We report here on novel technology improvements to high-throughput electron-optical imaging pipelines, developed by coauthors at Voxa and currently in use by coauthors at Princeton and elsewhere. Significant advances in array volume imaging continue to improve the fidelity and size of 3D anatomy visualizations for structural biology, and provide promise for high throughput characterization in other fields such as nanoparticle synthesis and industrial processing. In 3D reconstructions, sectioned tissue is imaged and the volume reconstructed in software from the imaged layers, where the raw section thicknesses range from micrometers (light-optical) down to tens of nanometers (electron-optical and super-resolution) [1–2]. While several varieties of optical and electron optical array methods are available, electron microscopy remains a gold standard. However, traditional EM is a slow and latency-filled process, and most commercial instruments average <<1 Mpix/s data collection rate during imaging and require automation. Traditional sample handling and exchange processes further reduce this effective speed by orders of magnitude.
Coauthors at Voxa have previously reported on novel pipelined tools, collectively called Blade™, for increasing transmission electron microscopy (TEM) throughput to enable extremely large 3D volume reconstructions [3–4]. A centerpiece of the pipeline is the GridStage™ sample handling system that enables high-volume low latency delivery and imaging of serial sequential samples under vacuum into the electron microscope. Supporting capabilities within the Blade pipeline include a) post-staining to apply stains after sectioning (Strider™), b) conductive layer deposition to control charging and stabilize deposited films (Glow™, previously referred to as Pupa), and c) cassette-based vacuum sample storage (Pod™) [3]. These tools have been deployed at imaging centers focusing on neuroscience, including at the Allen Institute for Brain Science and at the Princeton Neuroscience Institute (PNI), acquiring datasets with detail down to the synaptic level (sub-10 nm) in extremely large volumes exceeding 1 mm3 (Fig. 1). Together with dedicated imaging integration software synchronizing the microscope settings and high-speed CMOS camera, Blade’s sustained practical image data collection rates are ~120 MPix/s for large volume reconstructions using ~36 MPix tiles, and Blade has been able to sustain 24/7 operation for several months at a time.
Figure 1.
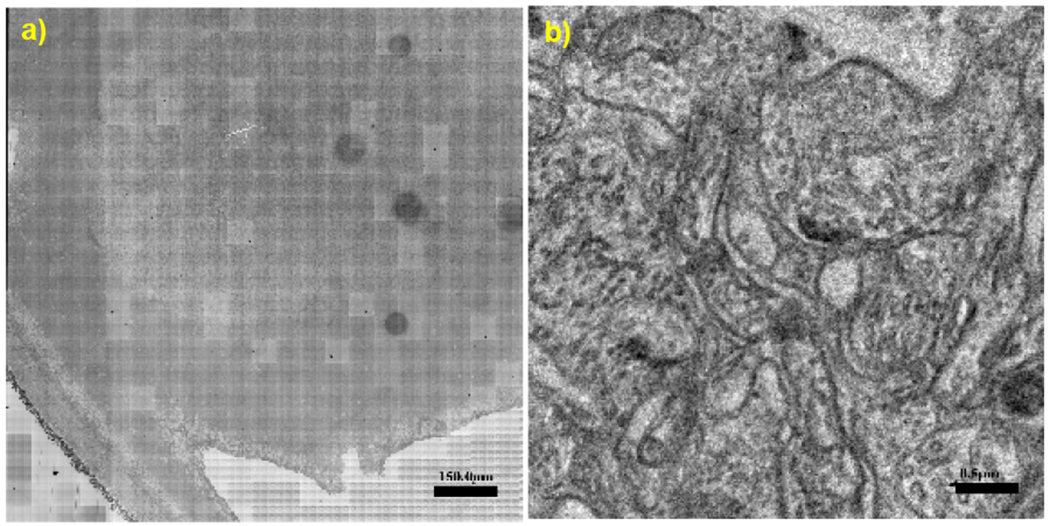
a) Aligned montage of mouse retinal tissue section acquired at 225 MPix/s on the Blade pipeline with Cricket (1.25 x 1.25 mm, 3.2 nm pixels, ~400 Gpix, <30 min acquisition). b) A 4 x 4 um region sampled from a), representative of image quality across the montage.
A new addition to the Blade pipeline, Cricket™, employs an innovative hybrid beam+stage scanning speed boost mechanism to further increase image data collection rate. Without Cricket, GridStage must make 10k mechanical sample translations to image about 1mm2. Consider that step-and-settle time (latency) of the sample stage remains a significant overhead even given short <100 ms translations offered by GridStage. Additionally, 10k steps per section means that stage motor life without Cricket can be as short as 3-6 months for 24/7 use. Both stage latency and motor longevity can be solved by Cricket reducing the number of mechanical translations by an order of magnitude.
Cricket jumps the electron beam around the current sample position in a tile pattern synchronously with a post-sample descan onto the TEM camera (Fig. 2a). The subtiles obtained with this “subscanning” technique are acquired and stitched together live on the acquisition computer to form a larger composite image “supertile” with ~10x fewer stage movements (Fig. 2b–c). Field aberrations and distortions are compensated to minimize error within the supertile to facilitate 2D stitching and 3D reconstruction. Supertiles can later be stitched together with significantly less computation, since each supertile will have already eliminated 20 edge-to-edge interfaces out of 24 total potential edges during acquisition. With Cricket, a large-area sample montage presenting a difficult puzzle with many small pieces becomes a child’s puzzle with 10x fewer, larger pieces comprising the same image.
Figure 2.
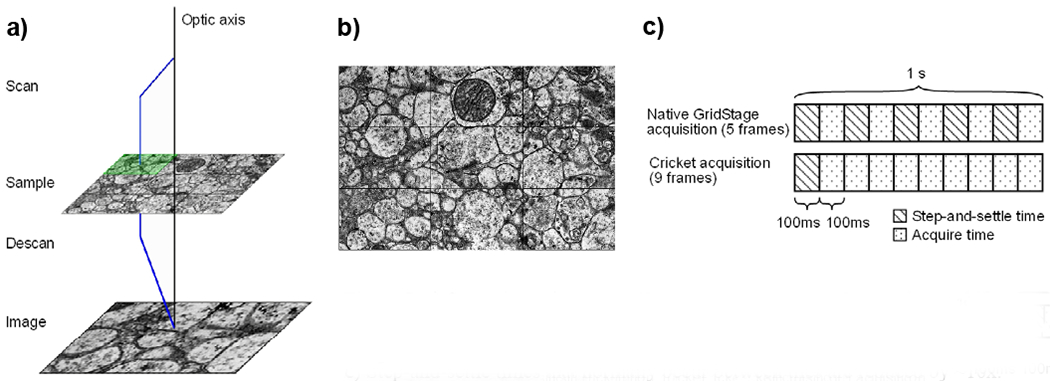
a) Operating principle of Voxa’s Cricket™. The electron beam shifts to illuminate areas of the sample while simultaneously descanning the post-sample beam onto the detector. b) Subtiles are stitched in real-time to form a “supertile”. c) Step-and-settle times that normally occur between tiles are reduced by ~10x.
The previously reported Blade pipeline takes image data acquisition speeds from <<1 Mpix/s with traditional EM workflow to ~120 MPix/s with negligible sample exchange time. The addition of Cricket further enhances Blade’s acquisition speed to >350 MPix/s. These resulting multi order-of-magnitude increases in practical speed provide—for the first time—capacity to create massive nanoscale-resolution 3D reconstructions in months rather than decades. [5]
Acknowledgments
This work was supported by Voxa, the Simons Foundation, the National Institutes of Health, and the CV Starr Fellowship of the Princeton Neuroscience Institute. Voxa innovations are patented and patent pending.
References:
- [1].Kubota Y, Sohn J, and Kawaguchi Y, Frontiers in Neural Circuits 12 (2018), Article 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Micheva KD, and Smith SJ, Neuron 55 (2007), 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Own CS, et al. Microscopy & Microanalysis 21 (2015), 9–10.26227685 [Google Scholar]
- [4].Own CS, et al. Microscopy & Microanalysis 23 (2017), 32–33. [Google Scholar]
- [5]. https://www.youtube.com/watch?v=3SMa45YAMK4.


