box were firstly found to be located in C2H2-ZFP proteins. Synteny analyses showed that only 4 pairs of C2H2 family genes in T. aestivum, 65 genes in B. distachyon, 66 genes in A. tauschii, 68 genes in rice, 9 genes in Arabidopsis, were syntenic relationships respectively. It indicated that TaZFPs were closely related to genes in Poaceae. From the published transcriptome data, totally 198 of 204 TaC2H2-ZF genes have expression data. Among them, 25 TaC2H2-ZF genes were certificated to be significantly differentially expressed in 5 different organs and 15 different development stages by quantitative RT-PCR. The 18 TaC2H2-ZF genes were verified in response to heat, drought, and heat & drought stresses. According to expression pattern analysis, several TaZFPs, like Traes_5BL_D53A846BE.1, were not only highly expressed in L2DAAs, RTLS, RMS, but also endowed tolerance to drought and heat stresses, making them good candidates for molecular breeding.
Conclusions
This study systematically characterized the TaC2H2-ZFPs and their potential roles in T. aestivum. Our findings provide new insights into the C2H2-ZF genes in T. aestivum as well as a foundation for further studies on the roles of TaC2H2-ZF genes in T. aestivum molecular breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03016-3.
Keywords: Triticum aestivum, C2H2-ZFP, Molecular structural analysis, Subet-specific motifs, Expression patterns
Background
The C2H2-type zinc finger proteins constitute a large family of transcription factors. The C2H2-ZF domain was defined as about 30 amino acids with two conserved Cysteine and two histidine residues which bound to one Zn2+ atom tetrahedrally and form a structure as C-X2-4-C-X3-F-X5-L-X2-H-X3-5-H [1]. According to the defined C2H2-ZF types, C2H2-ZFPs were classified into two subsets X-tandem-SF and X-isolated-SF. The X-tandem-SF contains two clearly subsets (X-t1-SF and X-t2-SF), and the X-isolated-SF contains ten clearly subsets (X-1i-Q-SF, X-1i-M-SF, X-1i-Z-SF, X-1i-D-SF, X-2i-Q-SF, X-2i-M-S and X-2i-Z-SF, X-2i-Mix-SF, X-3i-SF, and X-4i-SF), where X represents any species [2, 3]. Previous studies have shown that C2H2-ZF genes represent 0.8%, 2.3%, 3% of all genes in Saccharomyces cerevisiae, Diptera, mammals, respectively [4].
The C2H2-ZFP family, which have been reported in numerous plant genomes, significantly contribute to the plant growth development, and also participate in multiple biological processes by transcriptional regulation. For instance, genome-wide analyses of ZFP proteins in plants have been carried out in Arabidopsis (176), durum wheat (122), rice (189), maize (211), soybean (321), foxtail millet (124) and other species based on genome sequences [2]. In T. aestivum, the C1-2i Q-type and C3H zinc finger protein (ZFP) transcription factor subclass has been reported to play important roles in plant stress responses and physiological stages [5, 6]. In Arabidopsis, the AZF1, AZF2, AZF3, ZAT6, ZAT7, ZAT8, ZAT10, ZAT12 and ZAT18 have been shown to function in multiple cellular processes, including seed germination, drought, cold, high-salinity, and oxidative stress responses [7–14]. In rice, several members of the C2H2-type ZFPs, such as ZFP15, ZFP36, ZFP39, ZFP182, ZFP245, ZFP252, and ZFP179, have also been shown to be involved in the responses of drought, salinity, and oxidative stress [14–20]. In Poplar (Populus alba), PtrZFP2/19/95 showed high expression levels in leaves and/or roots under environmental stresses by genome-wide analysis, which provided a solid foundation for studying the biological roles of C2H2-ZF genes in Populus growth and development [20].
The conserved amino acid residues of these C2H2-ZF genes were consistent with previous studies of the C2H2 domain in other plant species [10]. These C2H2-ZF genes studies include main structural features in C2H2-ZFPs, such as the arrangement of C2H2-ZF domains (tandem or dispersed), the length of spacer between the ZFs, the number of C2H2-ZF domains and the “QALGGH” sequence, whereas yeast and animal C2H2-ZFPs do not have this motif [3]. Compared with other eukaryote C2H2-ZFPs, in multiple-fingered C2H2-ZFPs, the plant zinc-finger domains are separated by long spacers that vary in length and sequence (such as, the double zinc finger protein ZPT2-7 and ZPT2-11 in petunia (Petumia hybrida), the distance between its adjacent zinc finger structures is 19 and 65 amino acids, respectively), whereas the C2H2-ZFPs of yeast and animals are mostly clustered and separated by only six to eight amino acids [4, 21].
Moreover, the structures of C2H2-ZFPs also contain several non-zinc-finger motifs, such as EAR-motif R-box, PL-box, L-box, and EK-box in T. aestivum [22]. The EAR motif (DLNxxP or LxLxL) was showed to be a predominant transcriptional repressor motif in some plant ZFP proteins [22]. Overexpression of most DREB/ERF proteins with an EAR motif led to reduced expression levels of stress-related genes, and fusion of the EAR motif to a number of transcriptional activators was found to convert them to dominant repressors [23]. Thus, the different classifications or different types of C2H2-ZF domains provides some extrinsic of function information for the C2H2-ZF genes, might make C2H2-ZFP diverse multifunction in comprehensive physiological stages.
Bread wheat (Triticum aestivum) as a staple crop of the world plays essential roles to sustain food security [24, 25]. However, dissecting gene function in T. aestivum is not easy due to its hexaploid genome and highly redundancy of genes. In this study, the comprehensive analyses were carried out based on phylogenetic relationships, physicochemical properties, subcellular localization prediction, chromosomal locations, and conserved protein domain in bread wheat C2H2-ZF genes family. The results of this study will provide the foundation for further functional analyses of the C2H2-ZF genes families, important scientific significance and application value for our understanding of the genetics and the evolution of T. aestivum.
Results
Classification and distribution analysis of T. aestivum C2H2-ZFPs
The 457 putative C2H2-type Zinc Finger proteins (C2H2-ZFP) (including 253 splice variants), were identified from the Plant transcription factor database (PlantTFDB) and the iTAK database. Further identification was conducted through the HMM profile HMMER 3.0 program and SMART database search. After removing incomplete and duplicate sequences by using the CD-HIT software, a total of 204 non-redundant candidate genes were obtained to encode 457 proteins, of which 316 proteins in full length and 141 cases as partial in N- and/or C-terminal regions but containing complete C2H2-ZF domain. In addition, according to the retrieved of the PlantTFDB database, iTAK database and ExPASy server, the 204 T. aestivum C2H2-ZFPs sequences showed that their amino acid length, molecular weight (Mw), isoelectric points (pIs), subcellular localization predictions, for details about TaC2H2-ZFP, please refer to Additional file 1: Table S2.
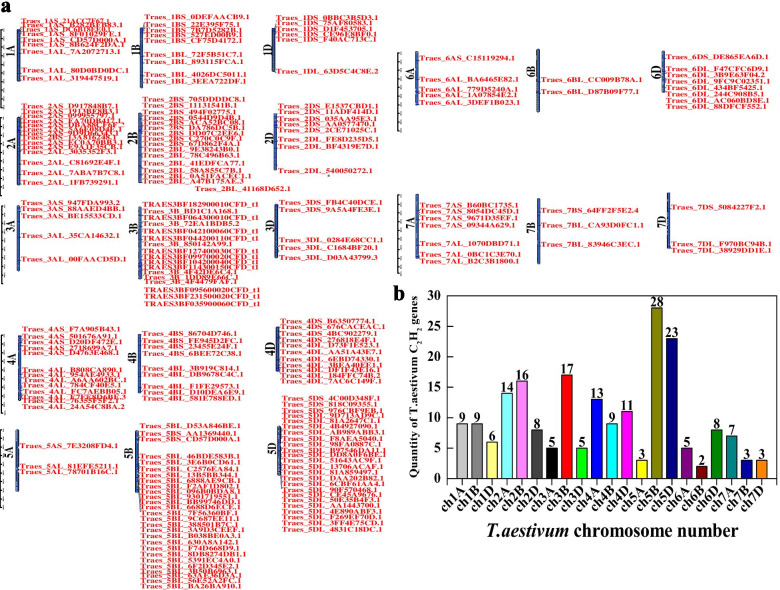
We identified 204 TaC2H2-ZF genes in T. aestivum, and revealed the distribution of the 204 TaC2H2-ZF genes on all the seven chromosomes in both A, B, and D sub-genomes (Additional file 1: Table S2; Fig. 1a). A maximum of 54 (26.5%) TaC2H2-ZF genes were located in chromosome 5, 48 on long arms and 6 on short arms, and 5B sub-genomes (26.47%) encoded a maximum number of TaC2H2-TaZF genes in the among of all sub-genomes. 38 (18.6%) TaC2H2-ZF genes were located in chromosome 2, 14 on long arms and 24 on short arms. The chromosome 7 encoded only 13 TaC2H2-ZF genes, and 7 out of 13 were distributed on long arms. 85 (41.67%) TaC2H2-ZF genes were encoded in the B sub-genome and 115 (56.37%) TaC2H2-ZF genes were distributed on long arms of the chromosomes, suggesting the members of each subsets were not distributed on each chromosome averagely. TaC2H2-ZF genes were also more identified on chromosomes 3 and 4, with 27 and 33 genes, respectively (Fig. 1b).
Fig. 1.
Chromosome localization of TaC2H2-ZFP members. a The TaC2H2-ZFP members were localized in bread wheat chromosomes (Chr1A, 1B, 1D–7A, 7B, 7D). b Numbers of TaZFP genes on each T. aestivum chromosome
Phylogeny analysis of C2H2-ZF domains
Furthermore, according to the variation of the plant-specific conserved amino acid sequence “QALGGH” and distances between metal ligands, the 457 C2H2-ZF domains of the T. aestivum were also classified into four categories (named Q, M, Z and D), the M and Z were further subdivided into M1, M2, M3, and M4, as well as Z1 and Z2 [2]. According to the classification results of the C2H2-ZF domain, 214 (46.8%) were classified as Ta-tandem-SF and 243 (53.2%) were classified as Ta-isolated-SF. 253 (55%) TaC2H2-ZF genes were different splice variants, while contain the same number C2H2-ZF domains for most TaC2H2-ZF genes, only the length of the amino acid sequence is different (Additional file 1: Tables S1 and S2). However, there are some TaC2H2-ZF splice variants were exceptions, which number and types of their domains have been reduced and changed. For instance, Traes_6DL_F47CFC6D9.1/2/3/4/5/6 have five C2H2 (M3;M4;M4;M4;Z1) domains, four splice variants Traes_6DL_F47CFC6D9.7/8/9/10 have three (M4;M4;Z1), the other three Traes_6DL_F47CFC6D9.11/12/13 only have two (M4;Z1) domains. And the C2H2 domains of Traes_3AS_BE15533CD.1/2 (Z1;D)-C2H2 domains are completely different from its variant Traes_3AS_BE15533CD.3 (M3-C2H2 domain). Undoubtedly, different splice variants of a TaC2H2-ZF gene have different number or type C2H2 domains, may perform different functions [26]. For the detailed information of the classified C2H2-ZFPs in T. aestivum, see Additional file 1: Table S1.
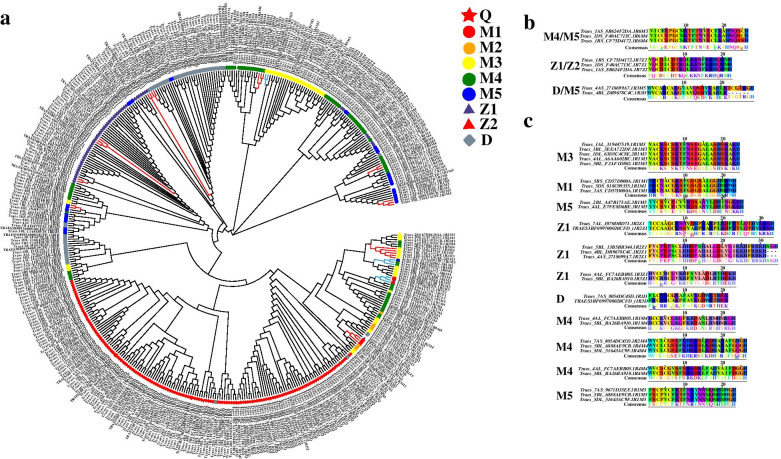
To evaluate the evolutionary relationships of the C2H2 gene family in T. aestivum, an unrooted phylogenetic tree was constructed with the neighbor-joining method based on the domain sequence alignment of 527 conserved ZFP domains (Fig. 2a). Through the analysis of domain sequence alignments, our analysis showed that most members were well separated. The majority of C2H2-ZF proteins in the same clade, particularly the most closely related proteins, typically shared common types (for example, Q-type, M1-type), indicating a similar potential function between these C2H2-ZF proteins. Within each subfamily member in a branch, the strong amino acid sequence conservation is evident from the short branch lengths at the tips of the trees, suggestion of strong evolutionary relationships among subfamily members (Fig. 2c). However, there are clear exceptions, and the different type subfamilies of C2H2-ZF proteins are found in the same clade (Fig. 2b, c). Our results cast a new light on classification of C2H2-ZF domain.
Fig. 2.
Phylogenetic tree showing the spread of C2H2 domain proteins of T. aestivum. The phylogenetic tree was constructed using the neighbor-joining method as implemented in MEGA 7.0 from TaC2H2-ZF protein sequences alignment. Bootstrap values from 1000 replicates are displayed at each node. The different C2H2 domain proteins on the tree can be divided by different colors
Gene structure and conserved motif analysis of C2H2-ZFPs
To further explore the evolutionary relationship among TaC2H2-ZF genes, an unrooted neighbor-joining phylogenetic tree of 204 TaC2H2-ZFPs full-length amino acid sequence was constructed. The gene structures and motif characteristics of TaC2H2-ZFPs were analyzed (Fig. 3a). The results of Gene Structure Display Server (GSDS) analysis showed that the number of introns in the TaC2H2-ZF genes contained from zero to ten introns, most of the genes had one to three introns (104 genes have no introns) (Fig. 3b), and most members contained typical C2H2 domains. The lengths of individual TaC2H2-ZFP were variable in intron length and it could partly reflect the length of different genes. For instance, the longest gene, Traes_5DS_976CBF9EB.1, with a size of 29.1 kb, was due mainly to the fact that it contained a longest intron with length of 21.9 kb.
Fig. 3.

Gene structure and motifs of TaC2H2-ZFP family. a The clustering of TaC2H2-ZF proteins based on Maximun Likelihoodphy phylogenetic tree. b The exon–intron structure of TaC2H2-ZF genes, the yellow boxes are the UTR region of TaC2H2-ZF genes, the green boxes are the exon of CDS, and the black line is the intron. c Schematic depiction of 15 conserved motifs in TaC2H2-ZF proteins. MEME online tool was used to identify the motifs of the TaC2H2-ZF proteins. Each motif is denoted by different colored blocks with respective numbers in the center of the motifs. The number in boxes (1–15) signifies motif 1–motif 15. The length and position of each colored box indicate the actual size of the motif. The full size and high resolution figure is available in Additional file 2
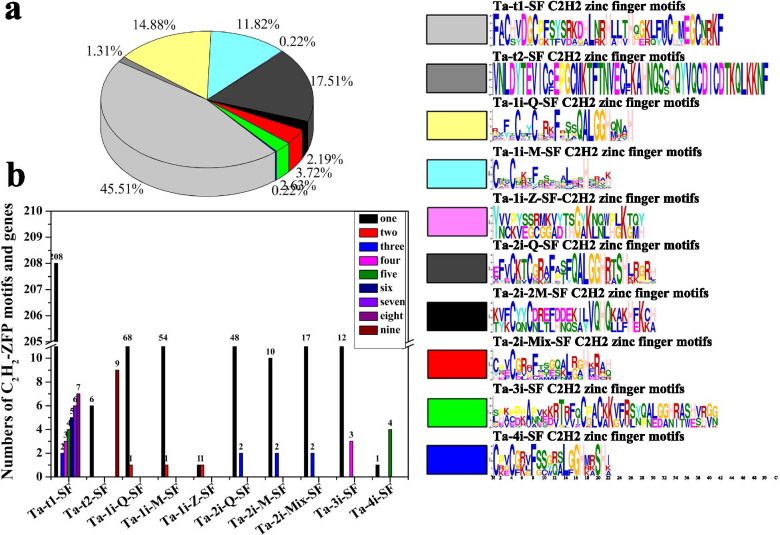
To identify common motifs among different groups of TaZFPs, we employed the MEME Version 5.1.1 [27] software to analyze the conserved motifs in amino acid sequences of 204 TaC2H2-ZFP genes in T. aestivum. Figure 3c shows the symbols of the motifs and the composition of the 204 TaC2H2-ZFP in the C2H2 family. Protein domain analysis showed that most members contained typical C2H2 domains. The C2H2 domains were annotated as the ZFP specific domains, in their N-terminal and C-terminal regions. Fifteen specific motifs were defined, with the ZFPs of subset Ta-t1-SF (Traes_6DL_3B9E63F04.2) containing the largest number of motifs. The TaC2H2-ZFPs belonging to the same subset have a similar motif composition (e.g. Traes_7AL_0BC1C3E70.1 and 7DL_38929DD1E.1, 1AS_21ACC7F67.1 and 6DL_F47CFC6D9.1). Also, some motifs appeared in only certain specific subsets. For example, motif 5 was unique to subset Ta-t1-SF and Ta-1i-M-SF, while L-box was specific to subset Ta-2i-Q-SF and Ta-3i-SF. However, it may be due to the divergence of the C2H2 motif, especially the diverse motif numbers and spacing in the amino acid sequences between Cys and Cys or Cys and His in each protein. Overall, aboved analysis suggested that the C2H2-ZFPs with only one and two Q-type C2H2 domains were most conservative in T. aestivum.
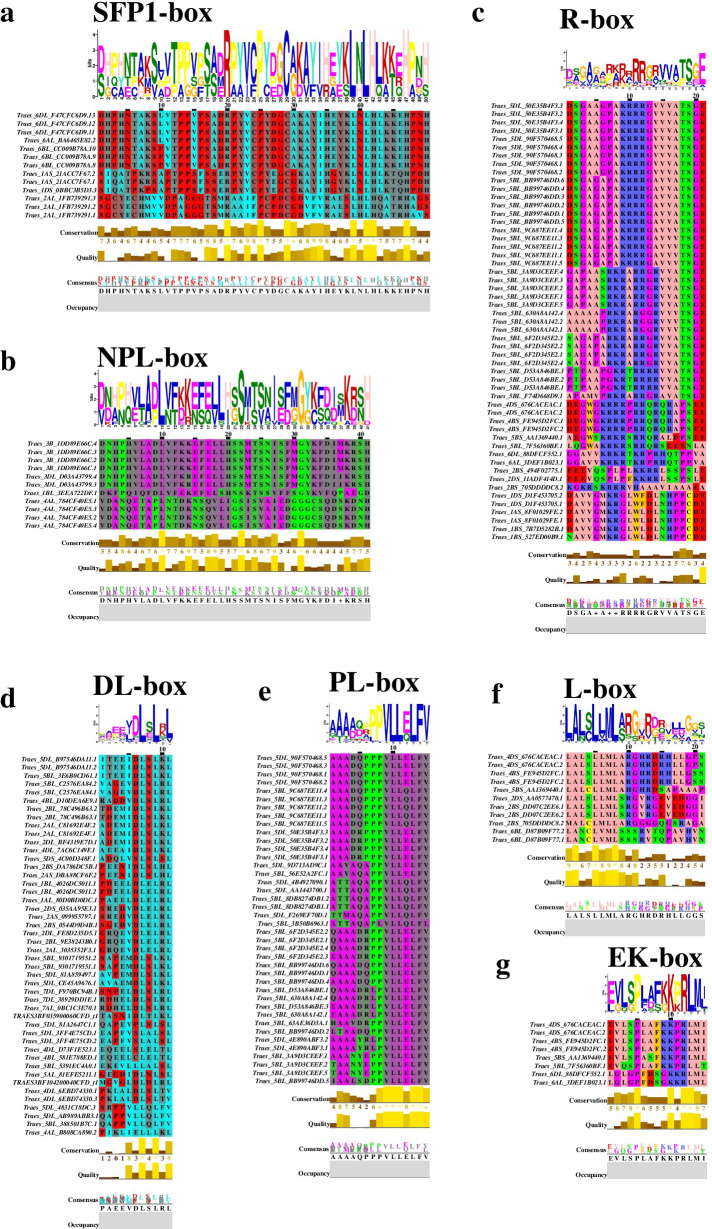
Discovery of conserved motifs specific of the C2H2-type zinc finger domain: SFP1-, DL-, R-, PL-, L-, EK-, and NPL-box
Several other conserved motifs except C2H2 subfamily members were detected by MEME algorithm and multiple sequence alignments (Fig. 4a-g). The C2H2 subset Ta-1i-M-SF is composed of ZF proteins that contain not only the C2H2-ZF domain but also a Nucleoplasmin-like (NPL) domain [28, 29]. Ta-2i-Mix-SF contains Schizosaccharomyces pombe finger protein 1 (SFP1) domain [30, 31]. Ta-1i-Q-SF contains DL-box. Ta-2i-Q-SF contain R-box, PL-box, L-box, and EK-box, respectively [23] (Fig. 4c-g). NPL-box (Ta-1i-M-SF) and L-box (Ta-2i-Q-SF) are located in the N-terminal region of C2H2-ZF domains. DL-box and R-box (Ta-1i-Q-SF) are located in the C-terminal region of C2H2-ZF domains. SFP1-box and R-box (Ta-2i-Q-SF) are located in the C-terminal region of the first C2H2-ZF domains. PL-box (Ta-2i-Q-SF) and EK-box are located in the C-terminal region of the second C2H2-ZF domains. Conservation motif of L-box, NPL-box and SFP1-box in some C2H2-type zinc fingered proteins have been showed in the previous studies [29, 31].
Fig. 4.
The conserved functional motifs and sequence logos of ten subsets proteins were predicted by MEME software. The highlights show the T. aestivum genome conservation of the C2H2 zinc finger motif. Numbers on the x-axis represent the sequence positions in zinc finger motifs. The y-axis represents the information content measured in bits
The DL-box is similar to the previously reported EAR motif (LDLSL) and may make the proteins contain transcriptional repressor functions [32]. Another promising finding was that different splice variants of many genes contained different motifs. For instance, two splice variants of Traes_5DL_50E35B4F3 contain R-box and PL-box, respectively. Traes_4DS_676CACEAC contain R-box, L-box, and EK-box in three splice variants. It indicated that the different subset members maybe contain a novel motif and execute different biological functions.
The C2H2 family genes in T. aestivum were more closely evolutionary relationship with Poaceae
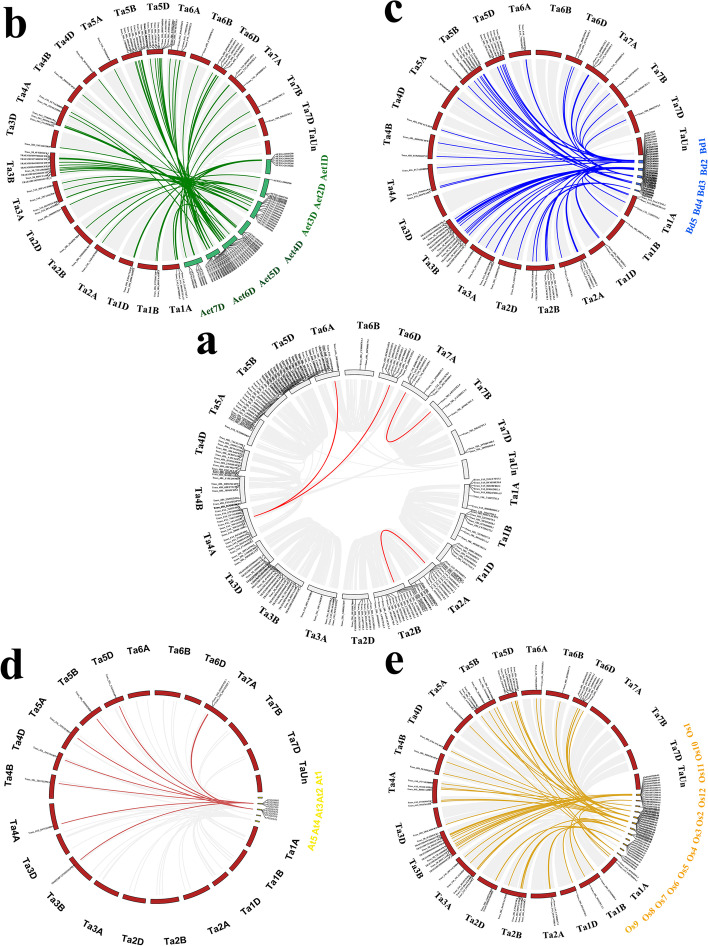
To further infer the phylogenetic mechanisms of the T. aestivum C2H2-ZF gene family and to better understand the origin, we constructed five comparative syntenic maps of T. aestivum associated with four representative species, including three monocots (A. tauschii, B. distachyon, and rice) and one dicot (Arabidopsis) (Fig. 5a-e). Only four pairs of C2H2 family genes in T. aestivum showed syntenic relationships. In four representative species, there was a syntenic relationship between C2H2 family genes and 65 genes in B. distachyon, 66 genes in A. tauschii, 68 genes in rice, 9 genes in Arabidopsis, which indicated that C2H2 family genes in T. aestivum were closely related to genes in the Poaceae family. More importantly, we found that some C2H2-ZF genes were associated with more gene pairs, such as Traes_2DS_2CE71025C.1 of the C2H2 gene family in T. aestivum was the syntenic relationship with KQK10684 and KQK06003. The Os05t0114400-01 is syntenic relationship with Traes_4AL_E7FE8D6BE.3, 7BL_83946C3EC.1 and 1BS_527ED00B9.1. The results indicated that these genes might have similar functions. Furthermore, there was a high degree of syntenic blocks among the T. aestivum, A. tauschii, B. distachyon, and rice as shown in Fig. 5a-e. In contrast, fewer gene pairs were located in syntenic blocks between T. aestivum and Arabidopsis, which may be related to the phylogenetic relationship between T. aestivum and three plant species.
Fig. 5.
The collinearity analysis of the C2H2-ZF gene family in T. aestivum. a Colinearity analysis of TaZFP genes. Duplicated gene pairs in the T. aestivum genome. b colinearity analysis of TaZFP genes with Aegilops tauschii. c Brachypodium distachyon. d Arabidopsis thaliana. e Oryza sativa Japonica (rice). Gray lines in the background indicate the collinear blocks within the T. aestivum and other plant genomes, while the dark lines highlight the syntenic TaZFP gene pairs
Interestingly, 33 TaC2H2-ZF genes in T. aestivum were found to be collinear with ZFP gene in A. tauschii, as well as with ZFP gene in B. distachyon and rice, but not found between T. aestivum and Arabidopsis, such as Traes_6DL_88DFCF552.1/KQK08112 and 4AL_24A54C8BA.2/Os05t0106000-01, which indicated that these orthologous pairs formed after the divergence of dicotyledonous and monocotyledonous plants. Additionally, one TaC2H2-ZF gene in T. aestivum was identified to be collinear with ZFP genes in A. tauschii, B. distachyon, rice, and Arabidopsis, indicating that these orthologous pairs may already exist before the ancestral divergence (Additional file 1: Table S3; Fig. 5a-e).
To better understand the evolutionary constraints acting on this gene family, we calculated the Ka/Ks ratios for the TaC2H2-ZF gene pairs. Generally, Ka/Ks > 1 indicates positive selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks < 1 indicates purification [33]. Additional file 1: Table S3 showed that the all Ka/Ks ratios were less than 1, suggesting that TaC2H2-ZF genes were undergone purifying selection, and it was speculated that their gene functions did not differentiate, which largely maintained the functional similarity of the members of the TaC2H2-ZFP family. The divergence time of duplication events were inferred by Ks (Additional file 1: Table S3). We predicted that the divergence time of C2H2-ZF gene pairs should be approximately 286 Mya (T. aestivum), 20 Mya (A. tauschii), 37 Mya (B. distachyon), 39 Mya (rice), 64 Mya (Arabidopsis) (Additional file 1: Table S3). The result indicated that TaC2H2-ZF gene family shared an intimate correlation with those in A. tauschii, B. distachyon, rice, and Arabidopsis.
Expression profiles of T. aestivum TaC2H2-ZF genes
To explore the expression patterns of T. aestivum C2H2-ZF genes during development and in response to abiotic stresses, we used publicly available RNA-seq data and mapped to the bread wheat genome by Borrill [34]. These samples included diverse developmental stages, tissues, and abiotic stress conditions. The RNA-seq data corresponded to 5 organs at 15 developmental stages [35].
To detect preferentially expressed TaC2H2-ZF genes in certain T. aestivum tissues and at certain stages, we found that totally 198 of 204 TaC2H2-ZF genes were expressed in all organs and development stages, and 37 of 204 TaC2H2-ZF genes were responded to heat, drought, and heat & drought abiotic stresses according to public transcriptome data.
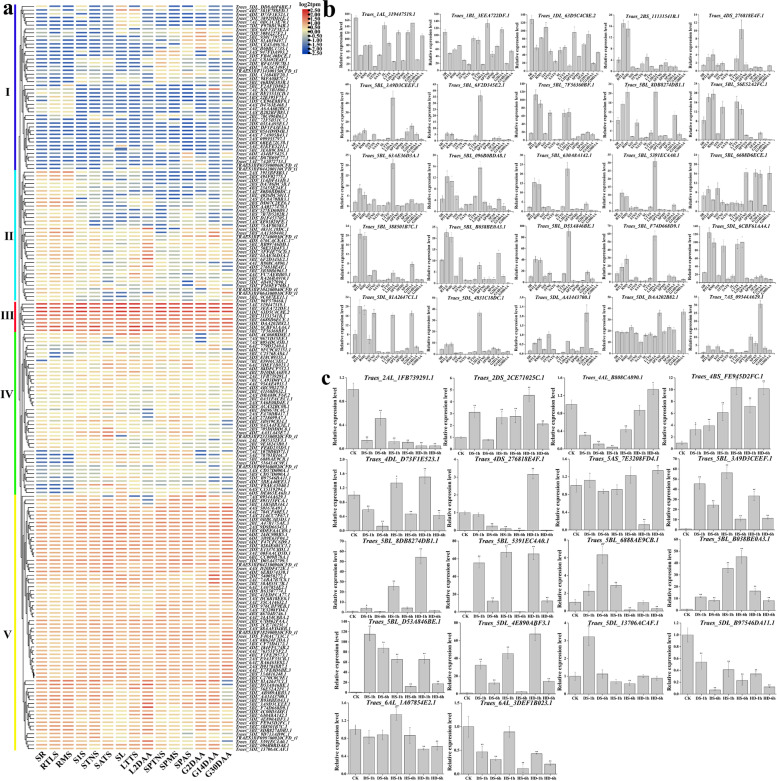
A heatmap showed the relative expression profile of 198 TaC2H2-ZF genes in 5 different organs and 15 different development stages (Additional file 1: Table S5; Fig. 6a). The 204 TaC2H2-ZF genes were clustered into five groups. The 44 genes (22.2%) of group I were lowly transcript accumulated in most organs and development stages. The 35 genes (17.7%) of group II were all preferentially moderately expression in the root. Among 35 genes, the 9 genes (25.7%) were moderately expression in stem, leaf, spike and grain, and 26 genes (74.3%) were lowly transcript accumulation in stem, leaf, spike and grain. The 8 genes (4.1%) of group III were highly transcript accumulation in all organs and development stages. The 43 genes (21.7%) of group IV were moderately expressed in most organs and development stages, but in which 12 genes (27.9%) were lowly transcript accumulation in root. The 74 genes (37.4%) of group V were moderately and consecutively expressed in all organs and development stages (Fig. 6a). We found that the Ta-t1-SF members were mainly distributed into the expression group V (44.6%), while the Ta-1i-Q-SF into group I (54.5%), the Ta-2i-Q-SF into group II (60%), the Ta-1i-M-SF into group III (50%) and IV (27.9%).
Fig. 6.
Transcriptome analysis of T. aestivum C2H2-ZF genes. a Expression profiles of 198 C2H2-ZF genes in different T. aestivum tissues. Hierarchical clustering of the relative transcript abundance profiles (log2 tpm) of genes. Different colors degrees represent different expression level. The scale bar indicates relative expression level as shown on the top right side. The different tissue types are shown on the bottom side: seedling roots (SR), roots at the three leaves (RTLS), roots at the meiosis (RMS), stems at the 1 cm spike (S1S), stems at the two nodes stems (STNS), stems at the anthesis stems (SATS), seedling leafs (SL), leafs at the three tillers (LTTS), leafs at the 2 days after anthesis (L2DAAs), spikes at the two nodes stems (SPTNS), spikes at the meiosis (SPMS), spikes at the anthesis stems (SPAS), grains at the 2 days after anthesis (G2DAAs), grains at the 14 days after anthesis (G14DAAs), grains at the 30 days after anthesis (G30DAAs). The individual gene names are indicated on the right side. b Expression analysis of 25 TaC2H2-ZF genes in different tissues and developmental stages by qRT-PCR, vertical bars indicate standard deviation. c Expression analysis of 18 TaC2H2-ZF genes in response to drought and heat stress by qRT-PCR, and vertical bars indicate standard deviation. The full size and high resolution figure is available in Additional file 3
To verify the expression profiles of TaC2H2-ZF genes obtained by the RNA-seq data analysis, qRT-PCR was performed for 43 selected genes (25 TaC2H2-ZF genes that showed a large change in different tissues expression, 18 TaC2H2-ZF genes is very significantly in response to drought and heat stresses). The gene-specific primers are listed in Additional file 1: Table S4. As shown in Fig. 6b and c, among the 25 genes, 3 genes showed the highest transcript accumulation in all development stages. 4 genes in the root (seedling, three leaves, and meiosis), stem (Spike at 1 cm and two nodes), leaf (seedling, Three tillers), grain (2 DAAs and 30 DAAs). The one genes showed moderately transcript accumulation in the root (three leaves and meiosis), stem (two nodes), leaf (Three tillers and 2 DAAs). The one gene showed moderately transcript accumulation in the root (three leaves and meiosis), leaf (2 DAAs). The 15 genes showed moderately transcript accumulation in the root (three leaves and meiosis), leaf (2 DAAs), spike (two nodes, meiosis, and anthesis), grain (2 DAAs and 30 DAAs), respectively. These results were almost consistent with those observed in the RNA-seq data analysis, while the levels of upregulation were slightly different (Fig. 6a, b). The expression of Traes_5DL_AA1443700.1 has almost no change at all stages, which is slightly different from the results of the above-mentioned RNA-seq data analysis (Fig. 6b).
Subsequently, the expression profiles of the 18 TaC2H2-ZF genes were further analyzed under drought and heat stresses, and most of which were well following microarray profiles. The qRT-PCR results were highly consistent with the RNA-seq data. Among the 18 TaZFPs, Traes_2AL_1FB739291.1 and 5DL_B97546DA11.1 were observed to be increased in response to drought and heat stress. The 4DS_276818E4F.1 was upregulated significantly at HD-1 h. The 4AL_B808CA890.1 was suppressed after DS-1 h, DS-6 h and HS-1 h treatments, whereas it was upregulated after HS-6 h, HD-1 h, and HD-6 h treatment (Fig. 6c). Notably, Traes_5BL_B038BE0A3.1, 5BL_3A9D3CEEF.1 and 5BL_D53A846BE.1 displayed significantly high expression levels at all treatment time points. The 5BL_5391EC4A0.1 and 5DL_4E890ABF3.1 displayed significantly high expression levels at DS-1 h, DS-6 h, HS-1 h, HD-1 h and HD-6 h treatment time points.
In summary, our results showed that these selected TaC2H2-ZF genes responded to one or more stresses. According to expression pattern analysis found, like Traes_5BL_D53A846BE.1, not only expressed highly in L2DAAs, RTLS, RMS, but also endowed tolerance to drought and heat stresses, making them good candidates for molecular breeding.
Discussion
T. aestivum C2H2-ZF proteins have a relatively conservative evolutionary process
C2H2-type zinc finger proteins belongs to one of the largest plant TF families and participates in multiple biological processes. Therefore, T. aestivum C2H2-ZF genes may be promising targets for crop breeding and improvement.
The 457 putative C2H2-type Zinc Finger proteins (C2H2-ZFP) (including 253 splice variants) were identified from T. aestivum. There are 179, 122, 189, 321 (including 135 pairs and 22 single genes), and 124 C2H2-ZFP members in Arabidopsis, durum wheat, rice, soybean, foxtail millet, respectively [1, 2] (Fig. 7a, b). Compared with the number of above species C2H2-ZFPs family members, T. aestivum have one main features in most of the plant C2H2-ZFPs due to its hexaploidy genome probably. in this study, T. aestivum C2H2-ZFPs were classified into four great categories (named Q, M, Z, and D) according to the variation of the conserved amino acid sequence “QALGGH” and distances between metal ligands, and 457 C2H2-ZFPs were further subdivided into ten different subsets, based on the arrangements, numbers, and types of C2H2-ZF domains [1] (Additional file 1: Table S1; Figs. 2a and 7a-b). This result was well identified by phylogenetic analysis (Fig. 2a). Previous studied have demonstrated that the molecular functions of the C2H2-ZFPs with tandem C2H2-ZF domain are generally different from the C2H2-ZFPs with dispersed fingers [36–38]. So, in this study, the TaC2H2-ZFPs with single or dispersed C2H2-ZF domains are generally classified separately from tandem C2H2-ZFP domain proteins. The TaC2H2-ZFPs were classified into two Ta-tandem-SF subsets and eight Ta-isolated-SF subsets (Additional file 1: Table S1; Fig. 7a, b). According on the statistical analysis results of linker lengths in T. aestivum C2H2-ZFPs, Ta-tandem-SF is different from the tandem ZFs in yeast and Arabidopsis (Additional file 1: Table S1) [36, 39]. Two Ta-tandem-SF subsets and eight Ta-isolated-SF were defined based on the numbers of C2H2-ZF domains according to the tandem-SF and isolated-SF in soybean and Brassica rapa L C2H2-ZFPs [2, 3]. In addition, two Ta-tandem-SF had no Q-type C2H2-ZF domain in these C2H2-ZFPs, which was similar to the yeast and animals [36]. Furthermore, the Q-type and M-type C2H2-ZF domains were similar, Z-type and D-type were completely different from the types (Additional file 1: Table S1), as these were defined in previous studies [2, 3, 40, 41].
Fig. 7.
Subset classification of C2H2-ZF genes in the T. aestivum genome. a Numbers of C2H2-ZFPs in the 10 different subsets. b Numbers of C2H2-ZFPs containing 1-7 and 9 C2H2-ZF domains
We also examine the C2H2-ZF domains type contained in the 11 subsets, and found that the Ta-1i-Q-SF, Ta-2i-Q-SF, Ta-2i-Mix-SF, Ta-3i-SF and Ta-4i-SF subsets of T. aestivum C2H2-ZFPs members contain one or two the QALGGH motif, and were identified as Q-type C2H2-ZFPs. (Additional file 1: Table S1; Fig. 7) [42]. However, the Q-type C2H2-ZFPs is considered as plant specific [43], animal and yeast lack this motif, which suggesting that Q-type C2H2-ZFPs might have evolved for as specific regulatory process unique in plants. And in the current study, the D, M1, M2, M3, M4, and M5 domains are generally generated is due to the simultaneous mutation of any first five amino acids in the conserved sequence QALGGH, respectively, leading to the different protein functions of TaC2H2-ZFPs [2, 3]. The complexity of these C2H2-ZF domains as well as the presence of other known domains in some C2H2-ZFPs indicates the functional diversity of these C2H2-ZFPs in plant growth and development. Our classifications of T. aestivum C2H2-ZFPs provided helpful information for further functional characterization of C2H2-ZFPs gene family among Poaceae.
In the current study, our analysis found that C2H2-ZFPs family genes in T. aestivum were closely related to genes in the Poaceae family. Most TaC2H2-ZF genes have orthologous genes in A. tauschii, B. distachyon, rice, and Arabidopsis (Fig. 4b-e, Additional file 1: Table S3), there are 66, 65, 68, and 9 C2H2-ZF gene pairs between T. aestivum and A. tauschii, B. distachyon, rice, and Arabidopsis, respectively. And they are 7.2 times, 7.4 times, and 7.6 times of number of the gene pairs between T. aestivum and Arabidopsis, respectively. From the results of gene replication, 33 TaC2H2-ZF gene pairs were duplicated genes and the high levels of collinear gene pairs were observed among A. tauschi, B. distachyon, and rice. The number of segmental duplication events was greater than that of tandem duplication events, indicating that segmental duplication contributed to TaC2H2-ZF genes expansion.
The analysis of syntenic and Ka/Ks values indicated that no positive selection occurred and underwent strong purifying selection in TaC2H2-ZF genes, and the results were consistent with the studies of C2H2-ZF genes in other plants [2, 3]. Overall, we inferred that the evolution of the TaC2H2-ZF gene family was similar to that in other plants. Compared with Arabidopsis, T. aestivum C2H2-ZF genes shared a strong relationship evolutionary with A. tauschi, B. distachyon, and rice C2H2-ZF genes.
Subfamily specific motifs (SFP1-, DL-, R-, PL-, L-, EK-, and NPL-box) of T. aestivum C2H2-ZF genes may contribute to the high adaptability of T. aestivum
Subsequently, sequence logo analysis to explore the sequence characteristics of the C2H2 domains from the ten major subsets were studied by the MEME motif search tool. The TaC2H2-ZF domains not only contain the C2H2 domain but also contain NPL, SFP1, DL-box, R-box, PL-box, L-box, and EK-box [29–32] (Fig. 4a-g). These motifs were previously reported in rice and Arabidopsis, [22].
The L-box have been shown in some 2-fingered Q-type ZFP proteins, such as AZF2 (Arabidopsis), ZFP15 (rice) and ZFP36 (rice) [19, 44, 45]. In the present study, all TaC2H2-ZFPs containing L-box also contained 2-fingered Q-type C2H2-ZF domain and belonged to Ta-2i-Q-SF subsets, suggesting L-box has a certain function with two Q-type ZF domain. For instance, Traes_6BL_D87B09F77.1 was homologous to rice ZFP15, both containing 2 Q-type C2H2-ZF domains and a L-box each. Traes_6BL_D87B09F77.1 expressed highest in the SPAS stages than other developmental stages. Meanwhile, transgenic rice ZFP15 showed it was important for pike development [45]. All these results suggested Traes_6BL_D87B09F77.1 has a certain role in spike development.
The DL-box is similar to the previously reported LDLNL-EAR motif suggesting it may function as transcriptional repressors [32]. Previous study showed that the EAR motif could reduce the transcriptional level of the reporter gene, and the transcriptional activation activity of some TFs, as well as negatively regulated genes involved in developmental, hormonal, and stress signaling pathways [46, 47]. For instance, overexpression of intact DREB/ERF proteins with an EAR motif led to the reduced expression levels of stress-related genes and decreased abiotic stress tolerance [23, 48, 49]. Furthermore, when EAR motif was tethered to transcriptional activators, the EAR motif functioned as dominant repressors [50]. In this study, we found that Traes_4AL_B808CA890.1, Traes_4DL_D73F1E523.1, and Traes_5DL_B97546DA11.1, all belonged to Ta-1i-Q-SF,contain LDLNL-EAR-motif-like domain. And through existing transcriptome data and real-time quantitative data analysis, it was found that the transcriptional levels of these three genes were decreased under drought and heat stress conditions. If a mutation occurs in this DLDLNL inhibitory domain, the protein's inhibitory function is abolished, For example, the EAR domain of TaRAP2.1L is responsible for a negative effect on growth and stress tolerance, while it became an activator after the EAR-motif was mutated to xAAAxxA from xDLNxxP [23, 51]. In briefly, both negative and/or positive regulators of gene expression containing the EAR motif domain play key roles in the regulation of plant stress responses.
The above analysis showed these TaC2H2-ZFPs had the high functional diversity in plant growth and development. This study cast a new light on multifunction of TaC2H2-ZF genes, and provided a certain reference value for people to understand the functions of the C2H2-ZF genes in T. aestivum. It is also worth discussing these significant facts revealed by the results of the above analysis.
Expression profiles revealed that TaZFPs comprehensively involved in development and abiotic stress responses
The RNA-seq analysis showed that 198 of 204 T. aestivum C2H2-ZF genes were expressed in 5 organs at 15 developmental stages. 134 (67.7%) of the TaC2H2-ZF genes were moderately and consecutively expressed in root and leaf (Fig. 6a). In plants, roots and leaves are the most important organs necessary for water and nutrient absorption, also crucial for water content regulation [52]. T. aestivum is widespread over the dryland world, and is grown in low rainfall condition, so drought/heat stress response are important for T. aestivum [48, 49]. Since dominant TaC2H2-ZFPs expressed in root and leaf, TaC2H2-ZFP transcription factors will is a major target for T. aestivum study breeding against drought stress mechanism. The study also indicated that T. aestivum C2H2-ZFPs played roles in root and leaf development more than other tissues or organs. There is considerable evidence that C2H2-ZFP transcription factors are involved widely in the development of multiple organs and tissues and abiotic stresses, such as seed maturation, floral development, spike development [1], salt stress tolerance [15, 53], drought stress tolerance [54].
In this study, we examined the expression patterns of 43 selected genes. The TaC2H2-ZFP genes of chromosome 5 account for 67.4% (Fig. 6a). Previous studies have shown that some C2H2-ZF genes located in chromosome 5 play important roles in the T. aestivum growth development stage and abiotic stress processes, such as WZF1, TaZFP15, TaZFP34 [55–57]. And most results were broadly consistent with the microarray profiles by real-time quantitative RT-PCR verification.
The Traes_5DL_6CBF61AA4.1 and 5BL_7F56360BF.1 were homologous to Arabidopsis AtZAT10 (AT1G27730.1, also called STZ). The three genes belong to the same subsets and all of them contain not only the Q-type C2H2-ZF domain but also a FDLNI (EAR-like motif) and an EK-box. Traes_5DL_6CBF61AA4.1 and 5BL_7F56360BF.1 were highly expressed in the SR, RTLS, RMS, S1S, STNS, LTTS and L2DAAs stages. Moreover, transgenic Arabidopsis overexpressing AtZAT10 showed growth retardation as demonstrated in a previous study [7, 22], suggesting that the functions of three homologous genes were probably conserved in both species.
Similarly, the Traes_4BS_FE945D2FC.1, homologous to Arabidopsis AT3G19580.2 (AZF2), contains 2 Q-type C2H2-ZF domains, a FDLNI (EAR-like-motif), an EK-box, and a L-box. Previous analysis showed that expression of AZF2 was strongly induced by drought stresses [7, 22]. The Traes_4BS_FE945D2FC.1 was significantly up-regulated at 6 h (up to tenfold) under heat & drought stresses, suggesting it had a similar function in drought stresses with AZF2. fThe Traes_5BL_D53A846BE.1 highly homologous to Arabidopsis AT2G37430.1 (ZAT11) and AT5G59820 (ZAT12), contains 2 Q-type C2H2-ZF domains. The Traes_5BL_D53A846BE.1, not only highly expressed in the RTLS, RMS and L2DAAs stages, but also responded to drought and heat stresses, making them good candidates for molecular breeding. While the ZAT11 is highly expressed in roots and particularly in root tips [58]. The ZAT12 can reduce heat-derived oxidative stress by activating multiple defense genes in tomato [59]. These results indicated that all the tested genes were more or less involved in development stages and stress response in T. aestivum, in a similar way as their orthologs in Arabidopsis. Further tissue expression and stress responsive expression analysis of a larger number of T. aestivum C2H2-ZF genes, including those lowly expressed genes, would allow to identify the most actives members in gramineae crops.
The above results indicated that these C2H2-ZF genes might be involved in the important regulatory networks or cross-talk triggered by different responses stresses. Overall, these findings provide insight into the potential functional roles of T. aestivum C2H2-ZF genes. The results provide a basis for further functional study of C2H2-ZF genes.
Conclusions
In the present study, we systematically analyzed the characterization of the TaZFP family. The 457 C2H2-ZF genes in the T. aestivum genome were identified (including 253 splice variants), according to defined C2H2-ZF domain types, and C2H2-ZFPs were classified into 10 distinct subsets. A total of 204 non-redundant candidate genes were obtained to encode 457 proteins. The 85 genes (41.67%) were encoded on the B sub-genome and 115 genes (56.37%) were distributed on long arms of the chromosomes. Synteny analysis showed that only 4 pairs of C2H2 family genes in T. aestivum, 65 genes in B. distachyon, 66 genes in A. tauschii, 68 genes in rice, 9 genes in Arabidopsis, were syntenic relationships respectively, indicating that TaZFPs were closely related to genes in the Poaceae. The specific conserved motifs of the C2H2-type zinc finger domain including SFP1-, DL- (EAR-like-motif), R-, PL-, L-, EK- and NPL-box were discovered, indicating the different subset members maybe contain a novel motif and execute different biological functions. According to public transcriptome data, totally 198 of 204 TaC2H2-ZF genes were found to have the expression data. The 25 TaC2H2-ZF genes were certificated to be significantly differentially expressed in 5 different organs and 15 different development stages by quantitative RT-PCR. The 18 TaC2H2-ZF genes were verified in response to heat, drought, and heat & drought stresses. Such as Traes_5BL_D53A846BE.1, it was not only highly expressed in L2DAAs, RTLS and RMS, but also endowed tolerance to drought and heat stresses, making them good candidates for molecular breeding. In summary, the present study provides the valuable reference information for further study of C2H2-ZF genes, and provides promising targets for further genetic engineering and genetic improvement in T. Aestivum.
Methods
Plant materials and abiotic stress treatments
Bread wheat (Triticum aestivum L. cv. Fielder) materials were acquired from Prof. Xue Gangping's lab in CSIRO Plant Industry. Seeds were germinated on wet paper towel and cultivated at 4℃ for 5 days, then placed at 12℃ for 5 day for germination. Germinated seedlings were grown at 22℃ in a greenhouse. Two-week-old wheat seedlings were treated by different stresses (heat, drought, and heat & drought). The abiotic stress treatments were performed by submerging T. aestivum seedling roots in solutions of 37℃ incubator, 25% PEG6000, respectively. T. aestivum seedlings treated with water were used as a mock control. The whole T. aestivum seedlings were sampled at different time points (0, 1, 6 h). Greenhouse-grown T. aestivum plants were used for measuring tissues specific expression patterns of 25 selected TaC2H2-ZF genes. Seedling roots (SR), roots at the three leaves (RTLS), roots at the meiosis (RMS), stems at the 1 cm spike (S1S), stems at the two nodes stems (STNS), stems at the anthesis stems (SATS), seedling leaves (SL), leaves at the three tillers (LTTS), leaves at the 2 days after anthesis (L2DAAs), spikes at the two nodes stems (SPTNS), spikes at the meiosis (SPMS), spikes at the anthesis stems (SPAS), grains at the 2 days after anthesis (G2DAAs), grains at the 14 days after anthesis (G14DAAs), grains at the 30 days after anthesis (G30DAAs) were collected. All collected tissue samples were immediately frozen in liquid nitrogen and stored at -80℃ for future analysis.
Data sets
The annotated genome sequences of A. tauschii(Aegilops tauschii), B. distachyon (Brachypodium distachyon), rice (Oryza sativa Japonica) and Arabidopsis(Arabidopsis thaliana) were downloaded from plantTFDBv5.0, (http://planttfdb.cbi.pku.edu.cn/phylo_tree.php?sp=Tae&fam=C2H2) [60], iTAK software v1.6 (http://itak.feilab.net/cgi-bin/itak/index.cgi) [61], Ensembl plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index). TAIR (Release 10, ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release) and JCVI, (Release 6.1, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1), respectively. The partially annotated genome sequences of T. aestivum and Oryza sativa were downloaded from Prosite (http://au.expasy.org/prosite/) and NCBI (https://www.ncbi.nlm.nih.gov/).
Identification and characterization of T. aestivum C2H2-ZFPs
For identification of T. aestivum C2H2-ZFPs family members, the information of T. aestivum C2H2-ZFPs sequences was downloaded from the plantTFDBv5.0, iTAK database, and the Ensembl plants. The C2H2-ZF domain was also queried for Hidden Markov Model (HMM) search through the HMMER 3.0 program. The TaC2H2-ZF domain was further determined using the SMART database (http://smart.embl-heidelberg.de/) [62].
The theoretical isoelectric point (pI), molecular weight (MW), atomic composition, instability index, aliphatic index, and grand average of hydropathicity (GRAVY) were downloaded from the ExPASy server (http://www.expasy.org/) [63]. The subcellular localization of each TaC2H2-ZF protein was predicted using the Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) webserver [64].
Classification and chromosomal location of T. aestivum C2H2-ZFPs
The identified 457 TaC2H2-ZFPs were further manually analyzed to search the numbers of C2H2-ZF domain, C2H2-ZF domain sequences, and the space length between C2H2-ZFPs using the SMART database (http://smart.embl-heidelberg.de/). We defined the C2H2 type for each identified T. aestivum C2H2 domain, according to the plant-specific amino acid residues and distances between two to nine C2H2-ZF domains, as have been previously adopted in soybean [3]. Tandem ZF was determined as ZF containing two to thirteen C2H2-ZF domains, and every two nearby domains are connected by < 12 amino acid residues, such as Tandem ZFs with two subsets assigned as Ta-t1-SF and Ta-t2-SF, respectively. ZF contains one ZF and/or two to four ZF domains each more than 11 amino acid residues between two nearby domains is considered to be scattered ZF. Also, different types of C2H2-ZFPs have been classified based on the variation of the plant-specific conserved domain “QALGGH” sequence and distances between metal ligands [1, 4, 54]. Generally, C2H2-ZFP were defined as four types: Q-type, M-type, Z-type, and D-type. C2H2-ZFP Q-type domains were defined as X2-C-X2-C-X7-QALGGH-X3-H. Those that have 1–5 amino acid degradations in the "QALGGH" domain and that there are certain modifications in the spacing between two cysteine and two histidines of Q type are defined as M-type. Those with more than 12 (Z1) and less than 12 (Z2) in their spacing between the second cysteine and the first histidine were defined as Z-type domains. However, compared with Q-type, M-type, and Z-type, the D-type does not include the second histidine in the C2H2-ZF domain. According to these defined C2H2-ZFP types, C2H2-ZFPs containing a single ZF domain were further classified into four clearly distinguishable subsets (Ta-1i-Q-SF, Ta-1i-M-SF, Ta-1i-Z-SF, and Ta-1i-D-SF). C2H2-ZFPs containing two Q-type or two M-type C2H2-ZF domains were defined as Ta-2i-Q-SF and Ta-2i-M-SF, respectively, while the C2H2-ZFPs containing two different types of C2H2-ZF domains were classified as Ta-2i-Mix-SF. All ZFPs containing three or four dispersed ZFs were classified into subsets of Ta-3i-SF or Ta-4i-SF, respectively [2, 3].
The TaC2H2-ZF genes approximate chromosomal location was determined by blastn search of cDNA sequences against chromosome sequences of T. aestivum available at Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast). The IWGSC bread wheat genome was assembled using the following parameters: E value ≤ 1e-10 and identity 85% [65].
The chromosomal location image of the TaC2H2-ZF genes was generated by TBtools software, according to the chromosomal position information provided in the EnsemblPlants database.
Analyses of phylogeny, gene structure and conserved motif
For the phylogenetic investigation, multiple alignments were performed with the domain sequences of the TaZFP family proteins by the neighbor-joining (NJ) method available via MEGA7 software [66] to construct an unrooted phylogenetic tree. Sequences were aligned using the MUSCLE alignment algorithm [66]. The tree obtained was the consensus of 1000 single trees provided by bootstraps, gaps/missing data treatment: partial deletion, model/method: LG model, rates among sites: gamma distributed with invariant sites (G + I).
For the TaC2H2-ZF gene exon–intron structure, we used the Gene Structure Display Server (GSDS 2.0) (http://gsds.cbi.pku.edu.cn) [67]. We analyzed the additional motifs outside the C2H2 domain of 204 C2H2-ZFP amino acid sequences using the Multiple Em for Motif Elicitation (MEME) suite Version 5.1.1 online program (http://meme-suite.org/tools/meme). The optimized parameters of MEME were employed as the following: maximum number of motifs, 10; minimum motif width, 6; and maximum motif width, 50.
Chromosomal distribution and synteny analysis of TaC2H2-ZF genes
The TaC2H2-ZF genes were mapped to the chromosome according to the chromosomal location given from the Ensemblplants database by using CIRCA (OMGenomics, http://omgenomics.com/circa/). Investigating the gene duplication events of having the Default parameters using the Multiple Collinearity Scan toolkit (MCScanX) [68].
To exhibit the syntenic relationship of the orthologous C2H2-ZF genes obtained from T. aestivum and A. tauschii, B. distachyon, Arabidopsis, O. Japonica the syntenic analysis maps were constructed using the Dual Systeny Plotter software (https://github.com/CJ-Chen/TBtools) [68]. Synonymous (Ks) and non-synonymous (Ka) substitution rates were calculated for each duplicated C2H2-ZF genes using a maximum likelihood model averaging in Ka/Ks Calculator [68]. For each gene pair, the divergence time of collinear gene pairs was calculated using the mean Ks values from T = Ks/(2λ × 10–6) Mya (λ = 6.5 × 10–9) [69].
Expression analysis of C2H2-ZF genes in T. aestivum
The expression profiles of 198 TaC2H2-ZF genes in different tissues (roots, stem, leaves, spikes, and grains) and different development stages were determined based on the RNA seq data available from Wheat Expression Browser powered by expVIP (http://www.wheat-expression.com/) [70]. Briefly, the raw data for RNA-Seq were downloaded, and subsequent data processing was performed to remove weakly expressed genes using the R software package. Finally, the gene expression heatmap was generated by using the TBtools view software.
RNA extraction and qRT-PCR analyses
Total RNA from different tissues (root, stem, leave, spike, grain) and various stress treated materials were extracted using Trizol reagent (Invitrogen) following the manufacturer’s instructions. Subsequently, RNA integrity and quality were checked by electrophoresis of RNA on 1.2% denaturing agarose gel. For qPCR analysis, first-strand cDNA synthesis was performed with the SuperScript II Reverse Transcriptase (Invitrogen) by the manufacturer's instructions. 25 tissue-specific TaC2H2-ZF genes expression patterns and 18 TaC2H2-ZF genes significantly in response to heat and drought stresses expression patterns were determined by qRT-PCR analysis. Gene-specific primer pairs in Additional file 1: Table S4. Accuracy and specificity of primers were checked by the Blast algorithm and their products were electrophoretically checked on 1.5% agarose gel for amplification accuracy. TaRP15 (Additional file 1: Table S4) was used as the internal reference gene when examining gene expression of 25 selected TaC2H2-ZF genes. the qRT-PCR analysis was performed to detect relative mRNA expression levels. During the qRT-PCR analysis, each qRT-PCR analysis was repeated three times, and data analyzed by analysis software based on the comparative 2−ΔΔCT method of relative gene quantification [3]. Relative expression of 25 selected TaC2H2-ZF genes and probability value (P) were also calculated using the qRT-PCR analysis software.
Supplementary Information
Additional file 1: Table S1. Classification of the 457 Triticum aestivum C2H2-ZFPs according to the organization of their contained C2H2 fingers. Table S2. List of 204 Triticum aestivum C2H2-ZFP genes and their related information. Table S3. One-to-one orthologous relationships between T.aestivum and T.aestivum, T.aestivum and A.tauschii, T.aestivum and B.distachyon, T.aestivum and Rice, T.aestivum and Arabidopsis. Table S4. Primers used for qRT-PCR. Table S5. Expression profiles of T. aestivum C2H2-ZFP genes.
Additional file 2: The full size and high resolution of Figure 3.
Additional file 3: The full size and high resolution of Figure 6.
Acknowledgements
The authors are grateful Dr. Shuai Hu, School of life sciences, Tsinghua University for critical review of this manuscript.
Abbreviations
- C2H2-ZFPs
C2H2-type zinc finger proteins
- T. Aestivum
Triticum aestivum
- A. tauschii
Aegilops tauschii
- B. distachyon
Brachypodium distachyon
- rice
Oryza sativa Japonica
- Arabidopsis
Arabidopsis thaliana
- CDS
Coding sequence
- pI
Isoelectric point
- MW
Molecular weight
- GRAVY
Grand average of hydropathicity
- NJ
Neighbor-joining
- GSDS
Gene Structure Display Server
- MEME
Multiple Em for Motif Elicitation
- MCScanX
Multiple Collinearity Scan toolkit
- SR
Seedling roots
- RTLS
Roots at the three leaves
- RMS
Roots at the meiosisstems
- S1S
At the 1 cm spike stems
- STNS
At the two nodes stems
- SATS
Stems at the anthesis stems
- SL
Seedling leafs
- LTTS
Leafs at the three tillers
- L2DAAs
Leafs at the 2 days after anthesis
- SPTNS
Spikes at the two nodes stems
- SPMS
Spikes at the meiosis
- SPAS
Spikes at the anthesis stems
- G2DAAs
Grains at the 2 days after anthesis
- G14DAAs
Grains at the 14 days after anthesis
- G30DAAs
Grains at the 30 days after anthesis
- qRT-PCR
Quantitative real-time PCR
- TFs
Transcription factors
Authors’ contributions
X.G. and M.Z. designed the experiments. Y.L. and X.G. wrote the main manuscript text. Y.L. and A.S. conducted the experiments. A.S., Q.W., X.Z., F.C., R.C. and H.X. collected and analysed phenotype data. Y.L., M.Z. and A.S. prepared Figures 1–7. All authors read and approved the manuscript.
Funding
This research was supported by grants from National Natural Science Foundation of China (31872866), Key Research & Development Project of Hunan Provincial Department of Science and Technology (2019NK2081), and National Key Research and Development Program of China (2017YFF0210301).
Availability of data and materials
The datasets analysed during the current study are available in the bread wheat Genome Project (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast), plantTFDBv5.0 (http://planttfdb.cbi.pku.edu.cn/phylo_tree.phpsp=Tae&fam=C2H2), iTAK software v1.6 (http://itak.feilab.net/cgi-bin/itak/index.cgi), Ensembl plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index). TAIR (Release 10, ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release) and JCVI (Release 6.1, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1) repository.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yongliang Li and Aolong Sun contributed equally to this work.
Contributor Information
Meng Zhang, Email: zhangmeng2019@hnu.edu.cn.
Xinhong Guo, Email: gxh@hnu.edu.cn.
References
- 1.Ming N, Ma N, Jiao B, et al. Genome wide identification of C2H2-Type Zinc finger proteins of tomato and expression analysis under different abiotic stresses. Plant Mol Biol Report. 2020;38(1):75–94. doi: 10.1007/s11105-019-01182-1. [DOI] [Google Scholar]
- 2.Intikhab Alam, Khadija, et al. Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L. PLoS One. 2019;14(5):e0216071. doi: 10.1371/journal.pone.0216071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan S, Li X, Li R, Wang L, Zhang C, Chen L, Hao Q, Zhang X, Chen H, Shan Z, Yang Z, Chen S, Qiu D, Ke D, Zhou X. Genome-wide identification and classification of soybean C2H2 Zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front Microbiol. 2018;9:126. doi: 10.3389/fmicb.2018.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D, He S, Song X, Zhai H, Liu N, Zhang D, et al. IbSIMT1, a novel salt-induced methyl transferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell Tissue Organ Cult. 2015;120:701–715. doi: 10.1007/s11240-014-0638-6. [DOI] [Google Scholar]
- 5.Cheuk A, Houde M. Genome wide identification of C1–2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol Genet Genomics. 2016;291(2):873–890. doi: 10.1007/s00438-015-1152-1. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Cao J, Gao C, Gao W, Yan S, Yao H, Xu K, Liu X, Xu D, Pan X, Lu J, Chang C, Zhang H, Ma C. Identification of the wheat C3H gene family and expression analysis of candidates associated with seed dormancy and germination. Plant Physiol Biochem. 2020;156:524–537. doi: 10.1016/j.plaphy.2020.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136(1):2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139(2):847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580(28–29):6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282(12):9260–9268. doi: 10.1074/jbc.M611093200. [DOI] [PubMed] [Google Scholar]
- 11.Devaiah BN, Nagarajan VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 2007;145(1):147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011;157(2):742–756. doi: 10.1104/pp.111.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin M, Wang Y, Zhang L, Li J, Quan W, Yang L, Wang Q, Chan Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J Exp Bot. 2017;68(11):2991–3005. doi: 10.1093/jxb/erx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Wang JF, Wang QH, Zhang HS. Identification of a rice zinc finger protein whose expression is transiently induced by drought, cold but not by salinity and abscisic acid. DNA Seq. 2005;16(2):130–136. doi: 10.1080/10425170500061590. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, Zhang HS. A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta. 2007;1769(4):220–227. doi: 10.1016/j.bbaexp.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009;23(15):1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.) FEBS Lett. 2008;582(7):1037–43. doi: 10.1016/j.febslet.2008.02.052. [DOI] [PubMed] [Google Scholar]
- 18.Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61(10):2807–2818. doi: 10.1093/jxb/erq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Liu Y, Wen F, Yao D, Wang L, Guo J, Ni L, Zhang A, Tan M, Jiang M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot. 2014;65(20):5795–5809. doi: 10.1093/jxb/eru313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.) BMC Plant Biol. 2015;15(1):1–20. doi: 10.1186/s12870-014-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takatsuji H. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54(6):582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kam J, Gresshoff PM, Shorter R, Xue GP. The Q-type C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress. Plant Mol Biol. 2008;67(3):305–322. doi: 10.1007/s11103-008-9319-3. [DOI] [PubMed] [Google Scholar]
- 23.Amalraj A, Luang S, Kumar MY, Sornaraj P, Eini O, Kovalchuk N, Bazanova N, Li Y, Yang N, Eliby S, Langridge P, Hrmova M, Lopato S. Change of function of the wheat stress-responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol J. 2016;14(2):820–32. doi: 10.1111/pbi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing Y, Liu J, Liu P, Ming D, Sun J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci Rep. 2019;9(1):1–15. doi: 10.1038/s41598-019-42177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Q, Zhang X, Zhang W, Zhang N, Song L, Liu L, Xue X, Liu G, Liu J, Meng D, Zhi L, Ji J, Zhao X, Yang C, Tong Y, Liu Z, Li J. QTL detection for kernel size and weight in bread wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci. 2018;9:1484. doi: 10.3389/fpls.2018.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danielli L, Li X, Tuler T, Daniel R. Quantifying the distribution of protein oligomerization degree reflects cellular information capacity. Sci Rep. 2020;10(1):17689. doi: 10.1038/s41598-020-74811-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 28.Kozowska M, Tarczewska A, Jakob M, Bystranowska D, Taube M, Kozak M, Czarnocki-Cieciura M, Dziembowski A, Orowski M, Tkocz K, Oyhar A. Nucleoplasmin-like domain of FKBP39 from Drosophila melanogaster forms a tetramer with partly disordered tentacle-like C-terminal segments. Sci Rep. 2017;7:40405. doi: 10.1038/srep40405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anselm E, Thomae AW, Jeyaprakash AA, Heun P. Oligomerization of Drosophila Nucleoplasmin-Like Protein is required for its centromere localization. Nucleic Acids Res. 2018;46(21):11274–11286. doi: 10.1093/nar/gky988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert B, Tomassetti S, Gloor Y, Dilg D, Mattarocci S, Kubik S, Hafner L, Shore D. Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 2019;33(5–6):288–293. doi: 10.1101/gad.322040.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SY, Chen HF, Yeh YC, Xue YP, Lan CY. The transcription factor Sfp1 regulates the oxidative stress response in Candida albicans. Microorganisms. 2019;7(5):131. doi: 10.3390/microorganisms7050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11(3):109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Yan H, Chen W, Liu J, Jiang C, Jiang H, Zhu S, Cheng B. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics. 2014;289(6):1061–1074. doi: 10.1007/s00438-014-0867-8. [DOI] [PubMed] [Google Scholar]
- 34.Borrill P, Harrington SA, Uauy C. Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploid wheat. G3 (Bethesda) 2017;7(9):3019–3029. doi: 10.1534/g3.117.043679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E, Leroy P, Mangenot S, Guilhot N, Le Gouis J, Balfourier F, Alaux M, Jamilloux V, Poulain J, Durand C, Bellec A, Gaspin C, Safar J, Dolezel J, Rogers J, Vandepoele K, Aury JM, Mayer K, Berges H, Quesneville H, Wincker P, Feuillet C. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345(6194):1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 36.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- 38.Xiong D, Wang Y, Deng C, Hu R, Tian C. Phylogenic analysis revealed an expanded C2H2-homeobox subfamily and expression profilesof C2H2zinc finger gene family in Verticillium dahliae. Gene. 2015;562:169–179. doi: 10.1016/j.gene.2015.02.063. [DOI] [PubMed] [Google Scholar]
- 39.Böhm S, Frishman D, Mewes HW. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997;25(12):2464–2469. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muthamilarasan M, Bonthala VS, Mishra AK, Khandelwal R, Khan Y, Roy R, Prasad M. C2H2-type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct Integr Genomics. 2014;14(3):531–543. doi: 10.1007/s10142-014-0383-2. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Liu D, Hu R, Hua C, Ali I, Zhang A, Liu B, Wu M, Huang L, Gan Y. AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem Biophys Res Commun. 2017;483(1):209–215. doi: 10.1016/j.bbrc.2016.12.164. [DOI] [PubMed] [Google Scholar]
- 42.Kubo K-I, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, et al. Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998;26:608–615. doi: 10.1093/nar/26.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Wang Z, Xu X, Zhang H, Li C. Genome-wide analysis of C2H2 Zinc-finger family transcription factors and their responses to abiotic stresses in Poplar (Populus trichocarpa) PLoS One. 2015;10(8):e0134753. doi: 10.1371/journal.pone.0134753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto H, Araki T, Meshi T, Iwabuchi M. Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene. 2000;248:23–32. doi: 10.1016/S0378-1119(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Wang J, Zhang H. Rice ZFP15 gene encoding for a novel C2H2-type zinc finger protein lacking DLN box, is regulated by spike development but not by abiotic stresses. Mol Biol Rep. 2005 Sep;32(3):177–83. 10.1007/s11033-005-2338-0. PMID: 16172918. [DOI] [PubMed]
- 46.Han G, Lu C, Guo J, Qiao Z, Sui N, Qiu N, Wang B. C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front Plant Sci. 2020 Feb;20(11):115. 10.3389/fpls.2020.00115. Erratum in Front Plant Sci 2020 Mar 18;11:298. PMID: 32153617; PMCID: PMC7044346. [DOI] [PMC free article] [PubMed]
- 47.Kulkarni M, Soolanayakanahally R, Ogawa S, Uga Y, Selvaraj MG, Kagale S. Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front Chem. 2017;5(5):106. doi: 10.3389/fchem.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng BH, Han YC, Xiao YY, Kuang JF, Fan ZQ, Chen JY, Lu WJ. The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J Exp Bot. 2016;67(8):2263–2275. doi: 10.1093/jxb/erw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan IC, Li CW, Su RC, Cheng CP, Lin CS, Chan MT. Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta. 2010;232(5):1075–1086. doi: 10.1007/s00425-010-1235-5. [DOI] [PubMed] [Google Scholar]
- 50.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13(8):1959–1968. doi: 10.1105/tpc.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 52.Cao H, Xu Y, Yuan L, Bian Y, Wang L, Zhen S, Hu Y, Yan Y. Molecular characterization of the 14–3–3 gene family in Brachypodium distachyon L. reveals high evolutionary conservation and diverse responses to abiotic stresses. Front Plant Sci. 2016;7:1099. doi: 10.3389/fpls.2016.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B, Zhao Y, Shi S, Yang M, Xiao K. TaZFP1, a C2H2 type-ZFP gene of T. aestivum, mediates salt stress tolerance of plants by modulating diverse stress-defensive physiological processes. Plant Physiol Biochem. 2019;136:127–142. doi: 10.1016/j.plaphy.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Zhang D, Tong J, Xu Z, Wei P, Xu L, Wan Q, Huang Y, He X, Yang J, Shao H, Ma H. Soybean C2H2-Type Zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis. Front Plant Sci. 2016;7:325. doi: 10.3389/fpls.2016.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto A, Minami M, Huh GH, Iwabuchi M. The putative zinc-finger protein WZF1 interacts with a cis-acting element of wheat histone genes. Eur J Biochem. 1993;217(3):1049–1056. doi: 10.1111/j.1432-1033.1993.tb18336.x. [DOI] [PubMed] [Google Scholar]
- 56.Sun Z-h, Ding C-h, Li X-j, Xiao K. Molecular characterization and expression analysis of TaZFP15, a C2H2-type zinc finger transcription factor gene in wheat (Triticum aestivum L.) J Integr Agric. 2012;11(01):31–42. doi: 10.1016/S1671-2927(12)60780-9. [DOI] [Google Scholar]
- 57.Chang H, Chen D, Kam J, Richardson T, Drenth J, Guo X, McIntyre CL, Chai S, Rae AL, Xue GP. Abiotic stress upregulated TaZFP34 represses the expression of type-B response regulator and SHY2 genes and enhances root to shoot ratio in wheat. Plant Sci. 2016;252:88–102. doi: 10.1016/j.plantsci.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 58.Liu XM, An J, Han HJ, Kim SH, Lim CO, Yun DJ, Chung WS. ZAT11, a zinc finger transcription factor, is a negative regulator of nickel ion tolerance in Arabidopsis. Plant Cell Rep. 2014;33(12):2015–2021. doi: 10.1007/s00299-014-1675-7. [DOI] [PubMed] [Google Scholar]
- 59.Shah K, Singh M, Rai AC. Effect of heat-shock induced oxidative stress is suppressed in BcZAT12 expressing drought tolerant tomato. Phytochemistry. 2013;95:109–117. doi: 10.1016/j.phytochem.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 60.Tian F, Yang DC, Meng YQ, Jin J, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):D1104–D1113. doi: 10.1093/nar/gkz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, Zhao PX, Rhee SY, Fei Z. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9(12):1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–60. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3(2):153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 65.Faraji S, Rasouli SH, Kazemitabar SK. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): insights into the roles in biological processes especially stress response. Biometals. 2018;31(6):1019–1042. doi: 10.1007/s10534-018-0146-y. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 69.Zhu T, Liu Y, Ma L, Wang X, Zhang D, Han Y, Ding Q, Ma L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020;20(1):420. doi: 10.1186/s12870-020-02576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okonechnikov K, Golosova O, Fursov M, UGENE team Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–7. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Classification of the 457 Triticum aestivum C2H2-ZFPs according to the organization of their contained C2H2 fingers. Table S2. List of 204 Triticum aestivum C2H2-ZFP genes and their related information. Table S3. One-to-one orthologous relationships between T.aestivum and T.aestivum, T.aestivum and A.tauschii, T.aestivum and B.distachyon, T.aestivum and Rice, T.aestivum and Arabidopsis. Table S4. Primers used for qRT-PCR. Table S5. Expression profiles of T. aestivum C2H2-ZFP genes.
Additional file 2: The full size and high resolution of Figure 3.
Additional file 3: The full size and high resolution of Figure 6.
Data Availability Statement
The datasets analysed during the current study are available in the bread wheat Genome Project (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast), plantTFDBv5.0 (http://planttfdb.cbi.pku.edu.cn/phylo_tree.phpsp=Tae&fam=C2H2), iTAK software v1.6 (http://itak.feilab.net/cgi-bin/itak/index.cgi), Ensembl plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index). TAIR (Release 10, ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release) and JCVI (Release 6.1, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1) repository.