Abstract
Background
Given the importance of neutralising antibodies in protection against SARS-CoV-2 infection, it is critical to assess neutralisation persistence long-term following recovery. This study investigated neutralisation titres against SARS-CoV-2 up to 6 months post-symptom onset in individuals with mild COVID-19.
Methods
Plasma neutralisation titres in convalescent COVID-19 individuals were determined at baseline and 6 months post-symptom onset using a cell culture infectious SARS-CoV-2 assay. Total SARS-CoV-2 spike-specific IgG and IgA binding was measured using a lectin capture ELISA and compared between timepoints and correlated to neutralising titres.
Findings
All 48 convalescent COVID-19 individuals were found to have detectable SARS-CoV-2 50% inhibitory dilution neutralisation titres (ID50) at baseline and 6 months post-symptom onset with mean ID50 of 1/943 and 1/411, respectively. SARS-CoV-2 neutralisation titres peaked within 1-2 months post-symptom onset. However, 50% of individuals showed comparable ID50 at baseline and 6 months post-symptom onset. Both SARS-CoV-2 spike-specific IgG and IgA levels correlated well with neutralising titres. IgG binding was found to be sustained up to 6 months post-symptom onset, whereas IgA levels declined.
Interpretation
This study demonstrates durability of SARS-CoV-2 spike-specific IgG and neutralisation responses following recovery from mild COVID-19. Thus, all subjects included in this study might potentially have protective levels of neutralising antibodies 6 months post-symptom onset. This study also demonstrates a relationship between spike-specific IgA and neutralisation decline, with implications for long-term protection against SARS-CoV-2 infection.
Funding
Novo Nordisk Foundation, Independent Research Fund Denmark and Danish Agency for Science and Higher Education.
Keywords: SARS-CoV-2, neutralising antibodies, neutralisation, COVID-19, IgG, IgA
Research in context.
Evidence before this study
Publications on longitudinal neutralisation in SARS-CoV-2 infection were searched in MEDLINE, PubMed, Embase and the WHO global research database using the search terms “severe acute respiratory syndrome coronavirus 2”, “SARS-CoV-2”, “COVID-19”, “longitudinal”, “neutralization/neutralisation”, “antibody”, which were published in English up to 31 March 2021. Publications in medRxiv and BioRxiv were not included due to lack of peer-review. A total of eight published studies were found, which assessed longitudinal neutralising antibodies following SARS-CoV-2 infection. Common limitations amongst these studies was the use of neutralisation assays using pseudotyped or surrogate models, limited follow up after SARS-CoV-2 infection and cohort size. Furthermore, most of these studies were limited to analysis of receptor binding domain antibodies and do not assess neutralisation targeting the whole spike protein or other SARS-CoV-2 proteins.
Added value of this study
This longitudinal study provides a biologically relevant analysis of neutralising antibody titres up to 6 months post-SARS-CoV-2 symptom onset, through the use of infectious whole SARS-CoV-2 neutralisation assays. All participants included in this study were found to have detectable neutralising titres 6 months post-symptom onset. Furthermore, IgG and IgA antibodies were assessed against whole SARS-CoV-2 spike protein and were found to correlate well with neutralising titres. While spike-specific IgG levels were sustained longitudinally, spike-specific IgA levels were found to wane and correlated with declining neutralising titres.
Implications of all the available evidence
Neutralising titres following SARS-CoV-2 infection follow dynamics similar to that of other acute viral infections, including that of other human coronavirus infections, and remain at significant levels 6 months post-symptom onset. These results would imply that immunity to SARS-CoV-2 does not rapidly wane, and individuals may be protected for at least 6 months post-symptom onset. These results also have important implications for longevity of vaccine-induced neutralising antibodies against SARS-CoV-2.
Alt-text: Unlabelled box
2. Introduction
The ongoing pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has affected over 100 million people and resulted in millions of deaths, worldwide [1]. The associated disease COVID-19 appears to have a broad spectrum of clinical syndromes, ranging from asymptomatic to mild flu-like symptoms to severe respiratory distress requiring respiratory support [2]. However, 85% of the infected individuals only report mild symptoms, which do not require hospitalization [2].
The SARS-CoV-2 RNA genome encodes 4 structural proteins, of which the spike (S) protein is the most immunodominant protein for neutralising antibody (nAb) responses [3]. These nAbs directed to the S protein are important for viral clearance and are a correlate for protection from infection/reinfection in animal challenge models and human vaccine trials [4], [5], [6], [7]. It is thought that most S-directed nAbs block the virus from interacting with the target host receptor, angiotensin-converting enzyme 2 (ACE2), particularly by interacting with the receptor binding domain (RBD) within the S protein [3]. For most typical acute viral infections, including that of closely related human coronaviruses SARS-CoV and Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV), nAb responses peak at 1-month post-symptom onset and wane to a level that is sustained longitudinally [8], [9], [10].
Limited studies on the longevity of antibody responses to SARS-CoV-2 indicate that, although overall antibody responses are maintained, nAbs follow the same pattern as typical acute viral infections [11], [12], [13], [14], [15], [16], [17], [18], [19]. Although advantageous due to the non-requirement of a high-level biosafety level facility, most of these studies are limited by the use of spike surrogate or pseudotyped neutralisation assays, which might not fully reflect neutralisation to the same extent as the assays performed with infectious virus [20], possibly due to absence of other nAb targets such as the nucleocapsid (N) [16], envelope (E) or membrane (M) proteins. Additionally, most of these studies have followed their participants for <6 months [12], [13], [14], [15],18], thus only representing a short time after symptom onset. Understanding the longevity of nAb responses is considered of high importance, especially considering that nAb induction is the primary goal of many of the vaccines currently being administered globally.
Here, we assessed neutralisation longitudinally up to 6 months post-symptom onset against a cultured SARS-CoV-2 isolate in 48 convalescent individuals with mild COVID-19. Concordant to this, total immunoglobulin G (IgG) and IgA levels were measured using an S protein-specific Enzyme-Linked Immunosorbent Assay (ELISA), which was compared longitudinally and to neutralising titres. This study offers important information regarding the longevity of antibody responses and virus neutralisation after SARS-CoV-2 infection in individuals with mild COVID-19, the most likely infection scenario, which may have important implications for protection against reinfection and for vaccine-induced nAbs.
3. Methods
3.1. Study cohort
From 15 April 2020 to 1 February 2021, individuals who recovered from mild SARS-CoV-2 infection (defined as the non-requirement of hospitalization), and healthy individuals were recruited into the Clinical, Virological and Immunological COVID-19 (CVIC) study at Department of Infectious Diseases, Copenhagen University Hospital, Hvidovre, Denmark. Subjects with mild COVID-19 (defined by the non-requirement of hospitalization or therapeutic intervention) were enrolled based on the inclusion criteria: ≥18 years of age, confirmed SARS-CoV-2 infection from routine polymerase chain reaction (PCR) and/or antibody testing, recovered from symptoms for ≥1week, and able to speak and read adequate Danish to provide written informed consent and to participate in the study interview. During the interview with a research nurse or a physician, included subjects were required to report on gender, year of birth, country of origin to determine ethnicity, possible way of SARS-CoV-2 transmission, date of symptom onset, duration (days) and type of COVID-19 symptoms. Only participants that had a 6-month sample collected (approximately 180 days after symptom onset) were selected for this study. The same criteria and interview were used for recruitment of healthy controls at Copenhagen University Hospital, Hvidovre, Denmark, with the exception that they had not exhibited COVID-19-like symptoms since March 2020. Only healthy participants that had been screened for SARS-CoV-2 antibodies through the WANTAI SARS-CoV-2 antibody ELISA (see below) were selected for this study. A summary of participant recruitment is shown in supplementary figure (S Fig) 1. Blood was collected in EDTA at baseline, and for individuals who recovered from mild SARS-CoV-2 infection also 6 months post-symptom onset, and processed using Ficoll density grade separation to isolate and store plasma and peripheral blood mononuclear cells (PBMCs) at -80°C and -150°C, respectively.
3.2. Ethics
The study was approved by the Regional Ethical Committee (H-20025872) and Data Protection Agency (P-2020-357), respectively, and was conducted in compliance with the Declaration of Helsinki guidelines. Written informed consent was provided by all subjects and study data were collected and managed using research electronic data capture (REDCap) tools hosted at Copenhagen University Hospital, Hvidovre, Denmark [21].
3.3. Neutralisation assay
The SARS-CoV-2 isolate used in Vero E6 cell-culture experiments was obtained from an individual presenting with COVID-19 at Copenhagen University Hospital, Hvidovre, Denmark in April 2020, as previously described [22]. The sequence of this virus can be found in GenBank (accession number MZ049597). Neutralisation experiments were performed by adding virus from Vero E6 cell-culture supernatants (multiplicity of infection [MOI] of 0.01 for 104 cells [virus titration shown in S Fig 2]) to serially diluted plasma (heat inactivated at 56°C for 30 min) from individuals with mild COVID-19 at a 1:1 ratio and incubated at room temperature for 1h. As a negative control, pooled plasma (heat inactivated at 56°C for 30 min) from 5 healthy individuals was included in each assay. A mouse derived SARS-CoV-2 spike neutralising antibody (Sino Biological #40592-MM57,RRID: AB_2857935) was used as a positive control. Following 1h incubation, plasma/virus and antibody/virus complexes were then added to Vero E6 cells (RRID: CVCL_0574) seeded the day before (104 cells/well; Corning white BioCoatTM Poly-D lysine coated plates, cat #: 354651) in quadruplicate. After 48 hours incubation at 37°C and 5% CO2, the cells were stained as described previously [22], but with the use of mouse-derived spike primary (Sino Biological #40592-MM57,RRID: AB_2857935) and GE Healthcare #NA931V (RRID: AB_772210) secondary antibodies. Spots representing virus infected cells were counted using an Immunospot series 5 UV analyser (Cellular Technologies). Single outliers of quadruplicates were calculated using a modified z-score system as previously described [23] and were removed from further analysis; thus a minimum of triplicates was used for all assays. Given that there was no significant difference between healthy and virus only controls (S Fig 2), the percentage neutralisation was calculated as:
Any overall neutralisation values (average of the triplicates/quadruplicates) that yielded higher than 100% was normalized to 100% and any overall neutralisation values that yielded lower than 0% was normalized to 0%.
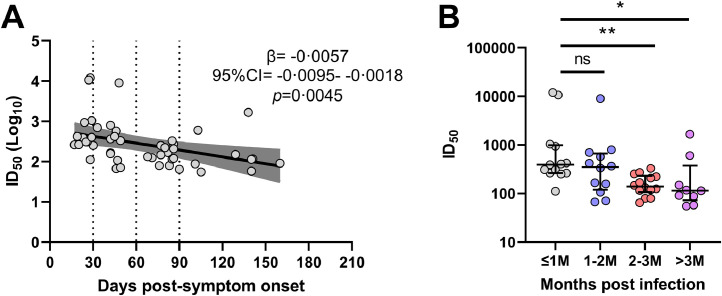
Fig. 2.
Neutralising titres against SARS-CoV-2 of baseline convalescent samples from 48 subjects with mild COVID-19. A) Linear regression analysis of baseline 50% inhibitory dilution neutralising titres (ID50), with a tendency for lower titres in samples collected further from symptom onset (β= -0•0057, 95% CI= -0•0095- -0•0018, p=0•0045). The dark grey shading indicates the 95% confidence interval of the beta estimate. B) Comparison of ID50 between stratified groups. Significantly lower titres were detected in group 2-3M (red; *p=0•012, Tukey's honest test) and group >3M (purple; *p=0•040, Tukey's honest test) when compared to group ≤1M (grey). No other significant differences were detected. Median and IQR are plotted. ns=not significant (p>0•05). See also legend to Figure 1 for definition of stratified groups.
The 50% inhibitory dilution neutralisation titres (ID50) of all included subjects with mild COVID-19 were first screened using three plasma dilutions (1/10, 1/100 and 1/1000) (S Fig 3), which helped set the parameters to perform at least 5 × 2-fold serial dilutions to identify an accurate ID50 value for each sample (S Fig 4).
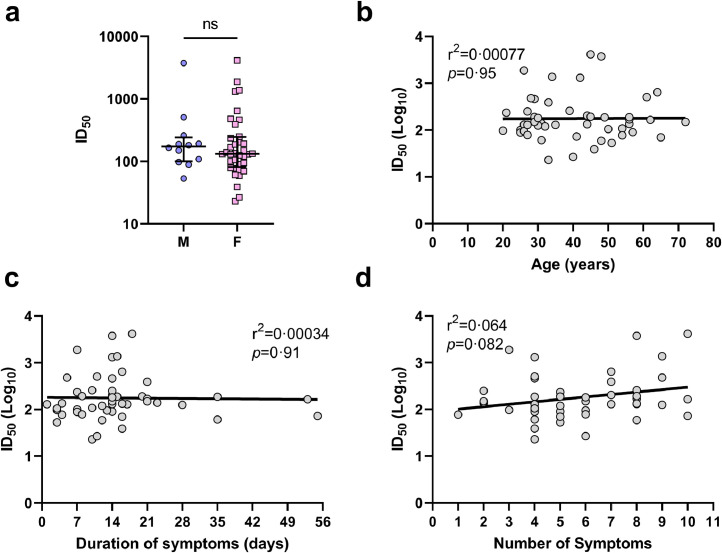
Fig. 3.
Time-matched analyses of 48 mild COVID-19 subject's demographic and clinical characteristics with neutralising titres at 6 months post-symptom onset. A) Comparison of 50% inhibitory dilution neutralising titres (ID50) between male (blue circles) and female (pink squares) at 6 months post-symptom onset (p=0•50, Mann-Whitney U test). Median and IQR plotted. B) Linear regression analysis of ID50 and age (p=0•95, r2=0•00077; simple linear regression analysis). C) Linear regression analysis of ID50 and the duration of symptoms reported (p=0•15, r2=0•00034; simple linear regression analysis). D) Linear regression analysis of ID50 and the number of symptoms reported (p=0•082, r2=0•064; simple linear regression analysis).
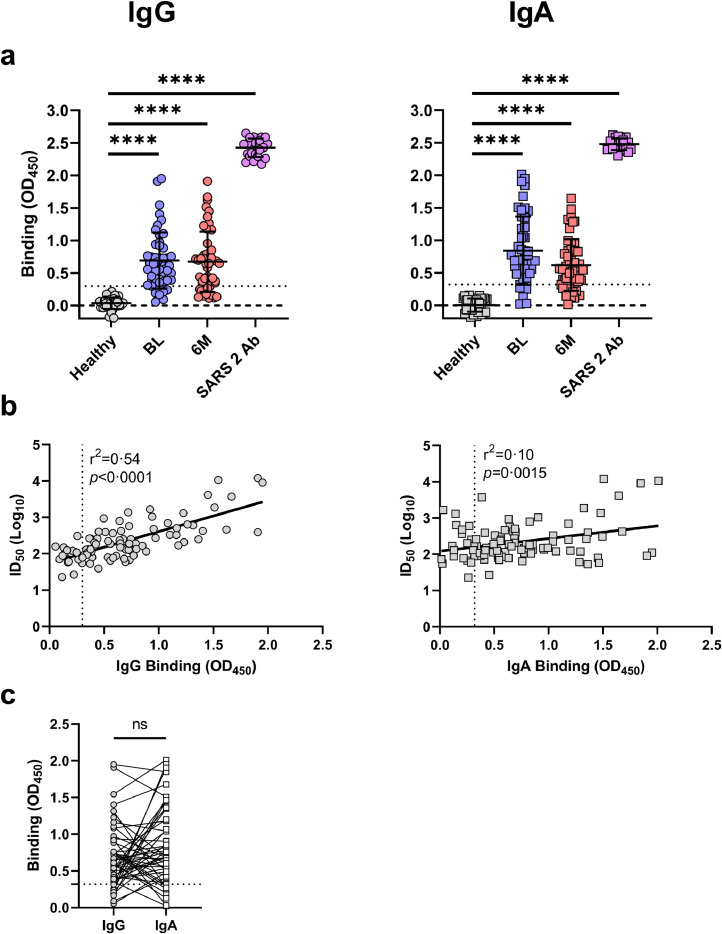
Fig. 4.
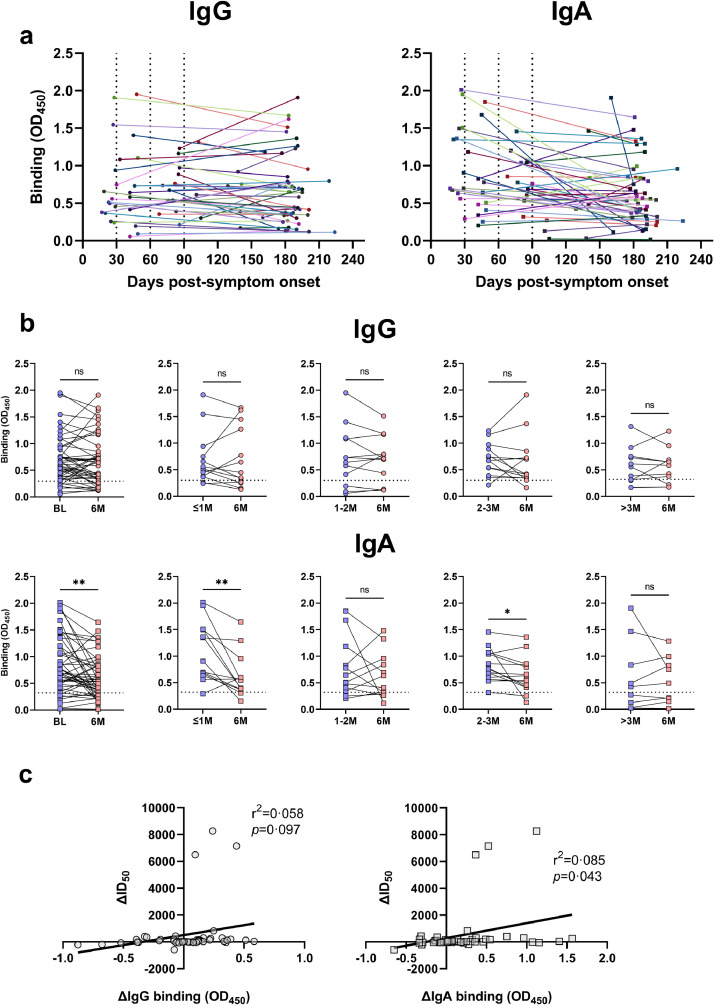
Total IgG and IgA binding (OD450) to the SARS-CoV-2 spike protein from 48 mild COVID-19 subject's plasma. A) Total binding of IgG (left panel; circles) and IgA (right panel; squares) at baseline (BL; blue) and 6-months (6M; red) for all subjects (n=48; 1/5 plasma dilution) compared to healthy controls (n=41; grey; 1/5 plasma dilution) and a SARS-CoV-2 spike-specific antibody (purple). The limit of detection is set to the mean +3SD of the healthy controls (IgG=0·3, IgA=0·32, dotted line). The overall binding of BL, 6M and SARS-CoV-2 antibody was significantly higher than healthy controls (****p<0·0001, unpaired parametric T tests). The mean and standard deviation are shown. The dashed line is plotted at 0. B) Correlation of 50% inhibitory dilution neutralising titres (ID50) against infectious culture derived SARS-CoV-2 to the spike-specific binding for IgG (left; circles; r2=0·54, p<0·0001; simple linear regression analysis) and IgA (right; squares; r2=0·10, p=0·0015; simple linear regression analysis). The dotted line on the X-axis represents the limit of detection for assays. C) Comparison of total IgG (circles) and IgA (squares) binding for the same time point in all subjects (n=96). The limit of detection (dotted line) has been set to the higher value of the two antibody isotypes (OD450 =0·32). No one subject is observed to have both undetectable IgG and IgA at any timepoint. No significant difference was observed between the overall binding of these isotypes (p=0·090, paired-parametric T test).
The change in titre between timepoints was calculated as:
3.4. Enzyme-linked immunosorbent assays
Qualitative assessment of the presence of SARS-CoV-2 RBD total antibody was done using the WANTAI SARS-CoV-2 antibody ELISA (Beijing Wantai, cat#: 256-WS-1096-96) according to the manufacturer's instructions. Undiluted and non-heat inactivated plasma was used for this assay. Specimens that gave an absorbance value greater than the cut off value (signal/noise ratio >1.0) were considered positive.
Quantitative assessment of SARS-CoV-2 S-specific antibodies was done using an in house developed ELISA. Spike protein was made by transfecting a pCG1 plasmid containing a codon-optimized spike sequence (plasmid kindly provided by Dr. Markus Hoffman [24]) into HEK293T cells (RRID: CVCL_0063) using a calcium phosphate transfection kit (Takara Bio, cat#: 631312) according to the manufacturer's instructions. Three days after transfection, the cells were collected, lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma/Aldrich, cat#: R0278) supplemented with a protease inhibitor cocktail (1:100 dilution; Sigma/Aldrich, cat#: P8340) and stored at -80°C. To capture the S protein in the cell lysate, NUNC Maxisorp plates (Sigma/Alrich, cat#: M9410) were coated with 250 ug/well of Galanthus nivalis (GNA) lectin (Medicago, cat#: L8275) and incubated at 37°C for 1.5 hours. The plates were washed with PBS tween 20 (0.05%; Sigma/Aldrich, cat#: P9416; PBS-T) and blocked overnight at 4°C with 5% non-fat dairy milk diluted in PBS-T. The following day, ELISA plates were washed and either S protein cell lysate or cell lysate without S protein was added (diluted 1:5 with 5% non-fat dairy milk). After a 1h incubation, the plates were washed and 2-fold serially diluted plasma (heat inactivated at 56°C for 30 min) was added in duplicate. A positive control SARS-CoV-2 spike-specific antibody (Sino Biological #40592-MM57, RRID: AB_2857935) was added to each plate in duplicate and a pool of plasma (heat inactivated at 56°C for 30 min) from 5 healthy individuals was used as a negative control. The plates were incubated for 1h, washed, and either, an anti-human IgG antibody (Jackson ImmunoResearch #209-035-088, RRID: AB_2339088; diluted 1:5000) or anti-human IgA antibody (Jackson ImmunoResearch #109-035-011, RRID: AB_2337580; diluted 1:5000) was added. For detection of the positive control antibody (mouse-derived), GE Healthcare #NA931V (RRID: AB_772210; diluted 1:5000) was added. Following a 1h incubation, the plates were washed and 3,3’,5,5’-tetramethylbenzidine (TMB; Sigma/Alrich, cat#: T0440) was added for 15 min. Following this, 1M hydrochloric acid (HCl) was added and absorbance was detected at 450nm using an ELx808 Ultra Microplate Reader (BioTek Instruments). The specific absorbance of antibody bound to S protein was calculated as:
The change in optical density at 450nm (∆OD450) between timepoints was calculated as:
3.5. Statistics
Neutralisation curves were constructed in GraphPad Prism (version 9.1.0.22) and the ID50 of plasma was calculated using non-linear regression (Log [inhibitor] vs normalized response [variable slope]). All statistical tests were performed in GraphPad Prism (9.1.0.22), with the exception of the multivariate linear regression analysis and the Tukey's honest tests, which were done in RStudio (RStudio Team (2020) Integrated Development for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/). The type of t test performed is indicated in the text and figure legends. In brief, parametric t test were performed on samples sizes greater than 20, non-parametric t tests were performed on sample sizes of 20 or less. The unparied non-parametric t test used was the Mann-Whitney U test and the paired non-parametric t test used was the Wilcoxon matched-pairs signed rank t test. Linear regression analyses were done in GraphPad Prism (9.1.0.22) using simple linear regression and where valid, the resulting beta estimate (β), 95% confidence interval (CI), the coefficient of determination (r2) and p value were reported. Statistical significance was defined as a p value less than 0·05.
3.6. Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3.7. Validation of cell lines, antibodies and reagents
All cell lines, antibodies and reagents included in this study were validated by the company or laboratory group from which they were purchased/gifted from. Sino Biological #40592-MM57 (RRID: AB_2857935) has been previously validated for use in neutralization and binding to S protein by others [25]. GE Healthcare #NA931V (RRID: AB_772210) has been validated for use as a secondary antibody by others [26]. Jackson ImmunoResearch #209-035-088 (RRID: AB_2339088) has been previously validated in ELISA [27]. Jackson ImmunoResearch #109-035-011 (RRID: AB_2337580) has been previously validated in ELISA by others [28]. These cell lines, antibodies and reagents were not further validated in our laboratory.
4. Results
4.1. Participant characteristics
The CVIC study has enrolled 102 individuals with mild COVID-19 and 97 healthy individuals. At the time of this study's initiation, 48/102 (47%) individuals with mild COVID-19 (45 [94%] diagnosed through PCR screening and 3 [6%] diagnosed by a rapid antibody test), who were enrolled during April through September 2020, had a baseline timepoint sample (BL; median time post-symptom onset=49 days [IQR=29-86]) and a follow-up timepoint sample at 6 months (6M; median time post-symptom onset=186 days [IQR=182-192]) collected, with a median of 127 days (IQR=101-154) between BL and 6M timepoints. Baseline timepoints were stratified into ≤1 month (≤1M; 14-30 days post-symptom onset), 1-2M (31-60 days), 2-3 M (61-90 days) and >3M (91-160 days). All reported COVID-19 symptom onset was between 24 March 2020 – 15 July 2020. The median duration of symptoms was 14 days (IQR=8-16). The most common symptoms were fatigue (n=35, 73%), fever (n=34, 71%) and headache (n=32, 67%) with a median of 5 (IQR=4-8) reported symptoms. Of these 48 subjects, 12 (25%) were males and 36 (75%) were females. The median age was 39 years (IQR=29-51) and all were Caucasian. Of the 97 healthy controls, 42 individuals, who were included between 28 February 2020 – 5 June 2020, were selected for inclusion in this study. Of these healthy individuals, 13 (31%) were males and 29 (69%) were females. Forty of the 42 (95%) were Caucasian, 1 (2·5%) was Hispanic and 1 (2·5%) was Middle Eastern and the median age was 34 (IQR=29-46). All included participants were screened using the WANTAI test, which is a highly sensitive ELISA that detects SARS-CoV-2 RBD-specific total antibody. Of all the included participants in this study, 48/48 (100%) with mild COVID-19 and 1/42 (2·5%) healthy control tested positive. This healthy control (H-40) was excluded from further analysis. A summary of subject data can be found in Table 1. More detailed summaries of SARS-CoV-2 infected, and healthy individuals can be found in Supplementary (S) Tables 1 and 2, respectively.
Table 1.
Summary of characteristics for 48 subjects with mild COVID-19 and 42 healthy controls
| Characteristic | Subjects with mild COVID-19 (n=48) | Healthy controls (n=42) |
|---|---|---|
| Age, median (IQR), years | 39 (29-51) | 34 (29-46) |
| Sex, n (%) | ||
| Male | 12 (25) | 13 (31) |
| Female | 36 (75) | 29 (69) |
| Ethnicity, n (%) | ||
| Caucasian | 48 (100) | 40 (95) |
| Hispanic | 1 (2•5) | |
| Middle Eastern | 1 (2•5) | |
| Infection confirmation, n positive (%) | ||
| Diagnostic PCR test | 45 (94) | |
| Diagnostic antibody test | 3 (6) | |
| WANTAI test | 48 (100) | 1 (2•5)# |
| Duration of symptoms, median (IQR), days | 14 (8-16) | |
| Baseline timepoint, median (IQR), days | 49 (29-86) | |
| 6-month timepoint, median (IQR), days | 186 (182-192) | |
| Days between timepoints, median (IQR), days | 127 (101-154) | |
| Symptoms, n (%) | ||
| Fatigue | 35 (73) | |
| Fever | 34 (71) | |
| Headache | 32 (67) | |
| Myalgia | 30 (63) | |
| Cough | 28 (58) | |
| Sore throat | 20 (42) | |
| Joint pain | 20 (42) | |
| Shortness of breath | 18 (38) | |
| Nasal congestion | 17 (35) | |
| Anosmia | 17 (35) | |
| Chest pain | 11 (23) | |
| Diarrhoea | 7 (15) | |
| Vomiting/Nausea | 2 (4) | |
| Total number of symptoms, median (IQR) | 5 (4-8) | |
| Transmission route, n (%) | ||
| Exposure at work | 26 (55) | |
| Unknown | 8 (17) | |
| Social activity | 5 (10) | |
| Choir activity | 5 (10) | |
| Household member | 4 (8) |
Excluded from further analysis (H-40).
4.2. Neutralisation titres against SARS-CoV-2 virus initially decrease and are then sustained longitudinally
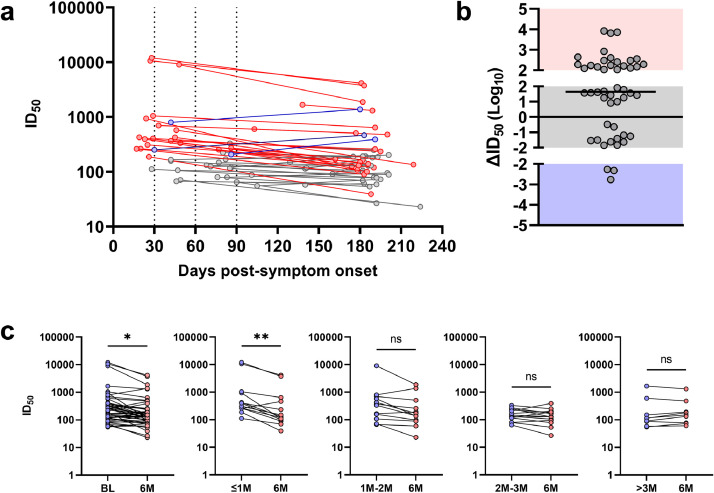
All subjects demonstrated neutralising activity at both timepoints (Figure 1a; mean BL ID50=1/943 and mean 6M ID50=1/411), with three subjects (6%; M-04, M-08 and M-30) demonstrating very high neutralisation titres (ID50>1/9000) at BL. Overall, the longitudinal nAb responses could be categorized into three distinct dynamic patterns i.e. decreased (defined as >100 ID50 decrease), unchanged (defined as ∆ID50 between 100 to -100) or increased neutralising titres (defined as >100 ID50 increase). These definitions were set based on the degree of variation between assays. Over the 6-month period, 21/48 (44%) individuals showed a loss of neutralising titres, 24/48 (50%) showed unchanged neutralising titres and 3/48 (6%) showed higher neutralising titres (Figure 1b). Of those that had decreased neutralising titres, 17/21 (81%) had their BL sample collected within 2 months of symptom onset. Of those that had unchanged neutralising titres, only 6/24 (25%) had their BL sample collected within 2 months of symptom onset. Upon follow up with the three subjects that showed boosted neutralisation titres (M-3, M-41 and M-57), all reported potential re-exposure to SARS-CoV-2 but were not confirmed to have re-infection in PCR testing.
Fig. 1.
Neutralising titres against SARS-CoV-2 of baseline and 6 months post-symptom onset convalescent samples from 48 subjects with mild COVID-19. A) 50% inhibitory dilution neutralising titres (ID50) against infectious cell-culture derived SARS-CoV-2 between baseline and 6-month time points, with decreased neutralising titres (red, n=21), unchanged neutralising titres (grey, n=24) and increased neutralising titres (blue, n=3) as defined in B. Each dotted line on the X-axis represents the time post-symptom onset according to group stratification, with the first section (14-30 days) comprising subjects with the baseline sample obtained ≤1M post-symptom onset, the second section (31-60 days) 1-2M, the third section (61-90 days) 2-3M and the last section (91-160 days) >3M post-symptom onset. B) The changes in neutralising titres (∆ID50) for all subjects from baseline to 6-months. Boosted titres (>100 ID50 increase) are shown in the blue shading, unchanged titres (∆ID50 between 100 and -100) are shown in the grey shading and decreased titres (>100 ID50 decrease) are shown in the red shading. The line in the grey shading represents the median change for all values. C) Comparisons of ID50 between baseline (BL; blue) and 6-month (6M; red) timepoints in all subjects (*p=0•045; Paired-parametric T test) and in stratified groups (Wilcoxon T tests; ≤1M to 6M **p=0•0046; others not significant [ns; p>0•05]).
When neutralising titres were compared between BL and 6M, there was an overall significant decrease at 6M (Figure 1c; p=0·045; paired parametric t test). However, when the subjects were stratified based on the collection time post-symptom onset (≤1M, 1-2M, 2-3M and >3M), it was only samples collected ≤1M that had a significant difference to their 6M timepoint (Figure 1c; p=0.0046, Wilcoxon T test). All other stratified comparisons did not reach significance when compared to their 6M timepoint (Figure 1c; p>0·05, Wilcoxon T tests). It is important to note that exclusion of the potentially re-exposed subject (M-3) in the 1-2M group renders the comparison to the 6M timepoint statistically significant (p=0·0049, Wilcoxon T test).
When BL samples were analysed, there was a significant trend for lower neutralisation titres with samples collected further from their symptom onset date (Figure 2a; β= -0·0057, 95% CI= -0·0095- -0·0018, p=0·0045). Even after removal of the three subjects that demonstrated high neutralising titres (M-04, M-08 and M-30), which were sampled closer to symptom onset, this trend remained significant (β= -0·0036, 95% CI= -0·0063- -0·00092, p=0·0096; data not shown). Stratification of subjects based on their collection time post-symptom onset showed that there were significant differences between ≤1M samples and 2-3M samples (Figure 2b; p=0·012; Tukey's honest test) and ≤1M samples and >3M samples (Figure 2b; p=0·040; Tukey's honest test). Other comparisons did not reach significance (p>0·05; Tukey's honest test).
4.3. Higher neutralising titres against SARS-CoV-2 at 6 months post-symptom onset may be associated with a more symptomatic disease
Given that collection of the 6M timepoint was time-matched for each participant, this permitted a time-matched analysis of neutralisation titres at 6M to other variables. Demographically, no significant differences were observed between 6M neutralisation titres and gender (Figure 3a; p>0·05, Mann-Whitney U test) or age (Figure 3b; p>0·05, r2=0·00077; simple linear regression analysis). Moreover, no correlation was observed between the 6M neutralisation titres and the duration of symptoms (Figure 3c; p>0·05, r2=0·00034; simple linear regression analysis). However, there was a tendency for a higher number of symptoms being associated with higher 6M neutralising titres (Figure 3d; p=0·082, r2=0·064; simple linear regression analysis). In a multivariate analysis accounting for gender, age and duration of symptoms (calculated in R statistical software; data not shown), this result became significant (β=0·15, SD=0·069, p=0·034).
4.4. Total spike-specific IgA but not IgG decreases longitudinally and is associated with changing neutralising titres
As determined by healthy controls (n=41), the limit of detection for the ELISA was set to the mean + 3SD for IgG (mean=0·032, SD=0·087, threshold set=0·3) and IgA (mean=0·0053, SD=0·10, threshold set=0·32). This threshold was used based on cut-off's set by others [29]. Based on these thresholds, 42/48 (88%) and 38/48 (79%) of samples had detectable levels of spike-specific IgG at BL and 6M, respectively (Figure 4a, S Fig 5). For IgA, 40/48 (83%) and 36/48 (75%) had detectable levels at BL and 6M, respectively (Figure 4a, S Fig 6). When the binding of spike-specific IgG and IgA was compared to the ID50 value for the same timepoint (n=96), a positive correlation was found for both isotypes (Figure 4b; IgG: p<0·0001, r2=0·54; IgA: p<0·0015, r2=0·10; simple linear regression analyses). All subjects were found to have either detectable IgG or IgA or both, indicating that the neutralisation observed is likely to originate from at least one or both isotypes (Figure 4c).
Fig. 5.
Longitudinal total binding (OD450) of IgG and IgA to the SARS-CoV-2 spike protein of 48 mild COVID-19 subjects. A) Total IgG binding (left) and IgA binding (right) between all baseline and 6-month time points, with each colour/shade representing a different subject. Each dotted line on the X-axis represents the time post-symptom onset stratification, as described in the Figure 1 legend. B) Comparisons of total binding IgG (top) and IgA (bottom) values between baseline (BL; blue) and 6-month (6M; red) timepoints in all subjects (Paired-parametric T tests) and in stratified groups (Wilcoxon T tests). Baseline (BL) and 6-month (6M) timepoints were significantly different for IgA (**p=0•0019). For the stratified groups, ≤1M and 2-3M were significantly higher than their 6M timepoint for IgA (**p=0•0051, *p=0•039). No other comparisons were statistically significant (ns; p>0•05). C) Correlation of the change in 50% inhibitory dilution neutralising titres (ID50) against SARS-CoV-2 to the change in IgG (left; circles; r2=0•058, p=0•097; simple linear regression analysis) and IgA (right; squares; r2=0•085, p=0•043; simple linear regression analysis) binding.
When compared longitudinally, no significant differences were found between BL and 6M timepoints in total IgG binding overall (p>0·05, paired parametric t test) or between stratified groups (i.e. ≤1M to 6M, 1-2M to 6M etc.; p>0.05, Wilcoxon T tests) (Figure 5a; p>0·05). For IgA, the 6M timepoint had significantly lower binding when compared to BL (Figure 5b; p=0·0019, paired parametric t test). After stratification, subjects in the ≤1M group were found to have the largest decline (Figure 5b; p=0·0051, Wilcoxon T test). Subjects in the 2-3M group also had a significant difference when compared to their 6M timepoint (Figure 5b; p=0·039, Wilcoxon T test). All other comparisons did not yield significance (p>0·05, Wilcoxon T tests).
To investigate if there was an association between the change in neutralising titres over time (∆ID50) and either the change in IgG or IgA binding over time (∆OD450), these variables were compared (Figure 5c). For IgG, no significant correlation was observed (p=0·097, r2=0·058; simple linear regression analysis). However, for IgA, a significant correlation was observed (p=0·043, r2=0·085; simple linear regression analysis), indicating that the difference in neutralising titres between timepoints may be accounted for by the levels of spike-specific IgA.
5. Discussion
This study demonstrated sustained neutralising titres up to 6 months post-symptom onset in individuals presenting with mild COVID-19, using a SARS-CoV-2 virus isolate and not pseudotyped or surrogate models. In addition, this study identified a concurrent decline in nAb titres and IgA levels, but not IgG levels, suggesting that waning neutralising titres associate with spike-specific IgA. Importantly, the kinetics of nAb responses seen here are concordant with that of other longitudinal SARS-CoV-2 and human coronavirus studies [11], [12], [13], [14], [15],[17], [18], [19], which may have been limited by either participant number [15,18] or the use of pseudotyped or surrogate models [11], [12], [13], [14], [15],17,19]. While this study, and others [11,30], show detectable neutralising titres 6 months post-symptom onset in all subjects, one study has reported some subjects with undetectable neutralisation at 6 months post-symptom onset [17], which may be somewhat artefactual, due to the use of a pseudotyped neutralisation assay with a truncated spike protein.
With mounting evidence of reduced SARS-CoV-2 cases due to the large rollout of current emergency use-approved (EUA) vaccines [31], it is clear that population-level protection can reduce the spread and burden of SARS-CoV-2 infection, which will contribute to controlling the ongoing pandemic. Currently, the minimum level of neutralising titres that are protective from SARS-CoV-2 infection/re-infection remains to be determined. For influenza, in vitro neutralising titres as low as 1/40 are considered high enough to be protective [32,33]. We observed that at the 6M timepoint post symptoms the neutralising titer was 1/59 (ID50; M-14) or higher among patients with mild COVID-19. However, one of the major challenges associated with protection from influenza infection, is the virus's capability of evolving through antigenic drift and shift, enabling evasion of previous established immunity and permitting re-infection [34]. While antigenic shift is not likely for human coronaviruses, there is increasing evidence of emerging antigenically drifted variants of SARS-CoV-2, which might have subverted neutralisation to natural or to vaccine-induced immunity [35,36]. Given that neutralisation to such variants was not measured in the present study, the estimated protection to the newly emerging SARS-CoV-2 variants could not be determined. Nevertheless, 3 subjects (M-3, M-41 and M-57) were observed to have boosted neutralising titres and IgG/IgA responses at 6 months post-symptom onset. Upon follow up with these individuals, all of whom are health care workers, they reported possible re-exposure to SARS-CoV-2 despite remaining PCR-negative following routine testing. While there is no definitive evidence of re-exposure, it is unlikely for individuals to have boosted immunity without re-exposure to the same antigen. Given that these individuals remained PCR-negative, this finding may suggest that they were protected from productive re-infection. With that said, the exact correlates of protective immunity to SARS-CoV-2 are not clear. In a recent study that followed up participants for less than 6 months, 5 reinfections of SARS-CoV-2 were documented from a cohort of 8758 people [37]. While all reported reinfections had relatively low neutralising titres (ID50 64 or less), the resulting severity of infection did not correlate with differences in their neutralising titres. Therefore, it is possible that in vitro neutralisation does not necessarily reflect protective immunity in vivo due to the requirement of other protective responses, such as T cell mediated immunity. While protection from SARS-CoV-2 infection specifically through T cell immunity has not been shown, the elicitation of long-lived memory-like T cells during primary infection would suggest they have a role in protective immunity against SARS-CoV-2 [38]. It is plausible that adequate protection from SARS-CoV-2 infection requires both robust antibody and T cell immunity to the challenging variant.
Since all the BL samples collected in this study were not time-matched, and neutralising titres change over time, correlates of neutralising titres in regard to clinical and demographic data were examined at the 6-month time-matched sample. From these analyses, a more symptomatic disease was found to be associated with higher neutralising titres. While a more symptomatic disease does not necessarily mean a more severe disease, it is interesting to note that higher neutralising titres have been reported in those with a more severe disease [15,18,20]. However, this study was limited by an uneven gender inclusion and a smaller sample size compared to most clinical cohorts. In turn, this limited demographic and clinical comparisons to neutralising data.
While the ELISA used in this study is not as sensitive as commercially available assays [39], there was clear evidence of spike-specific IgG and IgA in subjects with mild COVID-19 compared to healthy controls, which correlated well with neutralising titres. Similar to other studies [14,15,17,18], spike-specific IgG was not found to wane 6 months post-symptom onset and did not correlate with the overall declining neutralising titres. In contrast, spike-specific IgA was found to correlate with the overall decline in neutralising titres. This would suggest that spike-specific IgA responses are an important factor for waning neutralising titres, which may have implications for vaccine strategies.
An important strength in this study is the use of infectious SARS-CoV-2 virus for neutralisation assays, and not pseudotyped or surrogate models. However, a limitation is the use of highly permissive African green monkey kidney cells (Vero E6), which although being the most used cell line for these types of assays, and thus allowing more comparable results between studies, lack expression of transmembrane protease serine 2 (TMPRSS2), which is required for one of the entry pathways for SARS-CoV-2 [40]. Thus, the observed neutralisation might be limited to cathepsin-mediated ACE-2 entry only. Future studies beyond the scope of this study should address if there are any differences in neutralisation between Vero cells and lung epithelial cells [24].
In summary, this study shows that virus specific neutralising titres are sustained to detectable levels up to 6 months post-symptom onset in individuals with mild COVID-19. However, close monitoring of re-infection is warranted, especially given the significant propagation of emerging SARS-CoV-2 variants. This study also found that the change in neutralising titres correlated with the change of spike-specific IgA over time, highlighting the importance of this isotype for waning/boosted neutralising titres. In contrast, spike-specific IgG does not appear to wane following SARS-CoV-2 infection. Future studies using extended sampling beyond 6 months are required to assess the timepoint at which neutralising titres perhaps become undetectable. This will have implications for SARS-CoV-2 re-infection and indicate a time period for potential vaccine boosters.
Contributors
A.U., C.S., A.W., S.B., U.F., N.W. and J.B. conceived and designed the study; A.U., C.S., A.W., S.B., S.V., A-L.S. and N.W. collected and processed patient data and samples; A.U., C.F-A., L.S.M., S.F., R.L., A.F., S.R and J.B. generated assays and experimental data; A.U., C.S., N.W. and J.B. drafted the manuscript. Data was verified by C.F-A., S.B., S.F., U.F., N.W. and J.B. All authors critically revised the manuscript for important intellectual content and gave final approval for the submitted version.
Data sharing
Following publication, and in agreement with the Data Protection Agency, Denmark, the data generated in this study will be made available to researchers who provide a sound proposal. Proposals should be directed to jbukh@sund.ku.dk, and to gain access, data requestors will need to sign a data access agreement. Individual participant data will remain coded.
Declaration of Competing Interest
Nina Weis has been clinical investigator, lecturer or on advisory boards for Abbvie, Gilead, Glaxo Smith Kline and Merck Sharp Dohme and has received unrestricted grants for research from Abbvie and Gilead without relation to the present work and for the remaining authors there are no conflicts of interest.
Acknowledgements
We thank all the participants for their contribution to this study. We would also like to thank Susanne Ruszcycka and Greta Vizgirda, Department of Infectious Diseases, Copenhagen University Hospital, Hvidovre, Denmark, for sample processing. Thanks to Sofie Dikeledi Kold Jespersen and Caroline Nørløv Vinten and Magnus Illum Dalegaard, Department of Infectious Diseases, Copenhagen University Hospital, Hvidovre, Denmark, for blood sampling and REDCap data recording, respectively. We thank Markus Hoffman, University of Göttingen, Germany, for provision of the codon optimized cloned spike sequence. This work was supported by grants from The Capital Region of Denmark's Research Foundation [C.S., J.B.], the Novo Nordisk Foundation [N.W., J.B.], the Independent Research Fund Denmark [J.B.], the Candys Foundation (C.F-A., S.R., J.B.), The Danish Cancer Society (J.B.), Manufacturer Vilhelm Pedersen and wife's memorial scholarship (S.B), Master carpenter Jørgen Holm and wife Elisa. F. Hansen's memorial scholarship (S.B) and the Danish Agency for Science and Higher Education (S.R., J.B.). J.B. is the 2015 recipient of the Novo Nordisk Prize and the 2019 recipient of a Distinguished Investigator grant from the Novo Nordisk Foundation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103519.
Appendix. Supplementary materials
References
- 1.Organization WH. WHO Coronavirus Disease (COVID-19) situation reports. 2021 [Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed 31 March, 2021.
- 2.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortorici MA, Beltramello M, Lempp FA, Pinto D, Dang HV, Rosen LE. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370(6519):950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe PG, Perera R, Park WB, Song KH, Bang JH, Kim ES. MERS-CoV Antibody Responses 1 Year after Symptom Onset, South Korea, 2015. Emerg Infect Dis. 2017;23(7):1079–1084. doi: 10.3201/eid2307.170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mo H, Zeng G, Ren X, Li H, Ke C, Tan Y. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11(1):49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357(19):1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 11.L’Huillier AG, Meyer B, DO Andrey, Arm-Vernez I, Baggio S, Didierlaurent A. Antibody persistence in the first 6 months following SARS-CoV-2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021;27(5) doi: 10.1016/j.cmi.2021.01.005. 784-e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Long QX, Deng HJ, Hu J, Gao QZ, Zhang GJ. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muecksch F, Wise H, Batchelor B, Squires M, Semple E, Richardson C. Longitudinal Serological Analysis and Neutralizing Antibody Levels in Coronavirus Disease 2019 Convalescent Patients. J Infect Dis. 2021;223(3):389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford KHD, Dingens AS, Eguia R, Wolf CR, Wilcox N, Logue JK. Dynamics of Neutralizing Antibody Titers in the Months After Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J Infect Dis. 2021;223(2):197–205. doi: 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brochot E, Demey B, Touze A, Belouzard S, Dubuisson J, Schmit JL. Anti-spike, Anti-nucleocapsid and Neutralizing Antibodies in SARS-CoV-2 Inpatients and Asymptomatic Individuals. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.584251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abayasingam A, Balachandran H, Agapiou D, Hammoud M, Rodrigo C, Keoshkerian E. Long-term persistence of RBD-positive memory B cells encoding neutralising antibodies in SARS-CoV-2 infection. Cell Rep Med. 2021 doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamayoshi S, Yasuhara A, Ito M, Akasaka O, Nakamura M, Nakachi I. Antibody titers against SARS-CoV-2 decline, but do not disappear for several months. EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2021.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chia WN, Zhu F, Ong SWX, Young BE, Fong SW, Le Bert N. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Liu L, Nair MS, Yin MT, Luo Y, Wang Q. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect. 2020;9(1):2091–2093. doi: 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez S, Fernandez-Antunez C, Galli A, Underwood A, Pham LV, Ryberg LA. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrobial Agents and Chemotherapy. 2021;65 doi: 10.1128/AAC.00097-21. e00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iglewicz B, Hoaglin D. Volume 16: how to detect and handle outliers, The ASQC basic references in quality control: statistical techniques, Edward F. Mykytka, Ph.D., Editor; 1993.
- 24.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong J, Zhu C, Lai H, Feng C, Zhou D. Potent Neutralization Antibodies Induced by a Recombinant Trimeric Spike Protein Vaccine Candidate Containing PIKA Adjuvant for COVID-19. Vaccines (Basel) 2021;9(3) doi: 10.3390/vaccines9030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solis AG, Bielecki P, Steach HR, Sharma L, Harman CCD, Yun S. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature. 2019;573(7772):69–74. doi: 10.1038/s41586-019-1485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Underwood AP, Walker MR, Brasher NA, Eltahla AA, Maher L, Luciani F. Understanding the Determinants of BnAb Induction in Acute HCV Infection. Viruses. 2018;10(11):659. doi: 10.3390/v10110659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuen RR, Steiner D, Pihl RMF, Chavez E, Olson A, Smith EL. Novel ELISA Protocol Links Pre-Existing SARS-CoV-2 Reactive Antibodies With Endemic Coronavirus Immunity and Age and Reveals Improved Serologic Identification of Acute COVID-19 via Multi-Parameter Detection. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.614676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Algaissi A, Alfaleh MA, Hala S, Abujamel TS, Alamri SS, Almahboub SA. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci Rep. 2020;10(1):16561. doi: 10.1038/s41598-020-73491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temperton NJ, Chan PK, Simmons G, Zambon MC, Tedder RS, Takeuchi Y. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossman H, Shilo S, Meir T, Gorfine M, Shalit U, Segal E. COVID-19 dynamics after a national immunization program in Israel. Nat Med. 2021;27(6):1055–1061. doi: 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- 32.Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972;70(4):767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis. 2011;204(12):1879–1885. doi: 10.1093/infdis/jir661. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6(2):143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 35.Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 37.Dimeglio C, Herin F, Miedouge M, Martin-Blondel G, Soulat JM, Izopet J. Protection of healthcare workers against SARS-CoV-2 reinfection. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonifacius A, Tischer-Zimmermann S, Dragon AC, Gussarow D, Vogel A, Krettek U. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity. 2021;54(2):340–354. doi: 10.1016/j.immuni.2021.01.008. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padoan A, Bonfante F, Pagliari M, Bortolami A, Negrini D, Zuin S. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.