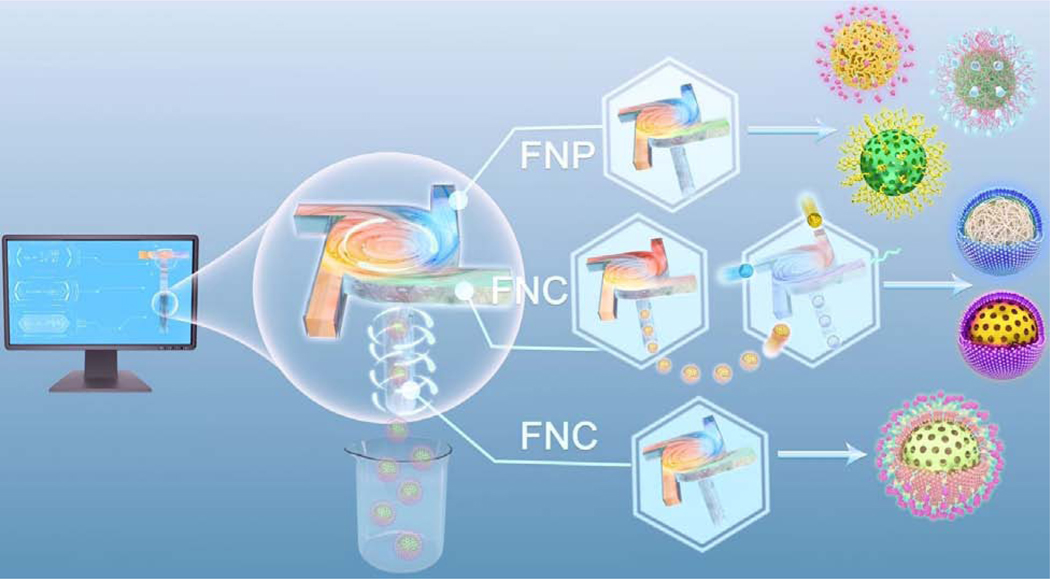
Abstract
Advances in nanoformulation have driven progress in biomedicine by producing nanoscale tools for biosensing, imaging, and drug delivery. Flash-based technology, the combination of rapid mixing technique with the self-assembly of macromolecules, is a new engine for the translational nanomedicine. Here, we review the state-of-the-art in flash-based self-assembly including theoretical and experimental principles, mixing device design, and applications. We highlight the fields of flash nanocomplexation (FNC) and flash nanoprecipitation (FNP), with an emphasis on biomedical applications of FNC, and discuss challenges and future directions for flash-based nanoformulation in biomedicine.
Keywords: flash nanoprecipitation, flash nanocomplexation, self-assembly, nanoformulation, drug delivery
Introduction
Nanoformulation, the construction of nanoparticles and nanocomplexes, has profoundly impacted the chemical, food, medical device, and pharmaceutical industries [1–4]. Advances in nanotechnology, materials science, and biotechnology are now converging in the exciting field of nanomedicine. However, nanomedicine can fulfill its potential only if it can be produced at scale with precise control of nanoparticle properties such as size, composition, and surface functionalization [5–7]. Self-assembly, in which disordered components autonomously form an organized structure, plays a key role in nanoformulation [8,9] and it has recent applications in antimicrobial treatment [10–12], cell-imaging [13–15], anti-inflammation [16], cancer therapies [17–20] and nanoparticle synthesis [21–23]. Self-assembly has been performed using a variety of techniques including layer-by-layer, bulk mixing, controlled growth, hydrodynamic focusing, and colloidal evaporation induction [24–26]. However, many of these nanoformulation methods cannot fully satisfy the requirements of biomanufacturing due to product heterogeneity, limited yield, and high batch-to-batch variation. Continuous, scalable, reproducible, and robust self-assembly technologies are needed. Flash technology, which involves rapid mixing in confined impingement jets mixers (CIJM) or multiple inlet vortex mixers (MIVM), facilitates self-assembly and formulation of nanocomplexes or nanoprecipitates in a low-cost, high-throughput, controllable manner [27]. The quality of the self-assembled nanoformulation can be maintained over the collection process in a manufacturing pipeline, which is vital for clinical translation.
In this review we describe the theoretical and experimental principles of flash-based self-assembly technology and its applications to biomedical nanoformulation. We begin by summarizing recent progress in the fabrication of nanomedicine using flash nanocomplexation (FNC) and flash nanoprecipitation (FNP). The first major theme of this review is that using theoretical models to optimize the dynamic flow mixing profile enables the design of more capable flash mixers. The second theme is that using flash-based self-assembly to generate nanoformulations via supersaturation or electrostatic complexation can be accomplished within dynamic mixers. We highlight differences between flash and conventional self-assembly approaches, and demonstrate the technical superiority of the rapid mixing methods. FNC can be used to produce nanomedicines (e.g. nanoparticles containing nucleic acids, proteins, and synthetic polymers for use as diagnostic or therapeutic agents) with uniform shape, controllable size, scalable yield, and high encapsulation efficiency. For example, FNC has been used to fabricate monodisperse, shape-controlled polyethylenimine (PEI) nanoparticles for gene delivery [28], and to produce size-tunable insulin/chitosan/tripolyphosphate (TPP) nanoparticles for oral insulin delivery in a scalable manner [29]. The insulin/chitosan/TPP nanoparticles exhibited narrow polydispersity, small size, and a high drug encapsulation rate, and were fabricated in a more reproducible, high-throughput, and controlled manner than when using conventional bulk mixing methods. FNC was also used to produce insulin/L-penetratin nanoparticles for oral insulin delivery that were coated with hyaluronic acid for better permeation into the intestinal mucus layer and increased transepithelial uptake. These intermediate-sized insulin nanoparticles were then encapsulated in enteric microcapsules using a multiple inlet vortex mixer to provide additional protection against the acidic environment of the stomach, allowing the controlled release of insulin in the small intestine.
Combining FNC techniques is a powerful approach to manipulating the structure of nanoformulations to improve their therapeutic efficacy. With intensive ongoing efforts to refine FNC techniques in the context of biomedical applications, flash technology is destined to have a fundamental impact on nanomedicine. This review will survey current developments in flash self-assembly, and will discuss challenges and opportunities in realizing the potential of flash-based nanoformulation in biomedicine.
Flash mixing devices
Flash self-assembly processes are conducted in microscale and milliscale confined mixers [30]. A variety of rapid mixers have been designed, some specifically for therapeutics/diagnostic formulation. Nanoformulation mixers provide a dynamic mixing environment with high Reynolds numbers, essential for precise control of nanoparticle properties [5,27,31]. Nanoparticle composition and surface modifications can be tailored by varying the configuration of the mixing chamber, flow rates of mixer inlets, and concentrations of solutions used.
Confined impinging jet mixer
Turbulent mixing is the key to achieving homogenous nanoformulations [32]. Self-assembly occurs at the molecular level during mixing, and manipulating the timescale of formation is crucial regardless of the physical forces driving self-assembly [33]. The dynamic collision of two or three jet flows in a mixing cavity reduces the timescale of nanoparticle nucleation and growth, enabling self-assembly within milliseconds. The advent of the confined impinging jet mixer (CIJM) was driven by demand for self-assembled polymer nanoformulation, and satisfied manufacturing requirements that could not be met using laminar flow devices.
In 2003, a highly dynamic turbulent mixer was developed by Prud’homme and co-workers at Princeton [32]. The CIJM was designed to have two or three fluidic inlets. For the two-inlet CIJM, two pathways lead to a small chamber where opposing streams containing solvent and non-solvent impinge to create turbulence. To maintain continuous flow, syringe pumps drive the streams from syringes into the mixing chamber. Design dimensions for the mixing chamber were specified by Johnson and Prud’homme [32]. The main body of the CIJM can be made of stainless steel or a solvent-compatible thermoplastic such as polyformaldehyde or high-density polyethylene [34].
The CIJM is a powerful tool for producing nanoformulations in the millisecond range through supersaturation and electrostatic complexation. However, CIJM geometry limits the ratio of solvent to non-solvent due to the requirement for equal momentum of the opposing streams in mixing [35]. This requirement restricts the use of CIJM to hydrophilic solutes and to a specific range of solvent-to-solute ratios.
Multiple inlet vortex mixer
To overcome the CIJM requirement of equal fluidic momentum, a multiple inlet vortex mixer (MIVM) was designed to allow unequal solvent-to-nonsolvent ratio mixing in a scalable manner. The concept of the MIVM is that momentum from each stream contributes independently to drive micromixing in the chamber [36]; therefore, it is possible to use larger differences in flow rate between flow inlets while maintaining dynamic micromixing.
Multiple inlet vortex mixers used in flash-based self-assembly generally consist of components manufactured using stainless steel and assembled with syringe adapters. MIVM are designed to be easily disassembled and cleaned. 3D printing can be used to fabricate MIVMs, and a 3D-printed MIVM was used to formulate lipid-polymer hybrid nanoparticles for biological applications [37]. The MIVM design allows the combination of immiscible solutions, providing an additional level of flexibility to the choice of materials for nanoformulations.
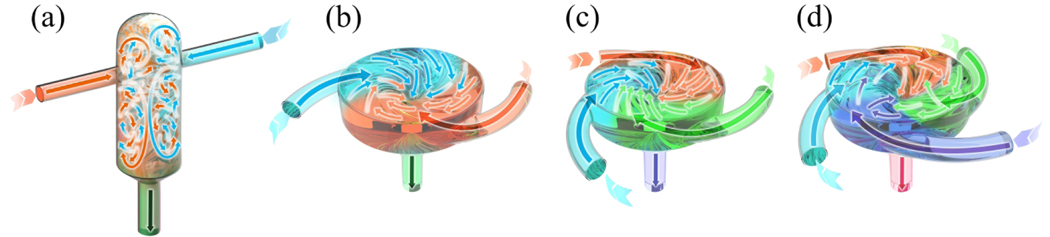
The geometries of CIJM and MIVM used in flash nanoformulation are illustrated in Fig. 1. Flash self-assembly involves creation of turbulent flow via dynamic mixing of multiple streams in a confined cavity. In addition to CIJM and MIVM, other mixer designs have been used to create turbulent mixing conditions, including coaxial turbulent jet mixers [6,25], K-M micromixers [38,39], double tubular counterflow micromixers (DTCF-MX) [40], staggered herringbone micromixers (SHM) [41,42], T connectors [43,44], and microfluidics-assisted confinement [7,45,46].
FIGURE 1.
Geometries of mixers used in flash nanoformulation processes. (a) CIJM, (b) two-inlet MIVM, (c) three-inlet MIVM, (d) four-inlet MIVM.
Principles of flow mixing
Reproducibly generating nanoparticles with well-controlled size and uniformity is critical for clinical translation [47]. Compared to batch-mode processing methods such as dialysis, emulsification, and slow precipitation, flash self-assembly provides better control of particle size and better reproducibility, scalability, and throughput capacity, by using continuous rapid mixing in a confined space. There are two major types of flash self-assembly: flash nanoprecipitation and flash nanocomplexation (Fig. 2). FNP uses dynamic mixing created by turbulent flow to stimulate nanoparticle nucleation and growth via solvent-induced supersaturation and polymer precipitation. FNC exploits polyelectrolyte complexation-induced phase separation. In FNC, nanocomposites undergo self-assembly via physical interactions such as electrostatic interactions and hydrogen bonding, and are formed within milliseconds or microseconds. The fluid dynamics of flash mixers in turbulent and laminar flow has been thoroughly studied in multiple fields using computational analysis and high-profile microscopy. This section discusses experimental characterization and computational simulation of flow behavior in FNP and FNC.
FIGURE 2.
Flash-based self-assembly, from computational simulations to nanoparticle formulation. FNP produces polymeric and inorganic nanoparticles via solvent supersaturation. FNC produces nanoformulations using polyelectrolyte complexation driven by physical interactions such as electrostatic interactions, hydrogen bonding, and metal-coordination, and often involves the use of multiple devices in parallel for nanoparticle formation and surface modification.
Flash nanoprecipitation
FNP is a scalable self-assembly process that uses rapid micromixing to establish homogeneous supersaturation conditions for controlled block copolymer self-assembly and precipitation of hydrophobic solutes [8, 22–25]. This process can be performed using a CIJM or MIVM to produce nanoformulations with controlled size distribution and high drug loading content.
There are several key timescales involved in FNP: mixing time (τmix), nanoformation time (τflash), copolymer aggregation time (τagg), and active organic nucleation and growth time (τng) [52,53]. Controlling τmix is crucial [54]. To attain homogeneous mixing free of mixing effects and a narrow particle size distribution, the generation of supersaturation by turbulent micromixing must be faster than the diffusion-limited aggregation that controls self-assembly [32,54–56]. In other words, τmix should be less than the copolymer aggregation time τagg for homogeneous mixing. When τagg is close to the nucleation and growth time τng, the block copolymer interacts with the active particle to alter nucleation and growth and causes colloidal stabilization. Liu et al. explored the optimal MIVM mixing time, inlet velocity, stream physical properties, and mixer geometry [35]. Their findings suggested that the Reynolds number (Re) plays a key role in mixing efficiency, and that Re >1600 allows sufficient mixing for sub-millisecond MIVM reactions.
Block copolymer selection is important in formulating stable nanoparticles using FNP. The hydrophobic and hydrophilic regions of block copolymers are balanced so that hydrophobic groups are located mainly in the nanoparticle core. The molecular weight of the hydrophobic polymer region does not affect the particle size distribution or stability [57,58]. Polymers with non-crystallizable hydrophobic blocks and a high glass transition temperature can facilitate long-term nanoparticle stability [59]. Increasing the molecular weight of the amphiphilic block copolymer reduces particle size and enhances colloidal stability [60–63]. Active ingredients involved in self-assembly should also be considered. For example, the amphiphilic stabilizer polyethylene glycol–polylactic acid (PEG-PLA), a block copolymer of hydrophilic PEG and hydrophobic PLA, was used to alter the molecular rearrangement of nanoparticles containing the antifungal itraconazole formulated using FNP. X-ray photoelectron spectroscopy revealed that higher PEG-to-PLA ratios resulted in disruption of the amphiphilic stabilizer by the drug, causing nanoformulation instability [64]. A separate study investigated the effect of the drug-to-polymer ratio (D/P) on the size and structure of drug-loaded nanoparticles formulated using FNP [65]. PEG-b-PLA nanoparticles loaded with the anti-cancer drug sorafenib were fabricated using a MIVM. A lower D/P (0.25) yielded nanoparticles ~140 nm with a densely packed core-shell structure, whereas a higher D/P (5) yielded larger products with a less dense, granular core structure. The drug release behavior and morphology of these therapeutic nanocarriers were thus strongly affected by D/P.
High drug loading is an advantage of FNP self-assembly versus slower mixing methods. The antioxidant β-carotene was used as a drug model to compare encapsulation efficiencies of FNP with a bulk mixing method; FNP demonstrated an encapsulation efficiency of 86.3%–93.6% β-carotene, versus ~20% for the bulk method [60]. When FNP was compared with dropwise addition for encapsulating sorafenib using PEG-b-PLA, nanoparticles made using FNP were smaller and contained more than twice the amount of sorafenib than nanoparticles made using dropwise addition [65]. Fast mixing (high Re) within a chamber provides an ideal niche for supersaturation-induced precipitation of drugs from organic solvent into anti-solvent within milliseconds. This process favors nucleation over particle growth, resulting in smaller and more homogenous nanoformulations. Particle growth is stopped by steric effects caused by the bulky polymeric stabilizer; almost all hydrophobic ingredients are encapsulated within the amphiphilic block copolymer shell. In contrast, bulk mixing reduces cargo loading efficiency due to unstable drug encapsulation and insufficient stabilizer coverage. For hydrophilic drug cargo mixed with PEG-PLA, fast mixing drives high drug loading and also entraps more polymeric (PEG) blocks in the core. Thus, the particle core grows larger but more hydrophilic cargo can be encapsulated [66].
To better understand the mechanisms of mixing and nanoprecipitation in FNP, computational fluid dynamics (CFD) and molecular dynamics (MD) [67–74] have been used to study mixing performance and reactions in CIJM and MIVM. Marchisio et al. used a CFD model to predict mixing in CIJM, and their predictions correlated well with experimental data at various Re for different inlet reactant concentrations and microreactor geometries [75,76]. Their work resulted in a reliable model for the design and scale-up of CIJM and MIVM. Olsen et al. proposed a delayed detached eddy simulation (DDES) model to simulate laminar and transitional flow in the complex geometry of the MIVM and to predict mean velocity and turbulent intensity, and validated the model using experimental data [77,78]. The DDES model provides insight into flow and mixing patterns in MIVM that were useful for optimizing the mixers.
Molecular dynamics simulations have been used to guide the design of complex polymeric Janus colloids with different shapes and coatings using amphiphilic block-copolymers and FNP processes [79]. MD was also used to study hydrophobic ion pairing (HIP) of an anionic drug and a cationic oleic acid (hydrophobic counterion) in solvent/anti-solvent solution [80]. Depending on the drug/counterion ratio, nanoparticle aggregation was prevented via electrostatic repulsion, indicating nanoparticle stability. MD has also been used to study the FNP-based fabrication of polymeric colloids containing inorganic nanoformulations [9]. The simulations revealed how the nanoparticle morphology depends on the interactions of growing nanoparticles with polymer and solvent, and how loading can be tuned by adjusting polymer and nanoparticle feed concentrations and their volume ratio. Diffusion-limited aggregation during turbulent mixing is key for forming stable nanoparticles within milliseconds. Whereas MD is useful in studying molecular interactions, larger-scale models such as coarse-grained simulations are helpful for studying nanoparticle-molecule interactions such as surface coating processes. A multiscale modeling approach combined MD with CFD and used a Smoluchowski population balance equation (PBE) to model FNP in CIJM [81]. The model described the FNP process at three size scales: molecular, nanoparticle, and macroscale, thus circumventing the standard nucleation and growth description and the need for parameter fitting.
Flash nanocomplexation
FNC is a kinetically controlled mixing process adapted from FNP. FNC can produce diverse nanoformulations with controlled size, high drug loading, and high encapsulation efficiency in a continuous and scalable manner [82–88]. To achieve uniform phase separation and nanoformation, a CIJM or a MIVM is used to facilitate rapid and efficient turbulent mixing of two or more polyelectrolyte solutions, similar to FNP [89]. FNC has significant advantages versus FNP. First, FNP uses organic solvents, and additional steps are required to remove these solvents [83,86]. The residual organic solvents are sometimes harmful to the environment and to humans. No organic solvents are involved in FNC, resulting in a simpler and less toxic preparation process. Second, FNC is more versatile than FNP for fabricating nanoformations, expanding the scope of applications. In FNP, the driving force for phase separation is typically solvent exchange, and nanoparticle fabrication is driven by hydrophobic interactions. However, in FNC, phase separation is induced by polyelectrolyte complexation, and many kinds of non-covalent and covalent interactions can be employed in fabrication, including ionic [86,90–92], hydrophobic [93], hydrogen bonding [16,83,94], metal coordination [85,93], and specific ligand or host-guest interactions [95]. Third, FNC can be used to assemble nanoformulations with diverse characteristics in a sequential process of nanoparticle formation followed by surface modification [87,88,96]. Thus, FNC allows rapid, scalable, and organic solvent-free fabrication of diverse nanoparticles with well-controlled structure, composition, size, and surface properties, with a more expansive range of potential applications.
Flash-based nanofabrication
The versatility and dynamic mixing of flash self-assembly technologies allows formulation of nanoparticles composed of organic and inorganic molecules for therapeutic, diagnostic, and imaging applications (Table 1). Cost-effectiveness is essential for clinical translation. Conventional nanofabrication methods developed in benchtop settings often have low reproducibility and high scale-up cost [97]. Flash nanofabrication methods offer lower cost and greater scalability, reproducibility, controllability, and product stability than bulk methods [98,99].
TABLE 1.
Nanoformulations constructed using flash-based self-assembly methods
| Organic nanoformulations | Inorganic and hybrid nanoformulations |
|---|---|
| β-carotene nanoparticles (NP) [60][97][100] | |
| Paclitaxel & vitamin E NP [101][102] | |
| Doxorubicin (DOX) NP [103] | |
| Cabazitaxel NP [104] | |
| α-lipoic acid & clozapine (antipsychotic) or cinnarizine (antihistamine) NP [105] | Silica NP, mesoporous silica nanoparticles (MSN) [22][108][109] |
| Polymyxin B / oleic acid NP [80] | |
| Cyclosporin or ibuprofen & mannitol or salbutamol sulfate NP [106] | Ammonium perchlorate NP [110] |
| Bifenthrin-loaded NP [107] | |
| NaCaS zein NP [26] | |
| Polyphenol-tocopherol polyethylene glycol 1000 succinate (TPGS) NP [16] | |
|
| |
| PEI/DNA NP [111][112] | (TA)-PEG-Au NP [113] |
| Tannic acid (TA)-interferon-α/CpG NP [94] | |
|
| |
| Chitosan/insulin/triphosphate (TPP) NP [114] | Polymeric NP decorated with Au, Pt, or Ag [115][116][117][118] |
|
| |
| Chitosan/antigen/adjuvant NP [119] | Lanthanide/gadolinium phosphate with PAA-PEG NP [120] |
|
| |
| Microgels & nanogels [121]; emulsion droplets [122] | Polymeric CdSe-ZnS quantum dots [123][124]; Polymeric PZn3 and iron oxide [125]; polymeric rare earth NP [126] |
|
| |
| Semiconducting polymeric dots (Pdots) [127] | Chitosan-Pb(NO3)2 NP [128] |
|
| |
| Amphiphilic polymersomes; bicontinuous nanospheres (BCN) [129] | Polymeric TiO2 or ZnO NP [130] |
|
| |
| Janus, Cerberus, disordered lamellar, and other nanocolloids [131][79] | Polymeric Janus nanocolloids decorated with Au [132] |
|
| |
| Non-spherical aggregation-induced emission (AIE) NP; nanorods & microrods [133] |
|
| Lipoplexes, lipo-polyplexes [134][135][136][137] | |
Scalable production of organic nanoformulations and nanocarriers
Brian Johnson and Robert Prud’homme were the first to develop methods for de novo fabrication of hydrophobic organic nanoparticles using FNP [97]. In their study, β-carotene was mixed with the amphiphilic copolymer polystyrene (PS)-b-PEO in an organic solvent stream, and the components were co-precipitated with water to form polymer-protected β-carotene-loaded nanoparticles. Subsequently, a variety of organic compounds were incorporated into stable nanoparticles with the aid of different amphiphilic copolymers using FNP [97][100]. These works introduce new techniques for compound loading in polymeric nanocarriers.
Hydrophobicity is a large determinant of particle stability. While selecting a cargo compound, cLogP values are used to quantify compound hydrophobicity. Compounds with high cLogP (>12) are hydrophobic, whereas compounds with small cLogP (<6) are prone to Ostwald ripening and aggregation [59]. A number of therapeutic compounds with low cLogP have been incorporated into stable nanoformulations using FNP. FNP was used to anchor the low cLogP anti-cancer drug paclitaxel with hydrophobic vitamin E succinate, enhancing particle stability and allowing controlled drug release [101][102]. The hydrophobic drug doxorubicin (DOX) (cLogP ~1.2) is often problematic in forming stable nanoparticles through precipitation using conventional mixing. By mixing DOX with PEG-b-PLA using a four-inlet MIVM, <100 nm nanoparticles were generated with good drug encapsulation efficiency and a pH-dependent drug-release profile [103]. In another example, the oncological drug cabazitaxel was conjugated with an amphiphilic polymer to form ~60 nm Cellax nanoparticles (Cellax-CBZ) which remain stable for over three months without post-processing; a broad therapeutic dosing window was also demonstrated [104]. FNP was also used to prepare stable nanoparticles containing the pesticide bifenthrin in a cost-effective manner [107]. Bifenthrin nanoparticles with a tunable loading rate of 50–91%, tunable size of 100–200 nm, and controlled drug release behavior were constructed by adjusting FNP parameters. An in situ salt-forming method using FNP was used to expand the scope of active pharmaceutical ingredients (APIs) that could be encapsulated into polymeric carriers [105]. Ionizable and low cLogP biologics such as α-lipoic acid combined with the antipsychotic clozapine or the antihistamine cinnarizine were stabilized in a polymeric carrier. Pharmaceutical compounds with distinctive solubilities such as cyclosporin or ibuprofen have been combined with mannitol or salbutamol sulfate and incorporated in polymeric nanoparticles using FNP [106]. Zein, an alcohol-soluble prolamin protein from corn with a variety of industrial and food uses, was incorporated into NaCaS zein nanoparticles and massively produced using FNP [26]. Using the charged groups of amino acid residues, a cornucopia of proteins can be prepared using this facile, continuous method. Therefore, FNP utilized solvent supersaturation to create majorly polymeric nanocarriers with diverse cargo loading.
FNC, unlike FNP, was initially focusing on fabricating polyelectrolyte nanoparticle complexes. FNC serves as an alternative platform for producing nanoparticles when compared to conventional bulk production. In one example, PEI/DNA nanocomplexes were prepared using a CIJM (Fig. 3a) [111][112]. The hydrodynamic size of the nanocomplex and plasmid loading was tuned by adjusting FNC parameters. Extended storage by lyophilization makes these polyplexes promising for clinical translation. The suitability of FNC for nanocomplex fabrication has been evaluated by comparing nanocomplexes produced using FNC and bulk mixing methods. Furthermore, nanocomplexes of chitosan, insulin, and TPP for use in treating type I diabetes were achieved by mixing positively-charged insulin and chitosan with negatively-charged TPP, and yielded smaller particles when using FNC than when using a dropwise-addition method (Fig. 3b) [114]. In another study, nanocomplexes of antigen and adjuvant were produced using FNC using chitosan as nanocarrier, resulting in production of size-controlled nanoparticles in a scalable manner [119]. In this study, 0.9 g of nanocomplexes containing ~30 mg of antigen was prepared in less than 1 h using a single device. The production can be multiplied by running several MIVM devices in parallel. Nanocomplex formulation using FNC to incorporate protein and antigen in nanocarriers is highly reproducible and promising for nanovaccine development at industrial scale.
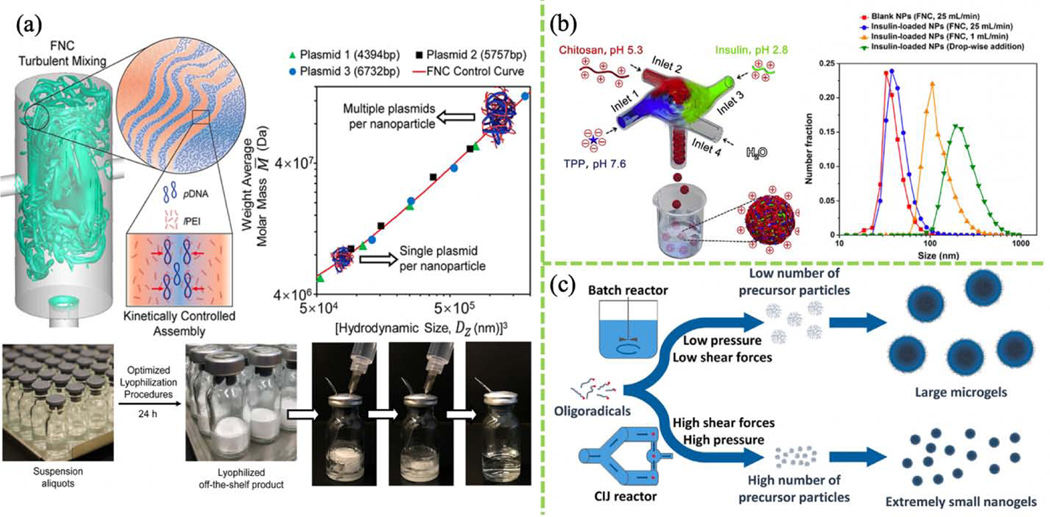
FIGURE 3.
(a) Turbulent mixing of pDNA and linear PEI solution in a CIJM for scalable fabrication of a plasmid nanocomplex. Transfection efficiency depends on nanocomplex size, which is closely related to the amount of DNA loaded. (Reprinted with permission from Ref. [112]. 2019, American Chemical Society.) (b) FNC system for preparing insulin/chitosan/TPP NPs. (Reprinted with permission from Ref. [114]. 2017, Elsevier.) (c) Synthesis of poly(N-vinylcaprolactam) (PVCL) nanogels can be achieved using surfactant-free FNP; batch mixing methods result in larger microgels. (Reprinted with permission from Ref. [121]. 2018, Elsevier.)
Microgels are crosslinked polymers with diverse applications. To fabricate microgels, oligoradicals are mixed in a batch reactor at low pressure, typically yielding a small number of precursor particles and large products. Small microgels (<100 nm) are useful in drug delivery. With the aid of FNP, 45-nm monodisperse poly(N-vinylcaprolactam) (PVCL) colloidal nanogels were created without using toxic surfactant [121]. This study involved the development of a high-pressure impinging jet reactor called a Microfluidizer (Fig. 3c), the first device to integrate a temperature sensor, intensifier pump, heat exchange chamber, and reaction chamber for scalable production of monodisperse nanogels.
Emulsification is another common industrial process. Emulsion using CIJM extends its potential by increasing throughput and reducing energy consumption. CIJM emulsification has be used to produce massive quantities (>176 g/min) of micron-size (~8 μm) oil-in-water droplets with high dispersed-phase content (80%) and good stability [122]. Again, flash-based techniques offer researchers versatility in constructing a great variety of nanoformulations.
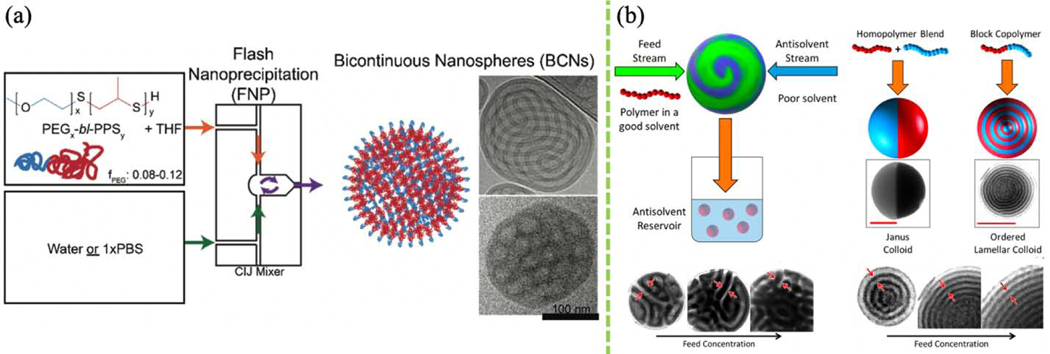
Polymeric nanocarriers are ubiquitous in nanomedicine. Self-assembled polymersomes (PS)—vesicles made using amphiphilic block copolymers—usually bear more shear stress than liposomes while maintaining water permeability during circulation, which is advantageous in drug delivery. These polymeric formulations have not yet achieved clinical transition partially due to challenges in large-scale fabrication. FNP was recently used to mass-produce PS with membrane-like structures. Gram-scale production of PS loaded with theranostic agents can be attained within minutes. Moreover, multilamellar, tubular, vesicular, and bicontinuous PEG-bl-poly(propylene sulfone) (PPS) PS have been developed, with either hydrophobic or hydrophilic lumens for loading a variety of molecules [129]. PS morphology depends on the hydrophobic/hydrophilic block ratio. Drug nanocarriers are subject to aggregation and cargo release once exposed to the oxidative cellular environment following uptake. Bicontinuous nanospheres (BCNs) are cubic PS that are morphologically similar to lipid cubosomes (Fig. 4a). BCNs are interconnected by labyrinthine narrow channels which efficiently trap payloads and have shown sustained drug release profiles, although some BCNs are prone to bursting and releasing their hydrophilic payloads. PEG-b-PPS BCNs and PS have been systemically evaluated and compared for their encapsulation efficiency, organ-level uptake, and drug delivery capabilities [129]. BCNs represent another level of the polymeric manufacturing technique that flash-based technique could offer.
FIGURE 4.
(a) Bicontinuous nanosphere formation using FNP. (Reprinted with permission from Ref. [129]. 2017, Elsevier.) (b) Janus colloid formation using FNP. (Reprinted with permission from Ref. [131]. 2019, American Chemical Society.)
FNP has also been used for the fast production of complex polymeric nanoparticles with Janus and patchy structures (Fig. 4b). Sosa and co-workers employed a rapid FNP process to manipulate soft colloidal particle surface properties and architecture. In a laboratory setting, a production rate of ~3 kg/day of polyisoprene-polystyrene Janus and Cerberus nanocolloids were demonstrated [131]. The authors alter the annealing schedule to control nanocolloid morphology. More complicated internal hierarchical structures have been generated by maneuvering phase separation and kinetic trapping, guided by MD simulations with explicit solvent [133]. Nanoparticles with non-spherical morphology have been fabricated at scale using FNP, including high aspect-ratio nanorods and microrods [138]. Small (~10 nm) semiconducting polymer dots (Pdots) for enhanced cell penetration have also been fabricated using FNP [127]. Compared to other bottom-up methods such as templating and self-organized precipitation, FNP offers a simple one-step, low-energy, scalable approach to fabricating stable and diverse colloidal nanoparticles.
Lipid-based nanoproducts have long been used in drug delivery. However, pure liposomal micelles are subject to cargo leakage, instability, and toxicity. The advent of lipid-based nanoparticle (LNP) systems addressed these issues. Lipo-complexes and lipo-polyplexes efficiently deliver multiple therapeutics or imaging agents. Effectively fabricating high-quality lipid-based nanoproducts merit both delivery-based therapeutics and diagnostics. In one study, a MIVM was used to prepare lipo-complexes and liposomal coating of the polyplex core through electrostatic interactions in one pot mixing. FNC not only provides versatility but flexibility and stability in preparing lipid-based nanoproducts. He et al. constructed thirteen groups of lipid-based nanoparticles with different lipid types (dipalmitoylphosphatidylcholine (DPPC) or 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) stabilized by cholesterol) and composition using two fabrication methods: FNC and rapid injection / manual mixing). The lipid-based nanoparticles fabricated using FNC showed more homogenous particle size across all tested formulations than rapid injection to produce blank DPPC/cholesterol liposomes and DOTAP/cholesterol/DNA lipo-complexes. In addition, purification and lyophilization systems can be combined with FNC to massively produce commercial-ready LNPs [134][135]. In another study, FNC was used to fabricate lipid-coated nanoparticles (lecithin/1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE)-PEG and poly(lactic-co-glycolic acid) (PLGA)) loaded with the anti-cancer drug doxorubicin. These hybrid nanoparticles were formulated using a 3D-printed herringbone-patterned MIVM. The herringbone structure inside the MIVM chamber was designed to promote formation of more uniform nanoparticles [136]. Lipid-polymer hybrid nanoparticles have also been fabricated using a MIVM. Distinct from lipo-polyplexes, lipid-polymer hybrids have a micellar configuration rather than a solid core-shell structure. Hybrid nanoparticles with small size, low polydispersity index (PDI), and high stability in serum have been mass-produced at >10 g/h [137]. Lipid-based surface modification will be discussed in a later section.
Scalable production of inorganic nanoparticles
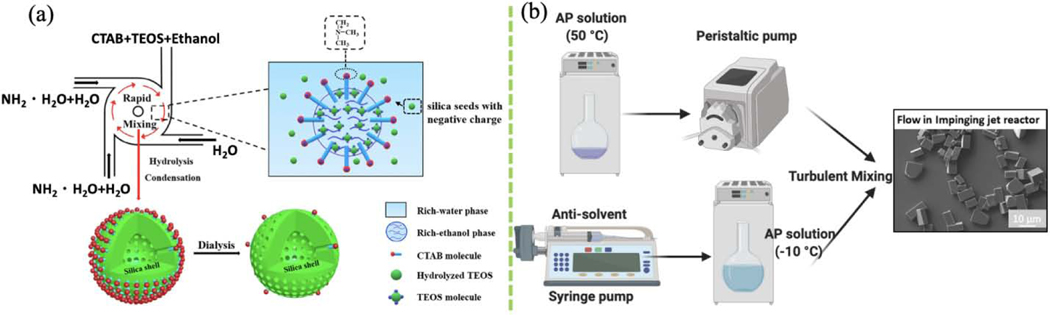
Inorganic nanoparticles are used in many fields. Solar cells, lubricants, batteries, and semiconductors often exploit the distinctive features of inorganic nanoparticles. Batch reactors remain the most common platform for wet synthesis of inorganic nanoformulations, but suffer from drawbacks such as heterogeneous nanoparticle size, low production rate, poor reproducibility due to insufficient mixing during particle assembly. Microfluidics systems such as capillaries, junctions, and hydrodynamic focusing were developed to overcome these drawbacks, but are limited to slow mixing and lamellar flow. To address slow mixing, microreactor system like interdigital microstructure mixer was developed. Mixing speed was improved by thinning the multilamellar flow by dividing and re-merging the inlet stream [108]. In speaking of synthesis, the Stöber process of synthesizing silica nanoparticles was also performed using this micromixing method. The effects of batch and micromixing methods on nanoparticle properties were compared. Narrower and smaller (<100 nm) silica nanoparticles were produced using micromixing due to the shorter nucleation time and uniform temperature and composition control in the micromixer [108]. In another study, a sequential FNP process was used to synthesize drug-loaded mesoporous silica nanoparticles (MSNs) (Fig. 5a). Formation of micelle-based cetyltrimethylammonium bromide (CTAB) templates followed by in situ MSN formation was achieved using two MIVMs in series. Importantly, silica shell thickness and cavity size were tunable by controlling the composition of reactant streams [109]. The intense mixing of the silicon precursor with the catalyst in the flash-based mixer with high shear stress shortens the preparation process. By tuning the composition and mixing parameters, cubic, golf-ball-like and hollow organosilica NPs can be produced [22]. MSN fabrication using FNP has the potential for scale-up production for industrial applications. Turbulent micromixing has also been favored to fabricate nanoparticles composed of the friction-reducing agent, molybdenum sulfide (MoS2) [139]. FNP has been used for the safe and rapid production of ammonium perchlorate (AP) particles. An integrated CIJM system linked to a temperature bath with a peristaltic pump was developed to produce <20 μm AP particles with minimum product clogging at the high rate of 3 kg/h (Fig. 5b) [110].
FIGURE 5.
(a) Preparation of mesoporous silica nanoparticles (MSN) by FNP. (Reprinted with permission from Ref. [109]. 2020, Elsevier.) (b) Integrated CIJ system for ammonium perchlorate (AP) particle production. (Reprinted with permission from Ref. [110]. 2019, Elsevier.) (Remade in BioRender.com).
Scalable production of organic-inorganic hybrid nanoparticles
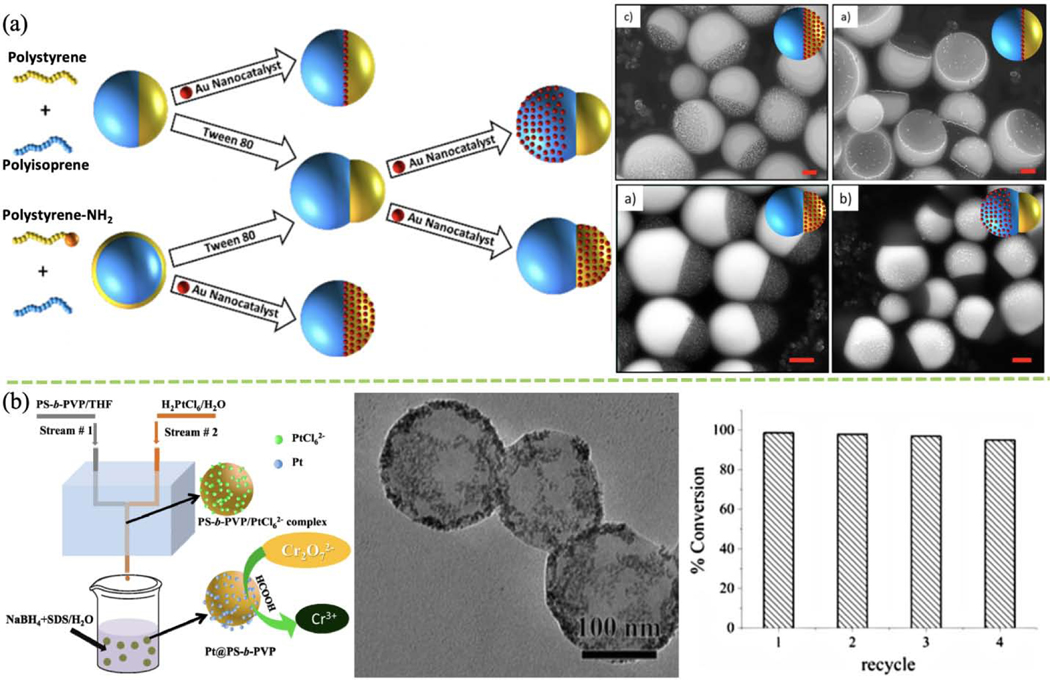
Organic-inorganic hybrid nanoparticles are versatile due to their combined properties. Lee and co-workers used FNP to decorate an Au nanocatalyst with polymeric Janus nanocolloids, causing the colloidal structure to metamorphose into a variety of structures (Fig. 6a). Moreover, Au nanocatalyst location can be manipulated by switching polymer functional groups. Adjusting total interfacial energy by using different surfactants and low MW block copolymers can transform the Janus structure from snowman-like or dumbbell to spherical [132]. Not only the morphology and configuration of nanocolloids can be controlled, size again can be easily manipulated with precision. Fabrication of tannic acid (TA)-PEG-Au nanoparticles with precise control of size over the range of 13–63 nm was achieved by Tang and co-workers [115]. In another study, hybrid Au/platinum (Pt) polystyrene-polyvinylpyrrolidone (PS-b-PVP) nanoparticles formed using FNP have shown high catalytic activity in p-nitrophenol reduction [116]. Similarly, Pt-embedded PS-b-PVP hybrid nanoparticles have shown excellent chromium reduction activity (Fig. 6b) [117]. Silver-decorated polymer nanocolloids with antibacterial and sensing abilities have also been fabricated using FNP [118].
FIGURE 6.
(a) Schematic and TEM images of FNP generation of Au-modified Janus and core-shell nano-hybrid. (Reprinted with permission from Ref. [132]. 2017, American Chemical Society.) (b) Schematic of FNP fabrication of Pt@PS-b-PVP nano-hybrid for Cr(VI) catalytic degradation to Cr(III); TEM image and recyclability test for reduction of Cr(VI) with four recycles. (Reprinted with permission from Ref. [117]. 2017, Springer Nature.)
Supramolecular assembly can be achieved using host-guest interactions in a MIVM [140]. Au-functionalized β-cyclodextrin (CD) (host) interacts with adamantyl-modified polypropylene dendrimers (Ad-PPI) via hydrophobic interactions to form kinetically labile complexes. These artificial networks can be used in bioimaging applications. Other interactions are also exploited for bio-imaging related hybrid nanocomposites preparation. For example, lanthanide and gadolinium phosphates stabilized by PAA-b-PEG were fabricated using ionic FNP (iFNP), which is driven by ionic interactions [120]. Ionic FNP proved to be highly effective for manufacturing hybrid nanoparticles for imaging applications. Metal-chelating hybrid nanoparticles for positron emission tomography (PET) were produced using FNP [141]. Multienzyme nanocomplexes were created using cadmium selenide (CdSe) quantum dots precipitated using FNP with Pluronic F127 polymer and labeled with enzymes, and were used to characterize enzyme colocalization using FRET [123]. Zhang et al. combined core-shell CdSe-ZnS quantum dots with a different polymer using FNP to form fluorescence nanoparticles for use in particle tracing, optical barcoding, and biosensors [124]. Furthermore, polymerized PZn3 and iron oxide nanocrystals have been formulated using FNP for T2-active MRI and NIR imaging, respectively [125]. The radionuclide 64Cu and the metal chelator phthalocyanine were co-encapsulated with polymeric stabilizers using FNP to form PET-active nanoparticles [141]. PEGylated GdPO4 inorganic and organic hybrid nanoparticles were fabricated using iFNP for use in multimodal imaging [120]. Quench iFNP (qiFNP) was developed to synthesize functional LnPO4-based nanoparticles in water for bioimaging [142]. Upconverting nanophosphors (UCNP) using the rare-earth phosphors NaYF4:Yb3+, and Er3+ together with PEG-b-PLA in photodynamic therapy (PDT) was also demonstrated [126]. In a different study, in situ reactive FNP (rFNP) was used to fabricate negative active material (NAM) nanoparticles using pH-sensitive chitosan with Pb(NO3)2 for use in a lead-acid battery application. The chitosan-based NAM shows a high discharge capacity and high recharging capacity (~700 cycles) [128]. Iron oxide, the antioxidant tannic acid, and PS-b-PEG were coprecipitated to form multifunctional nanoparticles of 50–265 nm diameter, with size tunable by adjusting the iron oxide-to-polymer ratio [143]. FNP has been used to fabricate sunscreen nanoformulations (inorganic (TiO2 or ZnO) and organic (Uvinul A Plus or T 150)) with spherical or rod-like shape that are stable for >80 days [130]. By exploring various nanomaterials and distinctive driving forces, flash-based self-assemblies were exploited to manifest its seminal potential in bioimaging applications.
Nanoparticle surface modification using flash technology
Surface modification of nanoparticles is used to improve cell uptake, targeted delivery, and particle stability. Surface modification uses hydrophobic interactions, hydrogen bonding, electrostatic interactions, π-bridges, and specific ligand interactions such as host–guest. Table 2 lists common polymeric and liposomal surface modifications that can be performed using flash technology.
TABLE 2.
Flash surface modifications
| Polymeric | Liposomal |
|---|---|
| PEG brushes [144] | DPPC/DMG-PEG-coated PEI/DNA polyplexes [147][134][135] |
| PEI-g-PEG mRNA-loaded NP [145] | |
| PEG-grafted chitosan NP [146] | |
|
| |
| PVP-coated paclitaxel NP [148] | DSPE-PEG-coated PLGA NP [136] |
| Lecithin DSPE-PEG-coated PLGA NP [149] | |
|
| |
| Double-layer-coated polyplexes (HA 1st layer, hypromellose phthalate 2rd layer) [29] | DOPE/DSPE-PEG-FA-coated drug nanocrystals |
| (MTX, DOX, CS) [150] | |
|
| |
| Eudragit (L100–55)-coated insulin polyplexes [151] | |
|
| |
| Chitosan-coated PCL NP [152] HA-coated HTCC NP [153] |
|
Polymeric surface modification using FNC and FNP
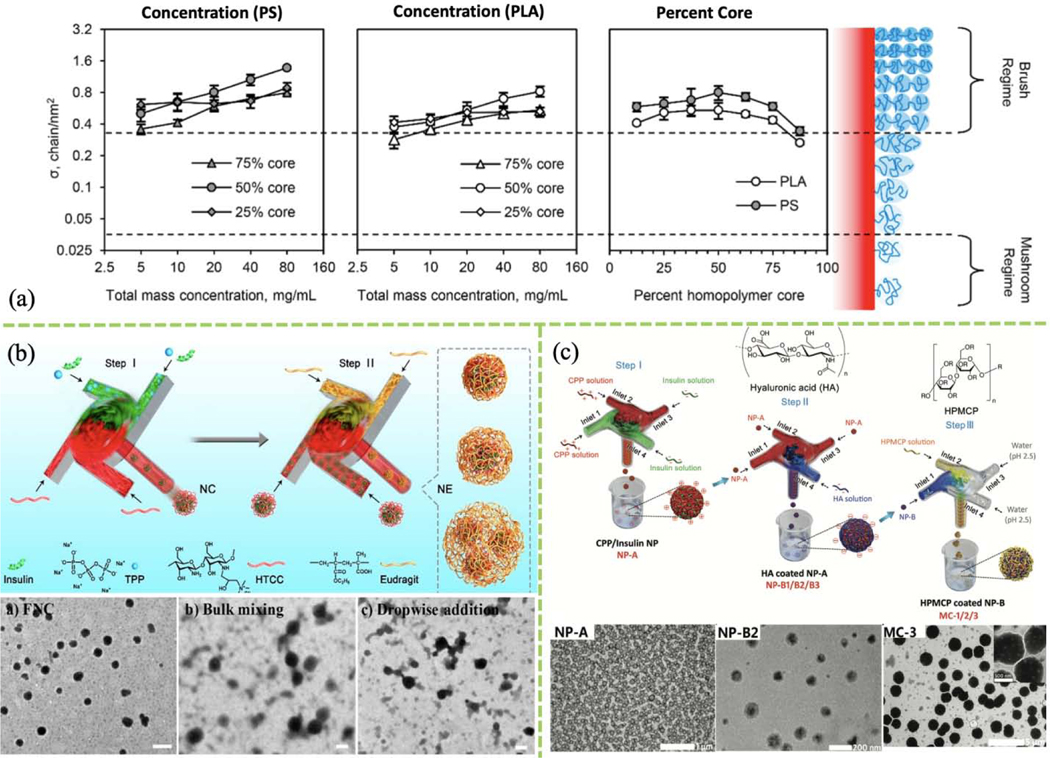
Natural and synthetic polymers are used to provide protection and stability for many therapeutic and diagnostic nanosystems. Most cargo-laden polymeric nanoparticles are fabricated by precipitation of organic molecules using anti-solvent. FNC-based self-assembly is attractive for drug delivery applications because an organic solvent is not required, which greatly reduces toxicity. Surface modification with polymers such as altering polymer brush density can enhance nanoparticle stability. The effects of polymer composition (PS and PLA), polymer mass concentration, and PEG graft surface density on polymer brush density were studied (Fig. 7a). PEG brush density was found increases with total brush mass concentration but decreases when the core is larger than 50% of the overall nanoparticle size [144]. However, polymeric brush surface density was weakly dependent on many FNP parameters, consistent with a study by García et al. in which surface PEG content was found to be independent of mixing time [66]. To take advantage of flash-based self-assembly for polymeric surface modification, other parameters such as turbulence, liquid viscosity, and temperature must be tested to obtain a model that can be used for therapeutic nanocarrier design. In a more recent study, messenger RNA (mRNA)-associated NPs were grafted with PEI-g-PEG through FNC. Those NP were used for pulmonary immune cells delivery and pulmonary immunomodulation. Authors found the PEGylated NPs with a PEG grafting ration about 0.5% have the highest transgene expression in the lung after i.v. administration. The lyophilized NPs are also stable for more than 4 months stored at sub-zero condition [145].
FIGURE 7.
(a) Effect of polymer concentration and core percentage on surface PEG brush density of polymeric core-shell NPs formed using FNP. (Reprinted with permission from Ref. [144]. 2018, American Chemical Society.) (b) Two-step FNC process preparing Eudragit-coated insulin-loaded HTCC/TPP nanocomplexes, and TEM images of nanocomplexes prepared by FNC, bulk mixing, or dropwise addition. (Reprinted with permission from Ref. [151]. 2019, American Chemical Society.) (c) Schematic of sequential FNC system for preparation of CPP/insulin NP (NP-A) core, hyaluronic acid-coated NPs (NP-B) and HPMCP-coated NP-B (MC); bottom: corresponding TEM images. (Reprinted with permission from Ref. [29]. 2018, Royal Society of Chemistry.)
Chitosan is widely used in oral drug delivery. Single-step rapid self-assembly of PEGylated chitosan nanoparticles was achieved using FNC with a two-inlet CIJM [146]. Intermolecular forces impact nanoparticle stability at different pH values, and adjusting the surface composition can be used to control stimuli-responsive cargo release. H-bonding, electrostatic interactions, and hydrophobic ion-pairing are important driving forces in FNC-based self-assembly. For example, PVP polymer was coated onto paclitaxel nanoparticles via H-bonding [148]. Furthermore, a double layer of polymeric coating was also reported using a three-step sequential FNC process (Fig. 7c). First, a MIVM was used to fabricate positively charged cell-penetrating peptide (CPP) complexes with negatively charged insulin. The resulting nanoparticles were coated with a layer of anionic HA in a second MIVM. Then, a third MIVM was involved to add another layer of hypromellose phthalate (HPMCP). All layers were formed through electrostatic interactions [29]. This sequential polymeric and polysaccharide surface modification technique adds flexibility to the development of therapeutics for oral drug delivery. Similarly, two MIVM were linked for the fabrication and subsequent coating of an insulin-loaded polymer-modified chitosan (HTCC)/TPP nanocomplex with a pH-responsive polymer (Eudragit L100–55) (Fig. 7b). The polymeric coating was also achieved through charge-charge interactions between the nanocomplex and Eudragit, providing stomach acid protection and site-specific release after oral administration [151].
Chemically-modified polymeric coatings contribute additional functions to drug delivery. High surface-charge and mucoadhesive chitosan-coated polycaprolactone (PCL) nanoparticles [152], dopamine-modified HA-coated metal-phenolic doxorubicin nanocomplexes [154], and thiolated HA-coated insulin-ammonium chloride-modified chitosan (HTCC) nanoparticles all demonstrate their uniqueness in drug delivery [153]. For example, FNC was used to produce insulin-lipid nanocomplexes through hydrophobic ion-pairing and to coat the surfaces with HA [155]. Targeting molecules can also be incorporated into the surface of nanoparticles. Breast cancer cell peptides and Luteinizing hormone were conjugated onto a PEG-PCL polymer, and the modified polymer was combined with a hydrophobic fluorophore and the paclitaxel to create surface functionalized-theranostic nanoparticles [156]. In addition to the aforementioned diabetic and cancer treatments, flash-based surface modifications are exploited in anti-bacterial applications. For example, mannose-targeting ligands for Mycobacterium tuberculosis were introduced onto polymeric nanocarrier surfaces using FNP; the surface density of the ligands could be precisely controlled to optimize nanoparticle uptake [157]. Magnetic iron oxide nanoparticles surface-modified with bacteria-targeting PEG stabilizer were developed for pathogen identification and separation [158]. Streptavidin-functionalized nanoparticle surfaces modified with a wide variety of biotinylated molecules or proteins such as antibodies were also demonstrated [61].
Liposomal surface modification using FNC
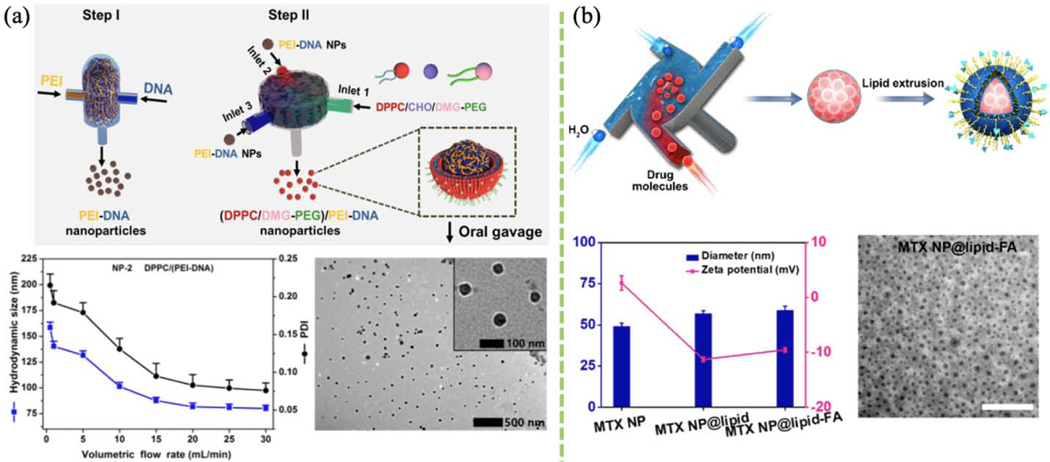
Liposomal systems are widely used in drug and gene delivery. Recent efforts have focused on harnessing lipids to optimize oral delivery. Oral gene delivery suffers from low transfection efficiency and bioavailability due to the secretion of negatively-charged mucin by the gut epithelia. Surface modification of naked plasmid nanoparticles with lipids enhances mucosa penetration and gene delivery efficacy. In a recent study, flash mixing of PEI and glucagon-like peptide 1 (GLP-1) forms PEI/DNA nanocomplexes via electrostatic interactions (Fig. 8a), and the resulting nanocomplexes express cationic surface property and can be easily trapped in the mucin layer. In addition, neutral DPPC/DMG-PEG was also utilized to modulate PEI/DNA polyplexes surface for other purposes. Both hydrophilic and neutral nanoparticle surface modification can be achieved using FNC [147][134][135]. Thus, FNC offers great flexibility in manipulating both the chemical and physical surface of nanocomplexes.
FIGURE 8.
(a) Schematic of lipid-coated DNA NPs prepared using FNC. NP size and PDI profile versus flow rate. TEM image of lipid-based PEI/DNA NPs. (Reprinted with permission from Ref. [159]. 2019, American Chemical Society.) (b) Schematic of the fabrication of lipid-stabilized solid drug NPs. Lipid-stabilized drug NP diameter and PDI. TEM image of lipid-coated MTX NPs. (Reprinted with permission from Ref. [150]. 2018, American Chemical Society.)
In recent studies, modified MIVMs were involved in constructing lipid-coated polymeric nanoparticles. For example, lipid-coated PLGA nanoparticles were formulated using a herringbone-patterned MIVM for drug testing in 2D and 3D tumor cultures. The lipid bilayers on the nanoparticle surface serve as protection and result in longer drug release [136]. DSPE-PEG lipid-coated PLGA nanoparticles were fabricated using a 3D-printed MIVM. In another study, the drugs myristic acid, rifampicin, and dexamethasone were encapsulated in a PLGA core, and the resulting DSPE-PEG coated nanoparticles showed two-week stability [149]. However, drug compounds loading often requires organic solvents so that the toxicity issue should be further addressed.
Conventional drug carriers are often limited by low loading, poor reproducibility, and short circulation time. Lipid-coated drug molecules have been developed to address these limitations. DOPE lipid, cholesteryl hemisuccinate, and targeting stabilizer DSPE-PEG-FA were combined in lipid-coated drug nanoparticles formed using FNP combined with manual extrusion processes (Fig. 8b) [150].
Biomedical applications
Flash-based self-assembly of a large variety of nanomaterials (organic, inorganic, and hybrid) has been used to fabricate innovative tools for small molecule delivery, macromolecule delivery, and imaging-related theranostic applications (Fig. 9).
FIGURE 9.
Flash technology has been used to incorporate small molecule drugs, macromolecules (such as nucleic acids, proteins, and synthetic polymers), and imaging agents into nanoparticles in a wide range of biomedical applications.
Small molecule delivery
Active pharmaceutical ingredients (APIs) are the essential entities in nanomedicine. However, the low solubility of many organic small molecule APIs leads to incomplete drug delivery and therapeutic failure. There is an increasing demand for effective small molecule delivery. Importantly, high-throughput techniques are needed for the fabrication of therapeutic nanoparticle systems that allow delivery of small molecule drugs with spatiotemporal control.
The chemotherapeutic drugs methotrexate (MTX), chlorambucil (CA), and doxorubicin (DOX) were involved in the flash system for improved antitumor activity in MCF-7 tumor-bearing mice. Lipids coated nanoparticles induce an enhanced permeability and retention (EPR) effect that the naked counterparts could not provide. Lipid-coated drug nanoparticles have shown better pharmacokinetic and biodistribution profiles than free drug molecules in vivo [150].
Oral chemotherapy for cancer treatment is favored due to high patient compliance, minimized invasiveness, and lower cost. However, the labile environment of the gastrointestinal (GI) tract poses a challenge to the functionality, pharmaceutical activity, and biological availability of orally administrated drug carriers [160]. Shen et al. used PLGA to encapsulate the cancer preventive agent SR12668 to produce stable nanocarriers with high drug loading. The nanoparticles showed better oral bioavailability and a better drug release profile in vivo than the commercial drug Labrasol [161]. In a different study, Le et al. created drug nanoparticles composed of tannic acid, paclitaxel, and PVP via H-bonding interactions that exhibit pH-dependent gut epithelial release, P-gp inhibitory function, and enhanced epithelial penetration after oral delivery in mice [148]. In another study, curcumin was encapsulated in a nanocomplex composed of TA and PVA and was used to inhibit prostate cancer PC3 cell growth. Interestingly, the nanocomplexes with the lowest curcumin loading elicited the most potent anti-proliferation effect among the groups tested, indicating the importance of drug loading on cancer cell uptake [162]. Despite all the promising results, further studies are still required to better understand the mechanism of anti-cancer nanoparticle uptake by cancer cells.
Besides cancer therapeutics, increasing bacterial antibiotic resistance has created an urgent need for anti-quorum-sensing therapies to regulate bacterial communication and the virulence cascade to combat the pathogens. One major challenge in translating quorum-sensing therapy is finding a suitable route and formulation for delivery. The autoinducer CAI-1 causes Vibrio cholerae to regulate its virulence level and manage antibiotic resistance through quorum sensing. However, oral delivery of water-soluble CAI-1 (cLogP ~ 4.6) fails to penetrate crypts of the upper small intestine. To address this, FNP formulated mucus-penetrating CAI-1 nanoparticles elicited a strong bacterial quorum-sensing response in a mouse model [163]. In another study, mucus-penetrating nanoparticles have also been used to treat infection by Pseudomonas aeruginosa, a gram-negative bacterium. The LasR inhibitor V-06–018 and vitamin E acetate were co-encapsulated in PEGylated amphiphilic polymers, and were used to achieve quorum-sensing inhibition, indicated by a reduction in the toxin pyocyanin [164]. Inhibition of M. smegmatis growth was also demonstrated using a rifampicin-loaded lipid-polymeric carrier [149].
Malaria is a deadly infectious disease caused by Plasmodium parasites. Current therapy relies on a combination of medicines and requires multiple dosages a day for a sustained period, which leads to drug resistance. To overcome this issue, the antimalarial drugs OZ439 [165] and lumefantrine (LMN) [166] were integrated into polymeric or liposomal nanoparticles to treat malaria, with improved drug bioavailability. In another study, abamectin (Abm), a nematicide for parasite inhibition, was encapsulated into MSNs, with loading efficiency greater than 90%. Abm release profiles were adjusted by modulating the CTAB template concentration, and 87% of the parasite Meloidogyne incognita was eliminated within 24 h using a slow Abm release profile [109]. Antibacterial and antiparasitic nanoparticles formulated using FNP are promising for combating drug resistance, with scalability that will allow future clinical translation.
Biomacromolecule delivery
Biopharmaceuticals such as proteins, peptides, and nucleic acids are important therapeutic components for combating diverse diseases such as cancer, infections, and autoimmune diseases. However, direct administration of these biomacromolecules is not feasible as nucleic acids and proteins are prone to degradation and clearance upon exposure to bodily fluids. Biofunctional molecules need to be protected until delivered to the target. Vehicles such as polymeric and peptide amphiphiles have been used as carriers in flash-based formulations for efficient and safe delivery of macromolecules.
Polymersomes have been used to load macromolecules for drug delivery [167]. However, lipophilic DiD and water-soluble FITC-labeled BSA-loaded BCNs have demonstrated higher encapsulation efficiency than their polymersome counterparts. Different molecular weight payloads such as calcein, ethyl eosin, 10 and 70 kDa dextran, and ovalbumin-TR were loaded in BCNs [168], and BCN accumulation in different organs was observed in a mouse model [169]. Reduced off-target retention in the liver and enhanced phagocytic target cell populations were reported [170]. Safety after intravenous administration was evaluated in a subsequent systematic toxicity study [171]. The BCNs did not elicit a significant inflammatory response in animal immune cells (Fig. 10a) [168]. These results establish that successful drug delivery using BCNs fabricated using flash-based self-assembly can be achieved with promising potential for clinical translation.
FIGURE 10.
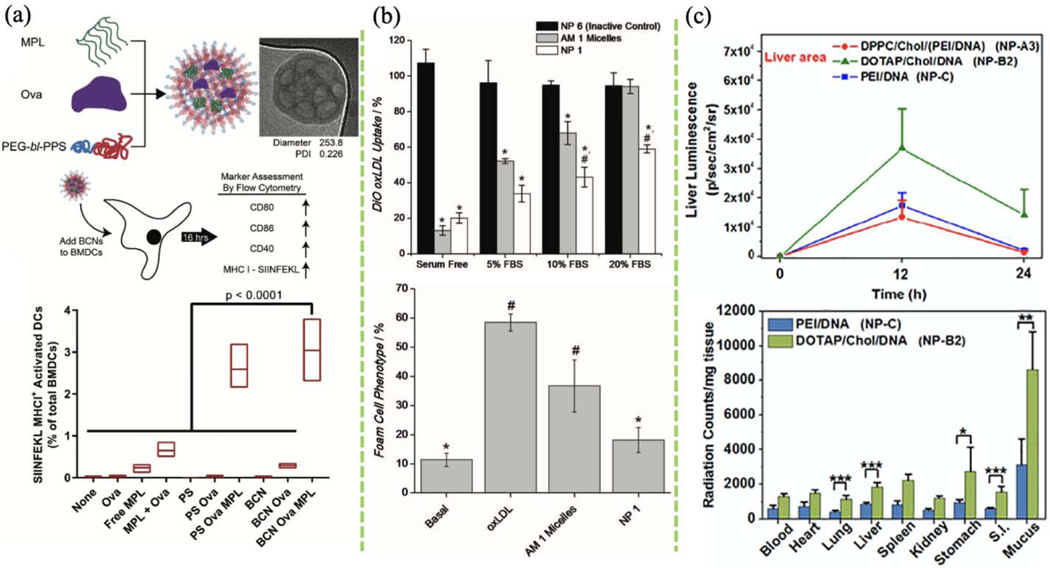
(a) In vitro antigen and adjuvant delivery to bone marrow-derived dendritic cells (BMDCs) using bicontinuous nanospheres (BCNs). TLR4 agonist monophosphoryl lipid A (MPL) with antigen ovalbumin (Ova) were co-loaded in BCNs via FNP. The effects of BCNs and polymersomes (PS) on BMDC activation were compared by comparing the expression of cell surface markers. (Reprinted with permission from Ref. [168]. 2018, Royal Society of Chemistry.) (b) Quantification of DiO-labeled oxidized low-density lipoprotein (oxLDL) uptake by human monocyte-derived macrophages (hMDMs) after 24 h incubation with FNP-based AM micelles or NP and quantification of foam cell phenotype after 48 h incubation. (Reprinted with permission from Ref. [172]. 2012, John Wiley and Sons.) (c) Top: transgene expression of plasmid in the liver and lung after single-dose delivery of different lipid-based NPs. Bottom: biodistribution of lipid-based NPs in different organs 6 h after administration. (Reprinted with permission from Ref. [134]. 2018, Elsevier.)
Bioactive amphiphilic macromolecule micelles are great delivery vehicles and were used to alleviate atherosclerosis by targeting inflammatory cells. Mucin acid-modified sugar-based amphiphilic micelles formulated by flash-based self-assembly exhibited excellent bioactivity in reducing the macrophage foam cell phenotype (Fig. 10b). This formulation promoted oxLDL internalization in macrophages, mitigated endothelial inflammation, and halted atherosclerosis progress [172]. In a subsequent study, Petersen and co-workers investigated the anti-atherogenic mechanism of the nanoformulation. They found that the mucin acid core-shell nanoparticles repress oxLDL internalization by macrophages via downregulation of surface expression of the scavenging receptors, SRA, and CD36, which are involved in oxLDL uptake [173].
Nanoparticle-based gene therapy has profound implications for clinical medicine. Utilizing PEI to form complexes with plasmids for gene delivery has been thoroughly studied due to its safety and high transfection efficiency. In one study, Santos et al. tested the bioactivity and transfection efficiency of (lPEI)/DNA polyplexes in BALB/c mice [111], and found that DNA samples prepared using high flow-rate FNC exhibited great gene transfection efficiency. In another example, oral DNA delivery using the lipoplex DOTAP/CHO/DNA showed better transfection efficiency than when using the naked PEI/DNA polyplexes (Fig. 10c) [134]. In a recent study, Hu and co-workers studied the fundamentals of the kinetic mixing in a CIJM for assembly of lPEI/pDNA. By controlling the FNC kinetic conditions, 1.3–21.8 pDNA were assembled into each nanocomplex, resulting in particle size of 35–130 nm. Nanocomplexes with intermediate plasmid payload (6–10 plasmids per particle) show the best transgene efficiency in the lungs of healthy and tumor-bearing murine models. Nanocomplexes with higher plasmid payloads lead to larger and more heterogeneous particles. Of note, lower toxicity in the liver was achieved without compromising transfection efficiency by reducing the amount of cationic lPEI in the nanocomplex [112]. Importantly, successful clinical translation of the flash-based nanoformulation depends on the prolonged and stable shelf-life and good reconstitution ability of the nanoformulation. The flash-based lPEI/pDNA nanocomplexes could be easily reconstituted in water without any sonication from lyophilization. The bioactivity studies show no statistical difference in terms of transfection efficacy between the lyophilized form and freshly-made ones. Again, all these studies confirm the versatility and flexibility of FNC-based technology on drug and gene delivery.
Nanocomplexes formulated by FNC also extend the scope of protein delivery. Oral delivery of insulin reduces cost and improves patient compliance. A sequential three-step FNC system was used to produce an HPMCP-protected HA-coated L-penetratin CPP/insulin nanocomplex. The outermost pH-sensitive HPMCP layer protects the insulin from the acidic stomach environment and disassociates at the higher pH of the small intestine. HA minimizes the interference of the mucosal layer and facilitates nanoparticle mucosa penetration. CPP aids the insulin in crossing the gut epithelia to guarantee insulin delivery into the bloodstream. These nanocomplexes exhibited high acid stability, epithelial penetration, and blood sugar control in the in vitro and in vivo and pharmacokinetic studies [29].
Moreover, nanoparticle vaccines have the advantages of lower risk, reduced side effects, and less immunogenicity versus free drugs. Immune modulation can be achieved by exploiting recombinant techniques to create subunit vaccines that optimize immune responses. Qiao et al. recently applied FNC to formulate a nanovaccine to prevent hand-foot-mouth disease, which is caused by the enterovirus antigen VP1. The adjuvants TNF-α and CpG were combined with chitosan and heparin to form nanovaccines with high size homogeneity, which prolongs lymph node retention. Strong Th1 and Th2 responses were illustrated after the adjuvant co-delivery. A lethal viral challenge following nanoparticle vaccination also showed comparable protection to that of a viral vaccine in a murine model [119]. In a more recent study, the tannic acid-associated antigen was used to complex with adjuvant (CpG or interferon-α) via hydrogen bonding through FNC. The formulated nanocomplexes serve as the vaccines to combat RBV-associated tumors [94]. Together, FNC provides researchers a promising platform in drug, protein and gene delivery with great multifunctionalities.
Disease imaging and theranostics
Effective diagnostic outcomes hinge upon the performance of bioimaging molecules. In the last five years, FNP has been used to formulate quantum dots [123][127], fluorescent molecules [174–180], and metal oxides [154,141] for diagnostic and imaging applications. Diverse fluorophore-loaded nanoparticles ranging from 30–800 nm with fluorescence emissions from 370–720 nm have been produced. For example, the hydrophobic fluorophore 6,13-bis(triisopropylsilylethynyl) (TIPS) pentacene was encapsulated into biodegradable PEO-b-PDLLA, and the nanoparticle surface was modified with streptavidin for targeted delivery and theranostics [61]. Binary organic nanoparticles consisted of two NIR-absorbing small molecules, quatterylene diimide and vanadyl phthalocyanine, were co-encapsulated in PS-b-PEG shells for photoacoustic imaging and photo-ablative therapeutic applications. Both ultrafast and delayed photoinduced charge transfer was also achieved due to the presence of the two NIR-absorbing molecules [174]. Co-delivery of imaging and therapeutic agents provides new strategies in theranostics. For example, coordination-driven FNC (cFNC) was introduced to rapidly fabricate metal-phenolic doxorubicin nanoparticles coated with a layer of HA showed good anti-cancer effect in MCF tumor-bearing mice. Paramagnetic ion iron (III) was used to coordinate doxorubicin-based metal-phenolic self-assembly to achieve pH-sensitive CD44+ receptor-targeted DOX delivery at a tumor site in a tumor theranostic [154]. Importantly, photothermal therapy (PTT) and photodynamic therapy (PDT) are recently developed nanomedical therapies whose efficacy and minimum invasiveness drive development of new biomaterials and fabrication methods. For example, inorganic materials such as ceramics can act as upconversion phosphors (UCPs) to generate infrared radiation and light excitation. Nanoparticles containing the rare-earth upconversion crystal NaYF4 :Yb3+, Er3+ showed anti-photobleaching properties under NIR exposure [176]. Effective HeLa cell killing within 45 min was observed using 134 W cm−2 NIR light (978 nm) using PEG-modified UCPs [126].
Nanoparticle size impacts cell uptake, the EPR effect, and stability, and thus therapeutic and diagnostic outcomes. Until recently, the “size gate” effect on lymph node (LN) drainage and trafficking remained elusive. Howard et al. elucidated the effect of nanoparticle size on LN trafficking and retention in mice [182]. Three sets of PEG-b-PLGA nanoparticles with sizes of 20, 40, and 100 nm were labeled with NIR dye. PLGA nanoparticles <20 nm showed the greatest penetration in both proximal and distal LNs. These small nanoparticles also tended to have longer retention in paracortex, and greater uptake by dendric cells (DC) post-s.c. administration. These results provide a reference for future design of vaccines and theranostics. Particle distribution is essential to understand particle uptake mechanisms and monitor particle profile in vivo. Quantitation of the biodistribution of nanoparticles can be performed using inductively-coupled plasma mass spectrometry (ICP-MS) of iodine as an alternative to the current gold standard, radiolabeling ex vivo bioimaging. In vivo nanoparticle tracking and monitoring requires stable iodine formulations. Di-iodination nanoparticles used in ICP-MS and X-ray contrast were produced to scale using FNP, and 45 wt% iodine with an ICP-MS sensitivity of 2 μg/mL was achieved. An attenuation rate of 4.8 Hu80 per mM matched commercially available agents [183]. Flash-based technology provides uniform imaging agents, which in turn is the key for high-quality nanoparticle tracking and hence leads to reproducible imaging outcomes.
Multienzyme colocalization and immobilization are useful for understanding and simulating catalytic mechanisms. In one study, PLGA coprecipitates with CdSe quantum dots to form micelles for enzyme colocalization. The enzymes horseradish peroxidase (HRP) and glucose oxidase (GOX) labeled with fluorescent dyes were then conjugated to the micelles. Nanometer-scale colocalization between the quantum dots and enzymes revealed by FRET illustrates the feasibility and versatility of multienzyme characterization [123].
Aggregation-induced emission (AIE) fluorophores are also attractive for bioimaging due to their unique light emission mechanism. AIE fluorophores emit almost no fluorescence unless aggregated; however, aggregation can also cause quenching. Thus, it is critical to precisely control the degree of AIE fluorophore aggregation. Traditional methods for AIE nanoparticle preparation rely on slow self-assembly processes which lead to broad particle size distributions. By using FNP, AIE nanoparticle size and fluorescence properties can be precisely controlled. AIE configuration can also be tuned to improve imaging outcomes, for example, to induce a redshift in fluorescence emission. In one study, AIE building blocks were designed to compose of tricyano-methylene-pyridine (TCM), which contains several donor-π-acceptor (D-π-A) dyes. This endows the AIE nanoparticles with excellent photostability, specific NIR fluorescence (656–768 nm), and a wide Stokes shift. Authors then demonstrated using saponin-based α-hederin nanoprecipitates with TCM-AIE fluorophore to achieve excellent biocompatibility and rapid HeLa cell imaging [180]. In another study, AIE-EDP nanoparticles were constructed with particle size corresponding to fluorescence wavelength [184].
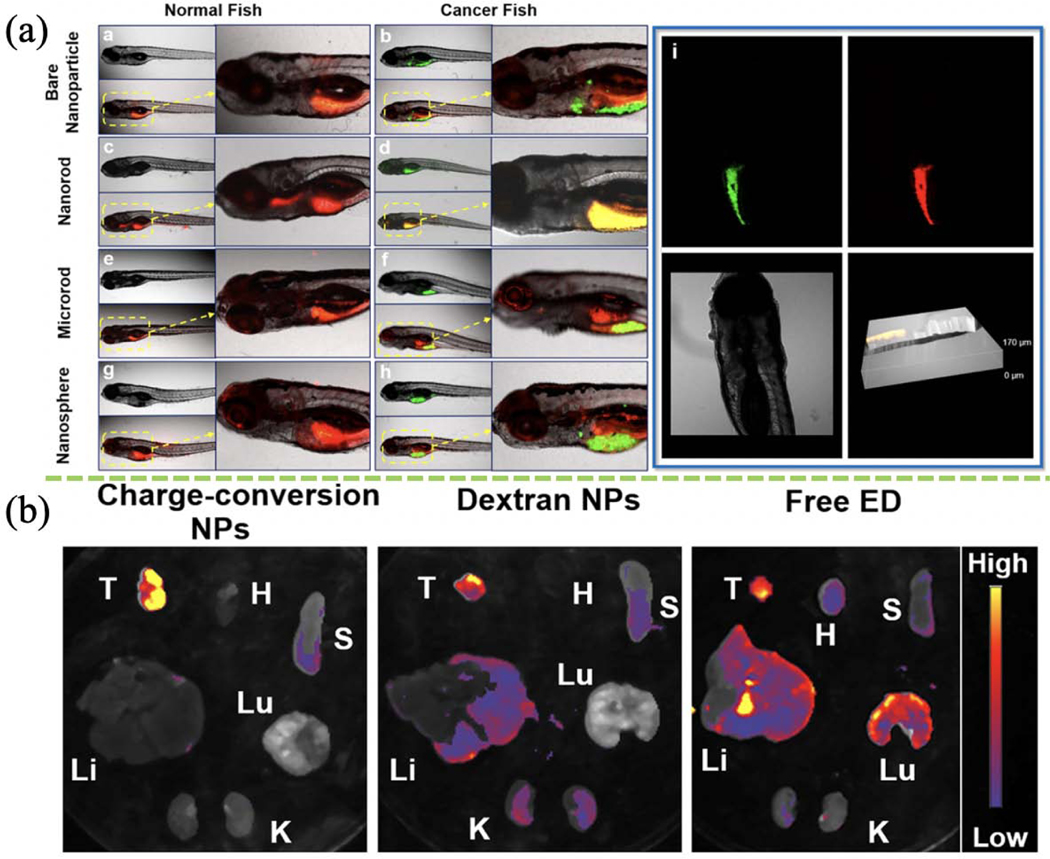
Imaging quality depends not only on the size of the nanoparticle, but also on nanoparticle morphology (e.g. aspect ratio and shape). Wang and colleagues produced a quinolinemalononitrile-encapsulated dextran-b-PLA nanoformulation with distinctive morphology for AIE cancer imaging [138]. Polymer-attached nanorods, microrods, nanospheres, and naked nanoparticles were evaluated for their in vivo tumor-targeting effect in a GFP-hepatocarcinoma zebrafish model (Fig. 11a). Nanorods with an aspect ratio of ~5 showed the best uptake and targeting performance in cancer cells. ED-encapsulated dextran-b-PLGA and PDMAEMA-b-PCL nanorods with the charge-conversion ability and a fluorescence emission spectrum of 570–620 nm showed enhanced cancer and organ imaging (Fig. 11b). The negative-to-positive charge-conversion of nanorods in the acidic tumor environment facilitates nanorod tumor uptake. The fluorescence intensity of nanorods was also pH-dependent. These findings are consistent with those of Wang et al. described above [181]. The flexibility of the self-assembly technique in producing nanoparticles can vastly benefit cellular imaging in many ways. Hereto, the flash-based technology offers excellent versatility in preparing bioimaging agents with different physicochemical properties, morphology, and consistent imaging outcomes.
FIGURE 11.
(a) Diagnostic effects of different nanoparticles fabricated using FNP on intact normal or tumor-bearing zebrafish. (Reprinted with permission from Ref. [138]. 2018, American Chemical Society.) (b) NPs formed using FNP-based charge-conversion enhance cancer imaging in tumor and other organs ex vivo (T: tumor, H: heart, Lu: lung, S: spleen, Li: liver, K: kidney). (Reprinted with permission from Ref. [181]. 2018, American Chemical Society.)
Importantly, the toxicity of many bright fluorescence probes used in vivo bioimaging is an unresolved problem. Fluorescent organic nanoparticles (FONs) were developed to reduce toxicity. Yan et al. combined FNP and freeze-drying with a plasticizing surfactant to manipulate FON crystallization, yielding stable nanocrystals with higher brightness and less toxicity for improved visualization in macrophages, mast cells, and vasculature in tumor sites in mouse models [185]. In another example, PEGylated multimodal composite nanocarriers (CNCs) prepared by FNP improve MRI and NIR visualization of cancer metastasis. CNCs encapsulating iron oxide-based nanocrystals and NIR fluorophores (PZn3) provide T2 contrast ability. The nanocarriers were modified with liver-targeting hydroxyl ligands and the in vivo timepoint of tumor metastasis was identified [125].
Of note, PET radionuclide imaging is a sensitive and low-threshold process for disease detection. Despite its high detection sensitivity, isobaric decay of erratic radioisotopes profoundly affects PET imaging. The nanoparticulate system vastly improves PET imaging stability and sensitivity through targeted delivery, prolonged circulation time, and the EPR effect. In one example, FNP was used to assemble PEGylated phthalocyanine-loaded nanoparticles without toxic radiotracer which greatly reduces the safety risk. Those phthalocyanine cores serve as reservoirs for 64Cu chelation, a common reagent in PET imaging. Enhanced radionuclide chelation (standard rate of 1845 M/h) was obtained [141]. Moreover, nanoparticle core composition, fluidity, and size all play roles in chelation kinetics and can be well-controlled in FNP processes. When designing a multifunctional imaging agent in the future, surface modification such as conjugating targeting ligands can also be considered for better imaging systems. Taken together, flash-based self-assembly is a great platform for encapsulating multiple types of imaging agents, and its versatility, flexibility, and consistency all render the desired imaging outcome of becoming more practical.
Concluding remarks
The rapid development of biomedicine requires the manufacturing of nanoparticle-based medicine in a designable and scalable manner. Flash-based self-assembly using rapid mixing technology provides a more versatile, scalable, reproducible, and robust approach to nanoformulation than traditional self-assembly techniques such as layer-by-layer, bulk mixing, controlled growth, and hydrodynamic focusing. Flash-based nanofabrication enables users to produce a variety of nanoproducts at scale with ease, changing the way materials manufacturing is approached in medicine. Nanoformulation optimization and screening typically requires copious amounts of therapeutics and biologics. Flash-based self-assembly reduces the amounts of reagents consumed while improving the homogeneity and stability of the product. Markwalter and Prud’homme developed the FNP strategy driven by phase inversion to fabricate nanoformulations with high loading, control of particle size, and multifunctional features. Mao et al. recently expanded the technique to FNC to fabricate nanoformulations driven by polyelectrolyte, reaping similar benefits of size control and compositional uniformity. FNP involves the mixing of solvent pairs, typically an organic solvent dissolving a hydrophobic polymer and hydrophobic drugs, and water as a poor solvent to induce the conversion of the continuous organic phase into a discrete phase containing drug-loaded nanoparticles. FNC typically involves charge interaction in an all aqueous system to form nanoparticles comprising a polar and charged carrier entrapping a nucleic acid and peptide cargo. Together, FNP and FNC pave the way toward manufacturing a wide variety of nanomedicines for use in bioimaging, diagnostics, and therapeutics.
Despite tremendous progress, challenges remain that have prevented the broader application of flash-based nanofabrication. For example, there is a need to better understand the relationship between flash-based processes and nanoformulation performance. Computational modeling and in silico experiments can also bring us trenchant insights on nanoformulation designing and particle interactions. Also, the development of a transparent device might be a useful tool for real-time visualization of the process. Such a design might contribute to developing large MIVM for scaling up production of small molecule drug-based nanoformulations for industrial translation, as well as for developing small MIVM for small scale production of personalized medicine and preserving precious reagents. New devices with different designs or connections that tailored to the self-assembly-based fabrication of specific nanomedicines are needed. This endeavor will require a better understanding of the driving forces during flash-based self-assembly. It is also critical to consider the cost factors at the front end of clinical translation, from material design to large-scale production. Other crucial factors such as efficiency, stability, and safety must also be considered. New flash-based nanomedicines fabricated using FNC via single and mixable driving forces are expected. More effective and scalable self-assembly strategies in this multidisciplinary field will lead to more flash-based nanomedicines emerging in the next decade.
Acknowledgments
The authors acknowledge funding support from NIH (UH3TR002151, UG3NS115598, RO1AR073935), USAMR W81XWH1910463, and HR00111920009 for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wang W, Zhou X, Bian Y, Wang S, Chai Q, Guo Z, Wang Z, Zhu P, Peng H, Yan X, Li W, Fu Y-X, Zhu M, Nat. Nanotechnol 15 (2020) 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shao D, Li M, Wang Z, Zheng X, Lao Y-H, Chang Z, Zhang F, Lu M, Yue J, Hu H, Yan H, Chen L, Dong W, Leong KW, Adv. Mater 30 (2018) 1801198. [DOI] [PubMed] [Google Scholar]
- [3].Merrifield DL, Shaw BJ, Harper GM, Saoud IP, Davies SJ, Handy RD, Henry TB, Environmental Pollution 174 (2013) 157–163. [DOI] [PubMed] [Google Scholar]
- [4].Yan H, Shao D, Lao Y, Li M, Hu H, Leong KW, Adv. Sci (2019) 1900605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hickey JW, Santos JL, Williford J-M, Mao H-Q, Journal of Controlled Release 219 (2015) 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lim J-M, Swami A, Gilson LM, Chopra S, Choi S, Wu J, Langer R, Karnik R, Farokhzad OC, ACS Nano 8 (2014) 6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ding S, Attia MF, Wallyn J, Taddei C, Serra CA, Anton N, Kassem M, Schmutz M, Er-Rafik M, Messaddeq N, Collard A, Yu W, Giordano M, Vandamme TF, Langmuir 34 (2018) 1981–1991. [DOI] [PubMed] [Google Scholar]
- [8].Shamay Y, Shah J, Işık M, Mizrachi A, Leibold J, Tschaharganeh DF, Roxbury D, Budhathoki-Uprety J, Nawaly K, Sugarman JL, Baut E, Neiman MR, Dacek M, Ganesh KS, Johnson DC, Sridharan R, Chu KL, Rajasekhar VK, Lowe SW, Chodera JD, Heller DA, Nature Mater 17 (2018) 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li N, Nikoubashman A, Panagiotopoulos AZ, Langmuir 35 (2019) 3780–3789. [DOI] [PubMed] [Google Scholar]
- [10].Song J, Yuan C, Jiao T, Xing R, Yang M, Adams DJ, Yan X, Small 16 (2020) 1907309. [DOI] [PubMed] [Google Scholar]
- [11].Ye Z, Zhu X, Acosta S, Kumar D, Sang T, Aparicio C, Nanoscale 11 (2019) 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lombardi L, Shi Y, Falanga A, Galdiero E, de Alteriis E, Franci G, Chourpa I, Azevedo HS, Galdiero S, Biomacromolecules 20 (2019) 1362–1374. [DOI] [PubMed] [Google Scholar]
- [13].Song J, Xing R, Jiao T, Peng Q, Yuan C, Möhwald H, Yan X, ACS Appl. Mater. Interfaces 10 (2018) 2368–2376. [DOI] [PubMed] [Google Scholar]
- [14].Yan R, Hu Y, Liu F, Wei S, Fang D, Shuhendler AJ, Liu H, Chen H-Y, Ye D, J. Am. Chem. Soc 141 (2019) 10331–10341. [DOI] [PubMed] [Google Scholar]
- [15].Yuan Y, Zhang J, Qi X, Li S, Liu G, Siddhanta S, Barman I, Song X, McMahon MT, Bulte JWM, Nat. Mater 18 (2019) 1376–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Le Z, Liu Z, Sun L, Liu L, Chen Y, ACS Appl. Bio Mater (2020) acsabm.0c00616. [DOI] [PubMed] [Google Scholar]
- [17].Ma K, Xing R, Jiao T, Shen G, Chen C, Li J, Yan X, ACS Appl. Mater. Interfaces 8 (2016) 30759–30767. [DOI] [PubMed] [Google Scholar]
- [18].Xing R, Liu K, Jiao T, Zhang N, Ma K, Zhang R, Zou Q, Ma G, Yan X, Adv. Mater 28 (2016) 3669–3676. [DOI] [PubMed] [Google Scholar]
- [19].Zhang R, Xing R, Jiao T, Ma K, Chen C, Ma G, Yan X, ACS Appl. Mater. Interfaces 8 (2016) 13262–13269. [DOI] [PubMed] [Google Scholar]
- [20].Zhu H, Wang H, Shi B, Shangguan L, Tong W, Yu G, Mao Z, Huang F, Nat Commun 10 (2019) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Le D, Dilger M, Pertici V, Diabaté S, Gigmes D, Weiss C, Delaittre G, Angew. Chem. Int. Ed 58 (2019) 4725–4731. [DOI] [PubMed] [Google Scholar]
- [22].Fu Z, Li L, Li F, Bhutto RA, Niu X, Liu D, Guo X, Ind. Eng. Chem. Res (2020) acs.iecr.0c02668. [Google Scholar]
- [23].Kang C, Honciuc A, Chem. Mater 31 (2019) 1688–1695. [Google Scholar]
- [24].Han U, Kim YJ, Kim W, Park JH, Hong J, Nanoscale 11 (2019) 13541–13551. [DOI] [PubMed] [Google Scholar]
- [25].Aubin J, Ferrando M, Jiricny V, Chemical Engineering Science 65 (2010) 2065–2093. [Google Scholar]
- [26].Li K-K, Zhang X, Huang Q, Yin S-W, Yang X-Q, Wen Q-B, Tang C-H, Lai F-R, Journal of Food Engineering 127 (2014) 103–110. [Google Scholar]
- [27].Saad WS, Prud’homme RK, Nano Today 11 (2016) 212–227. [Google Scholar]
- [28].Santos JL, Ren Y, Vandermark J, Archang MM, Williford J-M, Liu H-W, Lee J, Wang T-H, Mao H-Q, Small 12 (2016) 6214–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He Z, Liu Z, Tian H, Hu Y, Liu L, Leong KW, Mao H-Q, Chen Y, Nanoscale 10 (2018) 3307–3319. [DOI] [PubMed] [Google Scholar]
- [30].Ding S, Anton N, Vandamme TF, Serra CA, Expert Opinion on Drug Delivery 13 (2016) 1447–1460. [DOI] [PubMed] [Google Scholar]
- [31].Williford J-M, Santos JL, Shyam R, Mao H-Q, Biomater. Sci 3 (2015) 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Johnson BK, Prud’homme RK, AIChE J. 49 (2003) 2264–2282. [Google Scholar]
- [33].D’Addio SM, Prud’homme RK, Advanced Drug Delivery Reviews 63 (2011) 417–426. [DOI] [PubMed] [Google Scholar]
- [34].Han J, Zhu Z, Qian H, Wohl AR, Beaman CJ, Hoye TR, Macosko CW, Journal of Pharmaceutical Sciences 101 (2012) 4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu Y, Cheng C, Liu Y, Prud’homme RK, Fox RO, Chemical Engineering Science 63 (2008) 2829–2842. [Google Scholar]
- [36].Cheng JC, Olsen MG, Fox RO, Appl. Phys. Lett 94 (2009) 204104. [Google Scholar]
- [37].Le L, Bokare A, Erogbogbo F, Mater. Res. Express 6 (2018) 025403. [Google Scholar]
- [38].Nagasawa H, Aoki N, Mae K, Chem. Eng. Technol 28 (2005) 324–330. [Google Scholar]
- [39].Ding S, Serra CA, Anton N, Yu W, Vandamme TF, International Journal of Pharmaceutics 558 (2019) 1–8. [DOI] [PubMed] [Google Scholar]
- [40].Kamio E, Ono T, Yoshizawa H, Lab Chip 9 (2009) 1809. [DOI] [PubMed] [Google Scholar]
- [41].Belliveau NM, Huft J, Lin PJ, Chen S, Leung AK, Leaver TJ, Wild AW, Lee JB, Taylor RJ, Tam YK, Hansen CL, Cullis PR, Molecular Therapy - Nucleic Acids 1 (2012) e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cheung CCL, Al-Jamal WT, International Journal of Pharmaceutics 566 (2019) 687–696. [DOI] [PubMed] [Google Scholar]
- [43].Zelphati O, Nguyen C, Ferrari M, Felgner J, Tsai Y, Felgner P, Gene Ther 5 (1998) 1272–1282. [DOI] [PubMed] [Google Scholar]
- [44].Kasper JC, Schaffert D, Ogris M, Wagner E, Friess W, European Journal of Pharmaceutics and Biopharmaceutics 77 (2011) 182–185. [DOI] [PubMed] [Google Scholar]
- [45].Ho Y-P, Grigsby CL, Zhao F, Leong KW, Nano Lett. 11 (2011) 2178–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jeong GS, Chung S, Kim C-B, Lee S-H, Analyst 135 (2010) 460. [DOI] [PubMed] [Google Scholar]
- [47].Adjei IM, Peetla C, Labhasetwar V, Nanomedicine 9 (2014) 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu Y, Kathan K, Saad W, Prud’homme RK, Phys. Rev. Lett 98 (2007) 036102. [DOI] [PubMed] [Google Scholar]
- [49].Allen S, Journal of Controlled Release (2017) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grundy LS, Lee VE, Li N, Sosa C, Mulhearn WD, Liu R, Register RA, Nikoubashman A, Prud’homme RK, Panagiotopoulos AZ, Priestley RD, ACS Nano 12 (2018) 4660–4668. [DOI] [PubMed] [Google Scholar]
- [51].Zhu Z, Xu P, Fan G, Liu N, Xu S, Li X, Xue H, Shao C, Guo Y, Chemical Engineering Journal 356 (2019) 877–885. [Google Scholar]
- [52].Johnson BK, Prud’homme RK, Aust. J. Chem 56 (2003) 1021. [Google Scholar]
- [53].Shen H, Hong S, Prud’homme RK, Liu Y, J Nanopart Res 13 (2011) 4109–4120. [Google Scholar]
- [54].Johnson BK, Prud’homme RK, Phys. Rev. Lett 91 (2003) 118302. [DOI] [PubMed] [Google Scholar]
- [55].Pagels RF, Edelstein J, Tang C, Prud’homme RK, Nano Lett. 18 (2018) 1139–1144. [DOI] [PubMed] [Google Scholar]
- [56].Feng J, Markwalter CE, Tian C, Armstrong M, Prud’homme RK, J Transl Med 17 (2019) 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhu Z, Anacker JL, Ji S, Hoye TR, Macosko CW, Prud’homme RK, Langmuir 23 (2007) 10499–10504. [DOI] [PubMed] [Google Scholar]
- [58].Wang M, Xu Y, Wang J, Liu M, Yuan Z, Chen K, Li L, Prud’homme RK, Guo X, Chem. Lett 44 (2015) 1688–1690. [Google Scholar]
- [59].Zhu Z, Mol. Pharmaceutics 11 (2014) 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fu Z, Li L, Wang M, Guo X, Colloid Polym Sci 296 (2018) 935–940. [Google Scholar]
- [61].Jo A, Zhang R, Allen IC, Riffle JS, Davis RM, Langmuir 34 (2018) 15783–15794. [DOI] [PubMed] [Google Scholar]
- [62].Turino LN, Stella B, Dosio F, Luna JA, Barresi AA, Drug Development and Industrial Pharmacy 44 (2018) 934–941. [DOI] [PubMed] [Google Scholar]
- [63].Pagels RF, Edelstein J, Tang C, Prud’homme RK, Nano Lett. 18 (2018) 1139–1144. [DOI] [PubMed] [Google Scholar]
- [64].Wan KY, Wong KW, Chow AHL, Chow SF, International Journal of Pharmaceutics 542 (2018) 221–231. [DOI] [PubMed] [Google Scholar]
- [65].Wang M, Lin S, Wang J, Liu L, Zhou W, Ahmed RB, Hu A, Guo X, Cohen Stuart MA, Ind. Eng. Chem. Res 57 (2018) 11911–11919. [Google Scholar]
- [66].Rode García T, García Ac A, Lalloz A, Lacasse F-X, Hildgen P, Rabanel J-M, Banquy X, Langmuir 34 (2018) 5772–5780. [DOI] [PubMed] [Google Scholar]
- [67].Gavi E, Rivautella L, Marchisio DL, Vanni M, Barresi AA, Baldi G, Chemical Engineering Research and Design 85 (2007) 735–744. [Google Scholar]
- [68].Icardi M, Gavi E, Marchisio DL, Barresi AA, Olsen MG, Fox RO, Lakehal D, Chemical Engineering Journal 166 (2011) 294–305. [Google Scholar]
- [69].Cheng JC, Fox RO, Ind. Eng. Chem. Res 49 (2010) 10651–10662. [Google Scholar]
- [70].Di Pasquale N, Marchisio DL, Barresi AA, Chemical Engineering Science 84 (2012) 671–683. [Google Scholar]
- [71].Shi Y, Somashekar V, Fox RO, Olsen MG, J. Micromech. Microeng 21 (2011) 115006. [Google Scholar]
- [72].Cheng JC, Vigil RD, Fox RO, Journal of Colloid and Interface Science 351 (2010) 330–342. [DOI] [PubMed] [Google Scholar]
- [73].Nikoubashman A, Lee VE, Sosa C, Prud’homme RK, Priestley RD, Panagiotopoulos AZ, ACS Nano 10 (2016) 1425–1433. [DOI] [PubMed] [Google Scholar]
- [74].Keßler S, Drese K, Schmid F, Polymer 126 (2017) 9–18. [Google Scholar]
- [75].Marchisio DL, Computers & Chemical Engineering 33 (2009) 408–420. [Google Scholar]
- [76].Gavi E, Marchisio DL, Barresi AA, Chemical Engineering Science 62 (2007) 2228–2241. [Google Scholar]
- [77].Liu Z, Hill JC, Fox RO, Passalacqua A, Olsen MG, Chemical Engineering Science 193 (2019) 66–75. [Google Scholar]
- [78].Liu Z, Passalacqua A, Olsen MG, Fox RO, Hill JC, AIChE J. 62 (2016) 2570–2578. [Google Scholar]
- [79].Morozova TI, Lee VE, Bizmark N, Datta SS, Prud’homme RK, Nikoubashman A, Priestley RD, ACS Cent. Sci 6 (2020) 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kozuch DJ, Ristroph K, Prud’homme RK, Debenedetti PG, ACS Nano 14 (2020) 6097–6106. [DOI] [PubMed] [Google Scholar]
- [81].Lavino AD, Di Pasquale N, Carbone P, Marchisio DL, Chemical Engineering Science 171 (2017) 485–494. [Google Scholar]
- [82].Santos JL, Ren Y, Vandermark J, Archang MM, Williford J-M, Liu H-W, Lee J, Wang T-H, Mao H-Q, Small 12 (2016) 6214–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ke X, Tang H, Mao H-Q, International Journal of Pharmaceutics 564 (2019) 273–280. [DOI] [PubMed] [Google Scholar]
- [84].Qiao D, Liu L, Chen Y, Xue C, Gao Q, Mao H-Q, Leong KW, Chen Y, Nano Lett. 18 (2018) 3007–3016. [DOI] [PubMed] [Google Scholar]
- [85].Liu Z, Le Z, Lu L, Zhu Y, Yang C, Zhao P, Wang Z, Shen J, Liu L, Chen Y, Nanoscale 11 (2019) 9410–9421. [DOI] [PubMed] [Google Scholar]
- [86].He Z, Santos JL, Tian H, Huang H, Hu Y, Liu L, Leong KW, Chen Y, Mao H-Q, Biomaterials 130 (2017) 28–41. [DOI] [PubMed] [Google Scholar]
- [87].He Z, Liu Z, Tian H, Hu Y, Liu L, Leong KW, Mao H-Q, Chen Y, Nanoscale 10 (2018) 3307–3319. [DOI] [PubMed] [Google Scholar]
- [88].Tian H, He Z, Sun C, Yang C, Zhao P, Liu L, Leong KW, Mao H-Q, Liu Z, Chen Y, Adv. Healthcare Mater 7 (2018) 1800285. [DOI] [PubMed] [Google Scholar]
- [89].He Z, Hu Y, Nie T, Tang H, Zhu J, Chen K, Liu L, Leong KW, Chen Y, Mao H-Q, Acta Biomaterialia 81 (2018) 195–207. [DOI] [PubMed] [Google Scholar]
- [90].Sun L, Liu Z, Tian H, Le Z, Liu L, Leong KW, Mao H-Q, Chen Y, Biomacromolecules 20 (2019) 528–538. [DOI] [PubMed] [Google Scholar]
- [91].Hu Y, He Z, Hao Y, Gong L, Pang M, Howard GP, Ahn H-H, Brummet M, Chen K, Liu H, Ke X, Zhu J, Anderson CF, Cui H, Ullman CG, Carrington CA, Pomper MG, Seo J-H, Mittal R, Minn I, Mao H-Q, ACS Nano 13 (2019) 10161–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Sun L, Le Z, He S, Liu J, Liu L, Leong KW, Mao H-Q, Liu Z, Chen Y, Mol. Pharmaceutics 17 (2020) 757–768. [DOI] [PubMed] [Google Scholar]
- [93].He Z, Hu Y, Gui Z, Zhou Y, Nie T, Zhu J, Liu Z, Chen K, Liu L, Leong KW, Cao P, Chen Y, Mao H-Q, Journal of Controlled Release 301 (2019) 119–128. [DOI] [PubMed] [Google Scholar]
- [94].Liu H, Chen H, Liu Z, Le Z, Nie T, Qiao D, Su Y, Mai H, Chen Y, Liu L, Biomaterials 255 (2020) 120158. [DOI] [PubMed] [Google Scholar]
- [95].Mejia-Ariza R, Huskens J, J. Mater. Chem B 2 (2014) 210–216. [DOI] [PubMed] [Google Scholar]
- [96].Sun L, Liu Z, Tian H, Le Z, Liu L, Leong KW, Mao H-Q, Chen Y, Biomacromolecules 20 (2019) 528–538. [DOI] [PubMed] [Google Scholar]
- [97].Johnson BK, Prud’homme RK, Aust. J. Chem 56 (2003) 1021. [Google Scholar]
- [98].Feng J, Markwalter CE, Tian C, Armstrong M, Prud’homme RK, J Transl Med 17 (2019) 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Markwalter CE, Prud’homme RK, Journal of Pharmaceutical Sciences 107 (2018) 2465–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Shen H, Hong S, Prud’homme RK, Liu Y, J Nanopart Res 13 (2011) 4109–4120. [Google Scholar]
- [101].Prud’homme R, Saad W, Mayer L, 2 (2006) 3. [Google Scholar]
- [102].Pustulka KM, Wohl AR, Lee HS, Michel AR, Han J, Hoye TR, McCormick AV, Panyam J, Macosko CW, Mol. Pharmaceutics 10 (2013) 4367–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tam YT, To KKW, Chow AHL, Colloids and Surfaces B: Biointerfaces 139 (2016) 249–258. [DOI] [PubMed] [Google Scholar]
- [104].Bteich J, McManus SA, Ernsting MJ, Mohammed MZ, Prud’homme RK, Sokoll KK, Mol. Pharmaceutics 14 (2017) 3998–4007. [DOI] [PubMed] [Google Scholar]
- [105].Pinkerton NM, Grandeury A, Fisch A, Brozio J, Riebesehl BU, Prud’homme RK, Mol. Pharmaceutics 10 (2013) 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chiou H, Chan H-K, Prud’homme RK, Raper JA, Drug Development and Industrial Pharmacy 34 (2008) 59–64. [DOI] [PubMed] [Google Scholar]
- [107].Liu Y, Tong Z, Prud’homme RK, Pest. Manag. Sci 64 (2008) 808–812. [DOI] [PubMed] [Google Scholar]
- [108].Gutierrez L, Gomez L, Irusta S, Arruebo M, Santamaria J, Chemical Engineering Journal 171 (2011) 674–683. [Google Scholar]
- [109].Fu Z, Li L, Wang Y, Chen Q, Zhao F, Dai L, Chen Z, Liu D, Guo X, Chemical Engineering Journal 382 (2020) 122905. [Google Scholar]
- [110].Pal S, Madane K, Kulkarni AA, Chemical Engineering Journal 369 (2019) 1161–1171. [Google Scholar]
- [111].Santos JL, Ren Y, Vandermark J, Archang MM, Williford J-M, Liu H-W, Lee J, Wang T-H, Mao H-Q, Small 12 (2016) 6214–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hu Y, He Z, Hao Y, Gong L, Pang M, Howard GP, Ahn H-H, Brummet M, Chen K, Liu H, Ke X, Zhu J, Anderson CF, Cui H, Ullman CG, Carrington CA, Pomper MG, Seo J-H, Mittal R, Minn I, Mao H-Q, ACS Nano 13 (2019) 10161–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Tang C, Sosa CL, Pagels RF, Priestley RD, Prud’homme RK, J. Mater. Chem B 4 (2016) 4813–4817. [DOI] [PubMed] [Google Scholar]
- [114].He Z, Santos JL, Tian H, Huang H, Hu Y, Liu L, Leong KW, Chen Y, Mao H-Q, Biomaterials 130 (2017) 28–41. [DOI] [PubMed] [Google Scholar]
- [115].Tang C, Sosa CL, Pagels RF, Priestley RD, Prud’homme RK, J. Mater. Chem B 4 (2016) 4813–4817. [DOI] [PubMed] [Google Scholar]
- [116].Liu R, Sosa C, Yeh Y-W, Qu F, Yao N, Prud’homme RK, Priestley RD, J. Mater. Chem A 2 (2014) 17286–17290. [Google Scholar]
- [117].Zhang Z, Sun L, Liu R, Colloid Polym Sci 296 (2018) 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].He Y, Wang B, Hu X, Zhang X, Sun L, Priestley RD, Liu R, Colloid Polym Sci 295 (2017) 521–527. [Google Scholar]
- [119].Qiao D, Liu L, Chen Y, Xue C, Gao Q, Mao H-Q, Leong KW, Chen Y, Nano Lett. 18 (2018) 3007–3016. [DOI] [PubMed] [Google Scholar]
- [120].Pinkerton NM, Behar L, Hadri K, Amouroux B, Mingotaud C, Talham DR, Chassaing S, Marty J-D, Nanoscale 9 (2017) 1403–1408. [DOI] [PubMed] [Google Scholar]
- [121].Kather M, Ritter F, Pich A, Chemical Engineering Journal 344 (2018) 375–379. [Google Scholar]
- [122].Tripodi E, Lazidis A, Norton IT, Spyropoulos F, Ind. Eng. Chem. Res 58 (2019) 14859–14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Jia F, Zhang Y, Narasimhan B, Mallapragada SK, Langmuir 28 (2012) 17389–17395. [DOI] [PubMed] [Google Scholar]
- [124].Zhang Y, Clapp AR, RSC Adv. 4 (2014) 48399–48410. [Google Scholar]
- [125].Pinkerton NM, Gindy ME, Calero-DdelC VL, Wolfson T, Pagels RF, Adler D, Gao D, Li S, Wang R, Zevon M, Yao N, Pacheco C, Therien MJ, Rinaldi C, Sinko PJ, Prud’homme RK, Adv. Healthcare Mater 4 (2015) 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shan J, Budijono SJ, Hu G, Yao N, Kang Y, Ju Y, Prud’homme RK, Adv. Funct. Mater 21 (2011) 2488–2495. [Google Scholar]
- [127].He Y, Fan X, Sun J, Liu R, Fan Z, Zhang Z, Chang X, Wang B, Gao F, Wang L, Chem. Commun 56 (2020) 2594–2597. [DOI] [PubMed] [Google Scholar]
- [128].Zhu Z, Xu P, Fan G, Liu N, Xu S, Li X, Xue H, Shao C, Guo Y, Chemical Engineering Journal 356 (2019) 877–885. [Google Scholar]
- [129].Allen S, Osorio O, Liu Y-G, Scott E, Journal of Controlled Release 262 (2017) 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Shi L, Shan J, Ju Y, Aikens P, Prud’homme RK, Colloids and Surfaces A: Physicochemical and Engineering Aspects 396 (2012) 122–129. [Google Scholar]
- [131].Sosa C, Liu R, Tang C, Qu F, Niu S, Bazant MZ, Prud’homme RK, Priestley RD, Macromolecules 49 (2016) 3580–3585. [Google Scholar]
- [132].Lee VE, Sosa C, Liu R, Prud’homme RK, Priestley RD, Langmuir 33 (2017) 3444–3449. [DOI] [PubMed] [Google Scholar]
- [133].Grundy LS, Lee VE, Li N, Sosa C, Mulhearn WD, Liu R, Register RA, Nikoubashman A, Prud’homme RK, Panagiotopoulos AZ, Priestley RD, ACS Nano 12 (2018) 4660–4668. [DOI] [PubMed] [Google Scholar]
- [134].He Z, Hu Y, Nie T, Tang H, Zhu J, Chen K, Liu L, Leong KW, Chen Y, Mao H-Q, Acta Biomaterialia 81 (2018) 195–207. [DOI] [PubMed] [Google Scholar]
- [135].Nie T, He Z, Zhu J, Liu L, Chen Y, Adv. Therap (2020) 2000016. [Google Scholar]
- [136].Nimbalkar P, Tabada P, Bokare A, Chung J, Mousavi M, Simon M, Erogbogbo F, MRC 9 (2019) 1053–1059. [Google Scholar]
- [137].Fang RH, Chen KNH, Aryal S, Hu C-MJ, Zhang K, Zhang L, Langmuir 28 (2012) 13824–13829. [DOI] [PubMed] [Google Scholar]
- [138].Wang M, Xu Y, Liu Y, Gu K, Tan J, Shi P, Yang D, Guo Z, Zhu W, Guo X, Cohen Stuart MA, ACS Appl. Mater. Interfaces 10 (2018) 25186–25193. [DOI] [PubMed] [Google Scholar]
- [139].Santillo G, Deorsola FA, Bensaid S, Russo N, Fino D, Chemical Engineering Journal 207–208 (2012) 322–328. [Google Scholar]
- [140].Mejia-Ariza R, Huskens J, J. Mater. Chem B 2 (2014) 210–216. [DOI] [PubMed] [Google Scholar]
- [141].Lu HD, Wang LZ, Wilson BK, McManus SA, Jumai’an J, Padakanti PK, Alavi A, Mach RH, Prud’homme RK, ACS Appl. Mater. Interfaces 10 (2018) 3191–3199. [DOI] [PubMed] [Google Scholar]
- [142].Pinkerton NM, Hadri K, Amouroux B, Behar L, Mingotaud C, Destarac M, Kulai I, Mazières S, Chassaing S, Marty J-D, Chem. Commun 54 (2018) 9438–9441. [DOI] [PubMed] [Google Scholar]
- [143].Tang C, Amin D, Messersmith PB, Anthony JE, Prud’homme RK, Langmuir 31 (2015) 3612–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Pagels RF, Edelstein J, Tang C, Prud’homme RK, Nano Lett. 18 (2018) 1139–1144. [DOI] [PubMed] [Google Scholar]
- [145].Ke X, Shelton L, Hu Y, Zhu Y, Chow E, Tang H, Santos JL, Mao H-Q, ACS Appl. Mater. Interfaces (2020) acsami.0c08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Smith RA, Walker RC, Levit SL, Tang C, Polymers 11 (2019) 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Nie T, He Z, Zhou Y, Zhu J, Chen K, Liu L, Leong KW, Mao H-Q, Chen Y, ACS Appl. Mater. Interfaces 11 (2019) 29593–29603. [DOI] [PubMed] [Google Scholar]
- [148].Le Z, Chen Y, Han H, Tian H, Zhao P, Yang C, He Z, Liu L, Leong KW, Mao H-Q, Liu Z, Chen Y, ACS Appl. Mater. Interfaces 10 (2018) 42186–42197. [DOI] [PubMed] [Google Scholar]
- [149].Le L, Bokare A, Erogbogbo F, Mater. Res. Express 6 (2018) 025403. [Google Scholar]
- [150].Zeng Z, Zhao P, Liu L, Gao X, Mao H-Q, Chen Y, ACS Appl. Mater. Interfaces 10 (2018) 24969–24974. [DOI] [PubMed] [Google Scholar]
- [151].Sun L, Liu Z, Tian H, Le Z, Liu L, Leong KW, Mao H-Q, Chen Y, Biomacromolecules 20 (2019) 528–538. [DOI] [PubMed] [Google Scholar]
- [152].Zelenková T, Mora MJ, Barresi AA, Granero GE, Fissore D, Journal of Pharmaceutical Sciences 107 (2018) 1157–1166. [DOI] [PubMed] [Google Scholar]
- [153].Tian H, He Z, Sun C, Yang C, Zhao P, Liu L, Leong KW, Mao H-Q, Liu Z, Chen Y, Adv. Healthcare Mater 7 (2018) 1800285. [DOI] [PubMed] [Google Scholar]
- [154].Liu Z, Le Z, Lu L, Zhu Y, Yang C, Zhao P, Wang Z, Shen J, Liu L, Chen Y, Nanoscale 11 (2019) 9410–9421. [DOI] [PubMed] [Google Scholar]
- [155].Zeng Z, Dong C, Zhao P, Liu Z, Liu L, Mao H, Leong KW, Gao X, Chen Y, Adv. Healthcare Mater 8 (2019) 1801010. [DOI] [PubMed] [Google Scholar]
- [156].Akbulut M, Ginart P, Gindy ME, Theriault C, Chin KH, Soboyejo W, Prud’homme RK, Adv. Funct. Mater 19 (2009) 718–725. [Google Scholar]
- [157].D’Addio SM, Baldassano S, Shi L, Cheung L, Adamson DH, Bruzek M, Anthony JE, Laskin DL, Sinko PJ, Prud’homme RK, Journal of Controlled Release 168 (2013) 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Lu HD, Yang SS, Wilson BK, McManus SA, Chen CVH-H, Prud’homme RK, Appl Nanosci 7 (2017) 83–93. [Google Scholar]
- [159].Nie T, He Z, Zhou Y, Zhu J, Chen K, Liu L, Leong KW, Mao H-Q, Chen Y, ACS Appl. Mater. Interfaces 11 (2019) 29593–29603. [DOI] [PubMed] [Google Scholar]
- [160].Lin P-Y, Chiu Y-L, Huang J-H, Chuang E-Y, Mi F-L, Lin K-J, Juang J-H, Sung H-W, Leong KW, Adv. Sci 5 (2018) 1701079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Shen H, Banerjee AA, Mlynarska P, Hautman M, Hong S, Kapetanovic IM, Lyubimov AV, Liu Y, Journal of Pharmaceutical Sciences 101 (2012) 3877–3885. [DOI] [PubMed] [Google Scholar]
- [162].Ke X, Tang H, Mao H-Q, International Journal of Pharmaceutics 564 (2019) 273–280. [DOI] [PubMed] [Google Scholar]
- [163].Lu HD, Spiegel AC, Hurley A, Perez LJ, Maisel K, Ensign LM, Hanes J, Bassler BL, Semmelhack MF, Prud’homme RK, Nano Lett. 15 (2015) 2235–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lu HD, Pearson E, Ristroph KD, Duncan GA, Ensign LM, Suk JS, Hanes J, Prud’homme RK, International Journal of Pharmaceutics 544 (2018) 75–82. [DOI] [PubMed] [Google Scholar]
- [165].Lu HD, Ristroph KD, Dobrijevic ELK, Feng J, McManus SA, Zhang Y, Mulhearn WD, Ramachandruni H, Patel A, Prud’homme RK, ACS Infect. Dis 4 (2018) 970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Feng J, Zhang Y, McManus SA, Qian R, Ristroph KD, Ramachandruni H, Gong K, White CE, Rawal A, Prud’homme RK, Soft Matter 15 (2019) 2400–2410. [DOI] [PubMed] [Google Scholar]
- [167].Allen S, Osorio O, Liu Y-G, Scott E, Journal of Controlled Release 262 (2017) 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Bobbala S, Allen SD, Scott EA, Nanoscale 10 (2018) 5078–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Allen SD, Bobbala S, Karabin NB, Modak M, Scott EA, ACS Appl. Mater. Interfaces 10 (2018) 33857–33866. [DOI] [PubMed] [Google Scholar]
- [170].Yi S, Allen SD, Liu Y-G, Ouyang BZ, Li X, Augsornworawat P, Thorp EB, Scott EA, ACS Nano 10 (2016) 11290–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Allen SD, Liu Y-G, Bobbala S, Cai L, Hecker PI, Temel R, Scott EA, Nano Res. 11 (2018) 5689–5703. [Google Scholar]
- [172].York AW, Zablocki KR, Lewis DR, Gu L, Uhrich KE, Prud’homme RK, Moghe PV, Adv. Mater 24 (2012) 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Petersen LK, York AW, Lewis DR, Ahuja S, Uhrich KE, Prud’homme RK, Moghe PV, Mol. Pharmaceutics 11 (2014) 2815–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kudisch B, Maiuri M, Wang L, Lim T, Lu H, Lee V, Prud’homme RK, Scholes GD, Nanoscale 11 (2019) 2385–2392. [DOI] [PubMed] [Google Scholar]
- [175].Akbulut M, Ginart P, Gindy ME, Theriault C, Chin KH, Soboyejo W, Prud’homme RK, Adv. Funct. Mater 19 (2009) 718–725. [Google Scholar]
- [176].Ungun B, Prud’homme RK, Budijon SJ, Shan J, Lim SF, Ju Y, Austin R, Opt. Express 17 (2009) 80. [DOI] [PubMed] [Google Scholar]
- [177].Kumar V, Adamson DH, Prud’homme RK, Small 6 (2010) 2907–2914. [DOI] [PubMed] [Google Scholar]
- [178].Shan J, Budijono SJ, Hu G, Yao N, Kang Y, Ju Y, Prud’homme RK, Adv. Funct. Mater 21 (2011) 2488–2495. [Google Scholar]
- [179].Pinkerton NM, Gindy ME, Calero-DdelC VL, Wolfson T, Pagels RF, Adler D, Gao D, Li S, Wang R, Zevon M, Yao N, Pacheco C, Therien MJ, Rinaldi C, Sinko PJ, Prud’homme RK, Adv. Healthcare Mater 4 (2015) 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Zhang J, Wang Q, Liu J, Guo Z, Yang J, Li Q, Zhang S, Yan C, Zhu W-H, ACS Appl. Bio Mater 2 (2019) 943–951. [DOI] [PubMed] [Google Scholar]
- [181].Li M, Xu Y, Sun J, Wang M, Yang D, Guo X, Song H, Cao S, Yan Y, ACS Appl. Mater. Interfaces 10 (2018) 10752–10760. [DOI] [PubMed] [Google Scholar]
- [182].Howard GP, Verma G, Ke X, Thayer WM, Hamerly T, Baxter VK, Lee JE, Dinglasan RR, Mao H-Q, Nano Res. 12 (2019) 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Tang C, York AW, Mikitsh JL, Wright AC, Chacko A-M, Elias DR, Xu Y, Lim H-K, Prud’homme RK, Macromol. Chem. Phys 219 (2018) 1700592. [Google Scholar]
- [184].Wang M, Yang N, Guo Z, Gu K, Shao A, Zhu W, Xu Y, Wang J, Prud’homme RK, Guo X, Ind. Eng. Chem. Res 54 (2015) 4683–4688. [Google Scholar]
- [185].Yan X, Remond M, Zheng Z, Hoibian E, Soulage C, Chambert S, Andraud C, Van der Sanden B, Ganachaud F, Bretonnière Y, Bernard J, ACS Appl. Mater. Interfaces 10 (2018) 25154–25165. [DOI] [PubMed] [Google Scholar]