Abstract
Loss-of-function mutations in Na+/H + exchanger 6 (NHE6) (also termed SLC9A6) cause the X-linked neurogenetic disorder Christianson syndrome (CS). Using peripheral blood mononuclear cells, we developed induced pluripotent stem cell (iPSC) lines from a patient with the NHE6 nonsense mutation c.1569G > A (p. (W523X)) and diagnosed with CS and from a biologically-related control. Using CRISPR/Cas9 gene editing, we generated two isogenic control lines in which the c.1569G > A mutation was corrected. All lines were verified by DNA sequencing and for NHE6 protein expression, pluripotency, and differentiation potential. These lines will serve as a valuable resource for both basic and translational studies in CS.
Resource Table:
| Unique stem cell lines identifier | EMe-NH6W523S1 EMe-NH6W523XS7 EMe-NH6W523K5 EMe-NH6W523K17 |
| Alternative name(s) of stem cell lines | Ctl-404-S1 (EMe-NH6W523S1) CS-403-S7 (EMe-NH6W523XS7) 403-S7-KI-5 (EMe-NH6W523K5) 403-S7-KI-17 (EMe-NH6W523K17) |
| Institution | Cincinnati Children’s Hospital Medical Center Pluripotent Stem Cell Facility and Brown University |
| Contact information of the reported cell line distributor | Eric M. Morrow < eric_morrow@brown.edu> |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info | Sex: Male |
| Cell Source | Blood, PBMCs |
| Method of reprogramming | Sendai virus (CytoTune-iPS 2.0 Sendai Reprogramming Kit) |
| Clonality | Clonal |
| Evidence of the reprogramming transgene loss | N/A (non-integrating virus) |
| Cell culture system used | mTeSR Plus media, Matrigel-coated plates under feeder-free conditions |
| Type of Genetic Modification | |
| Disease-causing mutation present in iPSCs from patient and correction of mutation using CRISPR/Cas9 gene editing | |
| Associated disease | Christianson syndrome (OMIM 300243, phenotype; OMIM 300231, gene/locus) |
| Gene/locus | NHE6 (SLC9A6), Xq26.3 GRCh37/hg19 chrX:135,106,595 (modified nucleotide) |
| Method of modification/site-specific nuclease used | CRISPR/Cas9 |
| Site-specific nuclease (SSN) delivery method | RNP |
| All genetic material introduced into the cells | Sendai viral vectors for reprogramming, gRNA, ssODN |
| Analysis of the nuclease-targeted allele status | Sequencing of the targeted allele |
| Method of the off-target nuclease activity surveillance | N/A |
| Name of transgene | hOCT3/4, hSOX2, hc-MYC, hKLF4 |
| Eukaryotic selective agent resistance (including inducible/gene expressing cell-specific) | N/A |
| Inducible/constitutive system details | N/A |
| Date archived/stock date | N/A |
| Cell line repository/bank | NIMH Repository and Genomics Resource |
| Ethical/GMO work approvals | |
| Lifespan Healthcare - Rhode Island | |
| Hospital IRB 2, Study No.: 640,453 | |
| Addgene/public access repository recombinant DNA sources’ disclaimers (if applicable) | N/A |
1. Resource utility
Although Christianson syndrome (CS) is known to be caused by mutations in NHE6, the mechanisms underlying disease pathogenesis remain to be fully defined. These iPSC lines will provide a useful resource for basic cellular and molecular studies, as well as translational studies, in CS in a human stem cell model.
2. Resource details
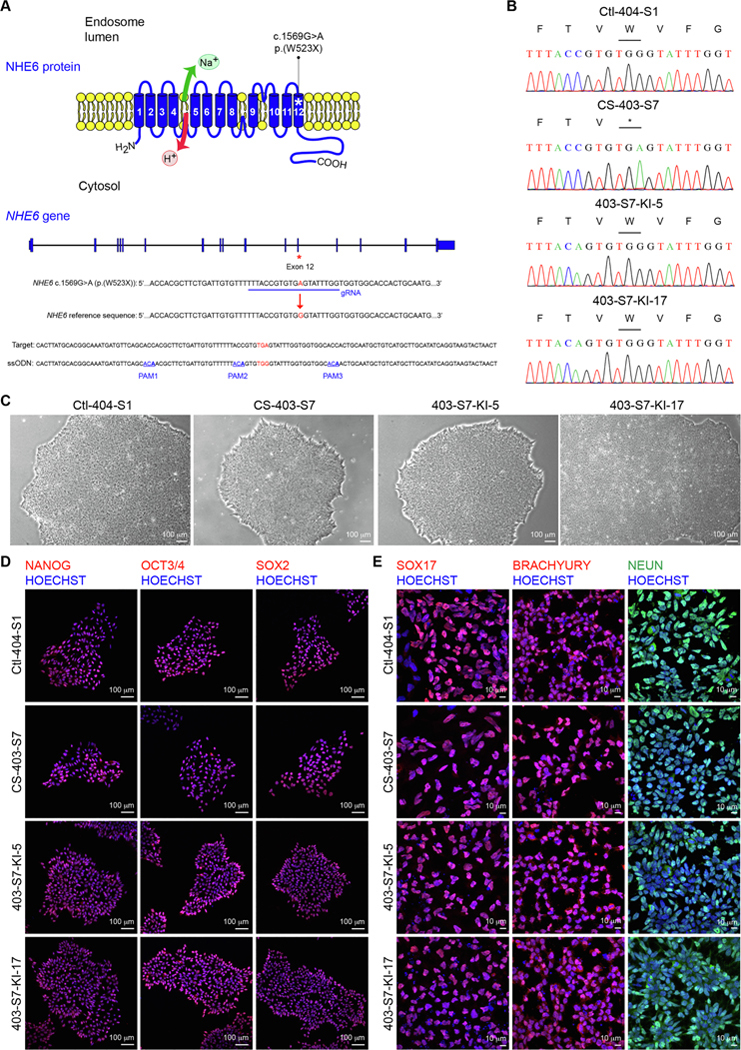
CS is an X-linked neurogenetic disorder with clinical features including intellectual disability, postnatal microcephaly, ataxia, epilepsy, hyperkinesis, nonverbal status, and cerebellar atrophy (Morrow and Pescosolido, 2018). CS is caused by pathogenic, loss-of-function mutations in the endosomal Na+/H+ exchanger 6 (NHE6) (also termed SLC9A6). Human induced pluripotent stem cells (iPSCs) have served as an important complement to animal models in translational studies targeting human genetic disease, including neurological and neuropsychiatric diseases. In order to establish a patient-derived cellular model for the study of CS, we reprogrammed peripheral blood mononuclear cells (PBMCs) from a male patient containing a c.1569G > A (p. (W523X)) mutation in NHE6 (NHE6 transcript NM_001042537.1 and NHE6 protein NP_001036002.1) (Fig. 1A & B) and a clinical diagnosis of CS into iPSCs. We also generated iPSCs from a genetically-related full brother who does not carry a pathogenic mutation in NHE6 (Fig. 1B). We then generated iPSCs wherein the c.1569G > A (p.(W523X)) mutation was corrected back to the reference sequence by using CRISPR/Cas9-mediated homology directed repair (HDR) knock-in methodology (Fig. 1A & B).
Fig. 1.
To be uploaded.
The c.1569G > A (p.(W523X)) mutation is located in exon 12 of NHE6 (Fig. 1A). CS iPSC line 403-S7 was randomly selected as template for gene editing. A gRNA was designed to target exon 12 of NHE6, and an ssODN with the targeted A > G mutation at the exact center was designed for HDR. To prevent re-targeting and recutting by Cas9, the ssODN was also designed to contain three silent mutations, each in a protospacer adjacent motif (PAM) (PAM1, PAM2, and PAM3) (Fig. 1A). A BtsIMutI restriction site (CAGTGTG) was introduced in PAM2 and served as a means for identification of successfully targeted clones. Following transfection of the ribonucleoprotein (RNP)/Cas9 complex together with ssODN and subsequent colony selection, Sanger sequencing was used to confirm correction of the c.1569G > A (p. (W523X)) mutation back to the NHE6 reference sequence (Fig. 1B). From 24 sequenced polyclonal colonies, two clones were identified showing the A > G correction, one of which was expanded so as to generate clones derived from a single cell. Overall, 14 clonal gene-corrected CS iPSC lines derived from a single cell were generated, two of which are characterized here (403-S7-KI-5 and 403-S7-KI-17).
Morphological analysis indicated that all four iPSC lines (biologically-related control, CS, and gene-corrected isogenic controls) displayed classic pluripotent stem cell morphology with a high nucleus to cytoplasm ratio and grew as high-density monolayers of tightly packed cells (Fig. 1C). The four iPSC lines also demonstrated expression of protein markers of pluripotency (Fig. 1D) and the ability to differentiate into the three germ layers (Fig. 1E). Results from SNP microarray analysis indicated that the two gene-corrected control iPSC lines contained a 1.39-Mb to 1.51-Mb copy number variant (CNV) gain at chromosome 20q11.21 in comparison to the parental CS iPSC line, with other reported results being generally comparable (Supplementary File 1). In addition to sequencing, western blot analysis confirmed expression of NHE6 protein (monomeric and dimeric forms) in the biological control and gene-corrected control iPSC lines, but not in the CS iPSC line (Supplementary Fig. 1A). No Mycoplasma was detected in cultures of the four iPSC lines (Supplementary Fig. 1B).
In summary, we report generation and characterization of a CS iPSC line containing a c.1569G > A (p.(W523X)) mutation in NHE6, an associated biologically-related control iPSC line, and two gene-corrected control iPSC lines (Table 1). These cell lines will serve as a valuable resource for both basic and translational studies in CS.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Typical pluripotent human stem cell morphology | Fig. 1 panel C |
| Pluripotency status evidence for the described cell line | Qualitative analysis (Immunocytochemistry) | Positive staining for pluripotency markers: NANOG, OCT3/4, and SOX2 | Fig. 1 panel D |
| Karyotype | Karyotype (G-banding) and higher-resolution, array-based assays (SNP) | Genotypes and CNVs reported Marker coverage: 843,390 spanning whole human genome CNV resolution: As low as 1 Kb |
Supplementary File 1 |
| Genotyping for the desired genomic alteration/allelic status of the gene of interest | PCR across the edited site or targeted allele-specific PCR | Confirmed correction of the NHE6 c.1569G > A (p.(W523X)) mutation by PCR + Sanger sequencing, with a hemizygous allelic status, in two clonal iPSC lines | Fig. 1 panel B |
| Verification of the absence of random plasmid integration events | PCR/Southern | N/A (non-integrating virus) | N/A |
| Parental and modified cell line genetic identity evidence | STR analysis, microsatellite PCR (mPCR) or specific (mutant) allele seq | Genotypes and CNVs reported, based on SNP microarray analysis | Supplementary File 1 |
| Mutagenesis / genetic modification outcome analysis | Sequencing (genomic DNA PCR or RT-PCR product) | Confirmed correction of the NHE6 c.1569G > A (p.(W523X)) mutation by PCR + Sanger sequencing in two clonal iPSC lines | Fig. 1 panel B |
| PCR-based analyses | N/A | N/A | |
| Southern Blot or WGS; western blotting | Confirmed expression of NHE6 protein by western blot in gene-corrected iPSC lines | Supplementary Fig. 1 panel A | |
| Off-target nuclease analysis (OPTIONAL) | PCR across top 5/10 predicted top likely off-target sites, whole genome/exome sequencing | N/A | N/A |
| Specific pathogen-free status | Mycoplasma | Negative | Supplementary Fig. 1 panel B |
| Multilineage differentiation potential | Embryoid body formation OR Teratoma formation OR Scorecard OR Directed differentiation | Demonstrated ability, by immunocytochemistry, of iPSC lines to differentiate into derivatives of all three germ layers | Fig. 1 panel E |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype - additional histocompatibility info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
3. Materials and methods
3.1. Generation and maintenance of iPSCs
The Institutional Review Boards at Brown University and Lifespan Healthcare approved the human subjects research protocol. Informed consent was obtained from all participants or guardians of participants. The protocol for generation of iPSCs from PBMCs was adapted from Kunisato et al. (2011), with Sendai virus used for transducing vectors, via spinoculation, allowing for expression of the reprogramming factors hOCT3/4, hSOX2, hc-MYC, and hKLF4 (CytoTune-iPS 2.0 Sendai Reprogramming Kit, ThermoFisher Scientific #A16568; Multiplicity of infection ratio of 5:5:3). The iPSC lines were cultured on Matrigel-coated plates (Corning #354277) under feeder-free conditions and maintained in mTeSR Plus media (StemCell Technologies #100–0276). Culturing medium was changed approximately daily. Enzyme-free passaging was performed using ReLeSR (StemCell Technologies #05872). Cell cultures were maintained at 37 ◦C in a humidified atmosphere of 95% air and 5% CO2.
3.2. CRISPR/Cas9-mediated gene editing
CRISPR/Cas9-based technology was used to revert CS iPSC line 403-S7 back to the reference sequence. The guide RNA (gRNA), chosen from three candidate gRNAs, was designed according to the CRISPOR web tool (http://crispor.tefor.net/) and was synthesized by Integrated DNA Technologies. A single-stranded oligodeoxyribonucleotide (ssODN) consisting of a 121-nucleotide ultramer (Integrated DNA Technologies) was used as template to repair the double-stranded break. The ssODN was designed to contain the desired reference NHE6 sequence, flanked with homology arms to the targeted genomic region. The ssODN was also designed to contain three silent mutations, each in a PAM (PAM1, PAM2, and PAM3), to prevent re-targeting and recutting by Cas9. Introduction of a BtsIMutI restriction site (CAGTGTG) in introducing one of the silent mutations served as a means for identification of successfully targeted clones.
For gene editing, ∼4 × 105 CS iPSCs containing a c.1569G > A (p. (W523X)) mutation were treated with Accutase (working dilution 1:3 in PBS; StemCell Technologies #07922). Cells were dissociated to generate a suspension of single cells, which were resuspended in mTeSR Plus media with 10 μM Rho-associated protein kinase inhibitor (ROCKi, Tocris Bioscience #Y-27632) for 24 h. Cells were then pelleted, resuspended in RNP/CRISPRMAX Cas9 Transfection Reagent (ThermoFisher Scientific #CMAX00003), and incubated for 5 min at room temperature before plating. For the RNP complex, 30 pmol gRNA and 15 pmol TrueCut Cas9 Protein (ThermoFisher Scientific #A36498) were combined for a ratio of 2:1, and 50% of the RNP complex solution was mixed with a minimum of 40 pmol ssODN for transfection.
To generate colonies derived from a single cell, the CS iPSCs transfected with the RNP/CRISPRMAX mix were diluted at ratios of 1:3,000 and 1:10,000, and plated on 10-cm dishes. Cells were grown on dishes, with medium of mTeSR Plus + ROCKi changed daily until colonies were visible. Pooled colonies were isolated and transferred to 96-well Matrigel-coated plates (Corning #354277) for culturing and subsequent DNA extraction and Sanger sequencing. Pooled colonies for which sequencing chromatograms showed a high peak demonstrating correction of the c.1569G > A (p.(W523X)) mutation were then dissociated and sorted so as to have a single cell per well of a 96-well plate. Colonies derived from a single cell were then further cultured, and correction of the c.1569G > A (p.(W523X)) mutation was confirmed by Sanger sequencing.
3.3. Genomic DNA extraction and Sanger sequencing
Genomic DNA was extracted from iPSC lines using Epicentre QuickExtract DNA Extraction Solution (Lucigen #QE09050), amplified by PCR using an Eppendorf Vapo.Protect Mastercycler Pro thermal cycler (Eppendorf), and sequenced using Sanger sequencing methods. Primers for PCR amplification of exon 12 of NHE6 are shown in Table 2.
Table 2.
Reagents details.
| Antibodies and stains used for immunocytochemistry/flow-cytometry/western blotting | |||
|---|---|---|---|
|
| |||
| Antibody | Dilution | Company Cat # and RRID | |
|
| |||
| Pluripotency markers | Mouse anti-SOX2 | 1:200 | R&D Systems #MAB2018, RRID:AB_358009 |
| Goat anti-NANOG | 1:200 | R&D Systems #AF1997-SP, RRID:AB_355097 | |
| Goat anti-OCT3/4 | 1:200 | R&D Systems #AF1759-SP, RRID:AB_354975 | |
| Differentiation markers | Goat anti-SOX17 | 1:200 | R&D Systems #AF1924-SP, RRID:AB_355060 |
| Goat anti-BRACHYURY | 1:200 | R&D Systems #AF2085-SP, RRID:AB_2200235 | |
| Rabbit anti-NEUN | 1:250 | Abcam #ab177487, RRID:AB_2532109 | |
| Secondary antibodies | Goat anti-mouse AlexaFluor 594 | 1:800 | ThermoFisher Scientific #A11032, RRID: AB_2534091 |
| Donkey anti-goat AlexaFluor 594 | 1:800 | ThermoFisher Scientific #A11058, RRID: AB_2534105 | |
| Goat anti-rabbit AlexaFluor 488 | 1:800 | ThermoFisher Scientific #A11008, RRID: AB_143165 | |
| Nuclear stain | Hoechst 33342 | 6.25 μg/mL | Invitrogen #33342 |
| Target protein | Rabbit anti-NHE6 | 1:1000 | Morrow laboratory (Ouyang et al., 2013) |
| Control protein | Mouse anti-α-TUBULIN | 1:5000 | Sigma-Aldrich #T6074, RRID:AB_477582 |
| Site-specific nuclease | |||
| Nuclease information | TrueCut Cas9 Protein | ThermoFisher Scientific #A36498 | |
| Delivery method | Lipofectamine CRISPRMAX Cas9 Transfection Reagent | ThermoFisher Scientific #CMAX00003 | |
| Selection/enrichment strategy | Restriction enzyme digestion and PCR analysis | ||
| Dilution of gene-corrected iPSCs to single cells and subsequent growth of clonal colonies | |||
| Primers and oligonucleotides used in this study | |||
|
| |||
| Target | Forward/Reverse primer (5′–3′) | ||
|
| |||
| Targeted mutation analysis | NHE6 exon 12 | Forward primer: AGAACGTGCTTTCTTGCTCTG Reverse primer: TGGAAAGATATCCCTCAAAGTC (Pescosolido et al., 2014) |
|
| gRNA oligonucleotide | GRCh37/hg19 chrX:135,106,584–135,106,603 | TTTACCGTGTGAGTATTTGG | |
| ssODN* | GRCh37/hg19 chrX:135,106,535–135,106,655 | CACTTATGCACGGCAAATGATGTTCAGCACAACGCTT CTGATTGTGTTTTTTACAGTGTGGGTATTTGGTGGTG GCACAACTGCAATGCTGTCATGCTTGCATATCAGGTA AGTACTAACT |
|
The three silenced PAMs are underlined and the silent mutations are in bold; the corrected “G” nucleotide is in bold and underlined.
3.4. Single nucleotide polymorphism (SNP) microarray analysis
Genomic DNA was extracted from iPSCs using the QIAamp DNA Mini Kit (Qiagen #51304) and analyzed using the Illumina CytoSNP-850K v1.2 BeadChip platform with BlueFuse Multi 4.5 Software (WiCell). Array design, genomic position, and chromosome banding are based on genome build GRCh37/hg19.
3.5. Immunoprecipitation and western blotting
IPSCs were harvested and then lysed in lysis buffer (50 mM Tris-HCl, pH 7.8, 137 mM NaCl, 1 mM NaF, 1 mM NaVO3, 1% Triton X-100, 0.2% Sarkosyl, 1 mM dithiothreitol, and 10% glycerol) supplemented with protease inhibitor cocktail and phosphatase inhibitor for 30 min on ice. Cell lysates were separated by centrifugation at 13,200 rpm for 15 min at 4 °C, and the remaining supernatants were removed for further processing. Protein concentration was measured by BCA assay using the Pierce BCA Kit (ThermoFisher Scientific #23225). For immunoprecipitation, aliquots of a custom-made rabbit anti-NHE6 antibody (Ouyang et al., 2013) were conjugated to Dynabeads Protein G (ThermoFisher Scientific #10004D) at room temperature for 2 h. Cell lysates were then incubated with anti-NHE6 antibody-conjugated beads overnight at 4 °C. The following day, the beads were gently pelleted, cell lysates were removed, and the beads were washed three times with PBS with 0.02% TWEEN 20 wash buffer. Pelleted beads were then boiled in sample buffer at 95 °C for 5 min before loading onto 4–12% SDS-PAGE gels (Novex #NP0321Box). Following separation of proteins by electrophoresis, gels were transferred to nitrocellulose membranes (Novex #LC2000). Western blots were performed using standard procedures (Lizarraga et al., 2021) and were analyzed with the Li-CoR Odyssey Imaging System. See Table 2 for antibodies and dilutions used.
3.6. Multilineage differentiation
IPSC lines were assayed for the ability to differentiate into the three germ layers (endoderm, mesoderm, and ectoderm) using the STEMDiff Trilineage Differentiation Kit (StemCell Technologies #05230). Differentiation into a specific germ layer was assessed by microscopy following processing of cells for immunofluorescence.
3.7. Immunofluorescent staining
For detection of NANOG, OCT3/4, SOX17, and BRACHYURY, iPSCs were fixed with 4% paraformaldehyde for 20 min at room temperature, washed with 1% BSA in PBS for 5 min, and permeabilized and blocked for 45 min at room temperature in PBS containing 0.3% Triton X-100, 1% BSA, and 10% normal donkey serum (Jackson ImmunoResearch #017–000-121). Cells were then incubated overnight at 4 °C with primary antibody diluted in blocking buffer, washed 3 × 5 min with 1% BSA in PBS, and incubated for 1 h at room temperature with secondary antibody diluted in 1% BSA in PBS. Nuclei were counterstained with Hoechst (1:1600 working dilution of 10 mg/mL stock; Invitrogen #33342). Cells were then washed 3 times with 1% BSA in PBS and mounted on slides with Fluoromount-G (SouthernBiotech #0100–01).
For detection of SOX2 and NEUN, iPSCs were fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized for 10 min in PBS containing 0.25% Triton X-100. Non-specific binding was blocked by incubation with 10% normal goat serum (Jackson ImmunoResearch #005–000-121) in PBS containing 0.1% TWEEN 20 (PBST) for 45 min. Cells were then incubated overnight at 4 °C with primary antibody diluted in PBST containing 2% normal donkey serum, washed 3 × 5 min with PBST, and incubated for 1 h at room temperature with secondary antibody diluted as for primary antibody. Nuclei were counterstained with Hoechst (1:1600 working dilution of 10 mg/mL stock; Invitrogen #33342). Cells were then washed 3 times with PBST and mounted on slides with Fluoromount-G (SouthernBiotech #0100–01). For detection of pluripotency marker expression, iPSCs were analyzed at the following passage numbers: 13 (Ctl-404-S1), 14 (CS-403-S7), and 34 (403-S7-KI-5, 403-S7-KI-17); for detection of germ layer marker expression, cells were analyzed at the following passage numbers: 12 (Ctl-404-S1), 13 (CS-403-S7), and 33 (403-S7-KI-5, 403-S7-KI-17). See Table 2 for antibodies and dilutions used.
3.8. Imaging of iPSCs
Brightfield microscopy images were captured using a Nikon Eclipse TS100 microscope equipped with a Q Imaging QIClick 1.4-MP CCD monochrome microscope camera. Fluorescence microscopy images were captured using an Olympus FV3000 confocal microscope.
3.9. Mycoplasma detection
Testing of iPSC culture supernatant for Mycoplasma was performed using the LookOut Mycoplasma PCR Detection Kit (Sigma #MP0035). Cultures were analyzed at the following passage numbers: 13 (Ctl-404-S1), 14 (CS-403-S7), and 34 (403-S7-KI-5, 403-S7-KI-17). PCR products were subjected to electrophoresis using a 1.2% agarose gel.
Supplementary Material
Acknowledgements
We thank the family who participated in this study. The study was supported by NIMH R01MH105442, NIMH R21MH115392, and NINDS/NIA R01NS113141 (E.M.M.). We thank the Brown University/Rhode Island Hospital Flow Cytometry and Sorting Facility and the Cincinnati Children’s Hospital Medical Center Pluripotent Stem Cell Facility for their services. We acknowledge Heather M. Thompson, Ph.D. of Brown University for her support in manuscript preparation and Li Li, Ph.D. of the Harvard Stem Cell Institute for technical advice and discussions.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102435.
References
- Kunisato A, Wakatsuki M, Shinba H, Ota T, Ishida I, Nagao K, 2011. Direct generation of induced pluripotent stem cells from human nonmobilized blood. Stem Cells Dev. 20, 159–168. 10.1089/scd.2010.0063. [DOI] [PubMed] [Google Scholar]
- Lizarraga SB, Ma L, Maguire AM, van Dyck LI, Wu Q, Ouyang Q, Kavanaugh BC, Nagda D, Livi LL, Pescosolido MF, Schmidt M, Alabi S, Cowen MH, Brito-Vargas P, Hoffman-Kim D, Gamsiz Uzun ED, Schlessinger A, Jones RN, Morrow EM, 2021. Human neurons from Christianson syndrome iPSCs reveal mutation-specific responses to rescue strategies. Sci. Transl. Med 13. 10.1126/scitranslmed.aaw0682. [DOI] [PMC free article] [PubMed]
- Morrow EM, Pescosolido MF, 2018. Christianson syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. (Eds.), GeneReviews((R)). University of Washington, Seattle: Seattle, WA: (1993–2018). [PubMed] [Google Scholar]
- Ouyang Q, Lizarraga S, Schmidt M, Yang U, Gong J, Ellisor D, Kauer J, Morrow E, 2013. Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron 80 (1), 97–112. 10.1016/j.neuron.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescosolido MF, Stein DM, Schmidt M, El Achkar CM, Sabbagh M, Rogg JM, Tantravahi U, McLean RL, Liu JS, Poduri A, Morrow EM, 2014. Genetic and phenotypic diversity of NHE6 mutations in Christianson syndrome. Ann. Neurol 76 (4), 581–593. 10.1002/ana.24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.