Abstract
Species of Diaporthe (syn. Phomopsis) are important endophytes, saprobes and pathogens, infecting a wide range of plants and resulting in important crop diseases. However, the species occurring on pear remain largely unresolved. In this study, a total of 453 Diaporthe isolates were obtained from branches of Pyrus plants (including P. bretschneideri, P. communis, P. pyrifolia and P. ussuriensis collected from 12 provinces in China) showing shoot canker symptoms. Phylogenetic analyses based on five loci (ITS, TEF, CAL, HIS, and TUB) coupled with morphology of 113 representative isolates revealed that 19 Diaporthe species were isolated, representing 13 known species (including D. caryae, D. cercidis, D. citrichinensis, D. eres, D. fusicola, D. ganjae, D. hongkongensis, D. padina, D. pescicola, D. sojae, D. taoicola, D. unshiuensis and D. velutina) and six new species described here as D. acuta, D. chongqingensis, D. fulvicolor, D. parvae, D. spinosa and D. zaobaisu. Although Koch’s postulates confirmed all species to be pathogenic, a high degree of variation in aggressiveness was observed. Moreover, these species have a high diversity, plasticity, and prevalence related to the geographical location and pear species involved.
Keywords: multi-gene phylogeny, pathogenicity, Pyrus, six new taxa, taxonomy
INTRODUCTION
Species of Diaporthe (asexual morph Phomopsis) are widely distributed, and infect a broad plant host range, e.g., fruit and forest trees, vegetables, and ornamental plants as endophytes, saprobes or pathogens (Santos & Phillips 2009, Santos et al. 2011, Udayanga et al. 2011, 2012, 2014a, b, Gomes et al. 2013, Gao et al. 2015, Marin-Felix et al. 2019). As plant pathogens Diaporthe spp. cause severe diseases, e.g., dieback, cankers, leaf spots, blights, decay or wilt of many economically important plants including Camellia, Citrus, Glycine, Helianthus, Persea, Vaccinium, and Vitis (Van Rensburg et al. 2006, Santos & Phillips 2009, Crous et al. 2011, 2016, Santos et al. 2011, Thompson et al. 2011, Grasso et al. 2012, Huang et al. 2013, Lombard et al. 2014, Gao et al. 2015, 2016, Udayanga et al. 2015, Guarnaccia & Crous 2017, 2018, Guarnaccia et al. 2018), resulting in major losses (Van Rensburg et al. 2006, Santos et al. 2011, Thompson et al. 2011). In recent years the taxonomy of Diaporthe species has been largely resolved based on multigene phylogenetic analyses including the rDNA internal transcribed spacer (ITS1, 5.8S, ITS2) region, partial translation elongation factor 1-alpha (TEF), beta-tubulin (TUB), histone H3 (HIS) and calmodulin (CAL) genes (Gomes et al. 2013, Marin-Felix et al. 2019). Based on this approach, Diaporthe species have been well characterised for those infecting grapevine and citrus in Europe (Guarnaccia & Crous 2017, Guarnaccia et al. 2018) and forest trees in China (Yang et al. 2018). Published results revealed numerous species infecting these crops, with four (D. bohemiae, D. celeris, D. hispaniae and D. hungariae), two (D. limonicola and D. melitensis spp. nov.) and 12 (D. acerigena, D. alangii, D. betulina, D. caryae, D. cercidis, D. chensiensis, D. cinnamomi, D. conica, D. fraxinicola, D. kadsurae, D. padina and D. ukurunduensis) from citrus, grapevine and forest trees, respectively (Guarnaccia & Crous 2017, Guarnaccia et al. 2018, Yang et al. 2018). Moreover, some Diaporthe taxa appear to be strictly host specific (Gomes et al. 2013). However, the Diaporthe spp. occurring on other economically important crops, such as Pyrus (pear), have been poorly studied.
Pear species represent the third most important temperate fruit crop after apple and grape worldwide. Pear originated in the Tertiary period in Western China, and is divided into two major groups: European and Asian pears, with Pyrus bretschneideri, P. communis, P. pyrifolia, P. sinkiangensis, and P. ussuriensis commercially cultivated (Silva et al. 2014, Ferradini et al. 2017). Three species, including P. bretschneideri, P. communis and P. pyrifolia are the major species cultivated in China, with a pear-cultivation area of 957 321 ha in 2017, producing 16.5 MT fruits, accounting for nearly 70 % of the global pear fruit yield (24.2 MT) (Wu et al. 2013, Zhao et al. 2016, FAO 2017).
Pear shoot canker is a devastating disease caused by Diaporthe spp. The disease was initially described on P. pyrifolia in Japan (Nasu et al. 1987), infecting pear branches, causing brown canker tissue around buds on the shoots, twigs, or large branches, and always killing the infected shoots or branches and the attached blossom and leaf buds. The disease has resulted in large losses to fruit production in China (Wang et al. 2011, Huang et al. 2014, Bai et al. 2015), and other countries, e.g., Japan and Korea (Tanaka & Endo 1930, Nasu et al. 1987). In our previous study, we preliminarily identified five Diaporthe species from pear samples collected from six provinces in China based on three loci including TEF, ACT and ITS sequences (Bai et al. 2015). However, these loci proved to be insufficiently robust to identify these species. Therefore, the species associated with pear shoot canker remain largely unresolved. The aims of the present study were thus as follows:
make an extensive survey of Diaporthe species associated with pear shoot canker in the major pear-cultivation provinces in China;
resolve the species identity based on multi-locus DNA sequence data;
characterise the morphology and evaluate the pathogenicity of the species involved; and
get insight into the diversity, incidence and biology of the Diaporthe species associated with pear shoot canker.
MATERIALS AND METHODS
Sampling and isolation
From May 2014 to December 2017, pear twigs, branches and trunks showing shoot canker symptoms were collected from 40 pear orchards in 15 provinces (including Chongqing, Fujian, Guizhou, Hebei, Henan, Hubei, Jiangsu, Jiangxi, Jilin, Liaoning, Shandong, Shanxi, Xinjiang, Yunnan and Zhejiang) of China. The pear species and varieties involved in the collection include P. pyrifolia cultivars (cvs.) Aigansui, Cuiyu, Cuiguan, Chuxialv, Huanghua, Hohsui, Jinqiu, Jinshui, Jinshui No. 2, Kousui, Minfu, Niitaka, Wanqiuhuang, Whangkeumbae, Yuanhuang and Yujing, P. bretschneideri cvs. Bayuesu, Dangshansu, Huangguan, Qingxiang, Wanyu, Yali and Zaobaisu, P. ussuriensis cv. Xiaonanguo, and P. communis cvs. Docteun Jule Guyot, Packham, J6, J23 and Winter decana.
The collected samples were subjected to fungal isolation as previously described (Bai et al. 2015). Briefly, infected tissues (4–5 mm2) were excised from the xylem or phloem under the canker lesions neighbouring the asymptomatic regions after surface-sterilised with 75 % ethanol for 45 s and 75 % NaClO for 45 s and then rinsed twice with sterilised water. The excised tissues were placed on potato dextrose agar (PDA, 20 % diced potatoes, 2 % glucose and 1.5 % agar) Petri dishes and incubated at 25 °C in the dark for 3–5 d. When colonies formed, each colony was transferred to a new PDA Petri dish and assigned a number. Each isolate was further purified by culturing a colony from a single conidium (Choi et al. 1999). The obtained isolates were stored in 25 % glycerol at −80 °C for later use. Type specimens of new species from this study were deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and ex-type living cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC), Beijing, China.
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from pure cultures using a modified cetyltrimethylammonium bromide (CTAB) protocol (Freeman et al. 1996), and subjected to PCR amplification of partial regions of five loci including partial ITS, TUB, TEF, CAL and HIS gene regions using corresponding primer pairs, e.g., ITS1/ITS4 (White et al. 1990), Bt2a/Bt2b (Glass & Donaldson 1995), EF1-728F/EF1-986R (Carbone & Kohn 1999), CAL-228F/CAL-737R (Carbone & Kohn 1999) and CYLH3F/H3-1b (Glass & Donaldson 1995, Crous et al. 2004), respectively. PCR parameters were initiated with 95 °C for 5 min, followed by 34 cycles of denaturation at 95 °C for 30 s, annealing at a suitable temperature for 30 s (56 °C for ITS, 52 °C for TEF, 54 °C for CAL, 57 °C for HIS and 60 °C for TUB), and extension at 72 °C for 30 s, and terminated with a final elongation step at 72 °C for 10 min. The PCR amplicons were purified and sequenced at the Sangon Biotech (Shanghai, China) Company, Ltd. The obtained sequences were analysed on DNAMAN (v. 9.0; Lynnon Biosoft), and deposited in GenBank (Table 1).
Table 1.
Collection details and GenBank accession numbers of isolates included in this study.
| Species | Culture no. | Host | Origin | GenBank accession number |
Mating type |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | CAL | HIS | TEF | TUB | MAT1 | MAT2 | ||||
| D. acuta | PSCG 045 | P. pyrifolia | Wuhan, Hubei | MK626956 | MK691123 | MK726160 | MK654809 | MK691223 | / | / |
| PSCG 046 | P. pyrifolia | Wuhan, Hubei | MK626958 | MK691124 | MK726162 | MK654803 | MK691224 | / | / | |
| PSCG 047 * | P. pyrifolia | Wuhan, Hubei | MK626957 | MK691125 | MK726161 | MK654802 | MK691225 | / | / | |
| D. caryae | PSCG 380 | P. pyrifolia | Nanjing, Jiangsu | MK626951 | MK691198 | MK726200 | MK654893 | MK691313 | – | + |
| PSCG 382 | P. pyrifolia | Nanjing, Jiangsu | MK626954 | MK691199 | MK726201 | MK654894 | MK691314 | – | + | |
| PSCG 520 | P. pyrifolia | Zhenjiang, Jiangsu | MK626952 | MK691200 | MK726202 | MK654895 | MK691315 | + | – | |
| PSCG 528 | P. pyrifolia | Zhenjiang, Jiangsu | MK626953 | MK691201 | MK726203 | MK654896 | MK691316 | – | + | |
| D. cercidis | PSCG 259 | P. pyrifolia | Yantai, Shandong | MK626847 | MK691170 | MK726154 | MK654795 | MK691218 | + | – |
| PSCG 273 | P. pyrifolia | Hangzhou, Zhejiang | MK626848 | MK691113 | MK726165 | MK654808 | MK691231 | – | + | |
| PSCG 275 | P. pyrifolia | Hangzhou, Zhejiang | MK626853 | MK691114 | MK726158 | MK654805 | MK691220 | + | + | |
| PSCG 439 | P. pyrifolia | Chongqing, China | MK626852 | MK691118 | MK726172 | MK654813 | MK691221 | – | + | |
| PSCG 513 | P. pyrifolia | Zhenjiang, Jiangsu | MK626850 | MK691117 | MK726223 | MK654815 | MK691219 | – | + | |
| PSCG 526 | P. pyrifolia | Zhenjiang, Jiangsu | MK626851 | MK691121 | MK726169 | MK654804 | MK691228 | + | – | |
| D. chongqingensis | PSCG 435 * | P. pyrifolia | Chongqing, China | MK626916 | MK691209 | MK726257 | MK654866 | MK691321 | – | + |
| PSCG 436 | P. pyrifolia | Chongqing, China | MK626917 | MK691208 | MK726256 | MK654867 | MK691322 | – | + | |
| D. citrichinensis | PSCG 462 | P. pyrifolia | Guiyang, Guizhou | MK626893 | MK691171 | MK726248 | MK654852 | MK691286 | + | – |
| D. eres | PSCG 007 | P. pyrifolia | Nanchang, Jiangxi | MK626884 | MK691157 | MK726216 | MK654835 | MK691278 | + | – |
| PSCG 017 | P. pyrifolia | Fuzhou, Jiangxi | MK626887 | MK691139 | MK726232 | MK654829 | MK691283 | – | + | |
| PSCG 023 | P. pyrifolia | Fuzhou, Jiangxi | MK626878 | MK691158 | MK726217 | MK654821 | MK691269 | + | – | |
| PSCG 041 | P. bretschneideri | Kunming, Yunnan | MK626880 | MK691144 | MK726219 | MK654840 | MK691265 | – | + | |
| PSCG 042 | P. bretschneideri | Kunming, Yunnan | MK626881 | MK691145 | MK726225 | MK654845 | MK691285 | – | + | |
| PSCG 043 | P. bretschneideri | Kunming, Yunnan | MK626879 | MK691146 | MK726229 | MK654844 | MK691266 | – | + | |
| PSCG 090 | P. communis | Yantai, Shandong | MK626872 | MK691159 | MK726236 | MK654828 | MK691281 | + | – | |
| PSCG 092 | P. communis | Yantai, Shandong | MK626896 | MK691147 | MK726227 | MK654823 | MK691264 | – | + | |
| PSCG 132 | P. pyrifolia | Sanming, Fujian | MK626891 | MK691133 | MK726212 | MK654816 | MK691250 | + | – | |
| PSCG 135 | P. pyrifolia | Sanming, Fujian | MK626873 | MK691160 | MK726213 | MK654837 | MK691251 | + | – | |
| PSCG 151 | P. pyrifolia | Sanming, Fujian | MK626876 | MK691161 | MK726239 | MK654820 | MK691262 | + | – | |
| PSCG 175 | P. pyrifolia | Yingtan, Jiangsu | MK626877 | MK691165 | MK726238 | MK654843 | MK691259 | – | + | |
| PSCG 202 | P. communis | Yantai, Shandong | MK626885 | MK691166 | MK726237 | MK654817 | MK691254 | – | + | |
| PSCG 245 | P. pyrifolia | Chongqing, China | MK626894 | MK691164 | MK726224 | MK654822 | MK691274 | + | – | |
| PSCG 250 | P. pyrifolia | Chongqing, China | MK626895 | MK691168 | MK726245 | MK654836 | MK691275 | – | + | |
| PSCG 261 | P. pyrifolia | Wuhan, Hubei | MK626904 | MK691141 | MK726241 | MK654826 | MK691252 | + | – | |
| PSCG 265 | P. pyrifolia | Wuhan, Hubei | MK626903 | MK691150 | MK726214 | MK654842 | MK691282 | + | – | |
| PSCG 276 | P. pyrifolia | Hangzhou, Zhejiang | MK626909 | MK691163 | MK726226 | MK654841 | MK691263 | + | – | |
| PSCG 299 | P. pyrifolia | Changli, Hebei | MK626900 | MK691154 | MK726246 | MK654818 | MK691255 | – | + | |
| PSCG 300 | P. pyrifolia | Changli, Hebei | MK626901 | MK691155 | MK726247 | MK654819 | MK691253 | – | + | |
| PSCG 306 | P. communis | Yantai, Shandong | MK626898 | MK691138 | MK726243 | MK654839 | MK691279 | – | + | |
| PSCG 321 | P. pyrifolia | Nanyang, Henan | MK626874 | MK691167 | MK726228 | MK654827 | MK691267 | – | + | |
| PSCG 322 | P. pyrifolia | Nanyang, Henan | MK626875 | MK691162 | MK726244 | MK654824 | MK691268 | – | + | |
| PSCG 324 | P. pyrifolia | Nanyang, Henan | MK626906 | MK691149 | MK726220 | MK654830 | MK691272 | – | + | |
| PSCG 325 | P. pyrifolia | Nanyang, Henan | MK626905 | MK691153 | MK726222 | MK654838 | MK691273 | – | + | |
| PSCG 346 | P. pyrifolia | Nanyang, Henan | MK626882 | MK691134 | MK726234 | MK654848 | MK691270 | – | + | |
| PSCG 358 | P. ussuriensis | Yingkou, Liaoning | MK626889 | MK691143 | MK726231 | MK654849 | MK691260 | – | + | |
| PSCG 362 | P. pyrifolia | Yingkou, Liaoning | MK626907 | MK691151 | MK726235 | MK654846 | MK691280 | + | – | |
| PSCG 376 | P. pyrifolia | Hangzhou, Zhejiang | MK626899 | MK691142 | MK726218 | MK654834 | MK691257 | – | + | |
| PSCG 377 | P. pyrifolia | Hangzhou, Zhejiang | MK626886 | MK691137 | MK726221 | MK654833 | MK691276 | + | – | |
| PSCG 381 | P. pyrifolia | Nanchang, Jiangxi | MK626897 | MK691148 | MK726215 | MK654847 | MK691277 | – | + | |
| PSCG 440 | P. pyrifolia | Wuhan, Hubei | MK626908 | MK691140 | MK726230 | MK654825 | MK691256 | + | + | |
| PSCG 512 | P. pyrifolia | Zhenjiang, Jiangsu | MK626883 | MK691135 | MK726240 | MK654832 | MK691271 | + | – | |
| PSCG 521 | P. pyrifolia | Zhenjiang, Jiangsu | MK626888 | MK691136 | MK726233 | MK654850 | MK691284 | – | + | |
| PSCG 529 | P. pyrifolia | Zhenjiang, Jiangsu | MK626902 | MK691156 | MK726242 | MK654831 | MK691258 | – | + | |
| D. fulvicolor | PSCG 051 * | P. pyrifolia | Wuhan, Hubei | MK626859 | MK691132 | MK726163 | MK654806 | MK691236 | – | + |
| PSCG 057 | P. pyrifolia | Wuhan, Hubei | MK626858 | MK691131 | MK726164 | MK654810 | MK691233 | – | + | |
| D. fusicola | PSCG 015 | P. pyrifolia | Fuzhou, Jiangxi | MK626915 | MK691210 | MK726254 | MK654861 | MK691320 | – | + |
| PSCG 030 | P. pyrifolia | Fuzhou, Jiangxi | MK626914 | MK691211 | MK726255 | MK654864 | MK691323 | – | + | |
| PSCG 118 | P. pyrifolia | Sanming, Fujian | MK626910 | MK691204 | MK726250 | MK654860 | MK691317 | – | + | |
| PSCG 178 | P. pyrifolia | Yingtan, Jiangxi | MK626913 | MK691206 | MK726251 | MK654862 | MK691324 | – | + | |
| PSCG 179 | P. pyrifolia | Yingtan, Jiangxi | MK626912 | MK691207 | MK726252 | MK654863 | MK691318 | – | + | |
| PSCG 371 | P. pyrifolia | Hangzhou, Zhejiang | MK626911 | MK691205 | MK726253 | MK654865 | MK691319 | – | + | |
| D. ganjae | PSCG 489 | P. pyrifolia | Guiyang, Guizhou | MK626955 | MK691202 | MK726204 | MK654897 | MK691287 | – | + |
| D. hongkongensis | PSCG 001 | P. pyrifolia | Nanchang, Jiangxi | MK626846 | MK691103 | MK726150 | MK654788 | MK691240 | + | – |
| PSCG 026 | P. pyrifolia | Fuzhou, Jiangxi | MK626861 | MK691106 | MK726153 | MK654789 | MK691241 | + | – | |
| PSCG 114 | P. pyrifolia | Sanming, Fujian | MK626867 | MK691104 | MK726146 | MK654785 | MK691212 | – | + | |
| PSCG 130 | P. pyrifolia | Sanming, Fujian | MK626862 | MK691105 | MK726151 | MK654786 | MK691239 | – | + | |
| PSCG 141 | P. pyrifolia | Sanming, Fujian | MK626854 | MK691110 | MK726147 | MK654787 | MK691213 | + | + | |
| PSCG 290 | P. pyrifolia | Hangzhou, Zhejiang | MK626870 | MK691107 | MK726152 | MK654794 | MK691214 | + | + | |
| PSCG 465 | P. pyrifolia | Sanming, Fujian | MK626863 | MK691109 | MK726148 | MK654790 | MK691242 | – | + | |
| PSCG 466 | P. pyrifolia | Sanming, Fujian | MK626864 | MK691111 | MK726149 | MK654792 | MK691217 | – | + | |
| PSCG 472 | P. pyrifolia | Sanming, Fujian | MK626865 | MK691108 | / | MK654793 | MK691215 | / | / | |
| PSCG 473 | P. pyrifolia | Sanming, Fujian | MK626866 | MK691112 | MK726187 | MK654791 | MK691216 | – | + | |
| D. padina | PSCG 160 | P. pyrifolia | Nanchang, Jiangxi | MK626892 | MK691172 | MK726249 | MK654851 | MK691261 | – | + |
| D. parvae | PSCG 034 * | P. bretschneideri | Kunming, Yunnan | MK626919 | / | MK726210 | MK654858 | MK691248 | + | – |
| PSCG 035 | P. bretschneideri | Kunming, Yunnan | MK626920 | MK691169 | MK726211 | MK654859 | MK691249 | + | – | |
| D. pescicola | PSCG 036 | P. bretschneideri | Kunming, Yunnan | MK626855 | MK691116 | MK726159 | MK654796 | MK691226 | + | – |
| PSCG 037 | P. bretschneideri | Kunming, Yunnan | MK626857 | MK691130 | MK726157 | MK654799 | MK691230 | – | + | |
| D. sojae | PSCG 177 | P. pyrifolia | Yingtan, Jiangxi | MK626940 | MK691188 | MK726189 | MK654882 | MK691302 | + | + |
| PSCG 283 | P. pyrifolia | Hangzhou, Zhejiang | MK626950 | MK691189 | MK726191 | MK654890 | MK691303 | + | + | |
| PSCG 481 | P. pyrifolia | Guiyang, Guizhou | MK626944 | MK691196 | MK726196 | MK654887 | MK691307 | + | + | |
| PSCG 486 | P. pyrifolia | Guiyang, Guizhou | MK626949 | MK691190 | MK726192 | MK654888 | MK691308 | + | + | |
| PSCG 488 | P. pyrifolia | Guiyang, Guizhou | MK626946 | MK691197 | MK726197 | MK654884 | MK691304 | + | + | |
| PSCG 490 | P. pyrifolia | Guiyang, Guizhou | MK626947 | MK691195 | MK726194 | MK654885 | MK691306 | + | + | |
| PSCG 492 | P. pyrifolia | Guiyang, Guizhou | MK626948 | MK691203 | MK726199 | MK654886 | MK691305 | + | + | |
| PSCG 502 | P. pyrifolia | Zhenjiang, Jiangsu | MK626941 | MK691191 | MK726193 | MK654891 | MK691309 | + | + | |
| PSCG 510 | P. pyrifolia | Zhenjiang, Jiangsu | MK626942 | MK691192 | MK726190 | MK654889 | MK691311 | + | + | |
| PSCG 518 | P. pyrifolia | Zhenjiang, Jiangsu | MK626945 | MK691192 | MK726198 | MK654883 | MK691312 | + | + | |
| PSCG 530 | P. pyrifolia | Zhenjiang, Jiangsu | MK626943 | MK691194 | MK726195 | MK654892 | MK691310 | + | + | |
| D. spinosa | PSCG 279 | P. pyrifolia | Hangzhou, Zhejiang | MK626925 | MK691126 | MK726155 | MK654801 | MK691235 | + | – |
| PSCG 383 * | P. pyrifolia | Nanjing, Jiangsu | MK626849 | MK691129 | MK726156 | MK654811 | MK691234 | – | + | |
| PSCG 388 | P. pyrifolia | Nanjing, Jiangsu | MK626860 | MK691128 | MK726171 | MK654798 | MK691229 | – | + | |
| PSCG 491 | P. pyrifolia | Guiyang, Guizhou | MK626856 | MK691127 | MK726170 | MK654807 | MK691237 | – | + | |
| D. taoicola | PSCG 292 | P. pyrifolia | Hangzhou, Zhejiang | MK626871 | MK691115 | MK726168 | MK654800 | MK691232 | – | + |
| PSCG 386 | P. pyrifolia | Nanjing, Jiangsu | MK626868 | MK691122 | MK726166 | MK654797 | MK691222 | + | – | |
| PSCG 413 | P. pyrifolia | Guiyang, Guizhou | MK626890 | MK691119 | MK726167 | MK654814 | MK691238 | – | + | |
| PSCG 485 | P. pyrifolia | Guiyang, Guizhou | MK626869 | MK691120 | MK726173 | MK654812 | MK691227 | – | + | |
| D. unshiuensis | PSCG 039 | P. bretschneideri | Kunming, Yunnan | MK626932 | MK691183 | MK726177 | MK654871 | MK691290 | + | – |
| PSCG 059 | P. pyrifolia | Wuhan, Hubei | MK626938 | MK691185 | MK726178 | MK654873 | MK691297 | + | – | |
| PSCG 060 | P. pyrifolia | Wuhan, Hubei | MK626929 | MK691179 | MK726185 | MK654875 | MK691292 | + | – | |
| PSCG 120 | P. pyrifolia | Sanming, Fujian | MK626926 | MK691174 | MK726174 | MK654868 | MK691288 | + | + | |
| PSCG 121 | P. pyrifolia | Sanming, Fujian | MK626936 | MK691175 | MK726180 | MK654876 | MK691289 | + | + | |
| PSCG 128 | P. pyrifolia | Sanming, Fujian | MK626927 | MK691184 | MK726175 | MK654880 | MK691295 | + | + | |
| PSCG 131 | P. pyrifolia | Sanming, Fujian | MK626934 | MK691176 | MK726176 | MK654869 | MK691293 | + | – | |
| PSCG 331 | P. pyrifolia | Sanming, Fujian | MK626937 | MK691186 | MK726182 | MK654870 | MK691291 | + | + | |
| PSCG 335 | P. pyrifolia | Sanming, Fujian | MK626933 | MK691177 | MK726186 | MK654881 | MK691299 | – | + | |
| PSCG 339 | P. pyrifolia | Sanming, Fujian | MK626928 | MK691181 | MK726188 | MK654879 | MK691300 | + | – | |
| PSCG 341 | P. pyrifolia | Sanming, Fujian | MK626935 | MK691182 | MK726183 | MK654878 | MK691296 | + | + | |
| PSCG 344 | P. pyrifolia | Sanming, Fujian | MK626931 | MK691187 | MK726181 | MK654874 | MK691298 | + | + | |
| PSCG 468 | P. pyrifolia | Sanming, Fujian | MK626939 | MK691180 | MK726184 | MK654872 | MK691301 | – | + | |
| PSCG 511 | P. pyrifolia | Zhenjiang, Jiangsu | MK626930 | MK691178 | MK726179 | MK654877 | MK691294 | – | + | |
| D. velutina | PSCG 134 | P. pyrifolia | Sanming, Fujian | MK626918 | MK691173 | MK726205 | MK654853 | MK691243 | + | – |
| PSCG 417 | P. pyrifolia | Guiyang, Guizhou | MK626921 | MK691152 | MK726206 | MK654854 | MK691244 | – | + | |
| D. zaobaisu | PSCG 031 * | P. bretschneideri | Kunming, Yunnan | MK626922 | / | MK726207 | MK654855 | MK691245 | + | – |
| PSCG 032 | P. bretschneideri | Kunming, Yunnan | MK626923 | / | MK726208 | MK654856 | MK691246 | + | – | |
| PSCG 033 | P. bretschneideri | Kunming, Yunnan | MK626924 | / | MK726209 | MK654857 | MK691247 | + | – | |
* = Ex-type culture. Newly described taxa and deposited sequences are in bold.
Phylogenetic analyses
New sequences generated in this study were blasted against the NCBIs GenBank nucleotide database to determine the closest relatives for a taxonomic framework of the studied isolates. Alignments of different gene regions, including sequences obtained from this study and sequences downloaded from GenBank (Table 2), were initially performed by using the MAFFT v. 7 online server (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh & Standley 2013) with default settings, and then manually adjusted in MEGA v. 7 (Kumar et al. 2016).
Table 2.
List of isolates of the Diaporthe species used in this study, with details about host/substrate, country, and GenBank accession numbers.
| Species | Culture1 | Host | Country | GenBank accession no. |
||||
|---|---|---|---|---|---|---|---|---|
| ITS | CAL | HIS | TEF | TUB | ||||
| D. acaciarum | CBS138862* | Acacia tortilis | Tanzania | KP004460 | – | KP004504 | – | KP004509 |
| D. alleghaniensis | CBS495.72 = ATCC 24097* | Betula alleghaniensis | Canada | KC343007 | KC343249 | KC343491 | KC343733 | KC343975 |
| D. alnea | CBS 146.46* | Alnus sp. | Netherlands | KC343008 | KC343250 | KC343492 | KC343734 | KC343976 |
| D. ampelina | CBS 114016* | Vitis vinifera | France | AF230751 | JX197443 | – | AY745056 | JX275452 |
| D. amygdali | CBS 126679* | Prunus dulcis | Portugal | KC343022 | KC343264 | KC343506 | KC343748 | KC343990 |
| CBS 115620 = FAU 1005 | Prunus persica | USA: | KC343020 | KC343262 | KC343504 | KC343746 | KC343988 | |
| D. anacardii | CBS 720.97* | Anacardium ocidentale | East Africa | KC343024 | KC343266 | KC343508 | KC343750 | KC343992 |
| D. angelicae | CBS 111592* | Heracleum sphondylium | Austria | KC343027 | KC343269 | KC343511 | KC343753 | KC343995 |
| D. apiculatum | CGMCC 3.17533* | Camellia sinensis | China | KP267896 | – | – | KP267970 | KP293476 |
| D. arctii | DP0482* | Arctium lappa | Austria | KJ590736 | KJ612133 | KJ659218 | KJ590776 | KJ610891 |
| D. arecae | CBS 161.64* | Areca catechu | India | KC343032 | KC343274 | KC343516 | KC343758 | KC344000 |
| ZJUD65 | Citrus sinensis | China | KJ490600 | – | KJ490542 | KJ490479 | KJ490421 | |
| ZJUD55 | Citrus sinensis | China | KJ490590 | – | KJ490532 | KJ490469 | KJ490411 | |
| CBS 535.75 | Citrus sp. | Suriname | KC343033 | KC343275 | KC343517 | KC343759 | KC344001 | |
| D. arengae | CBS 114979* | Arenga engleri | Hong Kong | KC343034 | KC343276 | KC343518 | KC343760 | KC344002 |
| D. baccae | CBS 136972* | Vaccinium corymbosum | Italy | KJ160565 | MG281695 | MF418264 | KJ160597 | MF418509 |
| D. batatas | CBS 122.21* | Ipomoea batatas | USA | KC343040 | KC343282 | KC343524 | KC343766 | KC344008 |
| D. beilharziae | BRIP 54792* | Indigofera australis | Australia | JX862529 | – | – | JX862535 | KF170921 |
| D. betulae | CFCC 50469* | Betula platyphylla | China | KT732950 | KT732997 | – | KT733016 | KT733020 |
| D. betulina | CFCC 52560* | Betula albosinensis | China | MH121495 | MH121419 | MH121455 | MH121537 | MH121577 |
| D. bicincta | CBS 121004* | Juglans sp. | USA | KC343134 | KC343376 | KC343618 | KC343860 | KC344102 |
| D. biguttusis | CGMCC 3.17081* | Lithocarpus glabra | China | KF576282 | – | – | KF576257 | KF576306 |
| D. camptothecicola | CFCC 51632 | Camptotheca acuminata | China | KY203726 | KY228877 | KY228881 | KY228887 | KY228893 |
| D. caryae | CFCC 52563* | Carya illinoensis | China | MH121498 | MH121422 | MH121458 | MH121540 | MH121580 |
| CFCC 52564 | Carya illinoensis | China | MH121499 | MH121423 | MH121459 | MH121541 | MH121581 | |
| D. castaneae | DNP 128* | Castanea mollissima | China | JF957786 | JX197430 | – | JX275401 | JX275438 |
| D. celastrina | CBS 139.27* | Celastrus sp. | USA | KC343047 | KC343289 | KC343531 | KC343773 | KC344015 |
| D. celeris | CPC 28262 | Vitis vinifera | Czech Republic | MG281017 | MG281712 | MG281363 | MG281538 | MG281190 |
| D. cercidis | CFCC 52565* | Cercis chinensis | China | MH121500 | MH121424 | MH121460 | MH121542 | MH121582 |
| CFCC 52566 | Cercis chinensis | China | MH121501 | MH121425 | MH121461 | MH121543 | MH121583 | |
| D. chamaeropis | CBS 454.81* | Chamaerops humilis | Greece | KC343048 | KC343290 | KC343532 | KC343774 | KC344016 |
| CBS 753.70 | Spartium junceum | Croatia | KC343049 | KC343291 | KC343533 | KC343775 | KC344017 | |
| D. charlesworthii | BRIP 54884m* | Rapistrum rugostrum | Australia | KJ197288 | – | – | KJ197250 | KJ197268 |
| D. chensiensis | CFCC 52567* | Abies chensiensis | China | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| D. citri | CBS 135422* | Citrus sp. | USA | KC843311 | KC843157 | KJ490523 | KC843071 | KC843187 |
| D. citrichinensis | ZJUD96 | Citrus sp. | China | KJ490631 | – | KJ490573 | KJ49051 | KJ490452 |
| D. convolvuli | CBS 124654 = DP0727* | Convolvulus arvensis | Turkey | KC343054 | KC343296 | KC343538 | KC343780 | KC344022 |
| D. cotoneastri | DP0667 | Juglans cinerea | USA | KC843328 | KC843155 | – | KC84312 | KC843229 |
| D. cuppatea | CBS 117499 = STE-U 5431* | Aspalathus linearis | South Africa | KC343057 | KC343299 | KC343541 | KC343783 | KC344025 |
| D. cytosporella | FAU461* | Citrus limon | Italy | KC843307 | KC843141 | MF418283 | KC843116 | KC843221 |
| D. dorycnii | MFLUCC 17-1015* | Dorycnium hirsutum | Italy | KY964215 | – | – | KY964171 | KY964099 |
| D. ellipicola | CGMCC 3.17084* | Lithocarpus glabra | China | KF576270 | – | – | KF576245 | KF576294 |
| D. endophytica | CBS 133811 = LGMF916* | Schinus terebinthifolius | Brazil | KC343065 | KC343307 | KC343549 | KC343791 | KC344033 |
| D. eres | AR5193* | Ulmus sp. | Germany | KJ210529 | KJ434999 | KJ420850 | KJ210550 | KJ420799 |
| CBS 101742 | Fraxinus sp. | Netherlands | KC343073 | KC343315 | KC343557 | KC343799 | KC344041 | |
| DLR12A | Vitis vinifera | France | KJ210518 | KJ434996 | KJ420833 | KJ210542 | KJ420783 | |
| DP0438 | Ulmus minor | Netherlands | KJ210532 | KJ435016 | KJ420886 | KJ210553 | KJ420816 | |
| FAU506 | Cornus florida | USA | KJ210526 | KJ435012 | KJ420842 | JQ807403 | KJ420792 | |
| D. eugeniae | CBS 444.82 | Eugenia aromatica | Indonesia | KC343098 | KC343340 | KC343582 | KC343824 | KC344066 |
| D. foeniculina | CBS 111553* | Foeniculum vulgare | Spain | KC343101 | KC343343 | KC343585 | KC343827 | KC344069 |
| FAU460 | Citrus limon | Spain | KC843304 | KC843138 | – | KC843113 | KC843218 | |
| AR5151 | Citrus latifolia | USA | KC843303 | KC843137 | – | KC843112 | KC843217 | |
| D. fraxini-angustifoliae | MFLUCC 15-0748 | Vitis vinifera | China | KT459428 | KT459462 | – | KT459446 | 960500551 |
| D. fukushii | MAFF 625034 | Pyrus pyrifolia | Japan | JQ807469 | – | – | JQ807418 | – |
| D. fusicola | CGMCC 3.17087* | Lithocarpus glabra | China | KF576281 | KF576233 | – | KF576256 | KF576305 |
| CGMCC 3.17088 | Lithocarpus glabra | China | KF576263 | KF576221 | – | KF576238 | KF576287 | |
| D. ganjae | CBS 180.91* | Cannabis sativa | USA | KC343112 | KC343354 | KC343596 | KC343838 | KC344080 |
| D. gulyae | BRIP 54025* | Helianthus annuus | Australia | JF431299 | – | – | JN645803 | – |
| D. helianthi | CBS 592.81* | Helianthus annuus | Serbia | KC343115 | KC343357 | KC343599 | KC343841 | KC344083 |
| D. helicis | AR5211= CBS 138596* | Hedera helix | France | KJ210538 | KJ435043 | KJ420875 | KJ210559 | KJ420828 |
| D. hongkongensis | CBS 115448* | Dichroa febrífuga | China | KC343119 | KC343361 | KC343603 | KC343845 | KC344087 |
| ZJUD74 | Citrus unshiu | China | KJ490609 | – | – | KJ490488 | KJ490430 | |
| D. incompleta | CGMCC 3.18288* | Camellia sinensis | China | KX986794 | KX999289 | KX999265 | KX999186 | KX999226 |
| D. inconspicua | CBS 133813* | Maytenus ilicifolia | Brazil | KC343123 | KC343365 | KC343607 | KC343849 | KC344091 |
| D. infecunda | LGMF912 = CPC 20288 | Schinus terebinthifolius | Brazil | KC343128 | KC343370 | KC343612 | KC343854 | KC344096 |
| D. juglandicola | CFCC 51134* | Juglans mandshurica | China | KU985101 | KX024616 | KX024622 | KX024628 | KX024634 |
| D. kadsurae | CFCC 52586* | Kadsura longipedunculata | China | MH121521 | MH121439 | MH121479 | MH121563 | MH121600 |
| CFCC 52587 | Kadsura longipedunculata | China | MH121522 | MH121440 | MH121480 | MH121564 | MH121601 | |
| D. kongii | BRIP 54031* | Helianthus annuus | Australia | JF431301 | – | – | JN645797 | – |
| D. limonicola | CPC 28200 = CBS 142549* | Citrus limon | Malta | MF418422 | MF418256 | MF418342 | MF418501 | MF418582 |
| D. litchicola | BRIP 54900* | Litchi chinensis | Australia | JX862533 | – | – | JX862539 | KF170925 |
| D. lithocarpus | CGMCC 3.15175* | Lithocarpus glabra | China | KC153104 | KF576235 | – | KC153095 | KF576311 |
| CGMCC 3.17098 | Lithocarpus glabra | China | KF576276 | KF576228 | – | KF576251 | KF576300 | |
| D. longicicola | CGMCC 3.17089* | Lithocarpus glabra | China | KF576267 | – | – | KF576242 | KF576291 |
| D. longicolla | FAU644 | Glycine max | USA | KJ590730 | KJ612126 | KJ659190 | KJ590769 | KJ610885 |
| FAU599 | Glycine max | USA | KJ590728 | KJ612124 | KJ659188 | KJ590767 | KJ610883 | |
| D. lusitanicae | CBS 123212* | Foeniculum vulgare | Portugal | KC343136 | KC343378 | KC343620 | KC343862 | KC344104 |
| D. mahothocarpus | CGMCC 3.15181* | Lithocarpus glabra | China | KC153096 | KT459461 | – | KC153087 | KF576312 |
| D. maritima | NB464-3A | Picea rubens | Canada | KU552027 | – | – | KU552022 | KU574616 |
| D. masirevicii | BRIP 57892a* | Helianthus annuus | Australia | KJ197277 | – | – | KJ197239 | KJ197257 |
| D. melitensis | CPC 27873 = CBS 142551 | Citrus limon | Malta | MF418424 | MF418258 | MF418344 | MF418503 | MF418584 |
| D. melonis | CBS 507.78* | Glycine soja | USA | KC343141 | KC343383 | KC343625 | KC343867 | KC344109 |
| D. middletonii | BRIP 54884e* | Rapistrum rugostrum | Australia | KJ197286 | – | – | KJ197248 | KJ197266 |
| D. miriciae | BRIP 54736j* | Helianthus annuus | Australia | KJ197282 | – | – | KJ197244 | KJ197262 |
| D. momicola | MFLUCC 16-0113 | Prunus persica | China | KU557563 | KU557611 | – | KU557631 | KU557587 |
| D. musigena | CBS 129519* | Musa sp. | Australia | KC343143 | KC343385 | KC343627 | KC343869 | KC344111 |
| D. neilliae | CBS 144. 27* | Spiraea sp. | USA | KC343144 | KC343386 | KC343628 | KC343870 | KC344112 |
| D. neoarctii | CBS 109490* | Ambrosia trifida | USA | KC343145 | KC343387 | KC343629 | KC343871 | KC344113 |
| D. neotheicola | CBS 123209 | Foeniculum vulgare | Portugal | GQ250192 | – | – | GQ250316 | – |
| D. nobilis | CBS 200.39 | Laurus nobilis | Germany | KC343151 | KC343393 | KC343635 | KC343877 | KC344119 |
| CBS 587.79 | Pinus pantepella | Japan | KC343153 | KC343395 | KC343637 | KC343879 | KC344121 | |
| D. novem | CBS 127270* | Glycine max, seed | Croatia | KC343156 | KC343398 | KC343640 | KC343882 | KC344124 |
| D. ovoicicola | CGMCC 3.17093* | Citrus sp. | China | KF576265 | KF576223 | – | KF576240 | KF576289 |
| D. padina | CFCC 52590* | Padus racemosa | China | MH121525 | MH121443 | MH121483 | MH121567 | MH121604 |
| D. pascoei | BRIP 54847* | Persea americana | Australia | JX862532 | – | – | JX862538 | KF170924 |
| D. passifloricola | CBS 141329* | Passiflora foetida | Malaysia | KX228292 | – | KX228367 | – | KX228387 |
| D. penetriteum | CGMCC 3.17532 | Camellia sinensis | China | KP267879 | – | KP293532 | KP267953 | KP293459 |
| D. perseae | CBS 151.73* | Persea gratissima | Netherlands | KC343173 | KC343415 | KC343657 | KC343899 | KC344141 |
| D. pescicola | MFLUCC 16-0105* | Prunus persica | China | KU557555 | KU557603 | – | KU557623 | KU557579 |
| MFLUCC 16-0106 | Prunus persica | China | KU557556 | KU557604 | – | KU557624 | KU557580 | |
| D. phaseolorum | CBS 116019 = STAM 30 | Caperonia palustris | USA | KC343175 | KC343417 | KC343659 | KC343901 | KC344143 |
| D. phragmitis | CBS 138897* | Phragmites australis | China | KP004445 | – | KP004503 | – | KP004507 |
| D. podocarpi-macrophylli | LC6200 | Podocarpus macrophyllus | China | KX986769 | KX999276 | KX999240 | KX999161 | KX999201 |
| D. pseudomangiferae | CBS 101339* | Mangifera indica | Dominican Republic | KC343181 | KC343423 | KC343665 | KC343907 | KC344149 |
| D. pseudophoenicicola | CBS 462.69* | Phoenix dactylifera | Spain | KC343183 | KC343425 | KC343667 | KC343909 | KC344151 |
| LC6150 | Phoenix canariensis | Uruguay | KY011891 | – | – | KY011902 | – | |
| D. pterocarpi | MFLUCC 10-0571* | Pterocarpus indicus | Thailand | JQ619899 | JX197451 | – | JX275416 | JX275460 |
| D. pterocarpicola | MFLUCC 10-0580a* | Pterocarpus indicus | Thailand | JQ619887 | JX197433 | – | JX275403 | JX275441 |
| D. pulla | CBS 338.89* | Hedera helix | Yugoslavia | KC343152 | KC343394 | KC343636 | KC343878 | KC344120 |
| D. ravennica | MFLUCC 15–0480 | Tamarix sp. | Italy | KU900336 | – | – | KX426703 | KX377688 |
| D. rhusicola | CBS 129528* | Rhus pendulina | South Africa | JF951146 | KC843124 | – | KC843100 | KC843205 |
| D. sackstonii | BRIP 54669b* | Helianthus annuus | Australia | KJ197287 | – | – | KJ197249 | KJ197267 |
| D. schini | CBS 133181* | Schinus terebinthifolius | Brazil | KC343191 | KC343433 | KC343675 | KC343917 | KC344159 |
| D. sennae | CFCC 51636* | Senna bicapsularis | China | KY203724 | KY228875 | KY228879 | KY228885 | KY228891 |
| D. sennicola | CFCC 51634* | Senna bicapsularis | China | KY203722 | – | KY228873 | KY228883 | KY228889 |
| D. serafiniae | BRIP 55665a* | Helianthus annuus | Australia | KJ197274 | – | – | KJ197236 | KJ197254 |
| D. sojae | FAU635* | Glycine max | USA | KJ590719 | KJ612116 | KJ659208 | KJ590762 | KJ610875 |
| FAU455 | Stokesia laevis | USA | KJ590712 | KJ612109 | KJ659201 | KJ590755 | KJ610868 | |
| DP0601 | Glycine max | USA | KJ590706 | KJ612103 | KJ659195 | KJ590749 | KJ610862 | |
| AR3602 | Cucumis melo | Japan | KJ590714 | KJ612111 | KJ659203 | KJ590757 | KJ610870 | |
| D. stewartii | CBS 193.36 | Cosmos bipinnatus | USA | FJ889448 | JX197415 | – | GQ250324 | JX275421 |
| D. subclavata | ZJUD95* | Citrus sp. | China | KJ490630 | – | KJ490572 | KJ490509 | KJ490451 |
| D. subordinaria | CBS 464.90* | Plantago lanceolata | New Zealand | KC343214 | KC343456 | KC343698 | KC343940 | KC344182 |
| D. taoicola | MFLUCC 16-0117* | Prunus persica | China | KU557567 | – | – | KU557635 | KU557591 |
| D. tectonendophytica | MFLUCC 13–0471* | Tectona grandis | China | KU712439 | KU749354 | KX999266 | KU749367 | KU743986 |
| D. tectonigena | LC6512 | Camellia sinensis | China | KX986782 | KX999284 | KX999254 | KX999174 | KX999215 |
| D. terebinthifolii | CBS 133180* | Schinus terebinthifolius | Brazil | KC343216 | KC343458 | KC343700 | KC343942 | KC344184 |
| D. thunbergiicola | MFLUCC 12–0033* | Thunbergia laurifolia | Thailand | KP715097 | – | – | KP715098 | – |
| D. ueckerae | FAU656* | Cucumis melo | USA | KJ590726 | KJ612122 | KJ659215 | KJ590747 | KJ610881 |
| D. unshiuensis | ZJUD52* | Citrus sp. | China | KJ490587 | – | KJ490529 | KJ490466 | KJ490408 |
| ZJUD49 | Citrus sp. | China | KJ490584. | – | KJ490526 | KJ490463 | KJ490405 | |
| CFCC 52595 | Carya illinoinensis | China | MH121530 | – | MH121488 | MH121572 | MH121607 | |
| D. vaccinii | CBS 160.32 = IFO 32646* | Oxycoccus macrocarpos | USA | KC343228 | KC343470 | KC343712 | KC343954 | KC344196 |
| D. velutina | CGMCC 3.18286 = LC 4421* | Neolitsea sp. | China | KX986790 | – | – | KX999182 | KX999223 |
| D. vexans | FAU597 | Solanum sp. | Dominican Republic | KJ590734 | KJ612131 | KJ659216 | KJ590774 | KJ610889 |
| D. virgiliae | CMW40748 | Virgilia oroboides | South Africa | KP247566 | – | – | – | KP247575 |
| Diaporthella corylina | CBS 121124* | Corylus sp. | China | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
1 AR, DP, FAU: Isolates in culture collection of Systematic Mycology and Microbiology Laboratory, USDA-ARS, Beltsville, Maryland, USA; BRIP: Queensland Plant Pathology herbarium/culture collection, Australia; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; CFCC: China Forestry Culture Collection Center, China; CGMCC: China General Microbiological Culture Collection; CMW: culture collection of the Forestry and Agricultural Biotechnology Institute; DNP: First author’s personal collection (deposited in MFLUCC); LC: Corresponding author’s personal collection (deposited in laboratory State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences); LGMF: Culture collection of Laboratory of Genetics of Microorganisms, Federal University of Parana, Curitiba, Brazil; MAFF: MAFF Genebank Project, Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; ZJUD: Zhejiang University.
*= Ex-type culture.
Three phylogenetic analyses were conducted based on concatenated loci for the D. eres species complex, D. sojae species complex and the remaining species. Of these, concatenated ITS, TEF, CAL, HIS and TUB were used for the D. sojae species complex and the remaining isolates except for the D. eres species complex, for which only TEF, CAL, HIS and TUB were analysed. Bayesian inference (BI) was used to construct phylogenies using MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003). The best-fit models of nucleotide substitution for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses (Table 3). Two analyses of four Markov Chain Monte Carlo (MCMC) chains were conducted from random trees with 15 × 106 generations for the D. eres species complex, 2 × 106 for the D. sojae species complex, and 15 × 106 generations for the remainder of the Diaporthe species. The analyses were sampled every 1 000 generations, which were stopped once the average standard deviation of split frequencies was below 0.01. The first 25 % of the trees were discarded as the burn-in phase of each analysis, and the remaining trees were summarised to calculate the posterior probabilities (PP) of each clade being monophyletic.
Table 3.
Nucleotide substitution models used in the phylogenetic analyses.
| Loci/Genes | Eres clade | Sojae clade | Arecae clade and other taxa |
|---|---|---|---|
| ITS | – | SYM+I+G | SYM+I+G |
| TEF | HKY+G | HKY+I+G | HKY+I+G |
| CAL | HKY+G | HKY+G | GTR+I+G |
| HIS | GTR+I+G | GTR+G | GTR+I+G |
| TUB | HKY+G | HKY+I+G | HKY+G |
Additionally, maximum parsimony analyses (MP) were performed on the multi-locus alignment using PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2002). Phylogenetic trees were generated using the heuristic search option with Tree Bisection Reconnection (TBR) branch swapping and 1 000 random sequence additions. Max trees were set up to 5 000, branches of zero length collapsed, and all multiple parsimonious trees were saved. Clade stability was assessed using a bootstrap analysis with 1 000 replicates. Afterwards, tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated. Furthermore, IQtree v. 1.6.8 was used for maximum likelihood (ML) analysis. The analysis was performed with a GTR site substitution model. The branch support was evaluated with a bootstrapping (BS) method of 1 000 replicates (Hillis & Bull 1993). Phylogenetic trees were visualised in FigTree v. 1.4.2 (Rambaut 2014). The alignments and phylogenetic trees were deposited in TreeBASE (Study 24313).
Morphological analyses
Fungal morphology was accessed by culturing a 4-d-old mycelial disc (5 mm diam) on a Petri dish containing PDA, oatmeal agar (OA; Crous et al. 2019), synthetic nutrient-poor agar medium (SNA; Nirenberg 1976), and 2 % tap water agar supplemented with sterile pine needles (PNA; Smith et al. 1996), wild fennel stems (Santos et al. 2010), and alfalfa stems (Udayanga et al. 2014a), respectively. Cultures were incubated at 25 °C with a 14/10 h fluorescent light/dark cycle. Growth rate (mm/d) was determined by similarly establishing each isolate on PDA and colony diameters were measured daily for 3 d. The colony morphologies were recorded after 14 d. Colony colours were rated according to Rayner (1970). Moreover, the shapes, colours and sizes of sporocarps, conidia, conidiophores, asci and ascospores were observed under a compound microscope (Olympus BX63 or Olympus SZX16, Japan), and 30–50 conidia or ascospores were measured to determine their sizes unless no or less spores were produced.
Prevalence
The prevalence of Diaporthe species in sampled provinces and the Pyrus spp. involved was calculated as previously described (Fu et al. 2019). The Isolation Rate (RI) was calculated for each species with the formula, RI % = (NS/NI) × 100, where NS was the number of isolates from the same species, and NI was the total number of isolates from each sample-collected region or Pyrus sp. (Fu et al. 2019).
Pathogenicity
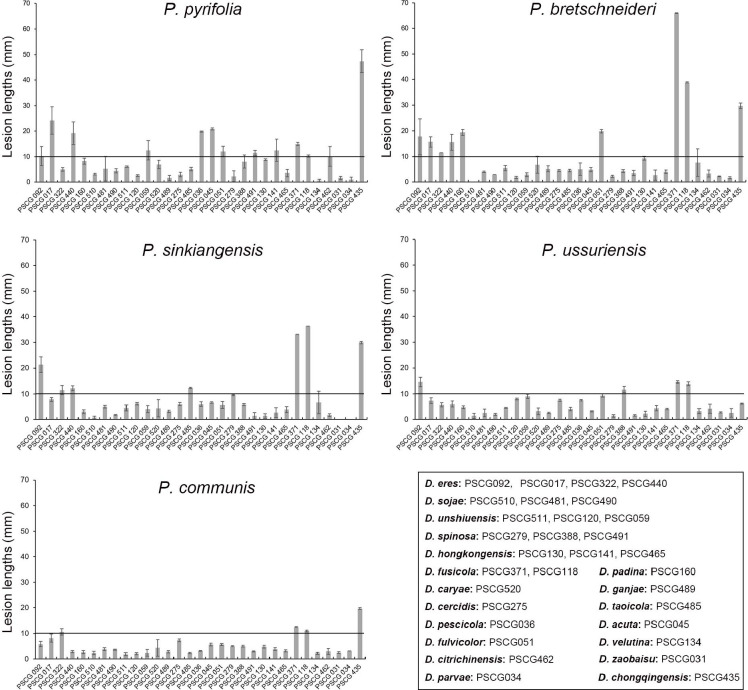
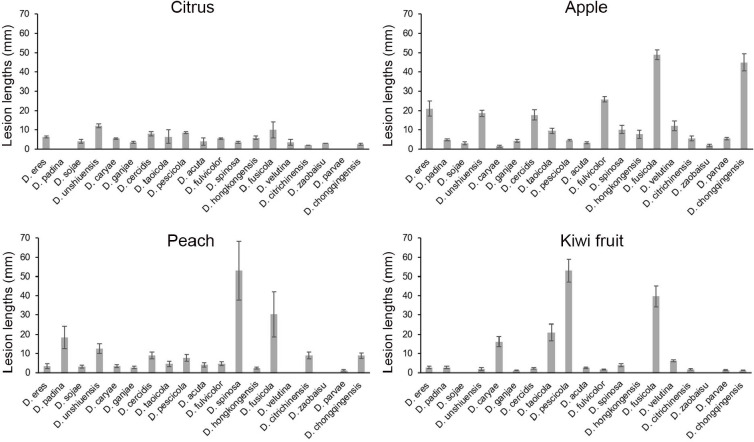
Host ranges were determined on detached shoots of P. pyrifolia cv. Hohsui, P. bretschneideri cv. Xuehua, P. ussuriensis cv. Hanxiang, P. communis cv. Docteun Jule Guyot, P. sinkiangensis cv. Kuerlexiangli, and other host plants, including Citrus reticulata cv. Rihui, Malus pumila cv. Hong Fushi, Prunus persica cv. Jinxiu, and Actinidia chinensis cv. Hongyang. Briefly, plant shoots 7.0 to 11.0 mm diam were disinfested with 75 % ethanol, and wounded between two of the closer buds with a punch (5 mm diam) on each shoot. Colonised PDA discs (5 mm diam) were excised from the colony margins after being cultured on PDA at 25 °C for 3 d, and inoculated in the hole of each shoot. Non-colonised PDA discs were used in parallel as controls. The inoculated shoots were incubated at 25 °C in plastic containers covered with a plastic film. Six branches were used for each inoculation treatment. A total of 31 isolates were used, namely: D. acuta (PSCG045), D. caryae (PSCG520), D. cercidis (PSCG275), D. chongqingensis (PSCG435), D. citrichinensis (PSCG462), D. eres (PSCG092, PSCG017, PSCG322, PSCG440), D. fulvicolor (PSCG051), D. fusicola (PSCG371, PSCG118), D. ganjae (PSCG489), D. hongkongensis (PSCG130, PSCG141, PSCG465), D. padina (PSCG160), D. parvae (PSCG034), D. pescicola (PSCG036), D. sojae (PSCG510, PSCG481, PSCG490), D. spinosa (PSCG279, PSCG388, PSCG491), D. taoicola (PSCG485), D. unshiuensis (PSCG511, PSCG120, PSCG059), D. velutina (PSCG134) and D. zaobaisu (PSCG031). The symptoms were recorded by taking photos, and the lesion lengths were measured at 8 dpi.
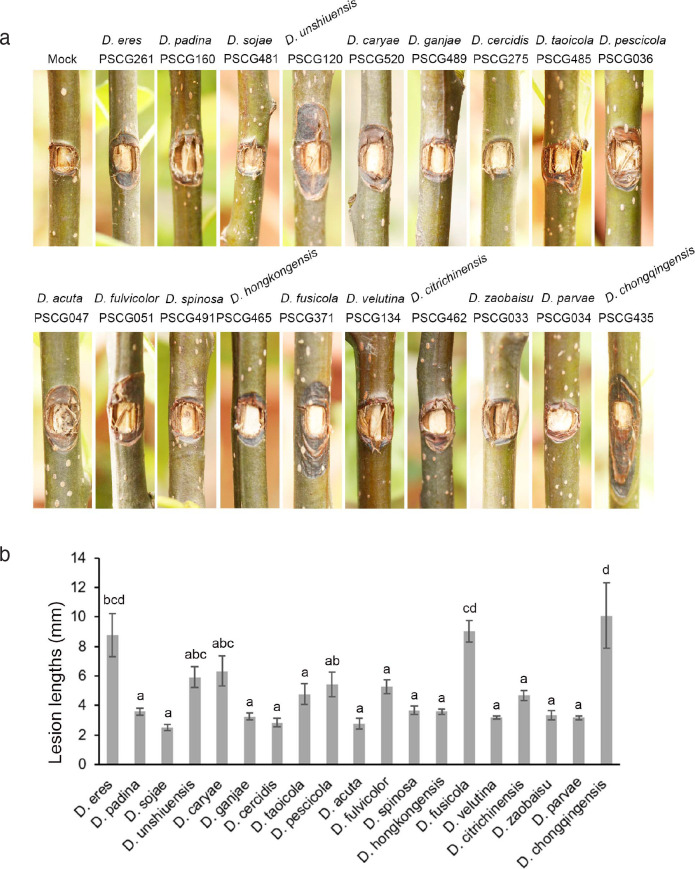
Pathogenicity tests were conducted by inoculating colonised PDA discs on intact shoots of 1-yr-old seedlings of P. pyrifolia cv. Cuiguan as described above. After inoculation, the seedlings were cultivated outdoors where the average daily lowest temperature was 15 °C and the highest temperature was 26 °C, with average humidity at 60 %. The tests were conducted in six repeats at two independent times. One representative isolate of each species was selected, namely: D. acuta (PSCG047), D. caryae (PSCG520), D. cercidis (PSCG275), D. chongqingensis (PSCG435), D. citrichinensis (PSCG462), D. eres (PSCG261), D. fulvicolor (PSCG051), D. fusicola (PSCG371), D. ganjae (PSCG489), D. hongkongensis (PSCG465), D. padina (PSCG160), D. parvae (PSCG034), D. pescicola (PSCG036), D. sojae (PSCG481), D. spinosa (PSCG491), D. taoicola (PSCG485), D. unshiuensis (PSCG120), D. velutina (PSCG134) and D. zaobaisu (PSCG033).
Mating-type test
The mating types (heterothallic or homothallic) were determined with a PCR-based mating type assay as previously described (Santos et al. 2010). The primers MAT1-1-1FW/MAT1-1-1RV were used for amplification of partial α1 box domain of the mating gene (MAT) MAT1-1-1, and primers MAT1-2-1FW/MAT1-2-1RV for amplification of partial HMG domain of the MAT1-2-1 gene.
RESULTS
Diaporthe isolates associated with pear shoot canker
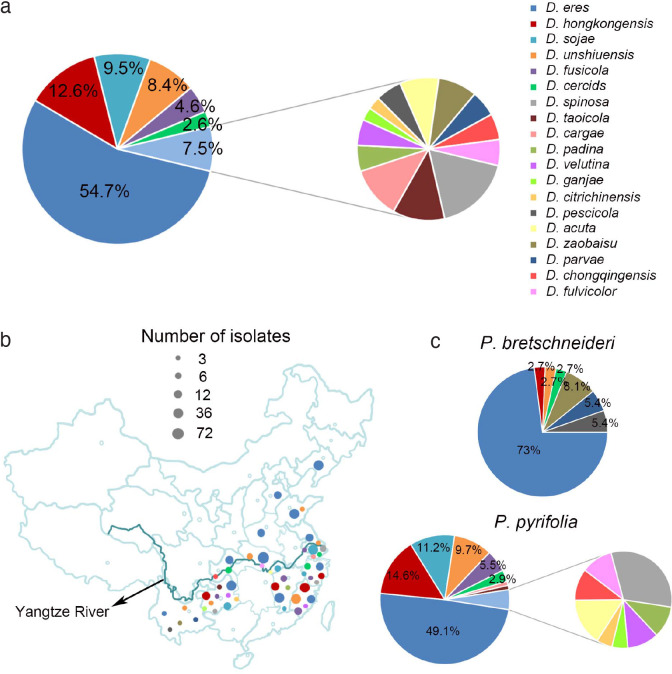
In the surveyed pear orchards, pear shoot canker showed symptoms including reddish brown canker lesions around buds (Fig. 1a, e), branch necrosis with oval or long cankers around branches (Fig. 1b–c), twig or branch cutting dieback (Fig. 1d), and curly white spore tendrils after rainfall in late summer (Fig. 1f). A total of 286 pear samples (shoots, branches, and twigs) affected by pear shoot canker collected from 12 provinces including Chongqing, Fujian, Guizhou, Hebei, Henan, Hubei, Jiangsu, Jiangxi, Liaoning, Shandong, Yunnan and Zhejiang provinces in China were subjected to fungal isolation, resulting in a total of 453 Diaporthe isolates identified based on morphology and ITS sequence data (see Appendix). However, no Diaporthe isolates were obtained from the samples collected from Jilin, Shanxi and Xinjiang provinces. A total of 113 representative isolates were chosen for further phylogenetic and taxonomic analyses (Table 1).
Fig. 1.

Representative symptoms of pear shoot canker on branches in the field. a. Newly developed reddish brown canker lesion around a bud of P. pyrifolia cv. Cuiguan; b–c. dieback symptoms resulting from lesion expansion around the branches of P. communis cv. Packham (b) and P. pyrifolia cv. Cuiguan (c); d. reddish brown necrosis at the cut of P. pyrifolia cv. Cuiguan; e. annular reddish brown lesion on branch of P. pyrifolia cv. Cuiguan; f. light-yellow spore tendrils released from pycnidia.
Phylogenetic analyses
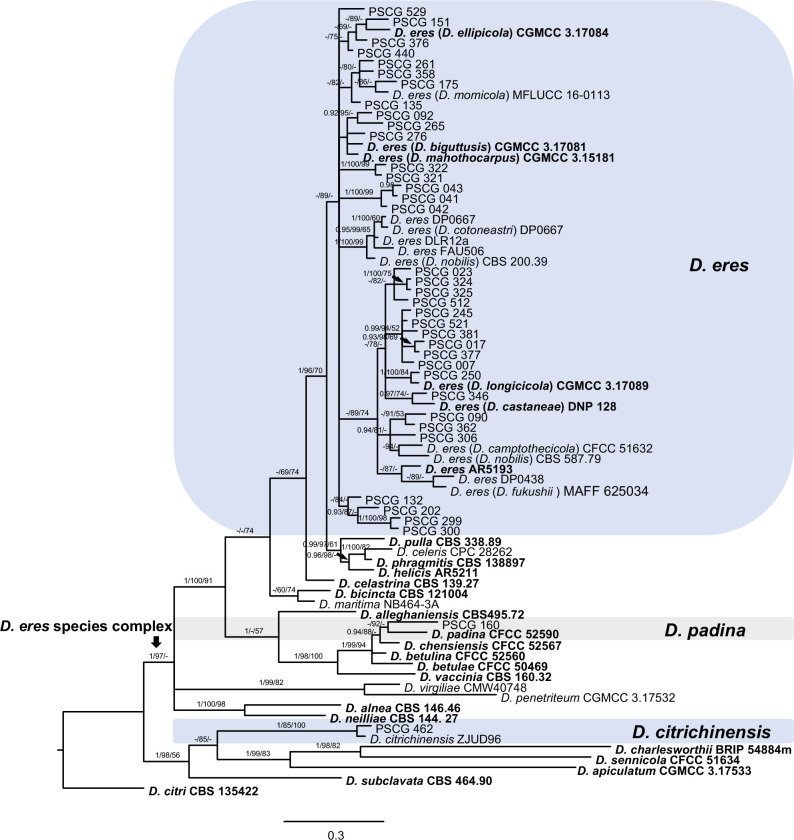
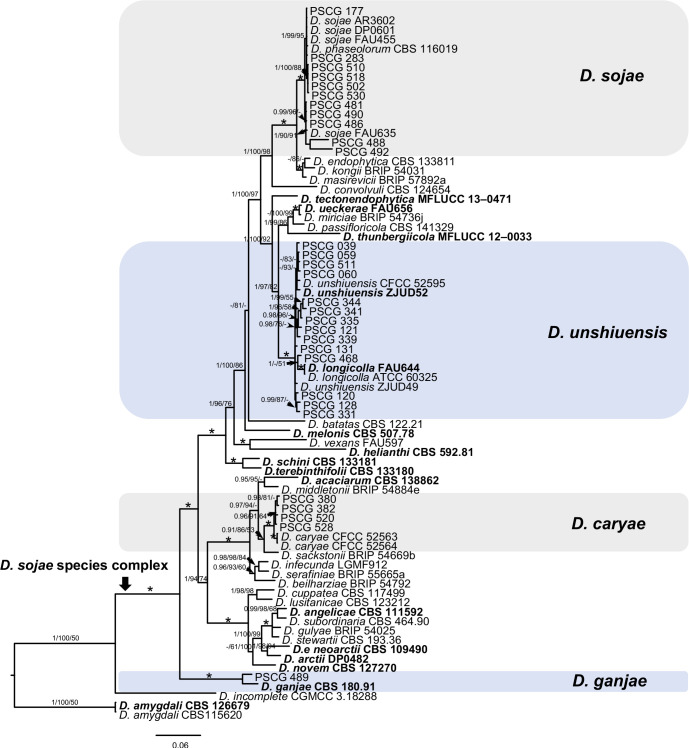
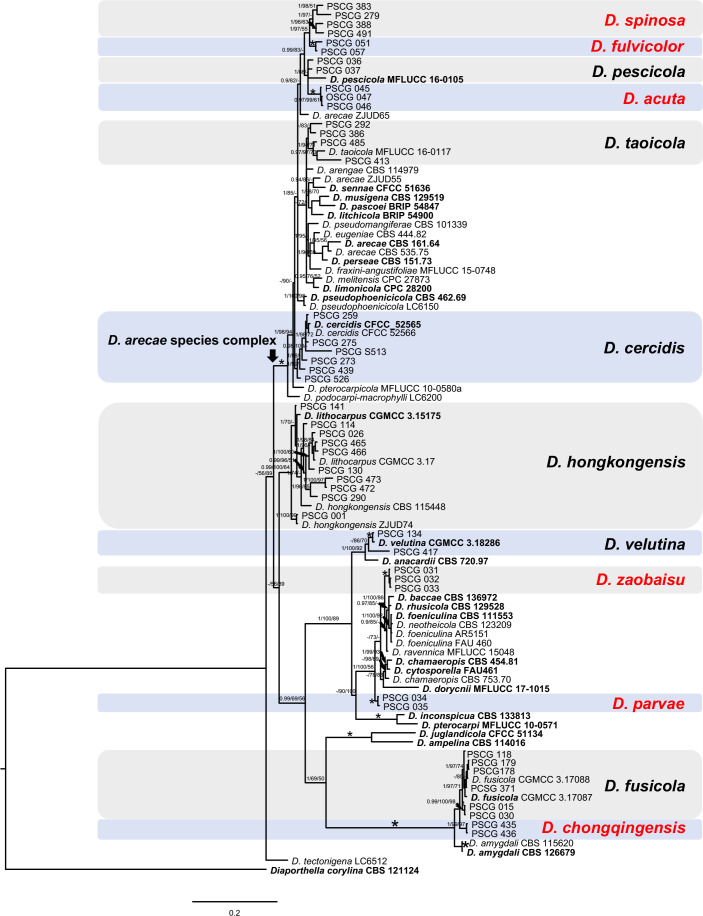
The 113 representative isolates (Table 1) were subjected to multi-locus phylogenetic analyses with concatenated ITS, TEF, CAL, HIS and TUB sequences together with 137 reference isolates from previously described species (Table 2). Results showed that these isolates clustered together with 19 species in three species complexes including D. eres (36 isolates), D. sojae (30) and D. arecae (21), and seven singleton species (26) (Fig. 2–4).
Fig. 2.
A Bayesian inference phylogenetic tree of 37 isolates in the D. eres species complex. The species D. citri (CBS 135422) was selected as an outgroup. The tree was built using concatenated sequences of the TEF, CAL, HIS and TUB genes. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %) and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/ML/MP). Ex-type strains were emphasized in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study. The scale bar indicates 0.3 expected changes per site.
Fig. 3.
A Bayesian inference phylogenetic tree of 30 isolates in the D. sojae species complex. The species D. amygdali (CBS 115620, CBS 126679) was selected as an outgroup. The tree was built using concatenated sequences of the ITS, TEF, CAL, HIS and TUB genes. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %) and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/ML/MP). The asterisk symbol (*) represents full support (1/100/100). Ex-type strains were emphasized in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study. The scale bar indicates 0.06 expected changes per site.
Fig. 4.
Phylogenetic tree generated by Bayesian analysis based on combined ITS, TEF, CAL, HIS and TUB sequence alignments of Diaporthe spp. The species Diaporthella corylina (CBS 121124) was selected as an outgroup. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %) and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/ML/MP). The asterisk symbol (*) represents full support (1/100/100). Ex-type strains were emphasized in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study. The scale bar indicates 0.2 expected changes per site.
In the phylogenetic tree constructed for the D. eres species complex, 37 isolates clustered in three clades corresponding to D. eres (35 isolates), D. padina (1) and D. citrichinensis (1) with a total of 1 504 characters including gaps (318 for TEF, 352 for CAL, 391 for HIS and 443 for TUB) included in the phylogenetic analysis (Fig. 2). Furthermore, D. biguttusis (CGMCC 3.17081), D. camptothecicola (CFCC 51632), D. ellipicola (CGMCC 3.17084), D. longicicola (CGMCC 3.17089), D. mahothocarpus (CGMCC 3.15181) and D. momicola (MFLUCC 16-0113) clustered together with D. eres, indicating that these species are synonyms of D. eres as previously proposed (Yang et al. 2018). In the D. sojae species complex, 30 isolates clustered into four clades corresponding to D. sojae (11 isolates), D. unshiuensis (14), D. caryae (4) and D. ganjae (1) (Fig. 3), with a total of 2 445 characters including gaps (480 for ITS, 380 for TEF, 560 for CAL, 539 for HIS and 482 for TUB) included in the phylogenetic analysis. In the D. arecae species complex, 12 isolates were assigned to three species, including D. cercidis (6), D. taoicola (4), D. pescicola (2), whereas nine isolates formed distinct clades with a highly supported subclade (1.00/100/100), which were identified as novel species and named D. spinosa (4), D. fulvicolor (2), and D. acuta (closely related to D. pescicola) (3), respectively. A total of 2 130 characters including gaps (510 for ITS, 296 for TEF, 437 for CAL, 465 for HIS, and 422 for TUB) were included in the multi-locus dataset. For the remaining isolates, 18 isolates were assigned to three species, including D. hongkongensis (10), D. fusicola (6) and D. velutina (2), whereas seven isolates formed distinct clades, and are identified as novel species, described as D. zaobaisu (3 isolates, closely related to D. ravennica), D. parvae (2) and D. chongqingensis (2, close to D. fusicola), respectively (Fig. 4).
TAXONOMY
Based on the morphology and multi-locus phylogeny, the 113 isolates were assigned to 19 species, including six newly described species. All species studied in culture are characterised below.
Diaporthe acuta Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830655; Fig. 5
Fig. 5.
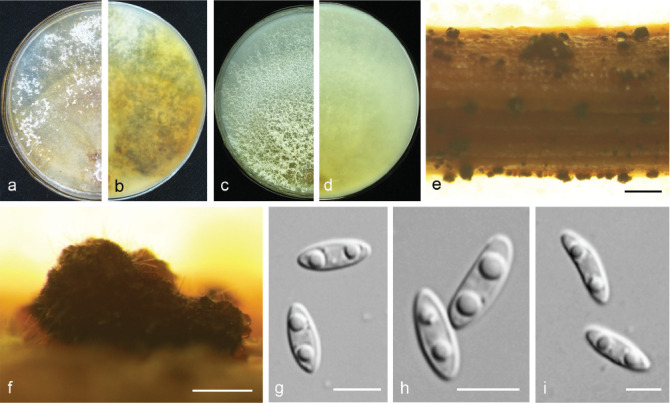
Diaporthe acuta (CGMCC 3.19600). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g–i. alpha conidia. — Scale bars: e = 1 mm; f = 200 μm; g–i = 5 μm.
Etymology. Named after the acute shape of both ends of its alpha conidia.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose or irregular, solitary or aggregated, exposed on the alfalfa stems surface, dark brown to black, 230–544 μm diam. Alpha conidia hyaline, aseptate, fusiform to oval, acutely round at both ends, bi- or multi-guttulate, 6–9.5 × 2–3 μm, mean ± SD = 7.8 ± 0.6 × 2.6 ± 0.2 μm, L/W ratio = 3 (n = 50). Beta and gamma conidia not observed.
Culture characteristics — Colonies on PDA with flattened mycelium, aerial mycelium scarce, flocculent scattered distribution, surface and reverse luteous. Colony diam 63–67 mm in 3 d at 28 °C. On OA with aerial mycelium white, fluffy, sulphur yellow pigment accumulation in the centre, pure white at the colony margin.
Materials examined. China, Hubei Province, Wuhan City, on branches of P. pyrifolia cv. Cuiguan, 1 Sept. 2014, Q. Bai (holotype HMAS 248147, culture ex-type CGMCC 3.19600 = PSCG 047); ibid., culture PSCG 045 and PSCG 046.
Notes — Three isolates were identified as D. acuta in a well-supported clade in the D. arecae species complex. This species is most closely related to D. pescicola, D. fulvicolor and D. spinosa, but easily distinguished from D. pescicola by 85 nucleotides difference in the concatenated alignment (40 in the ITS region, 6 TEF, 38 CAL and 1 TUB), from D. fulvicolor by 82 nucleotides difference (43 in the ITS region, 3 TEF, 17 CAL, 3 HIS and 16 TUB) and from D. spinosa by 24 nucleotides difference (13 in the ITS region, 7 CAL and 4 TUB). Moreover, D. acuta differs from D. pescicola in morphology, namely having smaller conidiomata (230–544 vs 637–881 μm), larger alpha conidia (6–9.5 × 2–3 vs 6–8 × 2–2.5 μm) (Table 4) and lacking beta conidia. However, its pycnidial conidiomata are larger than those of D. fulvicolor (230–544 vs 174–316 μm) and D. spinosa (230–544 vs 124–172 μm).
Table 4.
Conidial sizes of Diaporthe spp. studied.
| Species | Isolate No. | Conidia size ranges |
|||||
|---|---|---|---|---|---|---|---|
| Alpha conidia (μm) |
Beta conidia |
Means ± SD of conidia |
|||||
| Length (μm) | Width (μm) | Length (μm) | Width (μm) | Alpha conidia | Beta conidia | ||
| D. acuta | PSCG 047 | 6.14–9.53 | 2.20–2.94 | / | / | 7.76 ± 0.64 × 2.58 ± 0.17 | / |
| D. caryae | PSCG 528 | 5.23–7.07 | 2.16–3.00 | 24.36–30.82 | 0.99–1.50 | 6.17 ± 0.40 × 2.55 ± 0.19 | 27.56 ± 2.28 × 1.21 ± 0.15 |
| D. cercidis | PSCG 259 | 6.25–8.86 | 2.18–2.96 | / | / | 7.51 ± 0.67 × 2.50 ± 0.20 | / |
| D. chongqingensis | PSCG 435 | 5.27–7.69 | 2.08–2.94 | / | / | 6.39 ± 0.47 × 2.34 ± 0.18 | / |
| D. citrichinensis | PSCG 462 | 6.80–8.38 | 2.29–3.67 | 22.49–30.84 | 1.07–1.26 | 7.46 ± 0.42 × 2.74 ± 0.35 | 27.77 ± 4.60 × 1.17 ± 0.10 |
| D. eres | PSCG 321 | 5.14–7.15 | 2.00–2.89 | / | / | 6.23 ± 0.42 × 2.38 ± 0.18 | / |
| PSCG 377 | 6.22–8.11 | 2.28–3.39 | 21.58–39.28 | 1.03–1.65 | 7.07 ± 0.48 × 2.67 ± 0.24 | 32.98 ± 3.87 × 1.31 ± 0.17 | |
| PSCG 044 | 6.83–9.37 | 2.02–2.70 | 20.06–38.31 | 1.19–1.88 | 7.77 ± 0.58 × 2.38 ± 0.16 | 32.45 ± 5.31 × 1.43 ± 0.20 | |
| PSCG 250 | 5.43–8.27 | 1.92–2.78 | 30.34–37.31 | 1.10–1.40 | 6.49 ± 0.70 × 2.38 ± 0.21 | 33.45 ± 3.54 × 1.28 ± 0.16 | |
| PSCG 265 | / | / | 18.89–29.68 | 1.01–2.03 | / | 23.53 ± 2.69 × 1.51 ± 0.20 | |
| PSCG 276 | 6.08–8.68 | 2.58–3.37 | 21.50–30.34 | 1.08–1.86 | 7.46 ± 0.74 × 3.03 ± 0.32 | 26.14 ± 2.53 × 1.44 ± 0.16 | |
| PSCG 300 | 6.66–8.90 | 2.32–3.62 | 24.07–31.38 | 1.26–1.31 | 7.65 ± 0.54 × 3.05 ± 0.28 | 27.72 ± 5.16 × 1.29 ± 0.04 | |
| PSCG 325 | 6.58–7.92 | 2.22–3.04 | / | / | 7.14 ± 0.40 × 2.51 ± 0.18 | / | |
| PSCG 440 | 5.12–7.71 | 2.05–3.50 | 26.22–37.66 | 1.07–1.91 | 6.37 ± 0.69 × 2.62 ± 0.33 | 32.06 ± 2.93 × 1.32 ± 0.24 | |
| PSCG 529 | 5.74–7.51 | 2.11–2.90 | 24.96–36.81 | 1.13–1.57 | 6.41 ± 0.47 × 2.48 ± 0.22 | 29.95 ± 2.06 × 1.36 ± 0.12 | |
| PSCG 041 | 5.29–8.78 | 1.82–2.68 | 20.16–38.18 | 0.94–1.54 | 6.63 ± 0.67 × 2.25 ± 0.17 | 28.70 ± 3.83 × 1.29 ± 0.17 | |
| PSCG 092 | 7.06–9.13 | 2.48–3.63 | / | / | 8.10 ± 0.55 × 3.14 ± 0.26 | / | |
| PSCG 322 | 6.66–8.53 | 2.38–3.06 | / | / | 7.62 ± 0.46 × 2.69 ± 0.17 | / | |
| PSCG 358 | 5.96–7.17 | 2.25–2.83 | 28.94–39.48 | 1.05–1.60 | 6.58 ± 0.31 × 2.59 ± 0.15 | 33.84 ± 2.89 × 1.28 ± 0.18 | |
| PSCG 378 | 5.72–7.94 | 2.04–2.68 | 20.74–50.93 | 0.69–1.43 | 6.81 ± 0.48 × 2.34 ± 0.14 | 34.37 ± 8.27 × 1.20 ± 0.19 | |
| D. fulvicolor | PSCG 051 | 7.00–8.86 | 2.08–2.85 | / | / | 7.78 ± 0.44 × 2.52 ± 0.16 | / |
| D. fusicola | PSCG 015 | 5.18–7.15 | 1.76–2.44 | / | / | 6.20 ± 0.45 × 2.11 ± 0.16 | / |
| PSCG 118 | 4.86–6.89 | 1.76–3.17 | / | / | 5.83 ± 0.49 × 2.29 ± 0.27 | / | |
| PSCG 371 | 5.61–9.00 | 1.82–2.86 | / | / | 6.78 ± 0.68 × 2.22 ± 0.24 | / | |
| D. ganjae | PSCG 489 | 5.31–7.25 | 2.16–3.01 | / | / | 6.44 ± 0.41 × 2.62 ± 0.21 | / |
| D. hongkongensis | PSCG 465 | 5.44–8.32 | 1.89–2.69 | 14.01–22.64 | 0.93–1.46 | 6.88 ± 0.63 × 2.24 ± 0.17 | 16.75 ± 2.68 × 1.20 ± 0.18 |
| PSCG 466 | 6.06–8.98 | 1.79–2.87 | 14.67–23.92 | 0.80–1.35 | 7.15 ± 0.63 × 2.36 ± 0.22 | 19.20 ± 3.18 × 1.06 ± 0.17 | |
| PSCG 141 | 6.28–8.71 | 1.99–2.73 | 16.04–19.20 | 1.14–1.69 | 7.43 ± 0.63 × 2.29 ± 0.18 | 17.27 ± 1.42 × 1.41 ± 0.22 | |
| D. padina | PSCG 160 | 7.29–10.08 | 2.16–3.52 | 25.92–41.59 | 1.07–1.74 | 8.40 ± 0.63 × 2.86 ± 0.34 | 34.33 ± 3.32 × 1.33 ± 0.15 |
| D. pescicola | PSCG 036 | 6.05–7.77 | 1.93–2.75 | 21.17–30.63 | 1.12–1.74 | 6.99 ± 0.44 × 2.42 ± 0.17 | 24.99 ± 3.07 × 1.29 ± 0.21 |
| D. sojae | PSCG 486 | 6.29–7.83 | 2.32–3.20 | 14.58–23.09 | 1.09–1.81 | 7.00 ± 0.38 × 2.78 ± 0.19 | 18.78 ± 2.15 × 1.40 ± 0.17 |
| D. spinosa | PSCG 383 | 5.68–8.12 | 2.11–3.36 | 18.74–30.60 | 1.13–1.61 | 7.02 ± 0.64 × 2.58 ± 0.27 | 25.06 ± 2.76 × 1.34 ± 0.13 |
| PSCG 491 | 2.37 | 1.89–3.08 | 12.06–24.75 | 0.88–1.90 | 7.26 ± 0.85 × 2.78 ± 0.26 | 19.89 ± 3.25 × 1.41 ± 0.22 | |
| D. taoicola | PSCG 485 | 6.50–11.19 | 1.77–2.74 | / | / | 8.34 ± 0.94 × 2.31 ± 0.19 | / |
| D. unshiuensis | PSCG 120 | 5.48–6.72 | 2.12–2.61 | / | / | 5.94 ± 0.27 × 2.35 ± 0.13 | / |
| PSCG 128 | 4.22–6.84 | 2.18–2.83 | / | / | 5.44 ± 0.51 × 2.45 ± 0.15 | / | |
| PSCG 511 | 5.21–7.20 | 2.42–3.13 | / | / | 6.21 ± 0.52 × 2.81 ± 0.18 | / | |
| PSCG 468 | 5.08–7.01 | 2.25–2.83 | 21.07–32.33 | 1.16–1.43 | 5.92 ± 0.47 × 2.55 ± 0.15 | 27.56 ± 4.76 × 1.29 ± 0.13 | |
| PSCG 055 | 5.74–7.65 | 2.29–3.04 | / | / | 6.70 ± 0.53 × 2.62 ± 0.17 | / | |
| PSCG 059 | 4.53–6.35 | 2.01–2.77 | / | / | 5.53 ± 0.52 × 2.41 ± 0.20 | / | |
| D. velutina | PSCG 134 | 5.59–7.39 | 2.03–2.77 | / | / | 6.50 ± 0.43 × 2.41 ± 0.15 | / |
| D. zaobaisu | PSCG 032 | 5.23–6.90 | 2.12–2.58 | 21.43–28.16 | 0.86–1.44 | 5.96 ± 0.40 × 2.35 ± 0.09 | 24.52 ± 1.50 × 1.14 ± 0.14 |
| PSCG 033 | 5.38–8.45 | 1.89–2.90 | / | / | 6.83 ± 0.71 × 2.35 ± 0.27 | / | |
Diaporthe caryae C.M. Tian & Q. Yang, MycoKeys 39: 124. 2018 — Fig. 6
Fig. 6.
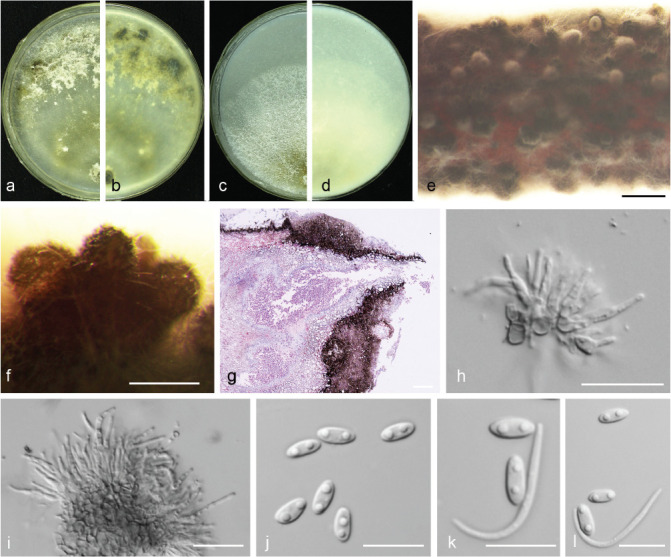
Diaporthe caryae (PSCG 528). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j. alpha conidia; k–l. alpha and beta conidia. — Scale bars: e = 1 mm; f–g = 200 μm; h–i = 20 μm; j–l =10 μm.
Description & Illustration — Yang et al. (2018).
Materials examined. China, Jiangsu Province, Nanjing City, on branches of P. pyrifolia cv. Cuiguan, 22 Aug. 2016, Y.S. Guo (culture PCSG 380, PCSG 382); Zhenjiang City, on branches of P. pyrifolia cv. Hohsui, 18 Nov. 2017, Y.S. Guo (culture PCSG 520, PCSG 528).
Notes — Diaporthe caryae was first reported on symptomatic twigs of Carya illinoensis in Jiangsu province, China (Yang et al. 2018). In this study, four isolates were identified as this species, and this is the first report of D. caryae responsible for pear shoot canker.
Pycnidial conidiomata of the isolate PSCG 528 are similar to the ex-type isolate CFCC 52563 (375–922 vs 450–836 μm). Alpha conidia of the isolate PSCG 528 are shorter than in isolate CFCC 52563 (5–7 × 2–3 vs 7–8.5 × 2–2.5 μm).
Diaporthe cercidis C.M. Tian & Q. Yang, MycoKeys 39: 124. 2018 — Fig. 7
Fig. 7.
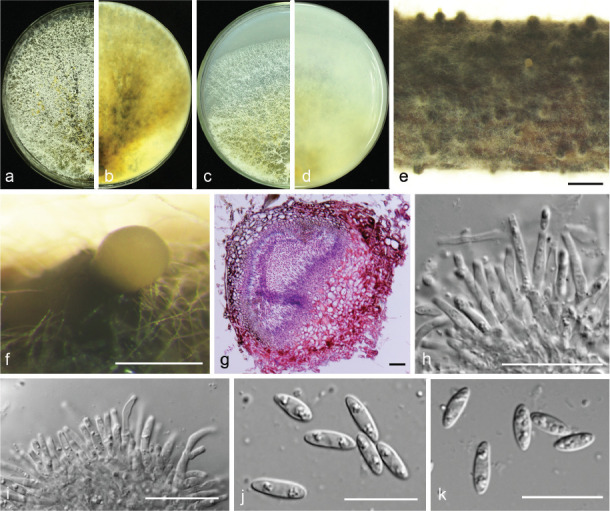
Diaporthe cercidis (PSCG 259). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j–k. alpha conidia. — Scale bars: e = 1 mm; f = 200 μm; g–h = 20 μm; j–k = 10 μm.
Description & Illustration — Yang et al. (2018).
Materials examined. China, Shandong Province, Yantai City, on branches of P. communis cv. Winter decana, 27 Nov. 2015, Y.S. Guo (culture PSCG 259); Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiyu, 7 Mar. 2016, Y.S. Guo (culture PSCG 273, PSCG 275); Chongqing City, on branches of P. pyrifolia cv. Huanghua, 29 Mar. 2017, Y.S. Guo (culture PSCG 439); Jiangsu Province, Zhenjiang City, on branches of P. pyrifolia cv. Aigansui, 18 Nov. 2017, Y.S. Guo (culture PCSG 513); ibid., on branches of P. pyrifolia cv. Hohsui, 18 Nov. 2017, Y.S. Guo (culture PCSG 526).
Notes — Diaporthe cercidis was first reported on twigs and branches of Cercis chinensis in Jiangsu province, China (Yang et al. 2018). In this study, six isolates were identified as belonging to this species, and this is the first report of D. cercidis responsible for pear shoot canker. The conidial size and morphology are similar to the ex-type isolate CFCC 52565, but the alpha conidia are multi-guttulate.
Diaporthe chongqingensis Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830656; Fig. 8
Fig. 8.
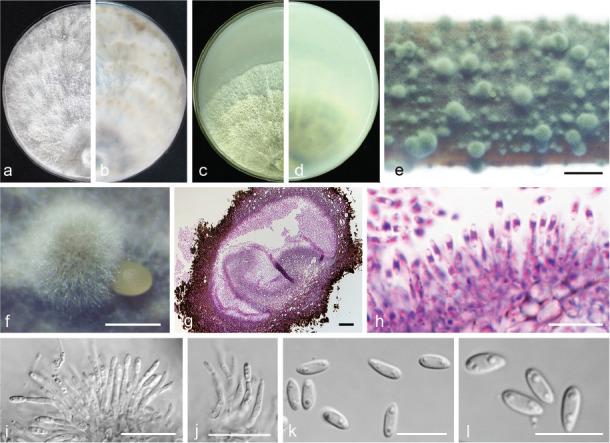
Diaporthe chongqingensis (CGMCC 3.19603). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–j. conidiophores; k–l. alpha conidia. — Scale bars: e = 2 mm; f = 500 μm; g = 50 μm; i–j = 20 μm; h, k–l = 10 μm.
Etymology. Referring to the city, Chongqing, where it was collected.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose, solitary or aggregated, wrapped in hyphae embedded in alfalfa stems surface, grey to black, 285–744 μm diam, yellowish translucent conidial drops exuded from the ostioles. Conidiophores hyaline, smooth, 1-septate, densely aggregated, unbranched, ampulliform, 6.5–12.5 × 2–6 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, straight, 14–26 × 1.5–2.5 μm, tapered towards the apex. Alpha conidia hyaline, aseptate, fusiform, biguttulate or multi-guttulate, acutely round at one end, 5.5–7.5 × 2–3 μm, mean ± SD = 6.4 ± 0.5 × 2.3 ± 0.2 μm, L/W ratio = 2.8 (n = 50). Beta and gamma conidia not observed.
Culture characteristics — Colony on PDA with flattened mycelium, white, smoke grey in the centre, reverse with smoke grey coloured pigments formed in the shape of a concentric ring pattern. Colony diam 40–49 mm in 3 d at 28 °C. On OA, colony with entire margin, grey olivaceous in the centre and white margin, reverse grey olivaceous pigments formed in the centre.
Materials examined. China, Chongqing City, on branches of P. pyrifolia cv. Huanghua, 29 Mar. 2017, Y.S. Guo (holotype HMAS 248148, culture ex-type CGMCC 3.19603 = PSCG 435); ibid., culture PSCG436.
Notes — Diaporthe chongqingensis is introduced based on the multi-locus phylogenetic analysis, with two isolates clustering separately in a well-supported clade (BI/ML/MP = 1/100/100). Diaporthe chongqingensis is most closely related to D. fusicola, but distinguished based on ITS and TEF loci from D. fusicola (96.6 % in ITS and 97 % in CAL) by 24 nucleotides in the concatenated alignment, in which 15 are distinct in the ITS region, six in the TEF region and three in the TUB region. Morphologically, D. chongqingensis differs from D. fusicola in its smaller alpha conidia (5.5–7.5 × 2–3 vs 5.5–9 × 2–3 μm).
Diaporthe citrichinensis F. Huang et al., Fungal Diversity 61: 247. 2013 — Fig. 9
Fig. 9.
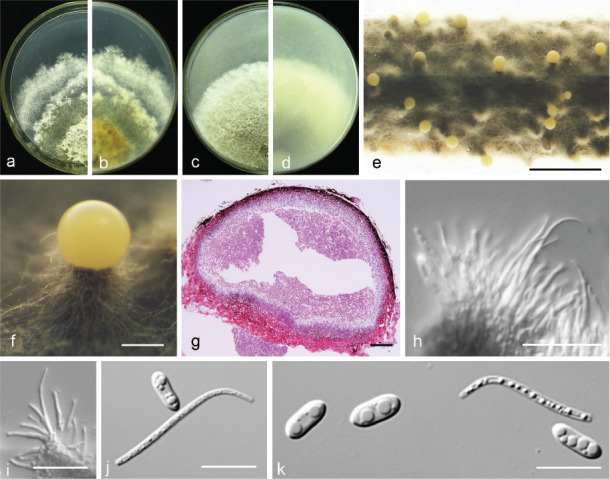
Diaporthe citrichinensis (PSCG 462). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j–k. alpha and beta conidia. — Scale bars: e = 2 mm; f = 200 μm; g = 50 μm; h–i = 20 μm; j–k = 10 μm.
Description & Illustration — Huang et al. (2013).
Materials examined. China, Guizhou Province, Guiyang City, on branches of P. pyrifolia cv. Jinqiu, 5 Mar. 2018, Y.S. Guo (culture PSCG 462).
Notes — Diaporthe citrichinensis was originally described from deadwood of Citrus unshiu in Shaanxi province, China (Huang et al. 2013). Isolate PSCG 462 clustered together with D. citrichinensis (ZJUD34) in the multi-locus phylogenetic tree. This is the first report of D. citrichinensis responsible for pear shoot canker. Pycnidial conidiomata of the ex-type isolate are slightly larger than those of the ex-type isolate ZJUD34 (375–922 vs 165–435 μm), and alpha and beta conidia of the ex-type are multi-guttulate.
Diaporthe eres Nitschke, Pyrenomyc. Germ. 2: 245. 1870 — Fig. 10
Fig. 10.
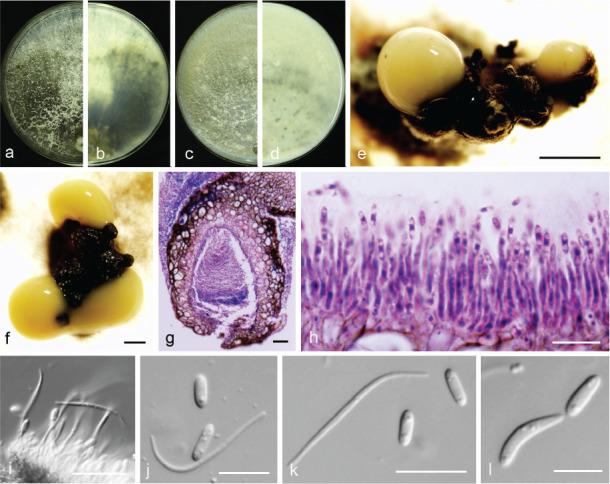
Diaporthe eres (PSCG 041). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e–f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j–k. alpha and beta conidia; l. alpha and gamma conidia. — Scale bars: e = 1 mm; f = 500 μm; g, i–j = 20 μm; h, k–l = 10 μm.
Synonym. Diaporthe nobilis Sacc. & Speg., Michelia 1(4): 386. 1878.
Description & Illustration — Udayanga et al. (2014b).
Materials examined. China, Henan Province, Nanyang City, on branches of P. pyrifolia cv. Wanqiuhuang, 17 Apr. 2016, Y.S. Guo (culture PCSG 321, PCSG 322, PCSG 325); Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiguan, 7 Mar. 2016, Y.S. Guo (PCSG 276); ibid., 22 Aug. 2016, Y.S. Guo (PCSG 377); Yunnan Province, Kunming City, on branches of P. bretschneideri cv. Zaobaisu, 17 Oct. 2014, Q. Bai (PCSG 041, PCSG 042); Chongqing City, on branches of P. pyrifolia cv. Huangguan, 27 Nov. 2016, Y.S. Guo (PCSG 250); Hubei Province, Wuhan City, on branches of P. pyrifolia cv. Jinshui, 27 Nov. 2016, Y.S. Guo (PCSG 265); ibid., on branches of P. pyrifolia cv. Yuanhuang, 10 Apr. 2017, Y.S. Guo (PCSG 440); Hebei Province, Cangzhou City, on branches of P. pyrifolia cv. Wanyu, 10 May 2016, Y.S. Guo (PCSG 300); Jiangsu Province, Zhenjiang City, on branches of P. pyrifolia cv. Hohsui, 18 Nov. 2017, Y.S. Guo (PCSG 529); Shandong Province, Yantai City, on branches of P. communis cv. Packham, 17 Oct. 2014, Q. Bai (PCSG 092); Liaoning Province, Yingkou City, on branches of P. pyrifolia cv. Huangjin, 29 June 2016, Y.S. Guo (PCSG 358).
Notes — Diaporthe eres is the type species of Diaporthe. It was described by Nitschke (1870) and collected from Ulmus sp. in Germany. It has a wide distribution and a broad host range as pathogen, endophyte or saprobe, and can cause a variety of plant diseases (Udayanga et al. 2014b). Recent studies indicated that D. biguttusis, D. camptothecicola, D. ellipicola, D. longicicola, D. mahothocarpus and D. momicola should be treated as synonyms of D. eres (Fan et al. 2018, Yang et al. 2018). The results of this study are consistent with the above. A large number of isolates clustered in D. eres. Bai et al. (2015) identified this species as responsible for pear shoot canker, and some of the isolates previously identified as P. fukushii were identified as D. eres in this study.
Diaporthe fulvicolor Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830657; Fig. 11
Fig. 11.
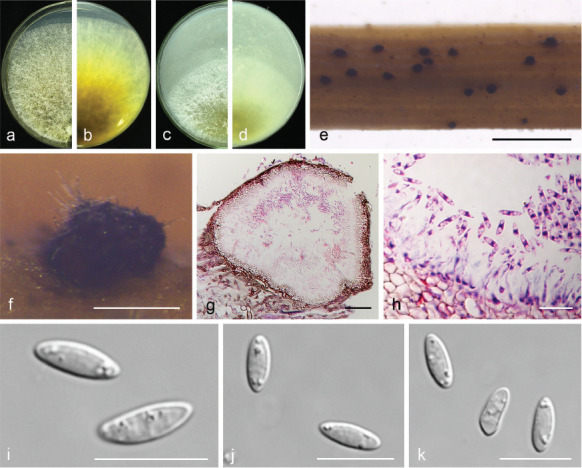
Diaporthe fulvicolor (CGMCC 3.19601). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h conidiophores; i–k. alpha conidia. — Scale bars: e = 2 mm; f = 200 μm; g = 50 μm; h–k = 10 μm.
Etymology. From Latin fulvi ‘tawny’, referring to tawny pigment accumulated in the centre of the colony.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose or irregular, solitary or aggregated, exposed on the alfalfa stems surface, dark brown to black, 174–316 μm diam. Conidiophores hyaline, smooth, 1-septate, densely aggregated, unbranched, cylindrical, straight, 5.5–8 × 2.5–3.5 μm. Conidiogenous cells phialidic, hyaline, terminal, ampulliform, 6.5–10 × 1.5–2.5 μm, tapered towards the apex. Alpha conidia hyaline, aseptate, fusiform to oval, acutely round at both ends, biguttulate or multi-guttulate, 7–9 × 2–3 μm, mean ± SD = 7.8 ± 0.4 × 2.5 ± 0.2 μm, L/W ratio = 3.1 (n = 50). Beta and gamma conidia not observed.
Culture characteristics — Colonies on PDA with aerial mycelium white, fluffy, reverse tawny pigment accumulation in the centre, surrounded by amber, pure white at the colony margin. Colony diam 52–55 mm in 3 d at 28 °C. On OA with entire margin, greyish yellow-green in the centre and white margin.
Materials examined. China, Hubei Province, Wuhan City, on branches of P. pyrifolia cv. Cuiguan, 1 Sept. 2014, Q. Bai (holotype HMAS 248149, culture ex-type CGMCC 3.19601 = PSCG 051); ibid., culture PSCG 057.
Notes — Diaporthe fulvicolor forms an independent clade in the D. arecae species complex (Fig. 4) and is phylogenetically distinct from D. pescicola and D. spinosa (described below). Diaporthe fulvicolor can be distinguished from D. pescicola in CAL and TUB loci by 57 nucleotide differences in concatenated alignment (40 in CAL and 17 in TUB), and from D. spinosa in CAL loci by 15 nucleotides (93 % in CAL). Moreover, D. fulvicolor differs from D. pescicola in having smaller conidiomata (174–316 vs 637–881 μm), and larger alpha conidia (7–9 × 2–3 vs 6–8 × 2–2.5 μm). Furthermore, D. fulvicolor differs from D. spinosa in its longer alpha conidia (7–9 × 2–3 vs 5.5–8 × 2–3.5 μm).
Diaporthe fusicola Y.H. Gao & L. Cai, Fungal Biol. 119: 300. 2015 — Fig. 12
Fig. 12.
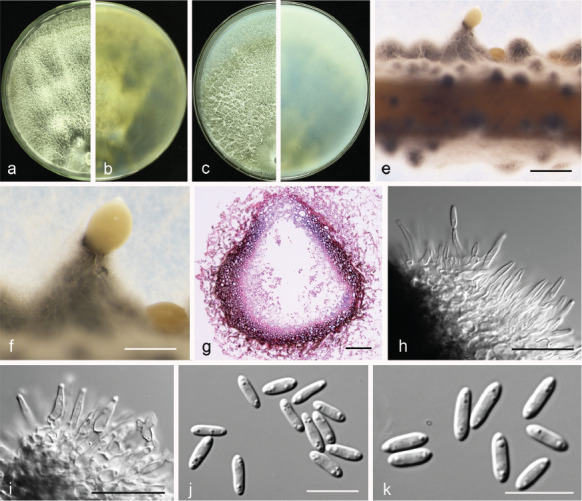
Diaporthe fusicola (PSCG 371). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j–k. alpha conidia. — Scale bars: e = 1 mm; f = 500 μm; g = 50 μm; h–i = 20 μm; j–k = 10 μm.
Description & Illustration — Gao et al. (2015).
Materials examined. China, Jiangxi Province, Fuzhou City, on branches of P. pyrifolia cv. Cuiyu, 2 Sept. 2014, Q. Bai (culture PSCG 015); Fujian Province, Sanming City, on branches of P. pyrifolia cv. Cuiyu, 10 Nov. 2014, Q. Bai (PSCG 118); Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiguan, 22 Aug. 2016, Y.S. Guo (PSCG 371).
Notes — Diaporthe fusicola was first described on leaves of Lithocarpus glabra in Zhejiang province, China (Gao et al. 2015). In this study, six isolates were identified as belonging to this species, and this is the first report of D. fusicola responsible for pear shoot canker. Bai et al. (2015) identified some of the isolates as P. amygdali, but they were identified as D. fusicola in this study.
Diaporthe ganjae R.R. Gomes et al., Persoonia 31: 22. 2013 — Fig. 13
Fig. 13.
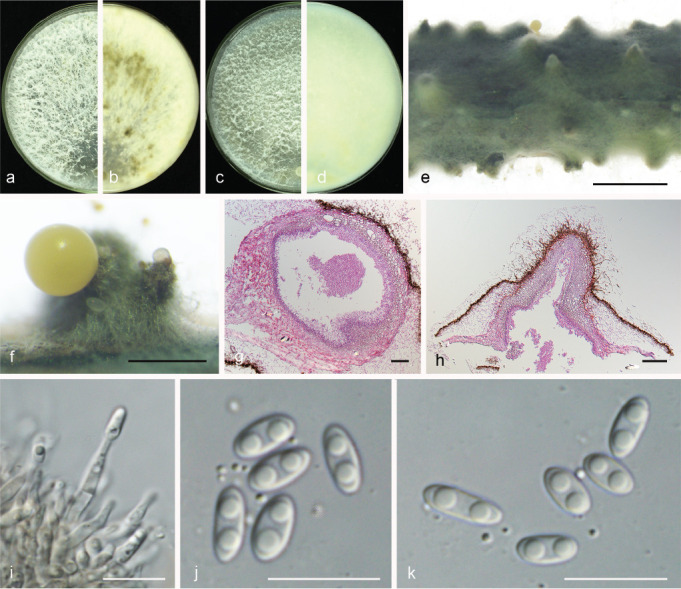
Diaporthe ganjae (PSCG 489). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g–h. section view of conidiomata; i. conidiophores; j–k. alpha conidia. — Scale bars: e = 2 mm; f = 500 μm; g = 50 μm; h = 100 μm; i–k = 10 μm.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose, conical or irregular, solitary or aggregated, exposed on the alfalfa stems surface, dark brown to black, 229–634 μm diam. Conidiophores hyaline, smooth, 1-septate, densely aggregated, unbranched, ampulliform, 5.5–7 × 2–4 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, 10.5–16 × 1.5–2.5 μm, tapered towards the apex. Alpha conidia hyaline, aseptate, fusiform to oval, obtuse rounded at both ends, biguttulate, 5.5–7.5 × 2–3 μm, mean ± SD = 6.4 ± 0.4 × 2.6 ± 0.2 μm, L/W ratio = 2.5 (n = 50). Beta and gamma conidia not observed.
Culture characteristics — Cultures on PDA with aerial mycelium white, fluffy, reverse with a mottled tawny pigment. Colony diam 79–81 mm in 3 d at 28 °C. On OA, colony with white aerial mycelium and lacking pigmentation.
Materials examined. China, Guizhou Province, Guiyang City, on branches of P. pyrifolia cv. Yuanhuang, 8 Nov. 2017, Y.S. Guo (culture PSCG 489).
Notes — Diaporthe ganjae was first reported from dead leaves of Cannabis sativa in Illinois, USA (Gomes et al. 2013). In this study, one isolate (PSCG 489) clustered together with the ex-type culture of D. ganjae (CBS 180.91) in the multi-locus phylogenetic tree (Fig. 3). This is the first description of its asexual morph and culture characteristics. Furthermore, this is the first report of D. ganjae responsible for pear shoot canker.
Diaporthe hongkongensis R.R. Gomes et al., Persoonia 31: 23. 2013 — Fig. 14
Fig. 14.
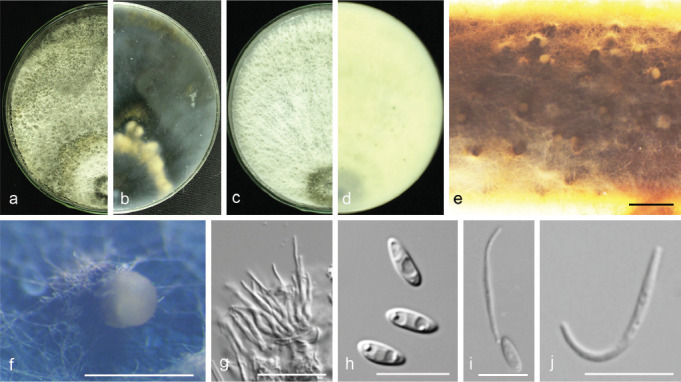
Diaporthe hongkongensis (PSCG 466). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. conidiophores; h. alpha conidia; i. alpha and beta conidia; j. beta conidia. — Scale bars: e = 1 mm; f = 200 μm; g = 20 μm; h–j = 10 μm.
Synonym. Diaporthe lithocarpi (Y.H. Gao et al.) Y.H. Gao & L. Cai, Fungal Biol. 119: 306. 2015. Nom. inval., Arts 41.1, F.5.1 (Shenzhen).
Description & Illustration — Gomes et al. (2013).
Materials examined. China, Fujian Province, Sanming City, on branches of P. pyrifolia cv. Cuiyu, 10 Nov. 2014, Q. Bai (PSCG 114); ibid., on branches of P. pyrifolia cv. Huanghua, 10 Nov. 2014, Q. Bai (culture PSCG 130, PSCG 141); Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiyu, 7 Mar. 2016, Y.S. Guo (culture PSCG 290); Fujian Province, Sanming City, on branches of P. pyrifolia cv. Cuiyu, 25 Nov. 2017, Y.S. Guo (PSCG 465, PSCG 466).
Notes — Diaporthe hongkongensis was first described from fruit of Dichroa febrifuga in Hong Kong, China (Gomes et al. 2013). This species often causes trunk diseases. In this study, 10 isolates were identified as belonging to this species, and this is the first report of D. hongkongensis responsible for pear shoot canker.
Diaporthe padina C.M. Tian & Q. Yang, MycoKeys 39: 137. 2018 — Fig. 15
Fig. 15.
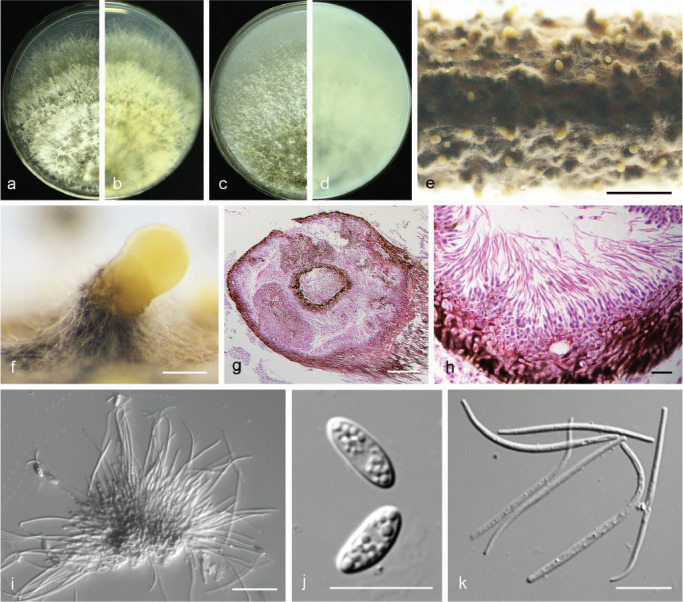
Diaporthe padina (PSCG 160). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j. alpha conidia; k. beta conidia. — Scale bars: e = 2 mm; f = 200 μm; g = 100 μm; i = 20 μm; h, j–k =10 μm.
Description & Illustration — Yang et al. (2018).
Materials examined. China, Jiangxi Province, Nanchang City, on branches of P. pyrifolia cv. Cuiguan, 27 Nov. 2014, Q. Bai (culture PSCG 160).
Notes — Diaporthe padina was first described from symptomatic twigs of Padus racemosa in Heilongjiang Province, China (Yang et al. 2018). In this study, one isolate was identified as belonging to this species, and this is the first report of D. padina responsible for pear shoot canker. Compared with the description of ex-type isolate CFCC 52590, pycnidial conidiomata of the isolate PSCG 160 are larger than CFCC 52590 (455–994 vs 330–520 μm), and conidiophores are longer (28–32 × 1–1.5 vs 5.5–12.5 × 1–1.5 μm). Alpha and beta conidia are both multi-guttulate, and longer than in isolate CFCC 52590 (alpha 7.5–10 × 2–3.5 vs 7–8 × 1.5–2 μm, beta 26–41.5 × 1–1.5 vs 21–24 × 1 μm).
Diaporthe parvae Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830658; Fig. 16
Fig. 16.
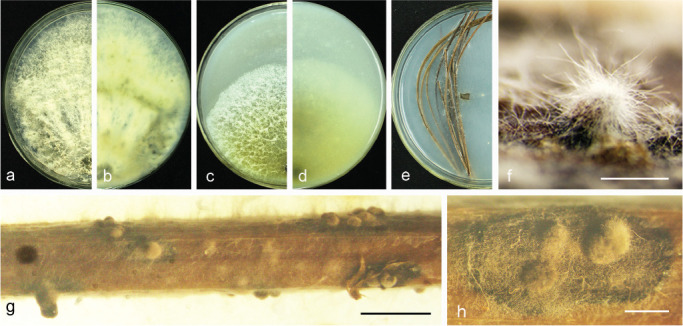
Diaporthe parvae (CGMCC 3.19599). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on PNA medium; f–h. conidiomata on alfalfa stems. — Scale bars: f = 100 μm; g = 5 mm; h = 1 mm.
Etymology. From Latin parva ‘small’, referring to smaller conidiomata.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose or irregular, solitary or aggregated, exposed on the alfalfa stems surface, dark brown to black, 253–455 μm diam. Alpha, beta and gamma conidia not observed.
Culture characteristics — Colony on PDA with flattened mycelium, white, reverse with non-uniform accumulation of citrine pigments. Colony 35.5–40 mm diam in 3 d at 28 °C. On OA with entire margin, aerial mycelium white, fluffy, citrine in the centre and white margin.
Materials examined. China, Yunnan Province, Kunming City, on branches of P. bretschneideri cv. Zaobaisu, 17 Oct. 2014, Q. Bai (holotype HMAS 248150, culture ex-type CGMCC 3.19599 = PSCG 034); ibid., culture PSCG 035.
Notes — Diaporthe parvae forms a distinct clade with high support (BI/ML/MP = 1/100/100), and differed with the closely related species (D. chamaeropis and D. cytosporella) on ITS and CAL loci (96 % in ITS and 83 % in CAL; and 98 % in ITS and 80 % in CAL, respectively). This species formed conidiomata-like structures, but remained sterile on various media including SNA, OA, PNA, fennel stems, alfalfa stems, pear stems and barleycorn at varied conditions, e.g., induced at black light and low temperatures, producing no conidiophores, conidiogenous cells and conidia.
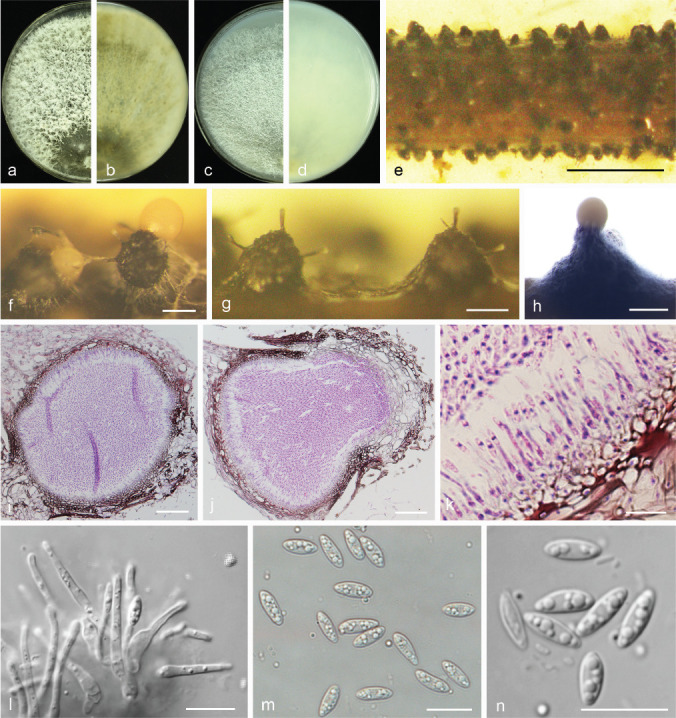
Diaporthe pescicola Dissanayake et al., Mycosphere 8: 542. 2017 — Fig. 17
Fig. 17.
Diaporthe pescicola (PSCG 036). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f–h. conidiomata; i–j. section view of conidiomata; k–l. conidiophores; m–n. alpha conidia. — Scale bars: e = 5 mm; f–g = 200 μm; h = 500 μm; i–j = 50 μm; k–n = 10 μm.
Description & Illustration — Dissanayake et al. (2017).
Materials examined. China, Shandong Province, Yantai City, on branches of P. bretschneideri cv. Zaobaisu, 17 Oct. 2014, Q. Bai (cultures PSCG 036, PSCG 037).
Notes — Diaporthe pescicola was first described from diseased shoots of Prunus persica in Hubei province, China (Dissanayake et al. 2017). In this study, two isolates (PSCG 036, PSCG 037) clustered together with the ex-type culture of D. pescicola (MFLUCC 16-0105) in the multi-locus phylogenetic tree (Fig. 4), and this is the first report of D. pescicola responsible for pear shoot canker.
Diaporthe sojae Lehman, Ann. Missouri Bot. Gard. 10: 128. 1923 — Fig. 18
Fig. 18.
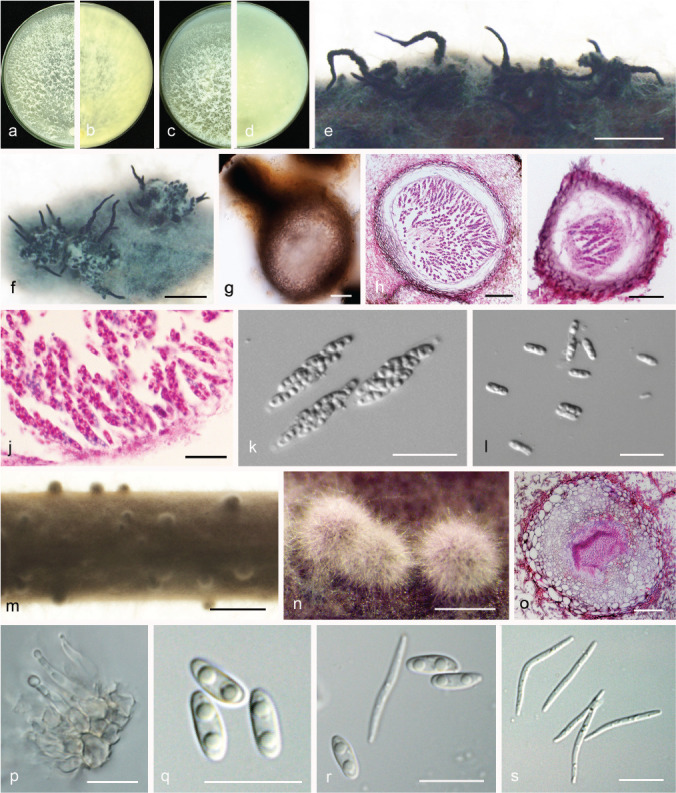
Diaporthe sojae (PSCG 486). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. ascomata on alfalfa stems; f. ascomata; g. ascoma; h–i. section view of ascoma; j–k. asci; l. ascospores; m. conidiomata on alfalfa stems; n. conidiomata; o. section view of conidiomata; p. conidiophores; q. alpha conidia; r. alpha and beta conidia; s. beta conidia. — Scale bars: e–f = 1 mm; g–h, o = 50 μm; i = 30 μm; j–l = 20 μm; m = 2 mm; n = 500 μm; p–s = 10 μm.
Description & Illustration — Udayanga et al. (2015).
Materials examined. China, Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiyu, 7 Mar. 2016, Y.S. Guo (culture PSCG 283); Guizhou Province, Guiyang City, on branches of P. pyrifolia cv. Yuanhuang, 8 Nov. 2017, Y.S. Guo (culture PSCG 481, PSCG 486, PSCG 488); Jiangsu Province, Zhenjiang City, on branches of P. pyrifolia cv. Hohsui, 18 Nov. 2017, Y.S. Guo (culture PCSG 502, PCSG 518); ibid., on branches of P. pyrifolia cv. Aigansui, 18 Nov. 2017, Y.S. Guo (culture PCSG 510); ibid., on branches of P. pyrifolia cv. Kousui, 18 Nov. 2017, Y.S. Guo (culture PCSG 530).
Notes — Diaporthe sojae was first reported on pods and stems of soybean, and subsequently reported on a wide range of hosts. It was also reported on some fruit trees in China, such as Vitis spp. (Dissanayake et al. 2015) and Citrus spp. (Huang et al. 2015). In this study, 11 isolates were identified as belonging to this species, and this is the first report of D. sojae responsible for pear shoot canker.
Compared with the description of the ex-type isolate FAU635, isolate PSCG 486 has shorter asci (33.5–39.5 × 6.5–9.5 vs 38.5–46.5 × 7–9 μm), slightly larger ascospores (10.5–13 × 3.5–4.5 vs 9.5–12 × 3–4 μm), and longer conidiogenous cells (8–14 vs 0.5–1 μm). Besides, beta conidia of isolate PSCG 486 were found to be hyaline, aseptate, multi-guttulate, filiform, curved, tapering towards both ends, 14.5–23 × 1–2 μm, mean ± SD = 18.8 ± 2.1 × 1.4 ± 0.2 μm, L/W ratio = 13.4.
Diaporthe spinosa Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830659; Fig. 19
Fig. 19.
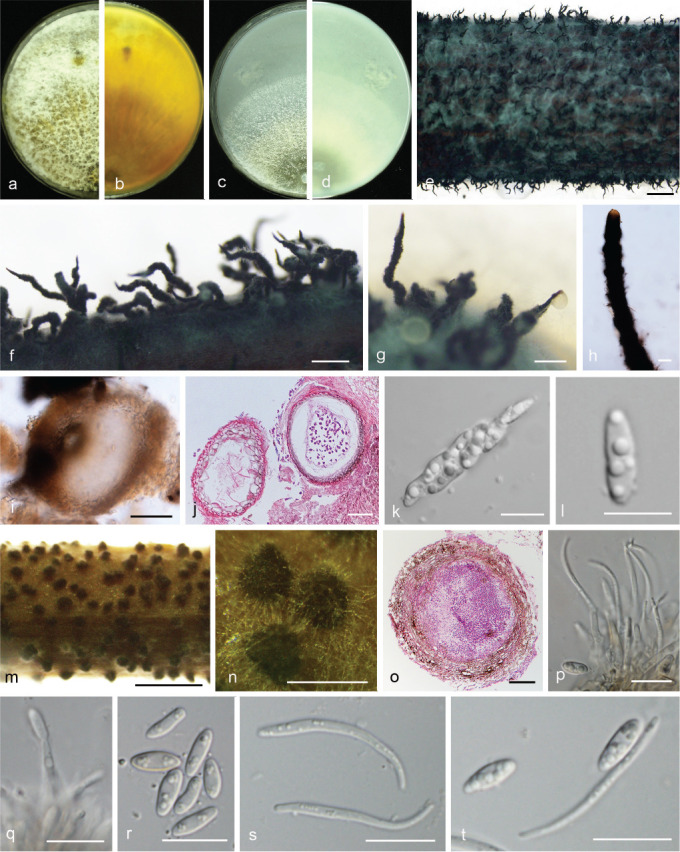
Diaporthe spinosa. a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. ascomata on alfalfa stems; f–g. ascomata; h. perithecial neck; i. ascoma; j. section view of ascoma; k. asci; l. ascospores; m. conidiomata on alfalfa stems; n. conidiomata; o. section view of conidiomata; p–q. conidiophores; r. alpha conidia; s. beta conidia; t. alpha and beta conidia (a–d, m–t. isolate PSCG 383; e–l. PSCG 491). — Scale bars: e, m = 2 mm; f–g, n = 500 μm; h–j, o = 50 μm; k–l, p–t = 10 μm.
Etymology. From Latin spinosus ‘spiny’, referring to its spiny perithecial necks.
Sexual morph on fennel stems. Ascomata black, deeply embedded in fennel stems surface, 702–1404 mm diam, densely clustered in groups, multiple tapering spiny perithecial necks protruding through substrata, 1235–1864 mm long. Perithecia oval to subglobose, dark brown, 67–215 μm, ostiolate. Asci fasciculate, unitunicate, 30.5–38.5 × 6–9 μm, 8-spored, sessile, elongate to clavate. Ascospores hyaline, two-celled, often biguttulate, elliptical to fusiform, 9.5–11.5 × 3–4 μm, mean ± SD = 10.5 ± 0.6 × 3.4 ± 0.3 μm, L/W ratio = 3.1 (n = 30). Asexual morph on alfalfa stems. Pycnidial conidiomata globose, solitary, exposed on the alfalfa stems surface, dark brown to black, 124–172 μm diam. Conidiophores hyaline, smooth, 1-septate, densely aggregated, unbranched, ampulliform, 6–9 × 3–4.5 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, straight, 8–29 × 1.5–2.5 μm, tapered towards the apex. Alpha conidia hyaline, aseptate, fusiform to oval, acutely round at both ends, biguttulate or multi-guttulate, 5.5–8 × 2–3.5 μm, mean ± SD = 7 ± 0.6 × 2.6 ± 0.3 μm, L/W ratio = 2.7 (n = 50). Beta conidia hyaline, aseptate, multi-guttulate, filiform, curved, tapering towards both ends, 18.5–30.5 × 1–1.5 μm, mean ± SD = 25.1 ± 2.8 × 1.3 ± 0.1 μm, L/W ratio = 19.3 (n = 38). Gamma conidia not observed.
Culture characteristics — Colony on PDA with fluffy mycelium, panniform, aerial mycelium white, reverse umber coloured, being darker at the centre and lighter at the edge. Colony diam 62.5–67.5 mm in 3 d at 28 °C. On OA, colony with entire margin, citrine green in the centre with a white margin.
Materials examined. China, Jiangsu Province, Nanjing City, on branches of P. pyrifolia cv. Cuiguan, 22 Aug. 2016, Y.S. Guo (holotype HMAS 248151, culture ex-type CGMCC 3.19602 = PCSG 383); ibid., culture PCSG 388; Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiguan, 7 Mar. 2016, Y.S. Guo (PCSG 279); Guizhou Province, Guizhou City, on branches of P. pyrifolia cv. Yuanhuang, 8 Nov. 2017, Y.S. Guo (PCSG 491).
Notes — Diaporthe spinosa forms a well-supported, independent clade in the D. arecae species complex (Fig. 4). It contains four isolates which are separated into two branches, with the former (PSCG 383, PSCG 279) differing from the latter (PSCG 388, PSCG 491) by unique fixed alleles in three loci including ITS positions 340 (C), 342 (G), 346 (A), 347 (A), 349 (G), 380 (T), CAL positions 368 (G), and HIS positions 162 (C), 163 (A), 191 (T), 193 (C), 194 (C), 195 (T), 205 (A), 213 (C), 404 (C), 417 (T), but without obvious differences in morphology of the asexual morph. Diaporthe spinosa is most closely related to D. pescicola and D. fulvicolor, but D. spinosa and D. pescicola can be clearly differentiated from the latter by 43 different unique fixed alleles in CAL loci, and 15 different unique fixed alleles in CAL loci can also distinguish D. spinosa from D. fulvicolor. This species differs from D. pescicola in its smaller conidiomata (124–172 vs 637–881 μm), and from D. fulvicolor in its shorter alpha conidia (5.5–8 × 2–3.5 vs 7–9 × 2–3 μm).
Diaporthe taoicola Dissanayake et al., Mycosphere 8: 543. 2017 — Fig. 20
Fig. 20.
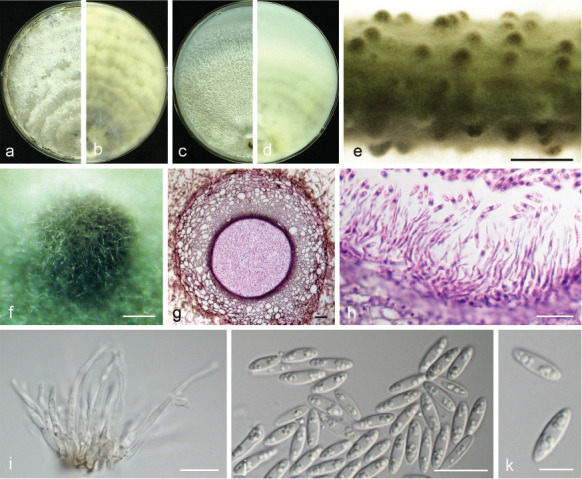
Diaporthe taoicola (PSCG 485). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. section view of conidiomata; h–i. conidiophores; j–k. alpha conidia. — Scale bars: e = 2 mm; f = 200 μm; g = 20 μm; h–j = 10 μm; k = 5 μm.
Description & Illustration — Dissanayake et al. (2017).
Materials examined. China, Zhejiang Province, Hangzhou City, on branches of P. pyrifolia cv. Cuiyu, 7 Mar. 2016, Y.S. Guo (culture PSCG 292); Guizhou Province, Guiyang City, on branches of P. pyrifolia cv. Jinqiu, 7 Mar. 2017, Y.S. Guo (culture PSCG 413); ibid., on branches of P. pyrifolia cv. Yuanhuang, 8 Nov. 2017, Y.S. Guo (culture PSCG 485).
Notes — Diaporthe taoicola was first described from diseased shoots of Prunus persica in Hubei province, China (Dissanayake et al. 2017). In this study, four isolates clustered together with the ex-type culture of D. taoicola (MFLUCC 16-0117) in the multi-locus phylogenetic tree (Fig. 4), and this is the first report of D. taoicola responsible for pear shoot canker.
Diaporthe unshiuensis F. Huang et al., Fungal Biol. 119: 344. 2015 — Fig. 21
Fig. 21.
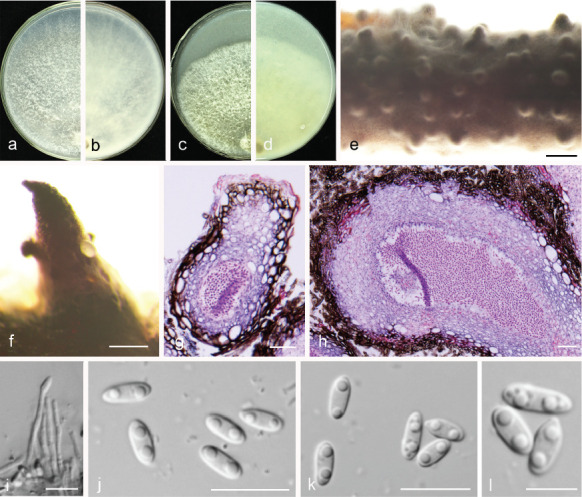
Diaporthe unshiuensis (PSCG 120). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g–h. section view of conidiomata; i. conidiophores; j–l. alpha conidia. — Scale bars: e = 1 mm; f = 200 μm; g–h = 20 μm; i–k = 10 μm; l = 5 μm.
Description & Illustration — Huang et al. (2015).
Materials examined. China, Fujian Province, Sanming City, on branches of P. pyrifolia cv. Minfu, 10 Nov. 2014, Q. Bai (culture PSCG 120); ibid., on branches of P. pyrifolia cv. Huanghua, 10 Nov. 2014, Q. Bai (PSCG 128); ibid., on branches of P. pyrifolia cv. Cuiyu, 25 Oct. 2017, Y.S. Guo (PSCG 468); Hubei Province, Wuhan City, on branches of P. pyrifolia cv. Cuiguan, 1 Sept. 2014, Q. Bai (PSCG 059); Jiangsu Province, Zhenjiang City, on branches of P. pyrifolia cv. Kousui, 18 Nov. 2017, Y.S. Guo (PSCG 511).
Notes — Diaporthe unshiuensis was initially described from twigs of asymptomatic Fortunella margarita in Zhejiang province, China (Huang et al. 2015). In this study, 14 isolates were identified as belonging to this species, and this is the first report of D. unshiuensis responsible for pear shoot canker. Bai et al. (2015) identified some of the isolates as P. longicolla, but they were re-identified as D. unshiuensis in this study.
Diaporthe velutina Y.H. Gao & L. Cai, IMA Fungus 8: 178. 2017 — Fig. 22
Fig. 22.
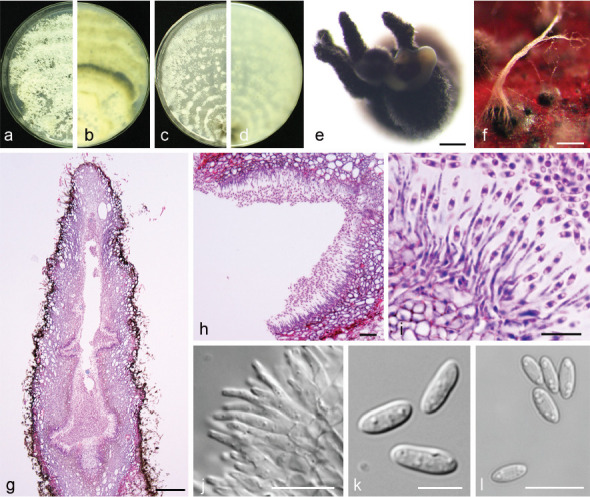
Diaporthe velutina (PSCG 134). a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e–f. conidiomata; g–h. section view of conidiomata; i–j. conidiophores; k–l. alpha conidia. — Scale bars: e–f = 200 μm; g = 100 μm; h = 20 μm; i–j, l = 10 μm; k = 5 μm.
Description & Illustration — Gao et al. (2017).
Materials examined. China, Fujian Province, Sanming City, on branches of P. pyrifolia cv. Huanghua, 10 Nov. 2014, Q. Bai (culture PSCG 134).
Notes — Diaporthe velutina was originally described from diseased leaves of Neolitsea sp. in Jiangxi province, China (Gao et al. 2017). In this study, one isolate (PSCG 134) clustered together with the ex-type culture of D. velutina (CGMCC 3.18286) in the multi-locus phylogenetic tree (Fig. 4), and this is the first report of D. velutina responsible for pear shoot canker. In this study, pycnidial conidiomata on alfalfa stems were globose, solitary or aggregated, exposed on the host surface, dark brown to black, 328–890 μm diam. Pycnidial conidiomata on PDA, OA or fennel stems were black, densely clustered in groups, with multiple tapering pycnidial necks protruding through substrata.
Diaporthe zaobaisu Y.S. Guo & G.P. Wang, sp. nov. — MycoBank MB830660; Fig. 23
Fig. 23.
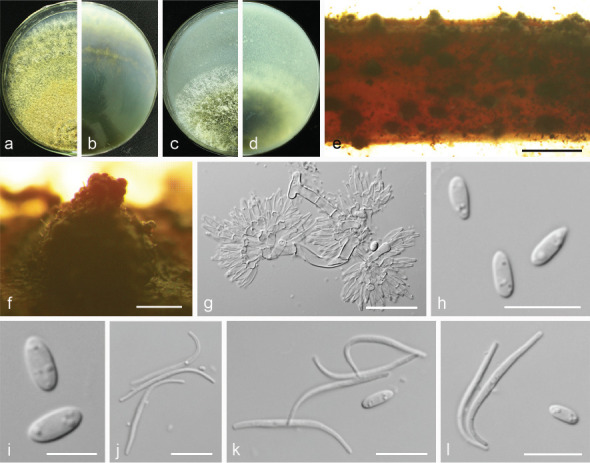
Diaporthe zaobaisu. a–d. Front and back view, respectively of colonies on PDA (a, b) and OA (c, d); e. conidiomata on alfalfa stems; f. conidiomata; g. conidiophores; h–i. alpha conidia; j. beta conidia; k–l. alpha and beta conidia (a–h. isolate PSCG 033; i–l. PSCG 032). — Scale bars: e = 2 mm; f = 200 μm; g = 20 μm; h, j–k = 10 μm; i = 5 μm.
Etymology. Referring to the host variety (P. bretschneideri cv. Zaobaisu), from which the fungus was isolated.
Sexual morph not observed. Asexual morph on alfalfa stems. Pycnidial conidiomata globose or irregular, solitary or aggregated, exposed on the alfalfa stems surface, dark brown to black, 235–445 μm diam. Conidiophores hyaline, smooth, 1-septate, densely aggregated, cylindrical, straight, 6–13 × 2.5–4 μm. Conidiogenous cells phialidic, hyaline, terminal, ampulliform, 8.5–12 × 2.5–3 μm, tapered towards the apex. Alpha conidia hyaline, aseptate, fusiform, biguttulate, 5.5–8.5 × 2–3 μm, mean ± SD = 6.4 ± 0.7 × 2.3 ± 0.2 μm, L/W ratio = 2.8 (n = 50). Beta conidia hyaline, aseptate, filiform, curved, tapering towards both ends, 21.5–28 × 1–1.4 μm, mean ± SD = 24.5 ± 1.5 × 1.1 ± 0.1 μm, L/W ratio = 22.3 (n = 41). Gamma conidia not observed.
Culture characteristics — Colonies on PDA flat with entire margin, colony honey in the centre with fluffy aerial mycelia and pale white margin; reverse with dull green pigment in the centre. Colony diam 40–44 mm in 3 d at 28 °C. On OA, colonies cottony, dense, greenish olivaceous in the centre; reverse dark herbage green.
Materials examined. China, Yunnan Province, Kunming City, on branches of P. bretschneideri cv. Zaobaisu, 17 Oct. 2014, Q. Bai (holotype HMAS 248152, culture ex-type CGMCC 3.19598 = PSCG 031); ibid., culture PSCG 032 and PSCG 033.
Notes — The three isolates studied form a well-supported independent clade distinct from known Diaporthe species. Diaporthe zaobaisu is most closely related to D. baccae, D. rhusicola, D. foeniculina, D. neotheicola and D. ravennica, but differentiated from them in ITS (9 different unique fixed alleles by D. baccae, 5 by D. rhusicola, 11 by D. foeniculina, 11 by D. neotheicola and 2 by D. ravennica) and TEF loci (21 different unique fixed alleles by D. baccae, 20 by D. rhusicola, 20 by D. foeniculina, 28 by D. neotheicola and 20 by D. ravennica). Moreover, D. zaobaisu differs from D. baccae in having shorter conidiophores (6–13 × 2.5–4 vs 20–57 × 2–3 μm) and conidiogenous cells (8.5–12 × 2.5–3 vs 9–23 × 1–2 μm) (Lombard et al. 2014). Alpha conidia are smaller than in D. foeniculina (5.5–8.5 × 2–3 vs 8.5–9 × 2–2.5 μm) and D. ravennica (5.5–8.5 × 2–3 vs 7–10.5 × 1.5–3 μm) (Udayanga et al. 2014a, Thambugala et al. 2016). Pycnidial conidiomata are smaller than in D. foeniculina (235–445 vs 400–700 μm) and D. neotheicola (235–445 vs 420–730 μm) (Santos & Phillips 2009, Udayanga et al. 2014a).
Prevalence of Diaporthe species
Prevalence analyses revealed that D. eres (248 isolates, 54.7 % of the total isolates) is the dominate species associated with pear shoot canker, followed by D. hongkongensis (57 isolates, 12.6 %, isolated from Guizhou, Jiangxi, Fujian and Zhejiang), D. sojae (43 isolates, 9.5 %, isolated from Guizhou, Hubei, Jiangsu, Jiangxi and Zhejiang), D. unshiuensis (38 isolates, 8.4 %, iso-lated from Guizhou, Hubei, Jiangsu, Shandong, Fujian and Yunnan), D. fusicola (21 isolates, 4.6 %, isolated from Guizhou, Jiangsu, Jiangxi, Fujian and Zhejiang), and D. cercidis (12 isolates, 2.6 %, isolated from Chongqing, Jiangsu and Zhejiang) (Fig. 24a). The remaining 13 species account for 7.5 % of the total isolates, with each less than 1 % prevalence (Fig. 24a).
Fig. 24.
The prevalence of Diaporthe species isolated from pear. a. Overall isolation rate (%) of Diaporthe species; b. distribution of Diaporthe species in China, each coloured circle represents one species, and the size of the circle indicates the number of isolates; c. isolation rate (%) of Diaporthe species from P. pyrifolia and P. bretschneideri, respectively.
Analysis of the abundance of Diaporthe species in the sampling areas revealed only two species identified from the north of the Yangtze River and 19 from the south, revealing obvious species diversity in the south (Fig. 24b). Analysis of the abundance of Diaporthe species on pear species revealed 15 species from P. pyrifolia and seven from P. bretschneideri, respectively (Fig. 24c), with only one species (D. eres) on the remaining pear species P. communis and P. ussuriensis. These findings might be due to the small samples obtained (with 20 and two samples collected in the field, respectively), since symptomatic branches were far less observed than these of P. pyrifolia and P. bretschneideri.
Pathogenicity and host range
The host range of the 19 Diaporthe species was accessed by inoculating mycelial discs onto detached shoots of five pear varieties (i.e., P. pyrifolia cv. Hohsui, P. bretschneideri cv. Xuehua, P. ussuriensis cv. Hanxiang, P. communis cv. Docteun Jule Guyot and P. sinkiangensis cv. Kuerlexiangli). At 11 d post inoculation (dpi), all Diaporthe isolates caused lesions on the inoculated shoots of P. pyrifolia, P. ussuriensis, P. communis, inducing reddish to black shoot canker symptoms, except for a D. sojae isolate (PSCG 510) inducing no lesions on P. bretschneideri, and a D. zaobaisu isolate (PSCG 031) and a D. parvae isolate (PSCG 034) on P. ussuriensis (Fig. 25). The lesion lengths varied significantly among the different isolates. Diaporthe fusicola and D. chongqingensis caused larger lesions (22–28 mm) on all the tested varieties, followed by the D. eres complex (7.6–14 mm), and the remaining isolates induced shorter lesions (1.5–10.5 mm). Most isolates induced longer lesions (longer than 10 mm) on the shoots of P. pyrifolia (13 isolates), P. bretschneideri (9) and P. sinkiangensis (7), while shorter lesions were observed on the shoots of P. communis (average 5 mm) and P. ussuriensis (5.6 mm). However, lesions longer than 10 mm were observed on P. ussuriensis (D. eres (PSCG092), D. spinosa (PSCG388) and D. fusicola (PSCG371, PSCG118)) and P. communis (D. eres (PSCG322), D. fusicola (PSCG371, PSCG118) and D. chongqingensis (PSCG435)) (Fig. 25). In parallel, no lesions developed on the twigs that were inoculated with PDA discs as control.
Fig. 25.
Lesion lengths on wounded pear twigs (P. pyrifolia cv. Hohsui, P. bretschneideri cv. Xuehua, P. ussuriensis cv. Hanxiang, P. communis cv. Docteun Jule Guyot and P. sinkiangensis cv. Kuerlexiangli) at 11 dpi induced by mycelia plugs of 31 representative isolates of 19 Diaporthe species.
One isolate of each species was further inoculated on intact pear seedlings (P. pyrifolia cv. Cuiguan) (Fig. 26). These results showed that all the isolates started to induce black lesions after 10 dpi. The lesions turned reddish and significant differences were evident among different species by 25 dpi (F = 8.735, P < 0.001). The induced symptoms matched the ones observed in the field. Diaporthe chongqingensis, D. fusicola and D. eres are highly aggressive (lesion lengths more than 8 mm). No lesions were induced in the control branches inoculated with PDA plugs. All branches showing canker symptoms induced by the inoculations were subjected to fungal isolation, and the results showed that the obtained colonies matched the inoculated ones in morphology and ITS sequence data.
Fig. 26.
Symptoms and lesion lengths induced by inoculation of wounded pear seedlings (P. pyrifolia cv. Cuiguan) at 25 dpi with mycelia plugs of representative isolates of 19 Diaporthe species. a. Representative symptoms as photographed at 25 days post inoculation (dpi); b. mean lesions lengths from six replicates of branches measured at 25 dpi. Statistical analysis was performed with SPSS Statistics 21.0 by one-way analysis of variance, and means were compared using Tukey’s test at a significance level of P = 0.05. Letters over the bars indicate the significant difference at the P = 0.05 level.
Host range was accessed on fruit trees including apple, peach, kiwifruit and citrus by inoculating the detached shoots with mycelium discs of one representative isolate from each Diaporthe species. The results showed that 13 species (including D. acuta, D. caryae, D. cercidis, D. chongqingensis, D. citrichinensis, D. eres, D. fulvicolor, D. fusicola, D. ganjae, D. pescicola, D. spinosa, D. taoicola and D. unshiuensis) infected all plants, resulting in lesions ranging from 1.5–49 mm on apple, 1.2–53 mm on peach, 1.2–53 mm on kiwifruit and 2–12 mm on citrus (Fig. 27). Of these, D. fusicola induced the longest lesions (32 mm) on four hosts compared to other species (less than 18.5 mm), as did D. spinosa (53 mm) on peach, D. pescicola (53 mm) on kiwifruit and D. chongqingensis (45 mm) on apple. Whereas D. padina and D. parvae infected all plants except for citrus, so did D. velutina except for peach, and D. sojae and D. hongkongensis except for kiwifruit. Diaporthe zaobaisu only infected citrus and apple, inducing lesions 3 and 2 mm long on their shoots, respectively.
Fig. 27.
Lesion lengths on wounded citrus, apple, peach and kiwifruit twigs at 11 dpi induced by mycelia plugs of representative isolates of 19 Diaporthe species.
Mating-type test
The mating-types of these 113 isolates were identified by PCR amplification of the mating genes (MAT1-2-1 and MAT1-1-1). These results showed that all D. sojae isolates are homothallic since both mating genes were detected in the same isolates; all the isolates of D. caryae, D. pescicola, D. spinosa, D. taoicola and D. velutina are heterothallic since only one of the mating genes was detected. For the remaining species (D. eres, D. unshiuensis, D. hongkongensis, D. cercidis), both mating genes were detected in some isolates while only one was detected in the remaining isolates of the same species, suggesting that they contain potentially homothallic as well as heterothallic isolates (Table 1).
DISCUSSION
Diaporthe species have been extensively investigated on several hosts (Gomes et al. 2013, Gao et al. 2017), but not yet on pear. Up to now, only eight species have been reported infecting pear, i.e., D. ambigua, D. infecunda, D. terebinthifolii, D. foeniculacea and D. oxe on P. communis, Phomopsis theicola and D. nobilis complex on P. pyrifolia and D. eres on P. communis (Smit 1996, Cloete et al. 2011, Santos et al. 2017b, Bertetti et al. 2018). In this study, we conducted extensive surveys of Diaporthe species associated with pear shoot canker in the major production provinces in China. Multi-locus phylogenetic and morphological analyses revealed 12 species (from 453 isolates) belonging to three Diaporthe species complexes, including the D. eres complex (D. eres and D. padina), D. sojae complex (D. caryae, D. ganjae, D. sojae and D. unshiuensis), and D. arecae complex (D. acuta, D. cercidis, D. fulvicolor, D. pescicola, D. spinosa and D. taoicola), and seven singleton species (D. chongqingensis, D. citrichinensis, D. fusicola, D. hongkongensis, D. parvae, D. velutina and D. zaobaisu). Of the 19 species, six species are newly described here, namely D. acuta, D. chongqingensis, D. fulvicolor, D. parvae, D. spinosa and D. zaobaisu. These species are all responsible for pear shoot canker, which could be confirmed following Koch’s postulates. To our knowledge, this is the first report that these species infecting pear are responsible for pear shoot canker besides D. eres.
Recently, Diaporthe species identification has been advanced by phylogenetic analysis based on multilocus DNA phylogeny including TEF, TUB, HIS and CAL genes (Santos et al. 2017a). Here, we resolved the Diaporthe species (P. fukushii, D. eres, P. amygdali, P. longicolla and D. neotheicola) that were previously identified based on phylogenetic analysis of TEF, ACT and ITS (Bai et al. 2015). Our results showed that these four species were incorrectly identified, and we reassigned isolates identified as P. fukushii to D. eres, P. amygdali to D. fusicola, P. longicolla to D. unshiuensis, and D. neotheicola to D. velutina (Fig. 2–4). Similarly, D. biguttusis, D. camptothecicola, D. ellipicola, D. longicicola, D. mahothocarpus and D. momicola clustered with D. eres (Fig. 2), suggesting that they are synonyms of D. eres, as previously proposed (Fan et al. 2018, Yang et al. 2018). Additionally, the ITS locus has been shown to be less optimal for closely related species (Farr et al. 2002, Gomes et al. 2013), especially in the D. eres complex (Santos et al. 2017a). Therefore, the ITS region was excluded from the phylogenetic analysis for the D. eres complex, which resulted in a well-supported phylogenetic tree (Fig. 2). However, for the D. sojae and D. arecae complexes, the phylogenetic analysis was still resolved with all these loci (Huang et al. 2013, Udayanga et al. 2014a, 2015). Furthermore, three new species (i.e., D. acuta, D. fulvicolor and D. spinosa) were identified as belonging to the D. arecae complex (Fig. 4).
Although the taxonomy of Diaporthe species has relied more heavily on molecular characteristics rather than on morphology (Castlebury et al. 2003, Crous & Groenewald 2005, Udayanga et al. 2012), we have noticed that most Diaporthe species exhibited morphological characteristics closely corresponding to their DNA phylogeny. For example, colonies of D. eres often secreted grey olivaceous pigments (Fig. 10, 15), D. arecae umber pigments (Fig. 5, 7, 11, 17, 19), while D. sojae lacked pigments (Fig. 13, 18, 21). Furthermore, their alpha conidial morphologies differed among these species complexes. Of those, most isolates in the D. eres complex exhibited short rod-like alpha conidia, D. sojae had oval conidia with obtusely rounded ends, and D. arecae had acutely rounded ends. In a previous study, gamma conidia were discovered for D. limonicola, which were hyaline, multiguttulate, fusiform to subcylindrical with an acute or rounded apex (Guarnaccia & Crous 2017). It is worthy to note that such conidia were also observed for D. eres (isolate PSCG 041) in this study (Fig. 10).
The prevalence analysis revealed that D. eres is the most prevalent species in China, which is consistent with observations made in our previous study (Bai et al. 2015), and corresponds to its biological trait of wide host range, since it infects many plants in the Rosaceae (Farr & Rossman 2018). Moreover, Diaporthe species are closely linked to the sampling area, with a higher diversity (19 species) in the south of the Yangtze River than that in the north (2). It might be due to the fact that the climate in the south is humid and warm, suitable for the survival and prevalence of Diaporthe species, while drought and extremely low temperatures in the north, especially in Gansu, Shanxi and Xinjiang, are unsuitable for Diaporthe. Moreover, P. pyrifolia trees are dominantly cultivated in the south, and are susceptive to infection by Diaporthe species. No Diaporthe species were detected from the pear samples collected in the north provinces including Gansu, Shanxi and Xinjiang. Instead, Botryosphaeria spp. were readily isolated from these samples, which induced stem canker following inoculation on pear stems, suggesting that these samples might be infected by pear stem canker instead of pear shoot canker.
Since Diaporthe spp. have an endophytic, saprobic or pathogenic lifestyle, we determined their pathogenicity to pear by inoculating colonised mycelial discs on shoots of five different pear species. These results showed that they are all pathogenic and responsible for pear shoot canker by fulfilling the Koch’s postulates. Moreover, these isolates showed significantly different virulence spectra related to species and host plants. For example, D. fusicola isolates were highly aggressive to P. bretschneideri, whereas D. parvae was only slightly aggressive on the same Pyrus species; D. chongqingensis isolates were aggressive to most of the tested Pyrus plants, but obviously less to P. ussuriensis. Additionally, the host ranges of these Diaporthe species also showed a clear diversity among them, exemplified by the fact that some infected all test plants, while others not. It is worth to note that most Diaporthe species have a wide host range, indicating that these species also pose threats to other fruit trees, as previously described (Gomes et al. 2013, Dissanayake et al. 2015). In fact, Diaporthe spp. have been reported infecting many plants resulting in severe diseases, e.g., seed decay of soybean (Sun et al. 2013), canker and twig dieback of jujube (Zhang et al. 2018), cordon dieback of kiwifruits (Díaz & Latorre 2018), and shoot canker diseases of citrus or grapevines (Van Niekerk et al. 2005, Huang et al. 2013), and of Rosaceae plants, e.g., peach (Dissanayake et al. 2017), apple (Abreo et al. 2012), blackberry (Vrandecic et al. 2011), and almond (Diogo et al. 2010).
In previous studies, 22 Diaporthe species have been characterised based on their mating type, revealing that most of the species are heterothallic except for D. ambigua which is homothallic, and D. viticola which is mixed (Santos et al. 2010). Recently, D. foeniculina, D. pyracanthae, D. malorum, and D. eres were also identified as being heterothallic (Santos et al. 2017b). Similarly, most of the species obtained in this study are heterothallic, with one species, D. sojae, being homothallic. Correspondingly, almost all of the obtained Diaporthe species were asexual, but D. sojae also produced ascomata with viable ascospores. It is worth to note that four species (D. unshiuensis, D. hongkongensis, D. cercidis and D. eres) were identified to be homo- as well as heterothallic, and the identification for D. eres differs from the previous report, which described D. eres as exclusively heterothallic (Santos et al. 2017b). For the heterothallic identification, we cannot exclude the possibility that one mating gene was undetected due to variation among isolates. For example, D. spinosa produced sexual sporocarps from single conidia, suggesting it to be homothallic, but only one mating type gene was detected (Table 1). Finally, the mating types detected by these primers need further confirmation since they might be inactive, or change due to mutation.
This study represents the most intensive investigation and the first resolution with multi-locus phylogenetic analysis of Diaporthe species infecting Pyrus plants, revealing six novel species that infect pear and are responsible for pear shoot canker. This study also characterises the taxonomic, morphological and biological diversity of Diaporthe spp. associated with different Pyrus spp. in China, with regards to geographical location, host range and mating type. As such it provides useful information to help understand the ecology of the Diaporthe spp. infecting pear, as well as for the control of pear shoot canker.
Acknowledgements
This study was financially supported by the fund of Integrated Research and Demonstration of Reduction of Fertilizer and Pesticide but Efficiency Enhancement for Pear Cultivation in the Yangtze River Basin (no. 2018YFD0201406), the earmarked fund for Pear Modern Agro-Industry Technology Research System (CARS-28-15) of the Chinese Ministry of Agriculture and the Fundamental Research Funds for the Central Universities (no. 2662016PY107). The authors thank Lei Cai for critical suggestions and Fang Liu for technical guidance in the morphological descriptions.
Appendix Number of samples and Diaporthe isolates collected from 12 regions in China.
| Province | Number of samples | Number of isolates |
|---|---|---|
| Chongqing | 11 | 16 |
| Fujian | 37 | 83 |
| Guizhou | 21 | 53 |
| Hebei | 18 | 10 |
| Henan | 11 | 10 |
| Hubei | 46 | 87 |
| Jiangsu | 35 | 47 |
| Jiangxi | 18 | 44 |
| Liaoning | 25 | 21 |
| Shandong | 27 | 40 |
| Yunnan | 8 | 12 |
| Zhejiang | 29 | 30 |
| Total | 286 | 453 |
REFERENCES
- Abreo E, Martínez S, Sessa L, et al. 2012. Phomopsis cotoneastri as a pathogen associated with trunk cankers and death of young apple trees cv. Cripps Pink. Journal of Phytopathology 160: 434–436. [Google Scholar]
- Bai Q, Wang GP, Zhai LF, et al. 2015. Biological and molecular characterization of five Phomopsis species associated with pear shoot canker in China. Plant Disease 99: 1704–1712. [DOI] [PubMed] [Google Scholar]
- Bertetti D, Guarnaccia V, Spadaro D, et al. 2018. First report of fruit rot in European pear caused by Diaporthe eres in Italy. Plant Disease 102: 1651. [Google Scholar]
- Carbone I, Kohn LM. 1999. Method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Castlebury LA, Farr DF, Rossman AY, et al. 2003. Diaporthe angelicae comb. nov., a modern description and placement of Diaporthopsis in Diaporthe. Mycoscience 44: 203–208. [Google Scholar]
- Choi YW, Hyde KD, Ho WH. 1999. Single spore isolation of fungi. Fungal Diversity 3: 29–38. [Google Scholar]
- Cloete M, Fourie PH, Damm U, et al. 2011. Fungi associated with die-back symptoms of apple and pear trees, a possible inoculum source of grapevine trunk disease pathogens. Phytopathologia Mediterranea 50: S176–S190. [Google Scholar]
- Crous PW, Groenewald JZ. 2005. Hosts, species and genotypes: opinions versus data. Australasian Plant Pathology 34: 463–470. [Google Scholar]
- Crous PW, Groenewald JZ, Risède J, et al. 2004. Calonectria species and their cylindrocladium anamorphs species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, et al. 2011. Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. (eds). 2019. Fungal Biodiversity. Westerdijk Laboratory Manual Series 1. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2016. Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz GA, Latorre BA. 2018. First report of cordon dieback of kiwifruits caused by Diaporthe ambigua and D. australafricana in Chile. Plant Disease 102: 446. [Google Scholar]
- Diogo ELF, Santos JM, Phillips AJL. 2010. Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. [Google Scholar]
- Dissanayake AJ, Liu M, Zhang W, et al. 2015. Morphological and molecular characterisation of Diaporthe species associated with grapevine trunk disease in China. Fungal Biology 119: 283–294. [DOI] [PubMed] [Google Scholar]
- Dissanayake AJ, Zhang W, Liu M, et al. 2017. Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere 8: 533–549. [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, et al. 2018. Diaporthe from walnut tree (Juglans regia) in China, with insight of the Diaporthe eres complex. Mycological Progress 17: 841–853. [Google Scholar]
- FAO – Food and Agricultural Organization of the United Nations, China . 2017. Pear fruits fresh and processed: annual statistics. http://www.fao.org/faostat/en/#data/QC. [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY. 2002. Morphological and molecular characterization of Phomopsis vaccinii and additional isolates of Phomopsis from blueberry and cranberry in the eastern United States. Mycologia 94: 494–504. [PubMed] [Google Scholar]
- Farr DF, Rossman A. 2018. Fungal databases, systematic mycology and microbiology laboratory. ARS, USDA. http://nt.ars-grin.gov/fungaldatabases/. [Google Scholar]
- Ferradini N, Lancioni H, Torricelli R, et al. 2017. Characterization and phylogenetic analysis of ancient Italian landraces of pear. Frontiers in Plant Science 8: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S, Katan T, Shabi E. 1996. Characterization of Colletotrichum gloeosporioides isolates from avocado and almond fruits with molecular and pathogenicity tests. Applied and Environmental Microbiology 62: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Crous PW, Bai Q, et al. 2019. Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 42: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Liu F, Cai L. 2016. Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. [Google Scholar]
- Gao YH, Liu F, Duan W, et al. 2017. Diaporthe is paraphyletic. IMA Fungus 8: 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Su YY, Sun W, et al. 2015. Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biology 119: 295–309. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. American Society for Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, et al. 2013. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso FM, Marini M, Vitale A, et al. 2012. Canker and dieback on Platanus × acerifolia caused by Diaporthe scabra. Forest Pathology 42: 510–513. [Google Scholar]
- Guarnaccia V, Crous PW. 2017. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. 2018. Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathologia Mediterranea 57: 307–319. [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, et al. 2018. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40: 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Huang DH, Zhou CH, Xu L, et al. 2014. New diseases of Jiangxi precocious pear shoot canker and its comprehensive prevention and control measures. Xiandai Horticulture 1: 67–68. [Google Scholar]
- Huang F, Hou X, Dewdney MM, et al. 2013. Diaporthe species occurring on citrus in China. Fungal Diversity 61: 237–250. [Google Scholar]
- Huang F, Udayanga D, Wang X, et al. 2015. Endophytic Diaporthe associated with Citrus: a phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Evolutionary Genetics Analysis 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Leeuwen GCM, Guarnaccia V, et al. 2014. Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, et al. 2019. Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92: 43–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu H, Hatamoto M, Date H, et al. 1987. Pear fruit rot caused by agents of Japanese pear canker, Phomopsis fukushii and blossom end rot of European pear phomopsis sp. Annals of the Phytopathological Society of Japan 53: 630–637. [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nitschke T. 1870. Pyrenomycetes Germanici 2. Eduard Trewendt, Breslau, 245. [Google Scholar]
- Nylander JAA. 2004. MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Rambaut A. 2014. FigTree, v. 1.4.2. Institute of evolutionary biology. University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute, Kew, UK. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Santos JM, Correia VG, Phillips AJ. 2010. Primers for mating-type diagnosis in Diaporthe and Phomopsis: their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114: 255–270. [DOI] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. 2009. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 109–123. [Google Scholar]
- Santos JM, Vrandecic K, Cosic J, et al. 2011. Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Alves A, Alves R. 2017a. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5:e3120. doi: 10.7717/peerj.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Phillips AJL, Crous PW, et al. 2017b. Diaporthe species on Rosaceae with descriptions of D. pyracanthae sp. nov. Mycosphere 8: 485–511. [Google Scholar]
- Silva GJ, Souza TM, Barbieri RL, et al. 2014. Origin, domestication, and dispersing of pear (Pyrus spp.). Advances in Agriculture 2014: 1–8. [Google Scholar]
- Smit WA. 1996. A new canker disease of apple, pear, and plum rootstocks – caused by Diaporthe ambigua in South Africa. Plant Disease 80: 1331–1335. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Sun S, Kim MY, Chaisan T, et al. 2013. Phomopsis (Diaporthe) species as the cause of soybean seed decay in Korea. Journal of Phytopathology 161: 131–134. [Google Scholar]
- Swofford D. 2002. PAUP 4.0 b10: Phylogenetic analysis using parsimony (*and other methods). Computer programme. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tanaka S, Endo S. 1930. Studies on a new canker disease of Japanese pear trees caused by Phomopsis fukushii n. sp. Transactions of the Tottori Society of Agricultural Sciences 2: 123–134. [Google Scholar]
- Thambugala KM, Daranagama DA, Phillips AJL, et al. 2016. Microfungi on tamarix. Fungal Diversity 82: 239–306. [Google Scholar]
- Thompson SM, Tan YP, Young AJ, et al. 2011. Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, et al. 2014a. Species limits in Diaporthe: molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 32: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, et al. 2014b. Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Diversity 67: 203–229. [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, et al. 2015. The Diaporthe sojae species complex: phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. [DOI] [PubMed] [Google Scholar]
- Udayanga D, Liu X, Crous PW, et al. 2012. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56: 157–171. [Google Scholar]
- Udayanga D, Liu X, McKenzie EHC, et al. 2011. The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. [Google Scholar]
- Van Niekerk JM, Groenewald JZ, Farr DF, et al. 2005. Reassessment of Phomopsis species on grapevines. Australasian Plant Pathology 34: 27–39. [Google Scholar]
- Van Rensburg JCJ, Lamprecht SC, Groenewald JZ, et al. 2006. Characterisation of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Studies in Mycology 55: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrandecic K, Jurkovic D, Cosic J, et al. 2011. First report of cane blight on blackberry caused by Diaporthe eres in Croatia. Plant Disease 95: 612–612. [DOI] [PubMed] [Google Scholar]
- Wang HY, Han XF, Gao JJ, et al. 2011. The cause of severe occurrence of pear shoot canker and its comprehensive control technology. Chinese Horticulture Abstracts 7: 164–165. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California. [Google Scholar]
- Wu J, Wang ZW, Shi ZB, et al. 2013. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Research 23: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, et al. 2018. High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QM, Yu CL, Li GF, et al. 2018. First report of Diaporthe eres causing twig canker on Zizyphus jujuba (Jujube) in China. Plant Disease 102: 1458. [Google Scholar]
- Zhao DY, Xu K, Yuan JC, et al. 2016. Analysis on the current situation of production and sales of world pear’s main country of origin and its development. China Fruits 2: 94–100. [Google Scholar]