Abstract
A rod-shaped, motile anaerobic bacterium, designated CCRI-22567T, was isolated from a vaginal sample of a woman diagnosed with bacterial vaginosis and subjected to a polyphasic taxonomic study. The novel strain was capable of growth at 30–42 °C (optimum, 42 °C), at pH 5.5–8.5 (optimum, pH 7.0–7.5) and in the presence of 0–1.5 % (w/v) NaCl (optimally at 0.5 % NaCl). The phylogenetic trees based on 16S rRNA gene sequences showed that strain CCRI-22567T forms a distinct evolutionary lineage independent of other taxa in the family Peptostreptococcaceae. Strain CCRI-22567T exhibited 90.1 % 16S rRNA gene sequence similarity to Peptoanaerobacter stomatis ACC19aT and 89.7 % to Eubacterium yurii subsp. schtitka ATCC 43716. The three closest organisms with an available whole genome were compared to strain CCRI-22567T for genomic relatedness assessment. The genomic average nucleotide identities (OrthoANIu) obtained with Peptoanaerobacter stomatis ACC19aT, Eubacterium yurii subsp. margaretiae ATCC 43715 and Filifactor alocis ATCC 35896T were 71.8, 70.3 and 69.6 %, respectively. Strain CCRI-22567T contained C18 : 1 ω9c and C18 : 1 ω9c DMA as the major fatty acids. The DNA G+C content of strain CCRI-22567T based on its genome sequence was 33.8 mol%. On the basis of the phylogenetic, chemotaxonomic and other phenotypic properties, strain CCRI-22567T is considered to represent a new genus and species within the family Peptostreptococcaceae, for which the name Criibacterium bergeronii gen. nov., sp. nov., is proposed. The type strain of Criibacterium bergeronii is CCRI-22567T (=LMG 31278T=DSM 107614T=CCUG 72594T).
Keywords: bacterial vaginosis, Criibacterium bergeronii, Peptostreptococcaceae, skin, vaginal flora
Strain CCRI-22567T was isolated from a vaginal clinical swab harvested from a woman diagnosed with bacterial vaginosis (BV; Nugent score of 9) in Spring 2014 at CHU de Québec-Université Laval (Quebec City, QC, Canada). Vaginal swabs requested by treating physicians are routinely submitted for BV testing using Nugent score at the clinical microbiology laboratory of the CHU de Quebec-Universite Laval (CHUL hospital). Swabs were streaked on sterile autoclaved glass slides so that swabs can be stored at 4 °C until the Nugent score report is generated. Anonymized swabs having Nugent scores between 7 and 10 were used for a research project to isolate BV-associated Mobiluncus species. This recovery of deidentified leftover clinical sample swabs was approved by the Research Ethics Committee of the ‘CHU de Québec-Université Laval’ (No. B13-08-1778, 26 September 2013). Swabs were used to inoculate Schaedler agar medium containing vitamin K and 5 % sheep blood. An isolated translucid round colony growing to <0.5 mm after 2 days of incubation at 35 °C under anaerobic conditions was subjected to 16S rRNA gene sequencing for species identification. Comparative 16S rRNA gene sequence analysis showed that strain CCRI-22567T represents a novel species of a new genus in the family Peptostreptococcaceae. The role of this new bacterium in BV remains unclear since Gardnerella vaginalis, Bacteroides fragilis and Mobiluncus curtisii (all species consistent with BV) were also isolated from the same vaginal swab. The comparison of the 16S rRNA gene sequence of strain CCRI-22567T with microbiome projects available in the GenBank NR database suggests that the same novel species is also present on sebaceous parts of the skin [1]. Furthermore, strain CCRI-24246 (=L9 284 001) was isolated in 2018 from pus cultivated from the left armpit of an 81-year-old woman, suggesting that these bacterial isolates most probably represent a commensal inhabitant of both skin and vaginal discharge. Anaerococcus prevotii, another member of the family Peptostreptococcaceae, is also well known to be an inhabitant of these sites [2]. To determine the taxonomic position of these isolates, they were subjected to further characterization in accordance with the recommendations outlined by Kämpfer et al. and Tindall et al. [3, 4].
Genomic DNA of strains CCRI-22567T and CCRI-24246 was extracted for 16S rRNA and whole genome sequencing as described previously [5]. PCR amplification of the 16S rRNA gene was performed with primers SSU27 and SSU1492 and sequencing using primers SSU27, SSU536, SSU685 and SSU926 as described by Sistek et al. [6]. 16S rRNA sequences were used for an initial blast against the GenBank NR database to identify the taxonomic neighbours of strains CCRI-22567T and CCRI-24246. Based on this analysis, the closest cultured relatives of strain CCRI-22567T were Peptoanaerobacter stomatis ACC19aT (90.1%), Eubacterium yurii subsp. schtitka ATCC 43716 (89.7 %) and Eubacterium yurii subsp. margaretiae ATCC 43 715 (89.6 %), respectively. These sequence identities were calculated based on nearly full-length sequences using the pairwise distance function of mega version 6.06 [7]. These identity values were below the accepted cut-off of 98.7 % for species delineation, and also under the cut-off of 95 % for the differentiation of a new genus [4, 8, 9]. Therefore, strains CCRI-22567T and CCRI-24246 meet the criteria of the description of a novel species and a new genus.
Subsequently, strains CCRI-22567T and CCRI-2424 16S rRNA sequences were aligned with all type strain sequence of every species of the family Peptostreptococcaceae according to LPSN [10] using muscle implemented in mega version 6.06 [7] to establish its relationship within the family Peptostreptococcaceae. Genera assigned to the families Peptostreptococcaceae and Clostridiaceae deemed to be closest to Criibacterium gen. nov. were discussed together here as inferred by Galperin et al. [11]. Organisms with names not yet validly published were also included to fully represent the family tree. The three subspecies of Eubacterium yurii were included in the analysis as they are members of Peptostreptococcaceae based on 16S rRNA phylogeny and are waiting to be transferred to another genus [12]. The 16S rRNA gene phylogenetic trees reconstructed using the maximum-likelihood method were carried out on a comparison of 1262 nucleotides using mega version 6.06 [7] (Fig. 1). Tree topology was confirmed using the minimum-evolution method and tree clustering structure with the neighbour-joining method (Fig. S1 and Fig. S2, available in the online version of this article). The maximum-likelihood tree reconstruction was similar to the one reported previously by Galperin et al. [11]. Strains CCRI-22567T and CCRI-24246 showed strong bootstrap values, supporting their relationship within the cluster consisting of Peptoanaerobacter stomatis, Eubacterium yurii and Filifactor species. That cluster is already known to form a deep branch within the family Peptostreptococcaceae [13]. While Filifactor villosus was isolated from a subcutaneous abcess, Peptoanaerobacter stomatis, Eubacterium yurii and Filifactor alocis are oral pathogens [14–17]
Fig. 1.
Maximum-likelihood phylogenetic tree showing the relationship of Criibacterium bergeronii within the family Peptostreptococcaceae. The 16S RNA sequences were aligned using muscle [37]. The phylogenetic tree was reconstructed using the maximum-likelihood method with the Tamura–Nei substitution model implemented in mega version 6.06 [7]. Only bootstrap values >50 % based on 1000 replications are shown at branching points. Organisms with names not validly published are presented with quotation marks. GenBank accession numbers for each 16S rRNA gene sequence are given in parentheses. Each bar indicates 0.02 substitutions per position. Clostridium botulinum ATCC 25763T and Clostridium butyricum DSM 10702T (Clostridium sensu stricto) were used as outgroups.
Whole-genome sequencing of strain CCRI-22567T was performed on an Illumina HiSeq 2500 using SBS version 4 to sequence a 126 bp paired-end library (Nextera XT, Illumina). A first genome assembly version using Ray software was described in Maheux et al. [5] (MBEW00000000.1). A second de novo assembly version (MBEW00000000.2) was performed using SPAdes software (v3.10.1) [18]. De novo assembly of the 7 787 976 reads yielded 79 contigs with an overall coverage of 450-fold. The genome length was 2 176 288 bp (N50 of 80 728 bp) and the G+C content was 33.8 mol%. The assembled genome was annotated using NCBI GenBank annotation pipeline (version 4.6) [19]. The genome of strain CCRI-22567T had a total of 2174 identified features, including six rRNAs, 40 tRNAs, four non-coding RNA and 2124 protein-coding sequences from which 436 were assigned as hypothetical proteins. Whole-genome sequencing of strain CCRI-24246 was performed on an Illumina Miseq using MiSeq Reagent Kit version 3 to sequence the 300 bp paired-end library. A total of 12 281 540 reads were assembled de novo in 139 contigs using SPAdes (version 3.13.0) [18] for an overall coverage of 400-fold. The total genome length was 2 611 389 bp (N50 of 115 916 bp) with an average G+C content of 32.9 mol%. The draft genome sequence was annotated using NCBI GenBank annotation pipeline (version 4.8) [19]. A total of 2554 features were identified, including seven rRNAs, 45 tRNAs, four non-coding RNA and 2497 putative coding sequences from which 516 were assigned as hypothetical proteins. As suggested by Chun et al. [20], 16S rRNA and recA genes sequences where extracted from genome assembly and compared to original Sanger sequences made on isolated strain CCRI-22567T. The individual 16S rRNA and recA genes sequences were 100 % identical (ambiguous bases were excluded) to sequences from the whole genome assembly, thus ensuring the authenticity of the whole genome data. Comparison of the proteins encoded by the two genomes using CD-HIT version 4.8.1 [21] revealed that strains CCRI-22567T and CCRI-24246 have 1654 protein clusters in common (at 95 % identity), while they have 441 and 820 unique protein clusters, respectively. Furthermore, the differences between the two genomes of strains CCRI-22567T and CCRI-24246 are mainly found in regions of higher G+C content. The genes annotated in those parts belong to bacteriophages or transposons and thus appear to be related to mobile genetic elements.
According to Chun et al. [20], the 16S rRNA gene comparison could have been sufficient to describe strains CCRI-22567T and CCRI-24246 as a new species/new genus with regard to the established cut-off [9, 20]. To further support this description, the genomic average nucleotide identity was also calculated using OrthoANIu software [22] with the strains CCRI-22567T and CCRI-24246 genome assemblies and the closest related whole genomes available at the time. The OrthoANIu identity obtained was 71.8 % with P. stomatis ACC19aT (NZ_AFZF00000000.2), 70.3 % with Eubacterium yurii subsp. margaretiae ATCC 43715 (NZ_AEES00000000.1) and 69.6 % with F. alocis ATCC 35896T (NC_016630.1). A predicted digital DNA–DNA hybridization (dDDH) value was also determined using the GGDC 2.1 web service using the same whole genomes (http://ggdc.dsmz.de) [23]. The returned dDDH values were 27.8, 25.6, and 25.4 %, respectively. These OrthoANIu and dDDH values were below the 95–96 and 70 % accepted threshold for species delineation, respectively, thus supporting the C. bergeronii new species description [20, 22, 24, 25]. The OrthoANIu and digital DDH values between the two strains of C. bergeronii were 98.5 and 88.7 %, respectively, indicating that these two strains were members of the same species according to the previously mentioned cut-off values.
The relationship of strains CCRI-22567T and CCRI-24246 within the family Peptostreptococcaceae was investigated using available genome sequence data and the Type (Strain) Genome Server (TYGS) platform [26]. First, the 16S rRNA sequence of CCRI-22567T was used to identify all close type strains with available genomes. Then, the whole genomes of CCRI-22567T and CCRI-24246 were used to determine intergenomic distances between them. A minimum-evolution genome-based tree, supported by strong bootstrap values, showed the same clustering of isolates CCRI-22567T and CCRI-24246 within the P. stomatis, Eubacterium yurii spp. and F. alocis cluster (Fig. S3). This strong branching also supporting a new genus and a new species identification. Moreover, the two strains, CCRI-22567T and CCRI-24246, were also defined as belonging to the same species using complete genomes on the TYGS platform. The same taxonomic assignment as a new genus and a new species was also supported using Genome Taxonomy Database (GTDB-Tk version 1.3.0) [27]
A more detailed phenotypic characterization was performed to support the species description. Strains CCRI-22567T and CCRI-24246 were stored at −80 °C in Brucella broth (BD) liquid medium containing 15 % glycerol until the characterization tests could be performed. The strains were sub-cultured three times on sheep blood agar. Culture using pre-reduced media and phenotypic identification tests were performed as described in the Wadsworth and VPI anaerobic manuals [28, 29].
Strain CCRI-22567T did not grow in thioglycolate and tryptic soy broths. Thus, physiological characterization was carried out on tryptic soy agar with 5 % sheep blood (blood agar) for atmosphere and temperature tests and tryptic soy agar (TSA) for pH and NaCl concentration tests, respectively. Growth was monitored by direct observation. Growth of the type strain was first tested on blood agar in an anaerobic chamber containing a gas mix of 10 % H2, 5 % CO2 and balance with N2, and microaerophilic conditions, in an anaerobic jar containing a Campy GasPak EZ (gas generating system pouch, Becton Dickinson and Company), as well as under aerobic conditions, with or without 5 % CO2. Strain CCRI-22567T grew only under anaerobic conditions. The determination of the temperature range for growth was tested at 25, 30, 35 and 42 °C on blood agar in an anaerobic atmosphere. Strain CCRI-22567T was capable of growth at 30–42 °C, with optimum growth at 42 °C. The pH range for growth was tested at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, 10.0 and 10.5, at 35 °C, on TSA using different buffers within their optimal range, in an anaerobic atmosphere. We used four different buffer systems: pH 4.0, 4.5, 5.0 and 5.5 with sodium acetate buffer; pH 6.0, 6.5, 7.0, 7.5 and 8.0 with sodium phosphate buffer; pH 8.5 and 9.0 with Bis-Tris propane buffer; pH 9.5, 10.0 and 10.5 with sodium carbonate buffer. Strains CCRI-22567T and CCRI-24246 were capable of growth at pH values from pH 5.5 to 8.5, with optimum growth at pH 7.0–7.5. The NaCl concentration range for growth was tested at 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.5 and 6.0 % (w/v), at 35 °C, on TSA and pH 7.0, in an anaerobic atmosphere. Strain CCRI-22567T was capable of growth at concentrations of 0–1.5 % (w/v) NaCl, with optimum growth at 0.5 % NaCl.
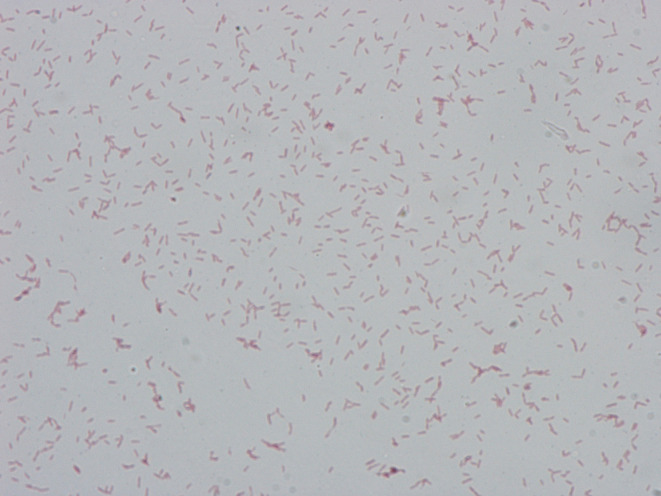
Morphological characteristics of colonies and cells of the type strain were determined using light microscopy (Leitz) and cryo electron tomography (cryo-ET). Cryo-ET is currently the only tool that preserves biological samples at or near the native, hydrated state. It allows for the three-dimensional imaging of whole cells to macromolecular resolution (3–4 nm). Thus, cryo-ET was the method used here to determine the cell envelope architecture [30]. Cryo-ET data was collected on frozen-hydrated bacterial samples. Briefly, cells were frozen on EM grids using a Vitrobot automated vitrification device (ThermoFisher Scientific). Plunge-frozen samples were imaged on a Titan Krios 300kV transmission electron microscope equipped with a Falcon 2 direct electron detector (ThermoFisher Scientific). Tilt series were recorded from −60° to +60° with 1° increment oscillations, and a defocus of −8 µm. The total final dose was 150 e-/Å [2]. Tilt series were recorded automatically using EPU Tomography software and tomographic reconstructions were calculated using the IMOD software package [31]. After 72 h of incubation on blood agar in an anaerobic chamber, colonies were grey-translucent, circular with entire edges, smooth and ~0.6 mm in diameter. Colonies were non-adherent and no haemolysis was observed. Gram staining was performed using a four-step Gram stain kit (BD) on cells from 72 h blood agar culture. Cells stained Gram-variable (CCRI-22567T stained Gram-negative, while CCRI-24246 stained Gram-variable, with red cells, purple cells and blue cells on the same glass slide) and both appeared as single cells or in pairs. Cells of strain CCRI-22567T were rod-shaped (~630 nm wide and ~1750 nm long) and non-spore-forming (Fig. 2). Although some genes of strains CCRI-22567T and CCRI-24246 have been annotated as having homologies with known sporulation proteins, no genes presenting homologies with the Spo0A regulator protein, which is essential to sporulation initiation, has been identified. Gelatin hydrolysis was performed as described by De la Cruz and Torres [32]. CCRI-22567T and CCRI-24246 did not grow on the nutrient gelatin medium. CCRI-22567T and CCRI-24246 showed motility in wet mount and flagella were observed with cryo-ET. When CCRI-22567T cells were imaged with cryo-ET (Fig. 3), they revealed a multi-layered cell envelope architecture reminiscent of typical Gram-negative bacteria with a relatively thin layer of peptidoglycan (~10 nm). A cytoplasmic membrane (CM) and an S-layer, forming a continuous mesh on the surface of cells, were clearly visible (Fig. 3). An outer layer (OL), similar in appearance to the CM was observed right underneath the S-layer, suggestive of a putative second membrane. A hidden Markov model search for outer membrane-related genes homologous to those of enterobacteria and Negativicutes failed to reveal any in the genomes of CCRI-22567T and CCRI-24246 (supplementary results). Therefore, characterization of this OL will require further investigations to determine its presence and composition in members of Peptostreptococcaceae. The genome annotation of CCRI-22567T revealed the presence of one gene annotated as S-layer protein and five S-layer homology domain-containing proteins. Additional ultrastructural features include the presence of flagella, flagellar motors and chemoreceptor arrays (Fig. 3b, d and e). Granules were also observed using cryo-ET (Fig. 3c), those are likely to be storage granules. Genome annotation revealed the presence of glgA, glgB and glgD genes known to code for glycogen storage granules as well as genes coding for polyphosphate storage ppk1 and ppx in both strains CCRI-22567T and CCRI-24246.
Fig. 2.
Gram stain of Criibacterium bergeronii CCRI-22567T after 96 h of incubation on sheep blood agar, under anaerobic atmosphere at 35 °C; ×1000 magnification.
Fig. 3.
Ultrastructure of Criibacterium bergeronii CCRI-22567 (n=7) revealed by cryo electron tomography. (a) A 20 nm thick tomographic slice reveals the presence of a cytoplasmic membrane (CM), thin peptidoglycan (PG) layer, an outer layer (OL) and a proteinaceous S-layer. The CM is also depicted in red while the OL is shown in blue. (b–e) Structural details reveal the presence of flagella and flagellar motors, chemoreceptor arrays and dense storage granules (likely polyphosphate). Scale bar, 200 nm.
Special-potency discs (Oxoid) containing colistin (10 µg), kanamycin (1000 µg) and vancomycin (5 µg) were used to group the anaerobic bacteria, as recommended in the Wadsworth manual [29]. Analysis using special potency discs showed that growth of strains CCRI-22567T and CCRI-24246 was resistant to 10 µg colistin and susceptible to 5 µg vancomycin and 1000 µg kanamycin.
The two novel isolates were examined (according to the manufacturer’s instructions) using Rapid ID 32A, RapID ANA II and API ZYM systems (bioMérieux). Under the rapid ID 32A system, strains CCRI-22567T and CCRI-24246 gave positive results for arginine dihydrolase, β-glucuronidase, indole production, arginine arylamidase, while it was negative for urease, β-galactosidase, β-galactosidase-6-phosphatase, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, mannose fermentation, raffinose fermentation, glutamic decarboxylase acid, α-fucosidase, nitrate reduction, proline arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase and serine arylamidase. Discordant results were observed for α-galactosidase, leucine arylamidase, and histidine arylamidase where strain CCRI-22567T gave positive results while strain CCRI-24246 gave negative results. Discordant results were also observed for alkaline phosphatase and glutamyl glutamic acid arylamidase where strain CCRI-22567T gave negative results while strain CCRI-24246 gave positive results. Numerical profiles obtained using the Rapid ID 32A system were 6040212001 for strain CCRI-22567T and 2040610000 for strain CCRI-24246, respectively.
Under the RapID ANA II system (Remel), both strains CCRI-22567T and CCRI-24246 gave positive results for arginine-β-naphthylamide and indole production from tryptophan, while they were negative for urea, p-nitrophenyl-β-d-disaccharide, p-nitrophenyl-α-l-arabinoside, α-nitrophenyl-β-d-galactoside, p-nitrophenyl-α-d-glucoside, p-nitrophenyl-β, d-glucoside, p-nitrophenyl-α-d-galactoside, p-nitrophenyl-α-l-fucoside, p-nitrophenyl-N-acetyl-β-d-glucosaminide, p-nitrophenylphosphate, leucyl-glycine-β-naphthylamide, glycine-β-naphthylamide, proline-β-naphthylamide, phenylalanine-β-naphthylamide, serine-β-naphthylamide and pyrroidonyl-β-naphthylamide. Microcode profiles obtained using the RapID ANA II system were 000044 for both strains.
Under the API ZYM system, both strains CCRI-22567T and CCRI-24246 gave positive results for phosphatase acid, naphthol-AS-BI-phosphohydrolase and β-glucuronidase, while they were negative for alkaline phosphatase, esterase, esterase lipase, lipase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Discordant results were observed for leucine arylamidase and α-galactosidase where strain CCRI-22567T gave positive results while strain CCRI-24246 gave negative results.
For the determination of the whole-cell fatty acid (CFA) composition, strains were extracted after 16 h growth at 35 °C in pre-reduced peptone–yeast–glucose (PYG) broth (Anaerobe Systems), using the Sherlock System (midi) as described previously [33] except that operating system version 4.5 was used. CFAs were identified using the peak-naming table found in midi (Moore 6.0). CFAs for strain CCRI-22567T were deemed to be qualitatively similar to other genera found in the families Clostridiaceae and Peptostreptococcaceae [34]. The main fatty acids in the cell membrane of strain CCRI-22567T comprised C18 : 1 ω9c and C18 : 1 ω9c DMA. The presence of a large quantity of these fatty acids appears to be specific to the type strain of this species. Furthermore, it appears that CFA C18 : 1 ω9c DMA (ECL 18.224) is not found in other taxa of the family Peptostreptococcaceae tested in this study (Table S1).
Scores from in-house generated mass spectrometry profiles [35] made using the two strains suggested that MALDI-TOF could be used to unambiguously identify C. bergeronii and had no significant match (<1.7) towards all other entries in the Bruker Biotyper library (version 7854). Genomic, morphological and chemotaxonomic characteristics of the 16 genera of Peptostreptococcaceae, including Criibacterium, are summarized in Table S2 [11, 36].
On the basis of the data from the polyphasic taxonomic analysis presented here, strain CCRI-22567T is considered to represent a new genus and species within the family Peptostreptococcaceae, for which the name Criibacterium bergeronii gen. nov., sp. nov., is proposed.
Description of Criibacterium gen. nov.
Criibacterium (Cri.i.bac.te′ri.um. N.L. neut. n. bacterium a small rod; N.L. neut. n. Criibacterium a small rod isolated at CRI-Centre de Recherche en Infectiologie.)
Members of the genus grow under anaerobic conditions. Cells are Gram-variable with a cytoplasmic membrane, a relatively thin layer of peptidoglycan, an additional outer layer of yet-uncharacterized composition and an S-layer (ultrastructural data from type strain only), rod-shaped with rounded ends, and non-spore-forming. Cells are motile. Colonies on blood agar are grey-translucent, circular with entire edges, smooth and raised. The predominant cellular fatty acids detected are C18 : 1 ω9c and C18 : 1 ω9c DMA. Phylogenetically, this genus is a member of the Peptostreptococcaceae and the type species is Criibacterium bergeronii.
Description of Criibacterium bergeronii sp. nov.
Criibacterium bergeronii [ber.ge.ro′ni.i. N.L. masc. gen. n. bergeronii of Bergeron, named after Professor Michel G. Bergeron (CM, OQ, M.D., FRCPC, FCAHS), infectious diseases specialist, serial entrepreneur, founder, and former director of Centre de Recherche en Infectiologie de l’Université Laval in Québec City (Québec), Canada].
C. bergeronii sp. nov. has the following characteristics in addition to those given for the genus. Cells are ~630 nm wide by ~1750 nm long and occur isolated or in pairs. Growth occurs on TSA with 5 % sheep blood under anaerobic conditions at 35 °C, but no growth occurs following subculture in thioglycolate and tryptic soy broths. After 72 h incubation under anaerobic conditions colonies are flat, translucent and <1.0 mm in diameter. Cells grow at 30–42 °C (optimum, 42 °C), at pH 5.5–8.5 (optimum, pH 7.0–7.5) and in the presence of 0–1.5 % (w/v) NaCl (optimally at 0.5 % NaCl). Using Rapid ID 32A, RapID ANA II and API ZYM systems, positive results are obtained for l-arginine, β-glucuronidase, indole production, arginine arylamidase, phosphatase acid, naphthol-AS-BI-phosphohydrolase and arginine-β-naphthylamide. Negative results are obtained for urea, β-galactosidase, β-galactosidase-6-phosphatase, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, mannose fermentation, raffinose fermentation, glutamic decarboxylase acid, α-fucosidase, α-mannosidase, nitrate reduction, proline arylamidase, leucyl glycine arylamidase, pyroglutamic arylamidase acid, tyrosine arylamidase, alanine arylamidase, cysteine arylamidase, glycine arylamidase, valine arylamidase, serine arylamidase, p-nitrophenyl-β-d-disaccharide, p-nitrophenyl-α-l-arabinoside, α-nitrophenyl-β-d-galactoside, p-nitrophenyl-α-d-glucoside, p-nitrophenyl-α-d-galactoside, p-nitrophenyl-α-l-fucoside, p-nitrophenyl-N-acetyl-β-d-glucosaminide, leucyl-glycine-β-naphthylamide, glycine-β-naphthylamide, proline-β-naphtylamide, phenylalanine-β-naphthylamide and pyrroidonyl-β-naphthylamide. The C. bergeronii type strain G+C content is 33.8 mol%, which is consistent with the other members of family Peptostreptococcaceae (Table S2) [11, 36]
The type strain, CCRI-22567T (=LMG 31278T=DSM 107614T=CCUG 72594T), was isolated from a woman diagnosed with bacterial vaginosis while strain CCRI-24246 was isolated from pus of the left armpit of an 81-year-old woman. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and draft whole genome sequences of strain CCRI-22567T are NR_164617.1 and MBEW00000000.2, respectively.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
The authors wish to thank Dr. Mark T. Riley (Department of Foreign Languages, California State University, Sacramento, CA, USA) and Mr. Hubert W. Hawkins (Manquin, VA, USA) for their help verifying the Latin name formation of the new species and genus, Dr. Luc Bissonnette for critical reading of the manuscript, as well as Julie Dunn and Patricia Corrado for phenotypic analysis of strain CCRI-24246. We acknowledge Dr. Kaustuv Basu and the Facility for Electron Microscopy Research (FEMR) at McGill University for assistance in data acquisition as well as Nancy Boucher and Pier-Luc Plante for their help in genome sequencing of strain CCRI-24246. We also thank Thomas Deschenes for his help in sequence analysis. We acknowledge the Bioimaging platform of the Infectious Disease Research Centre, funded by an equipment and infrastructure grant from the Canadian Foundation Innovation (CFI). F. R. was supported by a Mitacs post-doctoral fellowship. Computations were performed under the auspices of Calcul Québec and Compute Canada. The operations of Compute Canada are funded by the Canada Foundation for Innovation (CFI), the National Science and Engineering Research Council (NSERC), NanoQuébec and the Fonds Québécois de Recherche sur la Nature et les Technologies (FQRNT). J. C. is the recipient of the CIHR-Canada Research Chair in Medical Genomics (Tier 1 level). E. I. T. was supported by NSERC Discovery grant (GRPIN 04345) and is the recipient of the NSERC-Canada Research Chair in Microbial Ultrastructure (Tier 2level).
Author contributions
A. F. M., investigation, validation, formal analysis, data curation, writing - original draft preparation, writing - review and editing, supervision, project administration. D. K. B., investigation, validation, formal analysis, data curation, writing – review and editing. J. Y. A., investigation, validation, formal analysis, writing – review and editing. È. B., investigation, validation, formal analysis, data curation, writing – review and editing. S. B., investigation. K. A. B., investigation, formal analysis, resources, data curation, writing – review and editing. A. H., investigation, formal analysis. É. D., investigation, formal analysis. É. F. G., investigation. F. R., investigation, formal analysis. J. C., resources. M. D., investigation, resources, data curation. P. H. R., validation, writing - review and editing. M. B., validation, formal analysis, writing - review and editing, supervision, project administration. E. I. T., investigation, formal analysis, resources, data curation, writing – review and editing. R. F. O., resources, writing – review and editing, supervision, project administration.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: BV, bacterial vaginosis; CFA, cell fatty acid; CM, cytoplasmic membrane; Cryo-Et, cryo electron tomography; dDDH, digital DNA–DNA hybridization; EM, electron microscopy; OL, outer layer; PG, peptidoglycan; PYG, peptone yeast glucose; TSA, tryptic soy agar; TYGS, Type (Strain) Genome Server.
Two supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labutti K, Pukall R, Steenblock K, Glavina Del Rio T, Tice H, et al. Complete genome sequence of Anaerococcus prevotii type strain (PC1) Stand Genomic Sci. 2009;1:159–165. doi: 10.4056/sigs.24194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kämpfer P, Buczolits S, Albrecht A, Busse HJ, Stackebrandt E. Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int J Syst Evol Microbiol. 2003;53:893–896. doi: 10.1099/ijs.0.02710-0. [DOI] [PubMed] [Google Scholar]
- 4.Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 5.Maheux AF, Bérubé Ève, Boudreau DK, Raymond F, Corbeil J, et al. Draft genome sequence of Criibacterium bergeronii gen. nov., sp. nov., strain CCRI-22567T, isolated from a vaginal sample from a woman with bacterial vaginosis. Genome Announc. 2016;4:e00959–00916. doi: 10.1128/genomeA.00959-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sistek V, Maheux AF, Boissinot M, Bernard KA, Cantin P, et al. Enterococcus ureasiticus sp. nov. and Enterococcus quebecensis sp. nov., isolated from water. Int J Syst Evol Microbiol. 2012;62:1314–1320. doi: 10.1099/ijs.0.029033-0. [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stackebrandt E, Göker M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 9.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 10.Parte AC. LPSN - List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 11.Galperin MY, Brover V, Tolstoy I, Yutin N. Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov. Int J Syst Evol Microbiol. 2016;66:5506–5513. doi: 10.1099/ijsem.0.001548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade WG, et al. Family II. Eubacteriaceae, Genus I. Eubacterium. In: de Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, editors. Bergey’s Manual of Systematic Bacteriology, Volume 3: The Firmicutes. New York: Springer; 2009. pp. 865–891. [Google Scholar]
- 13.Slobodkin A. The Family Peptostreptococcaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer; 2014. pp. 291–302. [Google Scholar]
- 14.Sizova MV, Chilaka A, Earl AM, Doerfert SN, Muller PA, et al. High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae . Stand Genomic Sci. 2015;10:37. doi: 10.1186/s40793-015-0027-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margaret BS, Krywolap GN. Eubacterium yurii subsp. yurii sp. nov. and Eubacterium yurii subsp. margaretiae subsp. nov.: test tube brush bacteria from subgingival dental plaque. Int J Syst Bacteriol. 1986;36:145–149. doi: 10.1099/00207713-36-2-145. [DOI] [Google Scholar]
- 16.Cato EP, Moore LVH, Moore WEC. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the Human Gingival Sulcus. Int J Syst Bacteriol. 1985;35:475–477. doi: 10.1099/00207713-35-4-475. [DOI] [Google Scholar]
- 17.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 18.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 21.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon SH, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 25.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holdeman L, Cato E, Moor W. Anaerobe Laboratory Manual. 4th Edition. Blacksburg, VA: Virginia Polytechnic Institute and State University, Anaerobe Laboratory; 1977. [Google Scholar]
- 29.Jousimies-Somer HR, Summanen P, Citron DM, Baron EJ, Wexler HM. Wadsworth Anaerobic Bacteriology Manual. 6th ed. Belmont: Star Publishing; 2002. [Google Scholar]
- 30.Tocheva EI, López-Garrido J, Hughes HV, Fredlund J, Kuru E, et al. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 32.De la Cruz TE, Torres J. Gelatine hydrolysis test protocol. Vol. 8. Microbe (ASM press); 2013. pp. 1–9. [Google Scholar]
- 33.Bernard KA, Shuttleworth L, Munro C, Forbes-Faulkner JC, Pitt D, et al. Propionibacterium australiense sp. nov. derived from granulomatous bovine lesions. Anaerobe. 2002;8:41–47. doi: 10.1006/anae.2000.0408. [DOI] [Google Scholar]
- 34.Gerritsen J, Fuentes S, Grievink W, van Niftrik L, Tindall BJ, et al. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov. and Asaccharospora gen. nov. Int J Syst Evol Microbiol. 2014;64:1600–1616. doi: 10.1099/ijs.0.059543-0. [DOI] [PubMed] [Google Scholar]
- 35.Bernard KA, Pacheco AL, Loomer C, Burdz T, Wiebe D, et al. Corynebacterium lowii sp. nov. and Corynebacterium oculi sp. nov., derived from human clinical disease and an emended description of Corynebacterium mastitidis . Int J Syst Evol Microbiol. 2016;66:2803–2812. doi: 10.1099/ijsem.0.001059. [DOI] [PubMed] [Google Scholar]
- 36.Ezaki T, et al. Family VII. Peptostreptococcaceae fam. nov. In: de Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, editors. Bergey’s Manual of Systematic Bacteriology, Volume 3: The Firmicutes. New York: Springer; 2009. pp. 1008–1015. [Google Scholar]
- 37.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.