SUMMARY
Brain Machine Interfaces (BMIs) hold promise to restore impaired motor function and serve as powerful tools to study learned motor skill. While limb-based motor prosthetic systems have leveraged nonhuman primates as an important animal model,1–4 speech prostheses lack a similar animal model and are more limited in terms of neural interface technology, brain coverage, and behavioral study design.5–7 Songbirds are an attractive model for learned complex vocal behavior. Birdsong shares a number of unique similarities with human speech,8–10 and its study has yielded general insight into multiple mechanisms and circuits behind learning, execution, and maintenance of vocal motor skill.11–18 In addition, the biomechanics of song production bear similarity to those of humans and some nonhuman primates.19–23 Here, we demonstrate a vocal synthesizer for birdsong, realized by mapping neural population activity recorded from electrode arrays implanted in the premotor nucleus HVC onto low-dimensional compressed representations of song, using simple computational methods that are implementable in real time. Using a generative biomechanical model of the vocal organ (syrinx) as the low-dimensional target for these mappings allows for the synthesis of vocalizations that match the bird’s own song. These results provide proof of concept that high-dimensional, complex natural behaviors can be directly synthesized from ongoing neural activity. This may inspire similar approaches to prosthetics in other species by exploiting knowledge of the peripheral systems and the temporal structure of their output.
In brief
Songbirds, like humans, need to control a sophisticated vocal organ to produce rich vocal sequences. Arneodo et. al. use knowledge of the biomechanics of the vocal organ and the structure of the vocal sequence to synthesize birdsong from recorded premotor neural activity.
Graphical Abstract
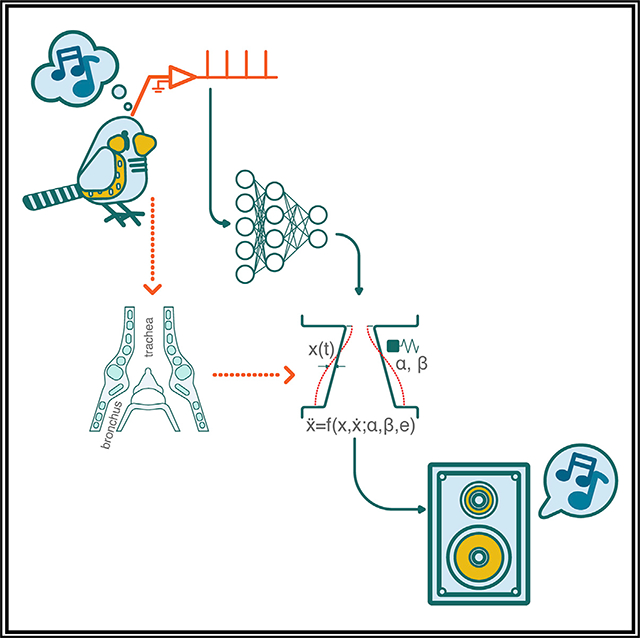
RESULTS AND DISCUSSION
We describe two methods for synthesizing realistic vocal signals from neural activity recorded in a premotor nucleus of zebra finches (Taeniopygia guttata). Each method exploits a different trait of the vocal-motor process. First, we leverage understanding of the biomechanics of birdsong production. We employ a biomechanical model of the vocal organ that captures much of the spectro-temporal complexity of song in a low-dimensional parameter space.24 This dimensionality reduction, compared to the full time-frequency representation of song, enables training of a shallow feedforward neural network (FFNN) that maps neural activity onto the model parameters. As a second synthesis method, we capitalize on predictive components in the temporal covariance between neural activity and song, which can be learned by a recurrent, long-short-term memory neural network (LSTM)25 trained directly on frequency domain representations (spectrograms) of the vocal output.
Neuronal input for each synthesis comes from the sensorymotor nucleus HVC, where neurons generate high-level commands that drive the production of learned song. Adult male zebra finches (Taeniopygia guttata) sing individually stereotyped motifs comprising a sequence of 3–10 syllables. Activity in multiple HVC neuronal subtypes is modulated during singing: projection neurons targeting area X and RA (HVCx/RA) evince short, precise, sparse activity bursts during the motif,15,17,26–30 while inhibitory interneurons (HVCI) display more tonic activity during singing.14,29,31,32 To obtain ensemble HVC activity and vocal output, we implanted 16- or 32-channel Si probes in male, adult (>120-day-old) zebra finches and recorded extracellular voltages simultaneously while each bird sang (n = 4 birds, 70–120 vocal motifs per session). Neural recordings were sorted automatically using Kilosort and manually curated to exclude noise.33 Non-noise clusters were classified as single- or multi-unit activity (SUA or MUA) based on the number of refractory period violations and putatively as projection or interneurons based on the sparseness of the activity during singing. The recordings were dominated by MUA clusters (n = 88) and HVC interneurons (HVCI; n = 29), with relatively few putative projection neurons (HVCx/RA; n = 15). Example song-aligned neural activity histograms are shown in Figure 1A. Example rasters with the numbers of clusters per bird are shown in Figure S1.
Figure 1. A neural-network-based decoder to synthesize birdsong from premotor neural activity.
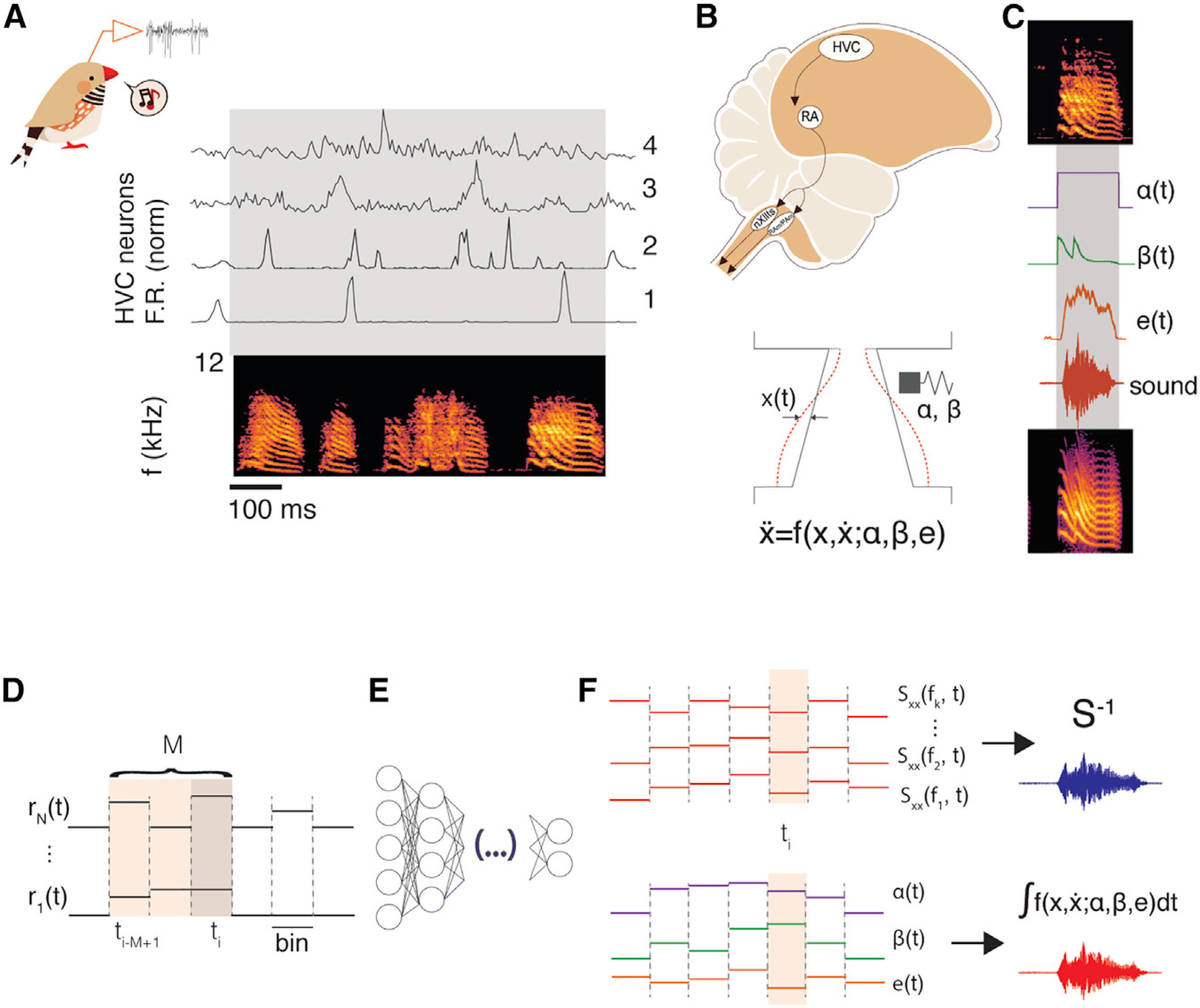
(A) Neural activity is collected from awake-singing animals. Sorted, extracellularly recorded single- and multi-units show different degrees of singing-related sparseness, robustness, and spiking precision (4 example clusters; top traces: normalized mean firing rate over 70 repetitions of the bird’s motif; below: spectrogram of the motif; see also Figure S1).
(B) Downstream of HVC, the posterior motor pathway leads into nuclei that control the muscles driving the sound production (nXII and RAm/PAm).34 Syringeal and respiratory muscles act coordinately to modulate the flow of air through sets of labia and produce sound.35 The complex labial motion is captured by the equations of a nonlinear oscillator;23 parameters that define acoustic properties of the sounds are surrogates of the activities of syringeal and respiratory muscles.36
(C) To reproduce a particular vocalization (top) from the biomechanical model, we fit the parameters (middle {α(t), β(t), e(t)}) such that, upon integration, the synthetic song (bottom) matches the pitch and spectral richness.
(D) The input of the neural network is an array with the values of a set of neural features (spike counts of sorted units/multi-units) over a window of M previous time steps.
(E) The hidden layer(s) of the network are composed either by a densely connected layer (FFNN) or two layers of LSTM cells.
(F) When training or reconstructing directly the spectral features of the song, the output of the network is a vector of powers across a range of frequency bands at a given time; the generated spectral slices are then inverted to produce synthetic song (top). When training or reconstructing via the biomechanical model, the output of the network at a given time is a 3-dimensional vector of parameters (as illustrated in C); the equations of the model are then integrated with these values to produce synthetic song (bottom). Illustrations were taken from Arneodo.37
Biomechanically meaningful compression enhances neurally driven synthesis
Synthesizing a complex motor sequence from neural activity requires mapping between two high-dimensional representations. To reduce the dimensionality of the problem, we leveraged a biomechanical model of the avian vocal organ that transforms neural activity to vocal output. The model accounts for the syrinx and the vocal tract.24,27,38 The syrinx contains labial folds that oscillate when induced by the sub-syringeal air sac pressure and modulate the airflow to produce sound (Figure 1B).35 The dynamics of the labia can be modeled after the motion equations of a nonlinear oscillator, in which the features of the sounds produced are determined by two time-varying parameters,23,24,36 representing physiological motor instructions (the sub-syringeal pressure and the activity of the muscles that tense the labia).24 In its simplest form, the syrinx model is computable in real time to produce synthetic vocalizations.38 We model the vocal tract (the trachea, the oropharyngeal-esophageal cavity, and the beak) as a passive acoustic filter that determines species-specific spectral traits, such as the timbre.24,27,39
To synthesize song from neural activity via the biomechanical model, we first fit the parameters of the model to produce a synthetic version of each vocalization.24,27,38 We searched for the parameters that produce, upon integration of the equations of the model, the closest match in pitch, spectral richness, and amplitude of the target vocalization. This effectively compresses each segment of a bird’s own song (BOS) into a time series in a 3D parameter space, which generates a corresponding segment of synthetic song (SYN) (Figure 1C).36,38 For each session, we randomly select 60% of the motifs for training, split each motif into 5-ms bins, and train a one-hidden-layer FFNN to predict the biomechanical model parameters corresponding to each bin independently from the neural activity in a 50-ms, immediately preceding time window. The neural activity was represented by the average firing rate of each cluster, split into 1-ms bins. To avoid introducing temporal correlations, we randomized the order in which each pair of neural activity window and target model parameters was presented to the network. After training, we predict the values of the biomechanical model parameters corresponding to a test set of neural activity and integrate the differential equations of the model to produce each bin of neurally driven synthetic song. This yields synthetic vocalizations that sound similar to the bird’s own. An example motif from each bird is illustrated in Figure 2 (and Audio S1, S2, S3, and S4).
Figure 2. Song synthesized from premotor neural activity via a biomechanical model of the vocal organ is similar to the recorded bird’s own song.
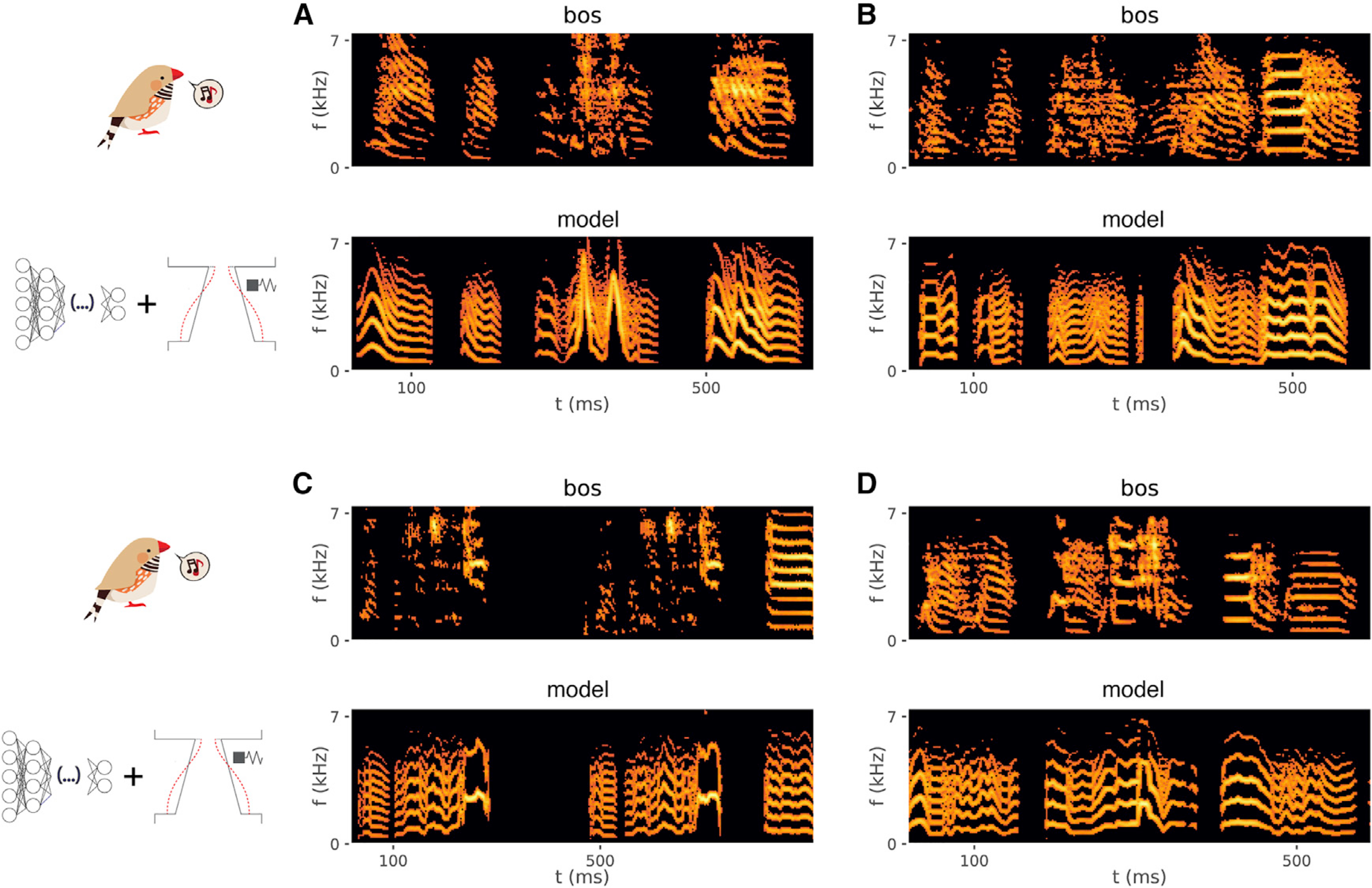
Spectrogram of one or two instances of a bird’s motif (BOS; upper) and corresponding song generated by inferring the biomechanical model parameters from neural activity using a shallow FFNN and integrating the model, for four different birds (z007, z017, z020, and z028, respectively; see also Audio S1, S2, S3, and S4, respectively, and Figures S2 and S4).
In contrast, implementing a FFNN to directly predict the spectro-temporal features of a song results in a low-quality synthesis. We trained a similar network as before but with the spectral components of the song, as represented by the power across 64 frequency bands, as the targets. Examples of songs synthesized in this way for each bird (Figure 3; Audio S1, S2, S3, and S4) show how the FFNN fails to produce well-defined harmonic stacks that are typical of the zebra finch song and to faithfully reproduce vocal onsets and offsets.
Figure 3. Direct spectrogram synthesis from neural activity.
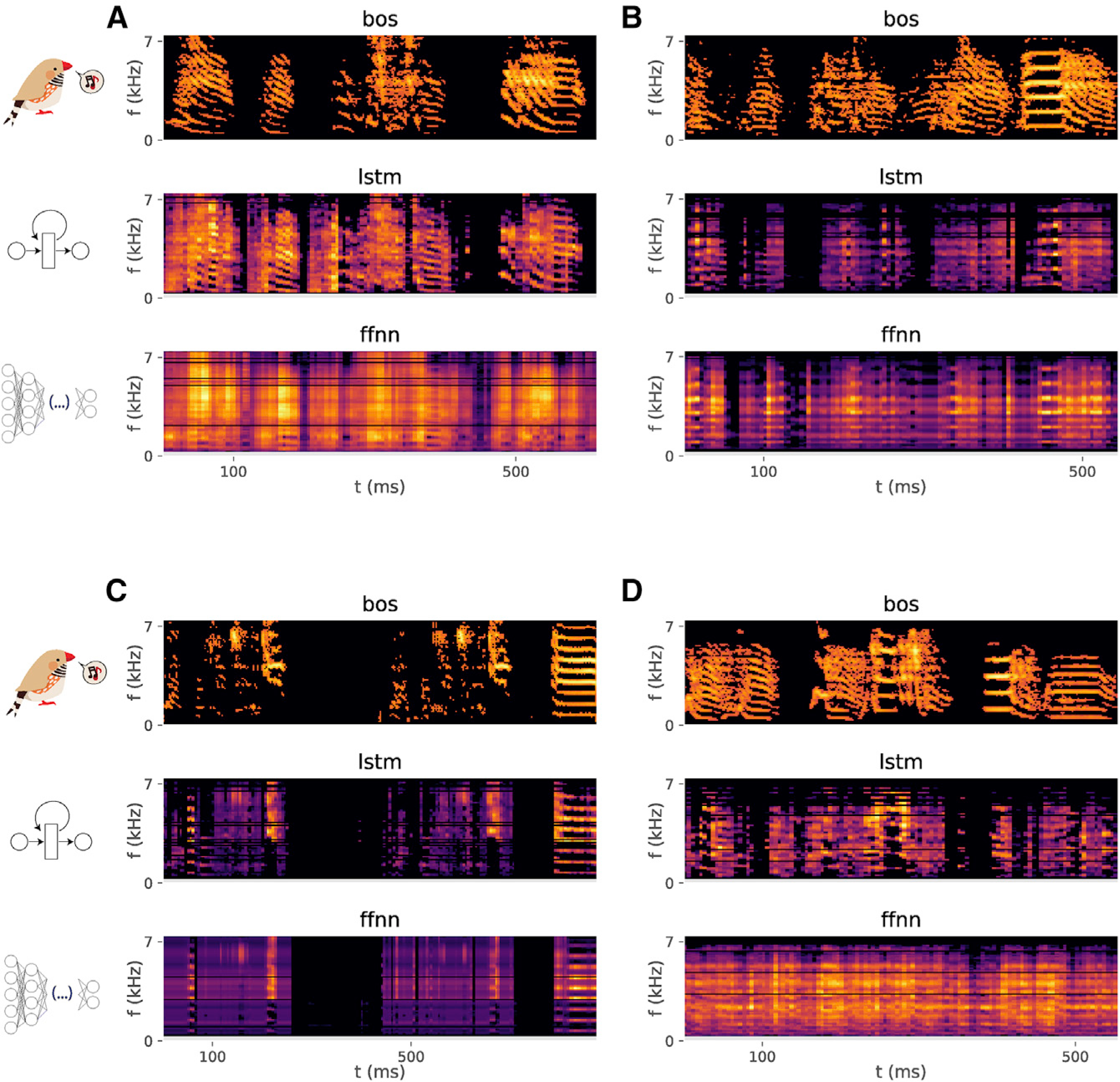
Spectrogram of a bird’s motif (BOS; upper), song generated by inferring the spectrogram directly from neural activity, after training an LSTM (middle), and song generated by feeding neural activity and spectrograms to a FFNN of similar architecture to the one used for Figure 2 (bird z007, z017, z020, and z028, respectively, for each panel. See also Audio S1, S2, S3, and S4, respectively, and Figure S4).
The differing capacities for the FFNN to predict model parameters compared to spectro-temporal coefficients (Figures 2, 3, and 4) suggest that reducing the dimensionality of the behavior enhances prediction. To confirm, we trained the FFNN to reproduce a different “compression” of the behavior, namely the first 3 principal components (PCs) of the spectrogram. The performance at predicting the values of the 3 PCs from the neural activity is similar to that at predicting the biomechanical model parameters (Figure S4A). The advantage of the latter is in their generative capacity to produce songs more similar to the BOS.
Figure 4. Performance comparisons aggregated for all birds.
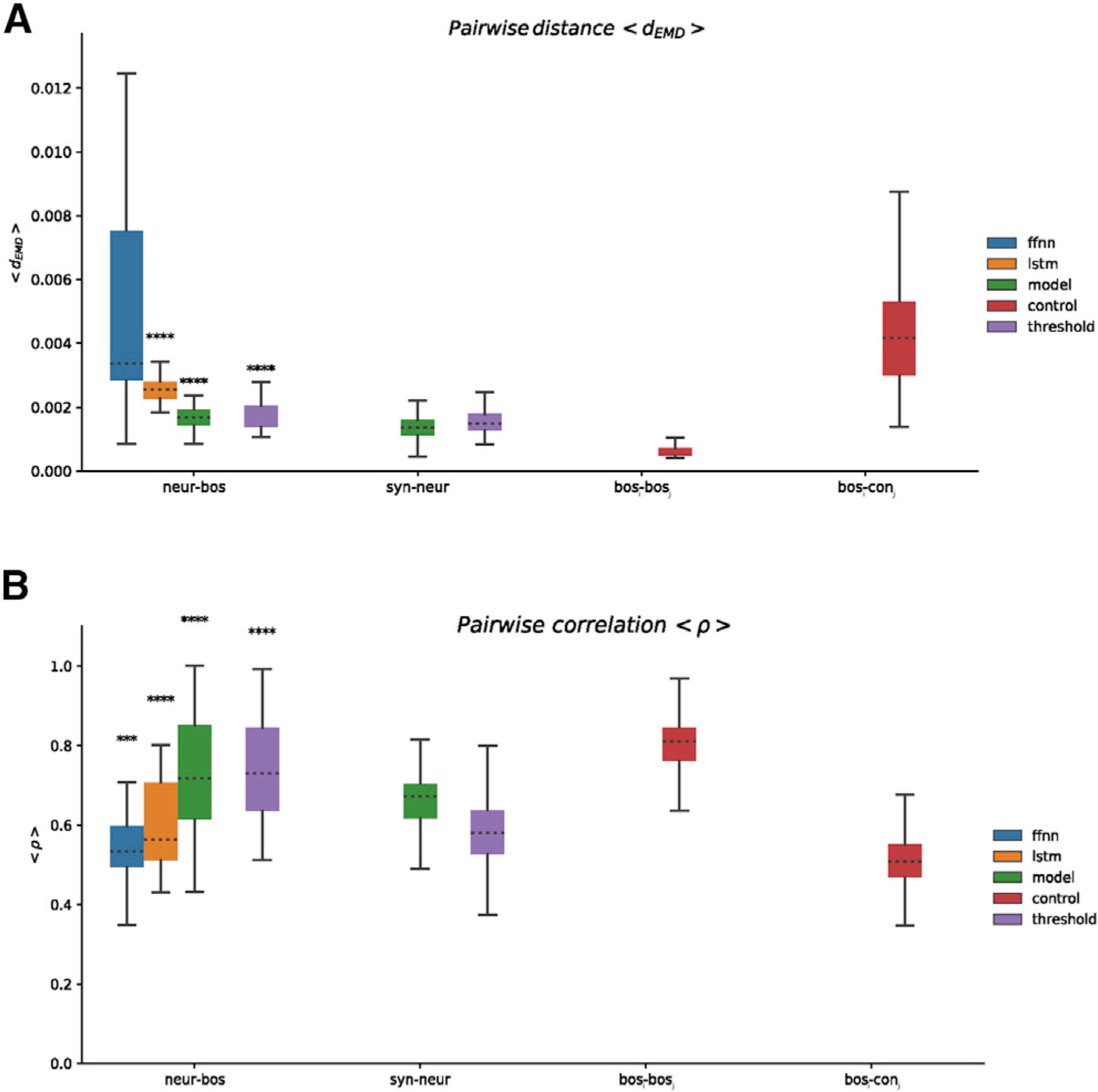
(A) Boxplots showing time-averaged earth mover’s distance (< dEMD >; lower = better). Neur-bos, distance between each pair of synthesized or target spectrograms in the test set. Ffnn and lstm indicate training directly with spectrogram via a FFNN and LSTM using sorted spikes, model indicates training or synthesis via the biomechanical model of the vocal organ using sorted spikes, and threshold indicates the same training or testing as in model, albeit using supra-threshold activity instead of sorted spikes. Syn-neur indicates the distance between the synthetic instance of each motif in the testing set (the one produced when fitting the parameters of the biomechanical model for a given motif) and the one synthesized from neural activity. Bosi-bosj indicates distance between each pair of motifs of BOS. Bosi-conj indicates distance between pairs of bos and songs from a pool of conspecific birds.
(B) Boxplots showing time-averaged spectral correlation (< ρ >; higher = better); same pairs as in (A) (***p < 0.001; ****p < 0.0001; Mann-Whitney U test, one sided against bosi-conj). Performance for each bird is shown in Figure S3.
The biophysical model can be integrated in real time to produce synthetic vocalizations online.38 This motivated us to skip the spike-sorting step and use a representation of the neural activity that requires cheaper computation and no manual curation. Instead of representing the neural activity with clustered spikes, we trained the FFNN to predict the biomechanical model parameters directly from supra-threshold events in each electrode signal.1,3,40–42 The results (Figures 4, S2, and S3) suggest that it is plausible to replace the spike sorting by a computationally cheaper representation of the neural activity without significant deterioration of the synthesis.
Exploiting temporal structure
The failure of the FFNN to accurately predict the spectral coefficients of a bird’s motif may reflect the inability of this model to capture more complex temporal dynamics across response clusters that precede specific vocalizations. To capture these dynamics, we trained an LSTM25 to predict the spectral components of the song (64 frequency bands) directly from the preceding 50 ms of neural activity, using the same input and output data as described in the previous section. Unlike the FFNN, the LSTM yields a neurally driven song synthesis that sounds similar to the intended bird’s own song (Figure 3; Audio S1, S2, S3, and S4).
Because zebra finch songs are highly stereotyped across renditions from the same singer, we wondered whether the LSTM might be capturing a trivial correspondence between two synchronized signals (the neuronal activity and the song). To rule out this possibility, we independently shuffled the spectral coefficients in each time slice of the spectrograms. This created a novel stereotyped “song,” in which the explicit relationship between neural activity and any given acoustic feature varied across time, but the temporal correlation between the average neural activity and the waveform envelope was unchanged. After normalizing the durations of all instances of the motif in a bird’s own song through dynamic time warping,43 we permuted the coefficients within each spectral time slice using the same pseudo-random mask for each motif. The quality of the LSTM synthesis dropped significantly for the pseudo-random stereotyped songs (Figure S4C), indicating a non-trivial relationship between temporal dynamics of HVC population responses and the spectro-temporal characteristics of natural song.
To assess the relative similarity of all our syntheses to BOS quantitatively, we compared the spectrograms of each synthesized and target motif pair using two different metrics. We computed the mean distance between spectrograms as the earth mover’s distance (dEMD)44 between each pair of corresponding spectral slices, averaged across time. Intuitively, dEMD measures the work required to transform one distribution of spectral power onto another and is sensitive to differences in spectral richness (harmonic stacks versus pure tones or broadband energy distribution) and vocalization onset or offset timing. Figure 4A displays a summary of pairwise mean distances < dEMD > aggregated for all birds (Figure S3 shows data grouped by bird). We show the distance between each motif of BOS and the corresponding synthesis achieved with each different strategy. For reference, we compute the distance between all pairs of motifs of BOS (bosi – bosj) and the distances between pairs of BOS and conspecific birds’ motifs (bosi – conj). We also computed the mean spectral correlation < ρ > between each synthetic motif and its target across the span of each motif. Results are presented in the same manner in Figures 4B and S3. Consistent with the intuition from Figures 2 and 3, both measures show that an FFNN trained to directly predict the spectrogram yields poorer song synthesis compared to the LSTM and the biomechanical model-aided network, even when trained with supra-threshold neural events.
We show that it is possible to synthesize a rich vocal behavior from neural ensemble activity recorded in singing songbirds, a well-established animal model for vocal communication. Similarity of the synthesis with respect to the bird’s intended vocalization is significantly enhanced by either compressing behavior into a low-dimensional parameter space or by exploiting the spectro-temporal correlation structure of song by the synthesis algorithm. Our results provide insight into how BMIs for complex behavior may be enhanced through detailed understanding of the underlying biomechanics of motor control and the statistical structure of the target behavior.
Introducing a biomechanical model of the vocal organ enables dimensionality reduction and generativity. Compressing the behavior into a few dimensions enables the use of cheap computations that can be implemented in real time38 with relatively small training sets. As a compressive model, the “latent” space provided by the time-varying parameters of the biomechanical model is attractive in that it is a proxy for muscular and respiratory activities.24 In principle, however, other low-dimensional representations of behavior should also be reconstructable from neural activity. Indeed, the 3 strongest PCs of the spectrogram can be predicted by a simple feedforward network almost as well as the parameters of the biomechanical model (Figures S4A and S4B), although these PCs yield poorer synthetic song (Figure S4A) compared to the biomechanical model (Figures 4A and S4B). As a generative model, song synthesized by the biomechanical model is similar to the BOS (Figure 2) and can evoke responses in neural circuits that are highly selective to the BOS.27,45–47 It remains to be tested whether the biomechanical model-generated song will be sufficient for sensory feedback in closed-loop experiments where neurally driven synthesis replaces BOS38 and whether the synthetic vocalizations can drive naturalistic responses in females, given the species’ ability to differentiate songs based on fine differences in spectro-temporal structure.39,48,49 Perhaps more detailed models are necessary to fulfill certain functions (BOS replacement and successful courtship). It is also likely that a similar approach can be translated to other species and motor behaviors that admit a low-dimensional, generative representation. This could be the case when the biomechanics are sufficiently understood6,22,41,50–56 or when there are enough examples of the behavior to enable data-driven dimensionality reductions.6,7,57
The limited repertoire of male zebra finches might suggest that a direct synthesis could be achieved by relatively simple means. Yet the FFNN trained to predict spectral coefficients, which, because of its loss function, is close to a regularized nonlinear regression, yielded poor-quality songs compared to all other methods (Figures 4 and S3). The reason for this is not entirely clear, but it may reflect the neuronal subtype compositions of our datasets. Unfortunately, the relatively low yield of well-isolated single units in our recordings (Figure S1) prevents us from examining the contributions of HVC interneurons and projection neurons directly. Most clusters in our datasets were MUA, which are likely dominated by interneuron activity.29 Although the precise function of HVC interneuron activity is not fully understood, it plays a prominent role in multiple models of HVC29,31,32 sequence generation, and fluctuations in the average firing rates of interneurons are closely timed to bursting in projection neurons.15 While one should not interpret our results to support the presence of an explicit “motor code” in HVC ensemble activity, employing a recurrent network that captures the temporal structure of our neural population activity, rather than a FFNN, nonetheless yields synthetic songs that are much closer matches to the birds own song.
We have demonstrated a BMI for a complex communication signal, using computation blocks that are implementable in real time in an established animal model for production and learning of complex vocal behavior. The strength of our approach lies in the ability to find a low-dimensional parameterization of the behavior in a manner that it can be driven with the activities recorded from relatively small samples (by tens) of neurons. Doing so with recordings from the superficially located nucleus HVC enables accessibility by less invasive micro-electrode arrays, capable of resolving not only LFP, which has been shown suitable for BMI,41 but also SUA and MUA.1,3,40–42,58,59 This provides a novel tool for probing the neural circuits underlying the production, acquisition, and maintenance of vocal communication signals and unlocks access to new models and experiments directed at understanding how neuronal activity is transformed into natural action and how peripheral effectors shape the neural basis of action.22,54 Our approach also provides a proving ground for vocal prosthetic strategies. While birdsong differs in important ways from human speech, the two vocal systems have many similarities, including features of the sequential organization and strategies for their acquisition,60,61 analogies in neuronal organization and function,10,12 genetic bases,9 and physical mechanisms of sound production.19,23 The experimental accessibility, relatively advanced understanding of the neural and peripheral systems, and status as a well-developed model for vocal production and learning make songbirds an attractive animal model to advance speech BMI, much like the nonhuman primate model for motor BMI.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed and will be fulfilled by the lead contact, Timothy Q. Gentner (tgentner@ucsd.edu).
Materials availability
Printable hardware and electronic designs developed for this work are available in the following github repository: https://github.com/singingfinch/bernardo.
Data and code availability
Data generated in this study have been deposited to https://doi.org/10.6084/m9.figshare.14502198.
Code for data acquisition, processing pipeline and analysis developed for this work is available in the following github repositories: https://github.com/zekearneodo/swissknife; https://github.com/kaichensh/curr_bio_2021.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Subjects
Electrophysiology data was collected from n = 4 adult (> 120 dph) male zebra finches.62 Birds were individually housed for the entire duration of the experiment and kept on a 14:10h light:dark cycle. The birds were not used in any other experiments.
Ethical note
All procedures were approved by the Institutional Animal Care and Use Committee of the University of California (protocol number S15027).
METHOD DETAILS
Neural and audio recordings
We used 4-shank, 16/32 site Si-Probes (Neuronexus A4×2-tet-5mm-150–200-121 -PEDOT coated, bird z007-; Buzsaki32 -pedot coated, bird z028-; A4×1-tet-3mm-150–121 -birds z017 & z020-). We mounted the probes on an in-house designed, printable microdrive and implanted them targeting nucleus HVC. Audio was registered with a microphone (Earthworks M30) connected to a preamplifier (ART Tube MP). Extracellular voltages and pre-amplified audio were amplified and digitized at 30kHz using an Intan RHD2000 acquisition system, Open ephys and custom software. Ref and gnd were shorted together via a 0 Ohm resistor on the headstage (Intan RHD2132/RHD2116).
Electrode implant
Animals were anesthetized with a gaseous mixture of Isoflurane/oxygen (1%–2.5%, 0.7 lpm) and placed in a stereotaxic frame. Analgesia was provided by means of a 2mg/kg dose of carprofen (Rimadyl) administered I.M. The scalp was partially removed and the upper layer of the skull over the y-sinus was uncovered. The probe was attached to the shaft of a microdrive of our design (https://github.com/singingfinch/bernardo/tree/master/hardware/printable_microdrive) which was printed in-house using a B9 Creator printer and the BR-9 resin. A craniotomy site was open 2400 μm lateral to the y-sinus (right/left hemispheres). The dura was removed, and the electrode array was lowered to a 300–500 μm depth. The opening was then covered with artificial dura (DOWSIL 3–4680 Silicone Gel Kit) and the microdrive was cemented to the skull using dental cement (C&B Metabond). A reference wire was made with a 0.5 mm segment of platinum-iridium wire (0.002”) soldered to a silver wire lead and inserted between the dura and the skull in through a craniotomy roughly 3mm medial (contralateral to the hemisphere where the electrode was inserted) and 1.5 mm anterior to the y-sinus. The reference electrode was cemented to the skull and the silver lead was soldered to the ref and gnd leads of the Neuronexus probe. Most of the open area, including the electrode and the microdrive, was covered with a chamber designed and 3D printed in house, which was cemented to the skull. The skin incision was sutured and glued to the chamber with superglue. The mass of the probe, microdrive and protective chamber was 1.2–1.4g. Upon returning to a single-housing cage, a weight reliever mechanism was set up: an end of a segment of thin nylon wire (fishing line) was attached to an ad hoc pin in the chamber; the other end routed through a set of pulleys and attached to a counterweight mass of ~1g.63
Dataset preparation
Song detection
A template matching filter written in python was used to find putative instances of the motif, and then curated manually to rule out false positives.
Spike sorting
Spikes were detected and sorted using Kilosort; details of the procedure can be found in Pachitariu et al.33 The number of clusters was initialized to 32/64 (twice the number of channels of the probe) and the algorithm was allowed to automatically merge similar clusters. In post hoc curation, we removed the clusters that were visibly noise (as per the waveform) and labeled units as putatively SUA/MUA depending on whether the fraction of refractory period (2ms) violations was below/above 3% respectively.
Single Unit type classification
SUA clusters were classified as putatively representing sparse firing projection neurons or tonically firing interneurons, based on their base firing rate and their bursting behavior. We labeled a SUA cluster a putative projection neuron if its mean, spontaneous firing rate was below 5Hz and it produced at most 4 bursts with a frequency of 100 Hz or higher during the motif.32
Supra-threshold event detection
We wrote scripts in Python to detect spiking events in each channel. First, the RMS of each channel was estimated using a running window, over a period of time that ranged from minutes to an hour. Then, events that deviated in absolute value more than a number of RMS (2.5–5.5) were detected using the package peakutils (min_distance = 0.5ms).
Neural activity features
With all clusters spike-sorted or supra-threshold events, we extracted spike counts within each motif and collapsed them into 1ms (30 samples at 30,000 samples/second) time bins.
Spectral features
When training the networks with spectral features, the target at each time step was a vector containing a spectrogram slice (in log power scale). We generated the spectral slices using the spectrogram function of the signal module in the scipy package.64 We used 5ms windows (150 samples) and kept the 64 first bands above 300 Hz.
Biomechanical model of the vocal organ
Model
A model of the zebra finch vocal organ has been previously introduced and it is explained in detail in Perl et al.36 and Arneodo et al.38 This model considers mainly a sound source and a vocal tract that further shapes the acoustics of the vocalizations.
The source (syrinx) comprises two sets of tissues or labia that can oscillate induced by the sub-syringeal pressure and modulate the airflow to produce sound.35 The motion of the labia is represented as a surface wave propagating in the direction of the airflow, that can be described in terms of the lateral displacement of the midpoint of the tissue.23 Its mathematical form is the motion equation of a nonlinear oscillator in which two parameters that determine the acoustic features of the solutions are controlled by the bird: the sub-syringeal air sac pressure and the stiffness of the restitution (through the activity of syringeal muscles). In order to integrate the model in real time, a set of equations was found that is computationally less expensive yet capable of displaying topologically equivalent sets of solutions as the parameters are varied:65
where x represents the departure of the midpoint position of the oscillating labia, γ is a time scaling factor, and the parameters α and β are functions of the air sac pressure and the activity of the ventral syringeal muscle, respectively.
The upper vocal tract further shapes the sound produced by the source, determining spectral properties such as the timbre. We used a model for the vocal that includes a tube, accounting for the trachea, followed by a Helmholtz resonator, accounting for the oropharyngeal-esophageal cavity (OEC)66,67 (see Figure 1A in Arneodo et al.38). The pressure at the input of the tube that represents the trachea is Pi(t) = ax(t) – r x(t – τ), where ax(t) is the contribution to the fluctuations by the modulated airflow, r is the reflection coefficient at the opposing end of the tube of length L and τ = 2L/c, with c the sound velocity. The pressure fluctuations at the output of the trachea force the air at the glottis, approximated by the neck of the Helmholtz resonator that represents the OEC. The mass of air at the glottis, forced into the cavity, is subject to a restitution force exerted by the larger mass of air in it.
In acoustics, it is common to write an analog electric computational model to describe a system of filters. The acoustic pressure is represented by an electric potential and the volume flow by the electric current.68 In this framework, short constrictions are inductors, and cavities (smaller than the wavelengths) are well represented by capacitors. The equations for the equivalent circuit of the post-tracheal part of the vocal tract, (see Figure 1B in Arneodo et al.38) read:
where the electric components relate to geometric parameters of acoustic elements, and are described in detail in Perl et al.36 and Arneodo et al.38 The pressure fluctuations at the glottal end of the trachea relate linearly to the electric tension Vext driving the circuit. Following the same scheme, the electrical potential at the resistor standing for the beak Vb = i3Rb is the analog of the pressure fluctuations at the output of the beak. In our model, this quantity is the sound radiated by the vocal organ.
Parameter fitting
In order to fit the parameter series that will lead to reconstruction of the song, we perform a procedure similar to that previously described.27,36 Timescale parameter is set to a value of 23,500; α is set to −0.15 during vocalization and 0.15 otherwise, and β is set in order to minimize the distance in the (pitch, spectral content) space between the synthesized and the recorded song segments;36 the envelope (e(t) in the main text) is obtained by rectifying and smoothing the recorded waveform; the parameters of the vocal tract were fixed, in the same values as in Perl et al.36 In order to extract the pitch of the song, we follow a modification of the automatic procedure presented in Boari et al.,69 and we add a layer of manual curation. When integrating the model, we apply the extracted envelope (e(t)) as an extra multiplicative factor when computing ax(t), since it recovers the amplitude fluctuations that were discarded when reducing the model to its normal form and driving it with the bi-valued parameter α. The parameters accounting for the geometry of the vocal tract are constants and are set to the same values as in Perl et al.24
Neural network training
Neural network -based decoders were implemented in python 3.6, using Tensorflow 2.0 and Keras. They were run on Ubuntu 16.04 and 18.04 PCs equipped with NVidia GPUs (Tesla k40c, Titan Xp, and Titan X Pascal). CUDA version was 10.2.
LSTM network architecture
The network has 2 layers of LSTM cells, with Nx5 cells in the first layer and N in the second, where N is the number of clusters in the neural data. The output layer has as many relu units as the target space (64 for the spectrogram bands). The input of the network is a Nx50 array that contains the spike count of each cluster, in each of 50, 1ms bins preceding the output bin. The output of the network is a 1×64 array containing the spectral bands corresponding to a 5ms bin. Both LSTM layers utilized 20% dropout and 0.001 L2 regularization during training to prevent overfitting.70
Feed-forward Network architecture
The network has 1 dense hidden layer of Nx25 relu units, where N is the number of clusters in the neural data. The output layer has as many relu units as the target space (p = 64 for the spectrogram bands, p = 3 for the biomechanical model parameters). The input of the network is a Nx50 vector that contains the spike count of each cluster, in each of 50, 1ms bins preceding the output bin. The output of the network is a 1xP array containing the spectral bands (p = 64) or the biomechanical model parameters (p = 3) corresponding to a 5ms bin. The hidden layer utilized 20% dropout and 0.001 L2 regularization during training
Training procedure
We utilized a gradient-based optimizer (Adam/rmsprop71) and mean square error (MSE) as a loss function for LSTM/FFNN. We used 40% of all the motifs for testing and the rest motifs for training. We made 3 passes using non-overlapping motifs as a testing set, in order to have as many decoded examples as the number of motifs in the session. In each pass, all of the neural-activity/decoder-target pairs (one per bin) were fed in random order to the network, both when training and decoding. We reserved 10% of the training set as a validation subset for early stopping, where the training session would be stopped if validation loss failed to decrease within 5/10 training epochs. Figure 4 shows the results of this motif -based training averaged across all birds. As an alternative training method, we masked a fraction of each motif (roughly 3.3%), trained on the complement, then generated the song corresponding to the masked fraction. We repeated this piece-wise procedure tiling the whole motif, and generated entire motifs using segments of data that were novel to the decoder. Figure S3 shows the results of both the motif-wise and piece-wise training for individual birds.
Song waveform generation
Spectrogram inversion
We used LSEE-STFTM algorithm to invert spectrograms back to audio waves,72 as implemented in the librosa python package.73 The algorithm iteratively estimates a signal from the short-time Fourier transform magnitude (STFTM), through minimizing the mean square error between the short-time Fourier transform (STFT) and the estimated STFT, and subsequently performs STFT on the estimated signal, the magnitude of which will be passed on to the next iteration.
Within each iteration, a signal was approximated using the equation below:
where x(n) denotes the estimated signal; w(n) denotes the analysis window used in STFT. The variable S is a positive integer, representing the sampling rate of the STFT. Here, yw(mS, n) is the target signal corresponding to Yw(mS, n), which denotes the target STFTM, in our case spectrogram powers. To calculate in each iteration, we used a sinusoidal window:72
where L represents the length of the window. Here, wr(n) is a rectangular window with an amplitude of within 0≤ n < L and zero anywhere outside. A modified Hamming window can be obtained by setting a = .54, b = − .46, . After obtaining an x(n) value within each iteration, the STFT of x(n) was calculated, which was used in place of Yw(mS, n) in the next iteration. The squared error between the target STFTM and the estimated STFTM is proven to decrease in each iteration of the algorithm.
Biomechanical model integration
Once the model parameters are predicted by the decoder, they are re-sampled and fed to an ordinary differential equation integrator. Resampling to 30 Khz is performed (with cubic interpolation). A fourth order runge-kutta ODE integrator (custom coded) integrates then the equations of the model with a time step of (900 KHz)−1.
Synthesis through principal components
PC decomposition was made using the PCA module in the scikit-learn package.74 We obtained the principal component decomposition of all the spectrograms of all the motifs sung by each bird during the length of the experiment. 512 frequency bins were used for the spectrograms, which were concatenated and projected onto the N principal components TN = SWN, where S is the 512-dimension spectrum time series and WN the transformation matrix (the matrix of the L eigenvectors of SST with the largest eigenvalues) via the pca.fit and pca.fit_transform methods. For reconstruction from principal components, the inverse transformation was applied by means of the pca.inverse_transform method.
Spectrum shuffle mask
Time warping
We adopted a simplified version of Dynamic Time Warping (DTW75) specific to zebra finch songs. Instead of segmenting the song into different syllables and matching each syllable to different syllable templates, we took advantage of the stereotypical nature of zebra finch songs and directly computed minimal distance matrices (D) between each song-level spectrogram and a spectrogram template. Starting at the first slice of each spectrogram,
Where i indexes the time frames of the input pattern, j indexes the time frames of a single template, l indexes the ordered steps along a specific path. d(i, j) is the local distance between slice i and slice j. wj(l) denotes the specific step at l in the space of j. Once a distance matrix D was calculated, we determined an optimal path with the lowest cumulative distance between the input and the template, and proceeded to stretch, delete or keep each input slice, depending on the path.
Masking
We applied a random yet consistent shuffling mask, P, to our entire warped spectrogram repertoire so that spectral consistency across time is disrupted while the temporal pattern within each motif remains. For the i-th spectrogram slice in each warped song, we shuffled all 64 spectral elements using the same shuffling pattern Pi. Treating all spectrograms with the same shuffling mask P enabled us to determine whether our model is decoding the spectral information within birdsongs or recreating the same pattern regardless of spectral consistency across time. In our shuffling training session, we used the shuffled spectrograms as output.
Reordering mask
After training, we tested our model on novel neural data, the target of which were also shuffled spectrograms. In order to visually compare our model’s performance with and without shuffling, we reordered the reconstructed shuffled spectrograms. We achieved this by applying a reordering mask, R, that traces and reverses all the shuffling done through the aforementioned shuffling mask P. For any spectrogram S, R(P(S)) = S.
QUANTIFICATION AND STATISTICAL ANALYSIS
Performance Evaluation
Root Mean Square Error (RMSE)
We used RMSE between each pair of original and predicted spectrogram magnitude as a metric to evaluate the performance of our models.
Spectral correlation
To obtain the spectral correlation across time for a pair of spectrograms, we first computed the pearson correlation coefficient between each corresponding pair of spectral slices that conform the two spectrograms (via the function pearsonr from the stats module of the scipy python package64). Then, we obtained the time-averaged value across the span of the motif.
Earth mover’s distance
To obtain the distance across time for a pair of spectrograms, we computed the earth mover’s distance (dEMD) or Wasserstein metric between each pair of spectral slices that conform the two spectrograms (via the function wasserstein_distance from the stats module of the scipy python package64). Prior comparison, each spectral slice was normalized such that the total area under the slice be 1; for silences, a value of 1 was assigned to the first bin of the spectrogram. Then, we obtained the time-averaged value across the span of the motif.
Spectrogram Normalization
In order to account for variations among motifs from different birds, we normalized spectrograms for each bird so that the collection of original spectrograms for each bird had a maximum power of 1 and minimum power of 0:
Where pi is the power of a point on either an original spectrogram or a predicted spectrogram before normalization, while is the normalized power of the corresponding point. pmax denotes the maximum power of the entire set of original spectrograms, while pmin represents the minimum power of the entire set of original spectrograms. With such normalization, we were able to account for variations among motifs from different birds while keeping the variations within motifs from the same bird.
Pairwise performance comparisons
We performed comparisons among and between different sets of songbirds (displayed in Figure 4 boxplots for instance). BOS-BOS: comparisons provide a baseline of the variability of the bird’s own motifs during the session: comparison across each pair of motifs. neur-bos: comparison across each pair of natural motifs and it’s corresponding one decoded from neural activity.
In order to provide an extra control reference, we also computed spectrogram comparisons against a set of 47 motifs from conspecific birds (other zebra finches; about half of them from our colony and half from other colonies). This produced the sets: BOS-CON: comparisons across each BOS motif and all of the conspecific (CON) motifs.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Deposited data | ||
| Original data | This paper, Figshare | https://doi.org/10.6084/m9.figshare.14502198 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| Code for acquisition, pipeline, analysis written in C/Python. | This paper | https://github.com/zekearneodo/swissknife |
| Code for analysis written in Python. | This paper | https://github.com/kaichensh/curr_bio_2021 |
| Other | ||
| Printable Microdrive and signal conditioning hardware designs. | This paper | https://github.com/singingfinch/bernardo |
| LIFE SCIENCE TABLE WITH EXAMPLES FOR AUTHOR REFERENCE | ||
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and virus strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children's Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350–13–2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433–59–5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124–87–8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14–486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical commercial assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | PerkinElmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202–1012 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09–0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207–1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106–5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
| PHYSICAL SCIENCE TABLE WITH EXAMPLES FOR AUTHOR REFERENCE | ||
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Chemicals, peptides, and recombinant proteins | ||
| QD605 streptavidin conjugated quantum dot | Thermo Fisher Scientific | Cat#Q10101MP |
| Platinum black | Sigma-Aldrich | Cat#205915 |
| Sodium formate BioUltra, ≥99.0% (NT) | Sigma-Aldrich | Cat#71359 |
| Chloramphenicol | Sigma-Aldrich | Cat#C0378 |
| Carbon dioxide (13C, 99%) (<2% 18O) | Cambridge Isotope Laboratories | CLM-185–5 |
| Poly(vinylidene fluoride-co-hexafluoropropylene) | Sigma-Aldrich | 427179 |
| PTFE Hydrophilic Membrane Filters, 0.22 μm, 90 mm | Scientificfilters.com/Tisch Scientific | SF13842 |
| Critical commercial assays | ||
| Folic Acid (FA) ELISA kit | Alpha Diagnostic International | Cat# 0365–0B9 |
| TMT10plex Isobaric Label Reagent Set | Thermo Fisher | A37725 |
| Surface Plasmon Resonance CM5 kit | GE Healthcare | Cat#29104988 |
| NanoBRET Target Engagement K-5 kit | Promega | Cat#N2500 |
| Deposited data | ||
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Structure of compound 5 | This paper; Cambridge Crystallographic Data Center | CCDC: 2016466 |
| Code for constraints-based modeling and analysis of autotrophic E. coli | This paper | https://gitlab.com/elad.noor/sloppy/tree/master/rubisco |
| Software and algorithms | ||
| Gaussian09 | Frish et al., 2013 | https://gaussian.com |
| Python version 2.7 | Python Software Foundation | https://www.python.org |
| ChemDraw Professional 18.0 | PerkinElmer | https://www.perkinelmer.com/category/chemdraw |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| Other | ||
| DASGIP MX4/4 Gas Mixing Module for 4 Vessels with a Mass Flow Controller | Eppendorf | Cat#76DGMX44 |
| Agilent 1200 series HPLC | Agilent Technologies | https://www.agilent.com/en/products/liquid-chromatography |
| PHI Quantera II XPS | ULVAC-PHI, Inc. | https://www.ulvac-phi.com/en/products/xps/phi-quantera-ii/ |
Highlights.
Songbirds sing rich, complex, learned songs
A model of their vocal organ can synthesize song with few control parameters
This allows neurally driven song synthesis via a simple neural network
Brain machine interfaces can be enhanced by understanding the biomechanics
ACKNOWLEDGMENTS
We thank K. Perks, B. Theilman, M. Thielk, and T. Sainburg for critical discussions and/or input on the manuscript. We acknowledge illustrations and artwork by Ondina Arneodo and Mariano Chena. This work was supported by the US National Institutes of Health (grant R01DC018446), the Kavli Institute for the Brain and Mind (IRG no. 2016-004), the Office of Naval Research (MURI N00014-13-1-0205), the US National Science Foundation (Graduate Research Fellowship Grant No. DGE-1650112 to D.E.B.), the University of California—Historically Black Colleges and Universities Initiative (D.E.B.), and a Pew Latin American Fellowship in the Biomedical Sciences (E.M.A.). The Tesla K40, Titan Z, and Titan X Pascal used for this research were donated by the NVIDIA Corporation.
Footnotes
DECLARATION OF INTERESTS
V.G. is a compensated consultant of Paradromics, Inc., a brain-computer interface company.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an under-represented minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.05.035.
REFERENCES
- 1.Gilja V, Nuyujukian P, Chestek CA, Cunningham JP, Yu BM, Fan JM, Churchland MM, Kaufman MT, Kao JC, Ryu SI, and Shenoy KV (2012). A high-performance neural prosthesis enabled by control algorithm design. Nat. Neurosci. 15, 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Velliste M, Perel S, Spalding MC, Whitford AS, and Schwartz AB (2008). Cortical control of a prosthetic arm for self-feeding. Nature 453, 1098–1101. [DOI] [PubMed] [Google Scholar]
- 3.Gilja V, Pandarinath C, Blabe CH, Nuyujukian P, Simeral JD, Sarma AA, Sorice BL, Perge JA, Jarosiewicz B, Hochberg LR, et al. (2015). Clinical translation of a high-performance neural prosthesis. Nat. Med. 21, 1142–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochberg LR, Bacher D, Jarosiewicz B, Masse NY, Simeral JD, Vogel J, Haddadin S, Liu J, Cash SS, van der Smagt P, and Donoghue JP (2012). Reach and grasp by people with tetraplegia using a neurally controlled robotic arm. Nature 485, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akbari H, Khalighinejad B, Herrero JL, Mehta AD, and Mesgarani N (2019). Towards reconstructing intelligible speech from the human auditory cortex. Sci. Rep. 9, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartier J, Anumanchipalli GK, Johnson K, and Chang EF (2018). Encoding of articulatory kinematic trajectories in human speech sensorimotor cortex. Neuron 98, 1042–1054.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anumanchipalli GK, Chartier J, and Chang EF (2019). Speech synthesis from neural decoding of spoken sentences. Nature 568, 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doupe AJ, and Kuhl PK (1999). Birdsong and human speech: common themes and mechanisms. Annu. Rev. Neurosci. 22, 567–631. [DOI] [PubMed] [Google Scholar]
- 9.Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, et al. (2014). Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis ED (2019). Evolution of vocal learning and spoken language. Science 366, 50–54. [DOI] [PubMed] [Google Scholar]
- 11.Brainard MS, and Doupe AJ (2000). Auditory feedback in learning and maintenance of vocal behaviour. Nat. Rev. Neurosci. 1, 31–40. [DOI] [PubMed] [Google Scholar]
- 12.Long MA, Katlowitz KA, Svirsky MA, Clary RC, Byun TM, Majaj N, Oya H, Howard MA 3rd, and Greenlee JDW (2016). Functional segregation of cortical regions underlying speech timing and articulation. Neuron 89, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadagkar V, Puzerey PA, Chen R, Baird-Daniel E, Farhang AR, and Goldberg JH (2016). Dopamine neurons encode performance error in singing birds. Science 354, 1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallentin D, Kosche G, Lipkind D, and Long MA (2016). Neural circuits. Inhibition protects acquired song segments during vocal learning in zebra finches. Science 351, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch GF, Okubo TS, Hanuschkin A, Hahnloser RHR, and Fee MS (2016). Rhythmic continuous-time coding in the songbird analog of vocal motor cortex. Neuron 90, 877–892. [DOI] [PubMed] [Google Scholar]
- 16.Roberts TF, Hisey E, Tanaka M, Kearney MG, Chattree G, Yang CF, Shah NM, and Mooney R (2017). Identification of a motor-to-auditory pathway important for vocal learning. Nat. Neurosci. 20, 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fee MS, Kozhevnikov AA, and Hahnloser RHR (2004). Neural mechanisms of vocal sequence generation in the songbird. Ann. N Y Acad. Sci. 1016, 153–170. [DOI] [PubMed] [Google Scholar]
- 18.Giret N, Kornfeld J, Ganguli S, and Hahnloser RHR (2014). Evidence for a causal inverse model in an avian cortico-basal ganglia circuit. Proc. Natl. Acad. Sci. USA 111, 6063–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elemans CPH, Rasmussen JH, Herbst CT, Düring DN, Zollinger SA, Brumm H, Srivastava K, Svane N, Ding M, Larsen ON, et al. (2015). Universal mechanisms of sound production and control in birds and mammals. Nat. Commun. 6, 8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava KH, Holmes CM, Vellema M, Pack AR, Elemans CPH, Nemenman I, and Sober SJ (2017). Motor control by precisely timed spike patterns. Proc. Natl. Acad. Sci. USA 114, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goller F, and Cooper BG (2004). Peripheral motor dynamics of song production in the zebra finch. Ann. N Y Acad. Sci. 1016, 130–152. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi DY, Fenley AR, Teramoto Y, Narayanan DZ, Borjon JI, Holmes P, and Ghazanfar AA (2015). Language development. The developmental dynamics of marmoset monkey vocal production. Science 349, 734–738. [DOI] [PubMed] [Google Scholar]
- 23.Titze IR (1988). The physics of small-amplitude oscillation of the vocal folds. J. Acoust. Soc. Am. 83, 1536–1552. [DOI] [PubMed] [Google Scholar]
- 24.Perl YS, Arneodo EM, Amador A, Goller F, and Mindlin GB (2011). Reconstruction of physiological instructions from zebra finch song. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 84, 051909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochreiter S, and Schmidhuber J (1997). Long short-term memory. Neural Comput. 9, 1735–1780. [DOI] [PubMed] [Google Scholar]
- 26.Leonardo A, and Fee MS (2005). Ensemble coding of vocal control in birdsong. J. Neurosci. 25, 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amador A, Perl YS, Mindlin GB, and Margoliash D (2013). Elemental gesture dynamics are encoded by song premotor cortical neurons. Nature 495, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picardo MA, Merel J, Katlowitz KA, Vallentin D, Okobi DE, Benezra SE, Clary RC, Pnevmatikakis EA, Paninski L, and Long MA (2016). Population-level representation of a temporal sequence underlying song production in the zebra finch. Neuron 90, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberti WA 3rd, Markowitz JE, Perkins LN, Liberti DC, Leman DP, Guitchounts G, Velho T, Kotton DN, Lois C, and Gardner TJ (2016). Unstable neurons underlie a stable learned behavior. Nat. Neurosci. 19, 1665–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahnloser RHR, Kozhevnikov AA, and Fee MS (2002). An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature 419, 65–70. [DOI] [PubMed] [Google Scholar]
- 31.Yu AC, and Margoliash D (1996). Temporal hierarchical control of singing in birds. Science 273, 1871–1875. [DOI] [PubMed] [Google Scholar]
- 32.Kozhevnikov AA, and Fee MS (2007). Singing-related activity of identified HVC neurons in the zebra finch. J. Neurophysiol. 97, 4271–4283. [DOI] [PubMed] [Google Scholar]
- 33.Pachitariu M, Steinmetz NA, Kadir SN, Carandini M, and Harris KD (2016). Fast and accurate spike sorting of high-channel count probes with KiloSort. In Advances in Neural Information Processing Systems 29, Lee DD, Sugiyama M, Luxburg UV, Guyon I, and Garnett R, eds. (Curran Associates; ), pp. 4448–4456. [Google Scholar]
- 34.Wild JM (1997). Neural pathways for the control of birdsong production. J. Neurobiol. 33, 653–670. [DOI] [PubMed] [Google Scholar]
- 35.Goller F, and Larsen ON (1997). A new mechanism of sound generation in songbirds. Proc. Natl. Acad. Sci. USA 94, 14787–14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perl YS, Arneodo EM, Amador A, and Mindlin GB (2012). Nonlinear dynamics and the synthesis of zebra finch song. Int. J. Bifurcat. Chaos 22, 1250235. [Google Scholar]
- 37.Arneodo O (2021). Zebra finch electrophysiology illustrations. Figshare. 10.6084/m9.figshare.14577156.v1. [DOI]
- 38.Arneodo EM, Perl YS, Goller F, and Mindlin GB (2012). Prosthetic avian vocal organ controlled by a freely behaving bird based on a low dimensional model of the biomechanical periphery. PLoS Comput. Biol. 8, e1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohr B, and Dooling RJ (1998). Detection of changes in timbre and harmonicity in complex sounds by zebra finches (Taeniopygia guttata) and budgerigars (Melopsittacus undulatus). J. Comp. Psychol. 112, 36–47. [DOI] [PubMed] [Google Scholar]
- 40.Christie BP, Tat DM, Irwin ZT, Gilja V, Nuyujukian P, Foster JD, Ryu SI, Shenoy KV, Thompson DE, and Chestek CA (2015). Comparison of spike sorting and thresholding of voltage waveforms for intracortical brain-machine interface performance. J. Neural Eng. 12, 016009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brincat S, Jia N, Salazar-Gómez AF, Panko M, Miller E, and Guenther F (2013). Which Neural Signals are Optimal for Brain-Computer Interface Control? (Verlag der Technischen Universität Graz). [Google Scholar]
- 42.Fraser GW, Chase SM, Whitford A, and Schwartz AB (2009). Control of a brain-computer interface without spike sorting. J. Neural Eng. 6, 055004. [DOI] [PubMed] [Google Scholar]
- 43.Kogan JA, and Margoliash D (1998). Automated recognition of bird song elements from continuous recordings using dynamic time warping and hidden Markov models: a comparative study. J. Acoust. Soc. Am. 103, 2185–2196. [DOI] [PubMed] [Google Scholar]
- 44.Peleg S, Werman M, and Rom H (1989). A unified approach to the change of resolution: space and gray-level. IEEE Trans. Pattern Anal. Mach. Intell. 11, 739–742. [Google Scholar]
- 45.Margoliash D, and Fortune ES (1992). Temporal and harmonic combination-sensitive neurons in the zebra finch’s HVc. J. Neurosci. 12, 4309–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doupe AJ, and Konishi M (1991). Song-selective auditory circuits in the vocal control system of the zebra finch. Proc. Natl. Acad. Sci. USA 88, 11339–11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dave AS, and Margoliash D (2000). Song replay during sleep and computational rules for sensorimotor vocal learning. Science 290, 812–816. [DOI] [PubMed] [Google Scholar]
- 48.Prior NH, Smith E, Lawson S, Ball GF, and Dooling RJ (2018). Acoustic fine structure may encode biologically relevant information for zebra finches. Sci. Rep. 8, 6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul A, McLendon H, Rally V, Sakata JT, and Woolley SC (2021). Behavioral discrimination and time-series phenotyping of birdsong performance. PLoS Comput. Biol. 17, e1008820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assaneo MF, Trevisan MA, and Mindlin GB (2013). Discrete motor coordinates for vowel production. PLoS ONE 8, e80373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tankus A, Fried I, and Shoham S (2012). Structured neuronal encoding and decoding of human speech features. Nat. Commun. 3, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.d’Avella A, Saltiel P, and Bizzi E (2003). Combinations of muscle synergies in the construction of a natural motor behavior. Nat. Neurosci. 6, 300–308. [DOI] [PubMed] [Google Scholar]
- 53.Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, and Datta SR (2015). Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YS, and Ghazanfar AA (2018). Vocal development through morphological computation. PLoS Biol. 16, e2003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tytell ED, Holmes P, and Cohen AH (2011). Spikes alone do not behavior make: why neuroscience needs biomechanics. Curr. Opin. Neurobiol. 21, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guenther FH, and Brumberg JS (2011). Brain-machine interfaces for real-time speech synthesis. Annu Int Conf IEEE Eng Med Biol Soc 2011, 5360–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinton GE, and Salakhutdinov RR (2006). Reducing the dimensionality of data with neural networks. Science 313, 504–507. [DOI] [PubMed] [Google Scholar]
- 58.Khodagholy D, Gelinas JN, Thesen T, Doyle W, Devinsky O, Malliaras GG, and Buzsáki G (2015). NeuroGrid: recording action potentials from the surface of the brain. Nat. Neurosci. 18, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hermiz J, Hossain L, Arneodo EM, Ganji M, Rogers N, Vahidi N, Halgren E, Gentner TQ, Dayeh SA, and Gilja V (2020). Stimulus driven single unit activity from micro-electrocorticography. Front. Neurosci. 14, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipkind D, Zai AT, Hanuschkin A, Marcus GF, Tchernichovski O, and Hahnloser RHR (2017). Songbirds work around computational complexity by learning song vocabulary independently of sequence. Nat. Commun. 8, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sainburg T, Theilman B, Thielk M, and Gentner TQ (2019). Parallels in the sequential organization of birdsong and human speech. Nat. Commun. 10, 3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arneodo E, and Gentner TQ (2021). Chronic recordings in HVC with silicon probe arrays in singing zebra finches. Figshare. [Google Scholar]
- 63.Yamahachi H, Zai AT, Tachibana RO, Stepien AE, Rodrigues DI, Cavé-Lopez S, Lorenz C, Arneodo EM, Giret N, and Hahnloser RHR (2020). Undirected singing rate as a non-invasive tool for welfare monitoring in isolated male zebra finches. PLoS ONE 15, e0236333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. ; SciPy 1.0 Contributors (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sitt JD, Amador A, Goller F, and Mindlin GB (2008). Dynamical origin of spectrally rich vocalizations in birdsong. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 78, 011905. [DOI] [PubMed] [Google Scholar]
- 66.Riede T, Suthers RA, Fletcher NH, and Blevins WE (2006). Songbirds tune their vocal tract to the fundamental frequency of their song. Proc. Natl. Acad. Sci. USA 103, 5543–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riede T, Schilling N, and Goller F (2013). The acoustic effect of vocal tract adjustments in zebra finches. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 199, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinsler LE, Frey AR, Coppens AB, and Sanders JV (1999). Fundamentals of Acoustics, Fourth Edition (Wiley; ). [Google Scholar]
- 69.Boari S, Perl YS, Amador A, Margoliash D, and Mindlin GB (2015). Automatic reconstruction of physiological gestures used in a model of birdsong production. J. Neurophysiol. 114, 2912–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, and Salakhutdinov R (2014). Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958. [Google Scholar]
- 71.Kingma DP, and Ba J (2014). Adam: a method for stochastic optimization. arXiv, arXiv:1412.6980v9. https://arxiv.org/abs/1412.6980. [Google Scholar]
- 72.Griffin D, and Lim J (1984). Signal estimation from modified short-time Fourier transform. IEEE Trans. Acoust. Speech Sig. Proc. 32, 236–243. [Google Scholar]
- 73.McFee B, Lostanlen V, Metsai A, McVicar M, Balke S, Thomé C, Raffel C, Zalkow F, Malek A, Lee K, et al. (2020). librosa/librosa: 0.8.0. Zenodo. [Google Scholar]
- 74.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830. [Google Scholar]
- 75.Anderson SE, Dave AS, and Margoliash D (1996). Template-based automatic recognition of birdsong syllables from continuous recordings. J. Acoust. Soc. Am. 100, 1209–1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study have been deposited to https://doi.org/10.6084/m9.figshare.14502198.
Code for data acquisition, processing pipeline and analysis developed for this work is available in the following github repositories: https://github.com/zekearneodo/swissknife; https://github.com/kaichensh/curr_bio_2021.


